Background: The androgen receptor (AR) plays a critical role in prostate cancer development and progression.
Results: Oncogenic PIM-1 kinases directly interact with AR and induce AR phosphorylation at multiple residues.
Conclusion: PIM-1 kinases differentially modulate AR activity via phosphorylation-dependent recruitment of distinct ubiquitin E3 ligases.
Significance: Our findings provide new insights into mechanisms by which AR activity may be regulated in prostate cancer cells.
Keywords: Androgen Receptor, Phosphorylation, Prostate, Protein Kinases, Ubiquitination, PIM1
Abstract
Androgen receptor (AR) plays a pivotal role in prostate cancer. Regulation of AR transcriptional activity by post-translational modifications, such as phosphorylation by multiple kinases, is well documented. Here, we report that two PIM-1 kinase isoforms which are up-regulated during prostate cancer progression, namely PIM-1S and PIM-1L, modulate AR stability and transcriptional activity through differentially phosphorylating AR at serine 213 (Ser-213) and threonine 850 (Thr-850). Although both kinases are capable of interacting with and phosphorylating AR at Ser-213, only PIM-1L could phosphorylate Thr-850. We also showed that PIM-1S induced Ser-213 phosphorylation destabilizes AR by recruiting the ubiquitin E3 ligase Mdm2 and promotes AR degradation in a cell cycle-dependent manner, while PIM-1L-induced Thr-850 phosphorylation stabilizes AR by recruiting the ubiquitin E3 ligase RNF6 and promotes AR-mediated transcription under low-androgen conditions. Furthermore, both PIM-1 isoforms could promote prostate cancer cell growth under low-androgen conditions. Our data suggest that these kinases regulate AR stability and transcriptional activity through recruitment of different functional partners in a phosphorylation-dependent manner. As AR turnover has been previously shown to be critical for cell cycle progression in prostate cancer cells, PIM-1 kinase isoforms may promote prostate cancer cell growth, at least in part, through modulating AR activity via distinct mechanisms.
Introduction
Hormone ablation therapy is the standard care for patients with advanced metastatic prostate cancer (1). Although most patients initially respond well to androgen deprivation therapy, many will relapse into an aggressive castration-resistant stage which is currently incurable. Better understanding mechanisms underlying castration resistance is critical for the development of more effective treatment (2).
The androgen receptor (AR)3 is a key regulator of survival and proliferation of prostatic cells. As a member of steroid hormone receptor family, it translocates to the nucleus upon binding to androgens and binds to the androgen response elements (AREs) in the regulatory regions of its targeted genes (3). Accumulating clinical data have demonstrated that a majority of castration-resistant prostate cancers still express AR and androgen-dependent genes, indicating that the AR-signaling pathway is functional under androgen-depleted conditions (4–6). Several independent studies also showed that AR is essential for both hormone sensitive and hormone refractory prostate cancer (7, 8). Deregulation of steroid biosynthesis enzymes, Mutations/amplification and alternative splicing of AR, alterations in protein kinases, growth factors, and nuclear receptor coactivators have been proposed to modulate AR signaling and may, therefore, contribute to castration-resistance (9–14). A substantial body of literature suggests that AR is regulated directly by phosphorylation (15, 16). We and others have showed that AR activity can also be regulated through tyrosine phosphorylation (17–20). In addition, MAPK, PKA, AKT/PKB, PKC, and CDKs are able to induce serine-threonine phosphorylation of AR (21–26). The mechanisms by which serine/threonine phosphorylation can regulate AR activity, especially under low androgen conditions, are poorly understood.
A newly emerging role of AR in regulating cell cycle progression is functioning as a component of DNA replication licensing factors, which are degraded during mitosis and allow relicensing only at the next cycle (27, 28). This process occurs primarily to ensure genetic stability by allowing replication to occur only once per cycle. A recent study showed that stabilizing AR in mitosis inhibits prostate cancer cell growth (29). Although it is known degradation of AR occurs primarily through a proteasome-dependent pathway, very little is known about how this process is regulated. In this study, we show that the decreased AR expression during mitosis was coincided with increased PIM-1 expression, suggesting that PIM-1 may play a role in regulation of AR turnover.
The human PIM1 gene was initially identified as a frequent integration site for Moloney murine leukemia virus (MuLV) (30). Several targets of PIM-1 regulate cell cycle progression including p21, p100, Cdc25A, Cdc25C, and HP1 (31–35). A number of studies have documented oncogenic synergy between c-Myc and PIM-1 (36–40). AKT/PKB shares a number of substrates with PIM kinases, including Bad and Mdm2 (42–44), which suggests that the two may regulate some overlapping targets in response to different extracellular stimuli. A consensus sequence for the PIM-1 preferred substrates, (K/R)3XS/TX(where X stands for any residue), has been derived from a screening on the peptide substrates and found in many known PIM-1 substrates (45).
PIM-1 overexpression has been found in human prostate cancer and mouse model prostate tumors (36, 46–48). PIM-1 was also identified as a potential diagnostic biomarker for prostate cancer (49). Both human and murine PIM1 genes encode two protein kinase isoforms, the 44 kDa PIM-1L and the 33 kDa PIM-1S, by virtue of an alternative upstream translation initiation site (50). PIM-1L possesses a unique proline-rich motif not present in PIM-1S, suggesting that it may interact with a different subset of proteins, such as those containing SH3 domains. We reported that the subcellular localization is different between the isoforms: PIM-1L resides mainly in the plasma membrane and cytosol while PIM-1S is predominantly in the nucleus (51). This raises the possibility that PIM-1 isoforms may serve divergent functions.
In the current study, we have shown that PIM-1L and PIM-1S can directly interact with AR and induce AR phosphorylation at different residues and recruit distinct ubiquitin E3 ligases to form complexes with AR in a phosphorylation-dependent manner. Our results suggest that PIM-1 kinases may promote prostate cancer cell growth, at least in part, through modulating AR activity via distinct mechanisms.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
Cell lines were purchased from American Tissue Culture Collections (ATCC) with the exception of CWR-R1 cells, which were provided by Drs. Gregory and Wilson of the University of North Carolina Chapel Hill. Cells were maintained in a 37 °C incubator at 5% CO2. 293T and COS-1 cell lines were cultured in DMEM (Mediatech Inc.) supplemented with 10% fetal bovine serum (FBS). LNCaP and CWR-R1 were cultured in RPMI 1640 medium (Mediatech Inc.) with 10% FBS. Cell proliferation was assayed using CCK-8 (Dojindo Molecular Technologies, Inc).
Constructs
PIM-1 constructs were cloned as previously described (51). All human PIM-1 constructs contain an N-terminal FLAG tag. PIM-1 LXXLL motif mutants (leucines 319, 322, and 323 mutated to alanines) were mutated using the QuickChange Mutagenesis Kit (Stratagene). All mutations were confirmed by sequencing. Expression constructs for FLAG-tagged AR, HA-RNF6, Probasin luciferase reporter ARR2-Luc, and specific shRNA constructs were described previously (17, 51, 52).
Transient Transfections
Cells were plated in 6-well plates at 1 × 105 cells/well for 24 h before transfection. 293T cells were transfected using a calcium phosphate transfection kit (Biological Mimetics, Inc). COS-1 cells were transfected using FugeneHD reagent (Roche). Lysates of cells were collected ∼48 h post-transfection.
Immunoprecipitation, Western Blotting, and Antibodies
For immunoprecipitations (IPs) and Western blots, cell lysates were collected using lysis/IP buffer as described previously (53). IPs were carried out by adding ∼1 μl of antibody per ml cell lysates and allowing incubation for ∼16 h at 4 °C. For detecting ubiquitinated proteins, denaturing conditions were used prior to IP (52). For Western blot analyses, cell lysates were separated on 8–10% SDS-polyacrylamide gels. Proteins were transferred to Immobilon-P polyvinylidene membranes (Millipore). Antibodies from Santa Cruz Biotechnology included AR (sc816 & sc7305), Pim-1 (sc13513), GAPDH (sc47724), actin (sc7210), pS213 (sc71773), Ub (sc8017), and Mdm2 (sc965). Other antibodies included tubulin (ABM G098), FLAG (Sigma F3165), pThr (Cell Signaling 9386S), pAKT (Ser-473, Cell Signaling 4051L), AKT (Cell Signaling 4691), and HA (Covance MMS-101P). The pART850 antibody was developed by immunizing rabbits with a synthetic peptide corresponding to phosphorylated threonine 850 of AR followed by two-round affinity purification using immunogen (Abgent).
GST Pull-down Assays
GST fusion proteins were expressed and purified as described previously (20, 52, 54). Briefly, immobilized GST fusion proteins were then incubated at 4 °C for 1 h with lysates of CWR-R1 and then separated by SDS-PAGE. Protein bands stained by Coomassie Blue dye were excised from the gel and submitted for mass spectrometry analysis by MALDI-TOF. For GST pull-down assay, immobilized GST fusion proteins were incubated with lysates from transfected 293T cells. The associated proteins were separated by PAGE and immunoblotted with anti-FLAG antibody.
In Vitro Kinase Assays
GST-PIM-1L, GST-PIM-1LKM, GST-PIM-1S, and GST-PIM-1SKM fusion proteins were purified and used for in in vitro kinase assays as described previously (54).
Quantitative Real-time RT-PCR
RNA was extracted using TRIzol reagent (Invitrogen). Total RNA was treated with DNase (Promega RQ1 kit) and transcribed into cDNA (Roche Transcriptor Reverse Transcriptase kit), both according to manufacturers' protocols. PCR was carried out using Roche FastStart High Fidelity PCR kit. Real-time PCR was carried out using Roche FastStart SYBR Green Master kit. 18S rRNA primers were TTGACGGAAGGGCACCACCAG (forward) and GCACCACCACCCACGGAATCG (reverse); AR primers were CTACTCCGGACCTTACGGGGACATGCG (forward) and GGGCTGACATTCATAGCCTTCAATGTGTGAC (reverse); AR target gene primers: PSA TCTGCGGCGGTGTTCTG (forward) and GCCGACCCAGCAAGATCA (reverse); KLK2 CATCCAGTCTCGGATTG (forward) and CTCATATT GTAGAGCGGGT (reverse); POV1 AGTGCTGT GTTCGCCTTG (forward) and CACCTCAGAGC CGCTAAG (reverse). The relative abundance of each transcript was quantified by using the ΔΔCt formula using 18S rRNA as an internal control.
Luciferase Assays
Approximately 2.5 × 104 COS-1 cells were seeded into each well of a 24-well plate 24 h prior to transfection. For serum starvation, cells were maintained in DMEM containing 0.1% FBS for 24 h post-transfection. The probasin promoter based ARR2-Luc construct was used a reporter and a promoter-less Renilla luciferase construct was used as an internal control. Experiments were carried out using Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol.
In Vitro Cell Proliferation Assays
Cells were plated in 100-mm plates at a density of ∼1 × 106 cells/plate 24 h prior to lentiviral infection. As vector-treated cells reached confluence, cells were fixed with 1% formaldehyde for 1 h, then stained with Coomassie Blue dye for 1 h. Densitometry was performed using a Bio-Rad Gel Imager.
Chromatin Immunoprecipitation (ChIP)
LNCaP cells were cultured and infected with lentiviruses as described above. Approximately 24 h postinfection, cells were serum-starved for 16 h, cross-linked with 1% formaldehyde, and then sonicated in 5-s intervals. Soluble chromatin was blocked with salmon sperm DNA and then immunoprecipitated with anti-AR (N20) antibody. Following washes with salt, LiCl, and TE buffers, crosslinks were reversed and DNA purified using Qiagen QIAquick PCR purification kit. Real time PCR was then carried out as described above. PSA Promoter ARE primers were AGGGATCAGGGAGTCTCACA (forward) and GCTAGCACTTGCTGTTCTGC (reverse); KLK2 Promoter ARE primers were GAGAATGCCTCCAGACTGAT (forward) and CTTGCCCTGTTGGCACCTA (reverse).
Phosphorylation Predictive Software
Publicly available NetPhos 2.0 software(55) was used to identify putative serine and threonine phosphorylation sites of AR. The resulting predicted sites were compared with the known PIM-1 target phosphorylation consensus sequence.
Statistical Analyses
Experiments were performed a minimum of three replicates. Statistical analyses were performed as a Student's t test using GraphPad Prism software (Version 5). p values of p < 0.05 were considered statistically significant.
RESULTS
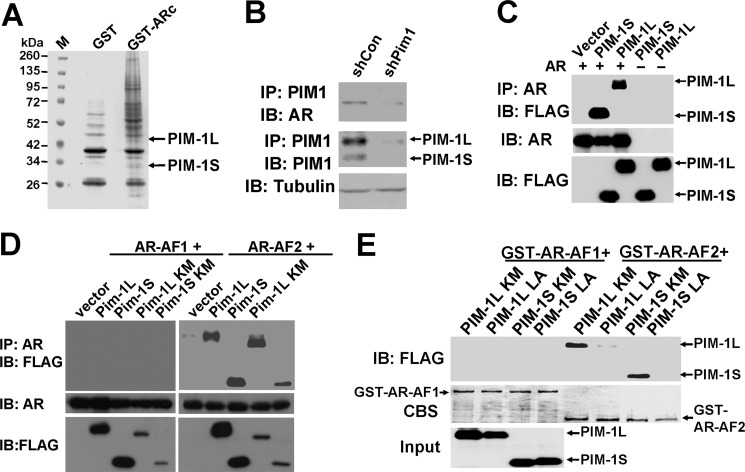
To identify AR-associated proteins in CWR-R1 cells, GST pull-down experiments were performed using the purified GST-tagged AR C terminus (amino acid 622–919) as described previously (52). Both PIM-1L and PIM-1S were among the proteins associated with AR (Fig. 1A). This interaction was validated by co-immunoprecipitation (co-IP) in a prostate cancer cell line CWR-R1 (Fig. 1B) and an overexpression system with AR and PIM-1 kinases in 293T cells (Fig. 1C). A subsequent co-IP experiment confirmed that PIM-1L and PIM-1S interacts with the C-terminal AF2 domain of AR, but not the N-terminal AF1 (Fig. 1D). This interaction appeared to be independent of PIM-1 kinase activity as the kinase-inactive mutants also pulled down the AF2 fragment. The direct binding of AF2 domain with PIM-1 kinases was supported by in vitro binding experiments using purified recombinant proteins (supplemental Fig. S1). It is noteworthy that the protein expression level of PIM-1 inactive mutants was considerably less than that of their kinase active counterparts. This phenomenon is likely due to the fact that PIM-1 can induce auto-phosphorylation (56), which may stabilize the kinase (57). Interestingly, PIM-1 kinases contain an LXXLL motif (X representing any amino acid, located between amino acids 319–323 in PIM-1L and amino acids 228–232 in PIM-1S), which is known to facilitate binding to nuclear receptors (58). We mutated all three leucine residues of kinase-inactive mutants to alanine and tested the effects on their binding to GST-tagged AF1 and AF2. As shown in Fig. 1E, neither PIM-1L-LA nor PIM-1S-LA mutants were able to associate with the AF2 domain of AR. Taken together, these observations suggest that both PIM-1 kinases can directly interact with the AF2 domain of AR through their LXXLL motif.
FIGURE 1.
PIM-1 kinases interact with AR C terminus via their LXXLL motif. A, GST pull-down identifies PIM-1 kinases as candidate AR-interacting proteins. Lysates from CWR-R1 cells were incubated with purified GST-tagged AR C terminus (GST-ARc) overnight and separated by PAGE. Marker (M) shows molecular weights with GST fusion proteins labeled. Bands from stained gel were excised and further analyzed by mass spectrometry. GST tag alone was used as a negative control. B, CWR-R1 cells were infected with lentivirus encoding the shRNA for PIM-1 or the control shRNA. At 48-h postinfection, cell lysates were immunoprecipitated with anti-PIM1 followed by Western blot analysis using the indicated antibodies. C, 293T cells were cotransfected with FLAG-tagged PIM-1 kinases and full-length AR followed by an IP with an anti-AR antibody. Control blots detected input levels of AR and PIM-1 kinase expression. D, 293T cells were cotransfected with AR AF1 (amino acids 1–564) or AF2 (amino acids 622–919) domains and FLAG-tagged PIM-1 constructs. Cell lysates were then immunoprecipitated with an anti-AR antibody followed by immunoblotting with an anti-FLAG antibody. Kinase-inactive mutants are indicated as KM. Total cell lysate levels of AR and FLAG-PIM1 were also immunoblotted as controls. E, 293T cells were transfected to overexpress FLAG-tagged PIM-1 kinase mutant (KM) or LXXLL mutant (LA) constructs. Lysates were incubated with purified GST-tagged AR-AF1 or -AF2 for GST pull-down assay. Western blots of pulldown or input samples were immunoblotted with anti-FLAG. Coomassie Blue-stained (CBS) membrane showed relative loading of GST AR AF constructs.
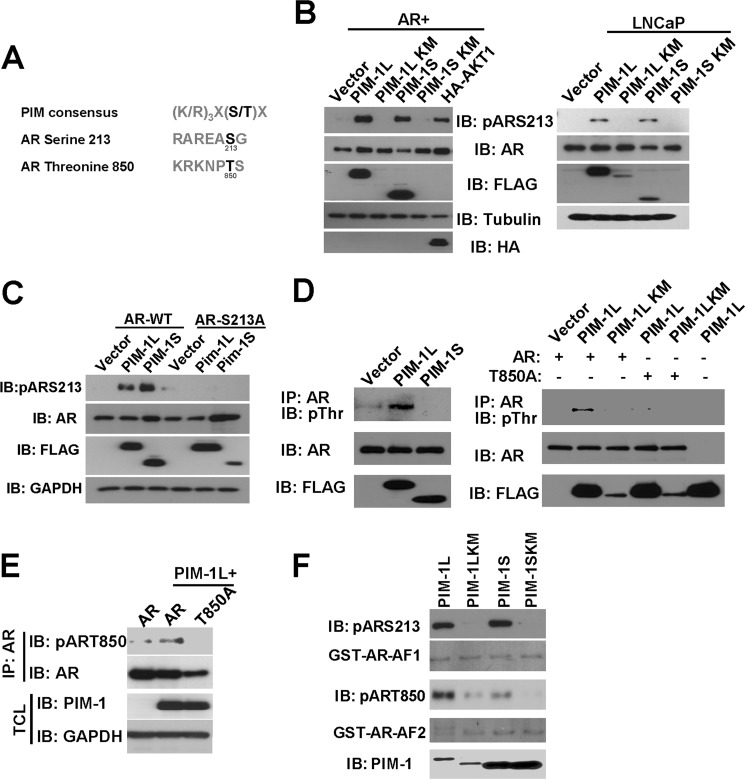
Using NetPhos predictive phosphorylation software, we were able to identify several serine and threonine residues in AR which could be potentially phosphorylated. Two potential phosphorylation sites serine 213 (Ser-213) and threonine 850 (Thr-850) are located in a sequence context closely resembling the known PIM-1 substrate sequence consensus (Fig. 2A).
FIGURE 2.
PIM-1 kinases phosphorylate AR at Ser-213 and Thr-850. A, putative PIM-1 phosphorylation sites in AR protein. B, PIM-1 kinases induce AR phosphorylation at Ser-213. FLAG-tagged PIM-1 kinases were transfected into COS-1 (left) or infected with lentiviruses in LNCaP cells (right). The level of AR Ser-213 phosphorylation in total cell lysates was detected by immunoblotting with phosphospecific antibody for Ser-213 of AR (pARS213). HA-tagged AKT1 served as a positive control for Ser-213 phosphorylation in COS-1 cells. AR, FLAG, tubulin, and HA blots were used as controls. C, COS-1 cells were cotransfected with wild-type AR (AR-WT) or the AR-S213A mutant with FLAG-tagged PIM-1 kinases. Total cell lysates were used for Western blot analysis as in B. D, PIM-1L induces AR phosphorylation at Thr-850. COS-1 cells were transfected with FLAG-tagged PIM-1 kinases and AR (left panel). Lysates were immunoprecipitated with anti-AR followed by immunoblotting with a phospho-threonine (pThr) antibody (right panel). AR and FLAG blots were used as controls. E, a similar IP experiment was performed along with the AR-T850A mutant to confirm specificity of pThr signal PIM-1L-induced AR Thr-850 phosphorylation was detected by an antibody specific for phosphorylated Thr-850 (pT850). F, in vitro kinase assays using purified GST-PIM-1S and -PIM-1L, AR phosphorylation was detected by using pARS213 and pART850 antibodies.
We first tested whether PIM-1 kinases could induce AR Ser-213 phosphorylation in an overexpression system. Both kinase-active PIM-1L and PIM-1S were able to induce AR phosphorylation detected by a phospho-S213 (pARS213) antibody (Fig. 2B, left panel). AKT was used as a positive control, as it has been shown to induce AR phosphorylation at Ser-213 (59). The induction of Ser-213 phosphorylation by PIM-1 kinases was also observed on endogenous AR in LNCaP cells as well (Fig. 2B, right panel). The specificity of the pARS213 antibody was confirmed by coexpression of the AR-S213A mutant with PIM-1 kinases (Fig. 2C).
To test whether PIM-1 kinases can also induce phosphorylation at threonine residues such as Thr-850, we overexpressed PIM-1L or PIM-1S with AR in COS-1 cells. Surprisingly, only PIM-1L was capable of inducing robust AR threonine phosphorylation (Fig. 2D). Coexpression of the T850A mutant with PIM-1L abrogated this signal, confirming that this was the major site responsible for the increased phosphothreonine. We then developed a phospho-specific antibody pART850 capable of detecting an increase in Thr-850 phosphorylation induced by PIM-1L (Fig. 2E). Mutation of Thr-850 diminished PIM-1L induced AR phosphorylation, confirming the specificity of the antibody and supporting previous experiments. In addition, we performed in vitro kinase assays and showed that purified PIM-1L, but not the kinase dead mutant, could phosphorylate both Ser-213 and Thr-850 very well (Fig. 2F). Meanwhile, PIM-1S displayed comparable activity toward Ser-213 but significantly less activity toward Thr-850 compared with PIM-1L. Taken together, these data suggest that PIM-1 kinases may differentially regulate AR phosphorylation in vivo.
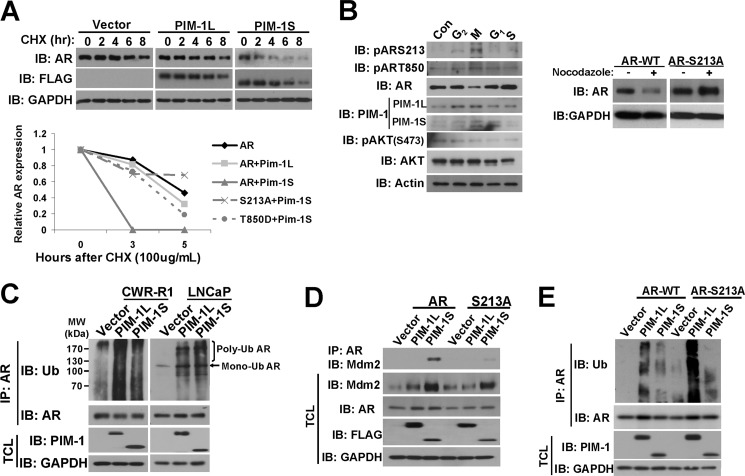
Because phosphorylation of AR at serine 213 induced by AKT has previously been shown to be associated with AR proteasomal degradation (60), we wondered whether PIM-1 kinases had the same effect. To measure the effects of PIM-1 kinases on AR protein stability, we treated cells with cycloheximide to block protein synthesis and measured the half-life of AR protein. As shown in the top panel of Fig. 3A, cotransfection of PIM-1S, but not PIM-1L, dramatically decreased the half-life of AR protein. In addition, the unphosphorylatable AR-S213A mutant is significantly more stable in the presence of PIM-1S while the phospho-mimic T850D mutant was partially resistant to PIM-1S induced destabilization (Fig. 3A, bottom panel). These data suggest that phosphorylation of Ser-213 is involved in the destabilization of AR and phosphorylation of Thr-850 induced by PIM-1L may protect AR from degradation.
FIGURE 3.
PIM-1S promotes AR turnover by recruitment of Mdm2. A, 293T cells were transfected with AR and FLAG-tagged PIM-1 kinases. At 16 h post-transfection, cells were treated with 10 μg/ml CHX for the times indicated. Total cell lysates were blotted for AR, while FLAG and GAPDH served as controls (top panel). The effects of S213A and T850D mutation on AR stability were also examined as above in cells treated with 100 μg/ml CHX. Quantification of AR levels was carried out using densitometry by normalizing AR to GAPDH expression (bottom panel). Values were set relative to AR expression at time 0. B, LNCaP cells were arrested in G2, M, G1, and S-phases after 16 h treatments of 2 μm etoposide, 200 ng/ml nocodazole, 15 μg/ml lovastatin, or 5 μg/ml aphidicolin, respectively (left panel). Total cell lysates were subjected to Western blot with the antibodies listed. Endogenous AR expression in LNCaP cells was knocked down by lentiviral shRNA and simultaneously replaced with the codon-switched AR-WT or AR-S213A mutant (right panel). Cells were then synchronized in M-phase using nocodazole treatment. AR expression was measured by Western blot with GAPDH serving as a loading control. C, FLAG-tagged PIM-1 kinases were overexpressed in CWR-R1 and LNCaP cells and then maintained in medium containing 0.1% FBS for 24 h. Lysates were subjected to denaturing conditions before IP with anti-AR as described under “Experimental Procedures.” Immunoprecipitates were immunoblotted with anti-Ub antibody while total cell lysate blots were used as controls. D, AR constructs and FLAG-tagged PIM-1 kinases were transfected into COS-1 cells followed by IP with anti-AR. Immunoprecipitates were immunoblotted with Mdm2 while total cell lysates were used for control blots. E, AR constructs and FLAG-tagged PIM-1 kinases were transfected into COS-1 cells followed by IP and Western blot as in C.
Previous studies in hematopoietic cell lines have shown that expression and activity of PIM-1S kinase is increased at G1-S and G2-M transitions (33, 34, 61, 62). AR protein has been reported to be destabilized during mitosis (28) and this degradation is considered necessary for AR to serve as a DNA replication licensing factor in prostate cancer cells as its stabilization inhibits proliferation (29). To determine whether decreased expression of AR coincided with PIM-1S activity, LNCaP cells were pharmacologically arrested at M, G1, mid-S, and G2 as previously described (23, 63). Fig. 3B shows that AR expression levels were significantly reduced in the M-phase in agreement with previously published studies (28). A noticeable increase in PIM-1L and PIM-1S expression was observed in G2 and M-phase arrested cells compared with others, which also coincided with elevated levels of phospho-S213 and phospho-T850 of AR. This confirmed that PIM-1 kinase expression levels and activity in prostate cancer cells are cell cycle-regulated. AKT activity was also determined by measuring serine 473 (Ser-473) phosphorylation which has been shown to be a critical regulator of full AKT activation (64, 65). We were unable to detect an increase of phospho-AKT in G2 and M phases, suggesting the increased phospho-S213 signal we observed was mainly due to elevated PIM-1 activity. Because increased phosphorylation at Ser-213 coincided with decreased AR expression, we hypothesized that this specific modification may play a role in regulating AR stability during mitosis. To test this, we utilized the AR replacement approach described previously (17) to knock down endogenous AR in LNCaP cells while simultaneously re-expressing either wild-type AR or the S213A mutant. Cells were then synchronized in M-phase with nocodazole. The S213A mutant was considerably more stable than the wild-type AR (Fig. 3B, right panel). These findings demonstrated that S213 phosphorylation plays an important role in AR stability during cell cycle, particularly during M-phase.
To identify a mechanism for PIM-1 induced changes on AR stability, we performed coimmunoprecipitation (co-IP) experiments to examine AR ubiquitination. As shown in Fig. 3C, both PIM-1L and PIM-1S could dramatically increase mono and polyubiquitination of endogenous AR in CWR-R1 and LNCaP cells. Since both isoforms could induce ubiquitination of AR yet only PIM-1S could cause AR degradation, we speculated that PIM-1 kinases promoted AR ubiquitination via different E3 ubiquitin ligase.
Since Mdm2 has been shown to bind to and ubiquitinate AR thus targeting it for proteolysis (60, 66) and Mdm2 is phosphorylated by AKT (42, 67–69) and PIM-1 (44, 70), we therefore tested whether PIM-1 kinases could modulate interaction between AR and Mdm2. As shown in Fig. 3D, only PIM-1S could enhance the interaction between endogenous Mdm2 and AR even though both isoforms could increase total endogenous Mdm2 levels. Furthermore, S213A mutation abrogated the binding of AR to Mdm2 promoted by PIM-1S. As a result, PIM-1S-induced AR polyubiquitination was also compromised (Fig. 3E). This suggested that phosphorylation of Ser-213 is required for PIM-1S promoted Mdm2 and AR interaction and Mdm2-mediated polyubiquitination of AR.
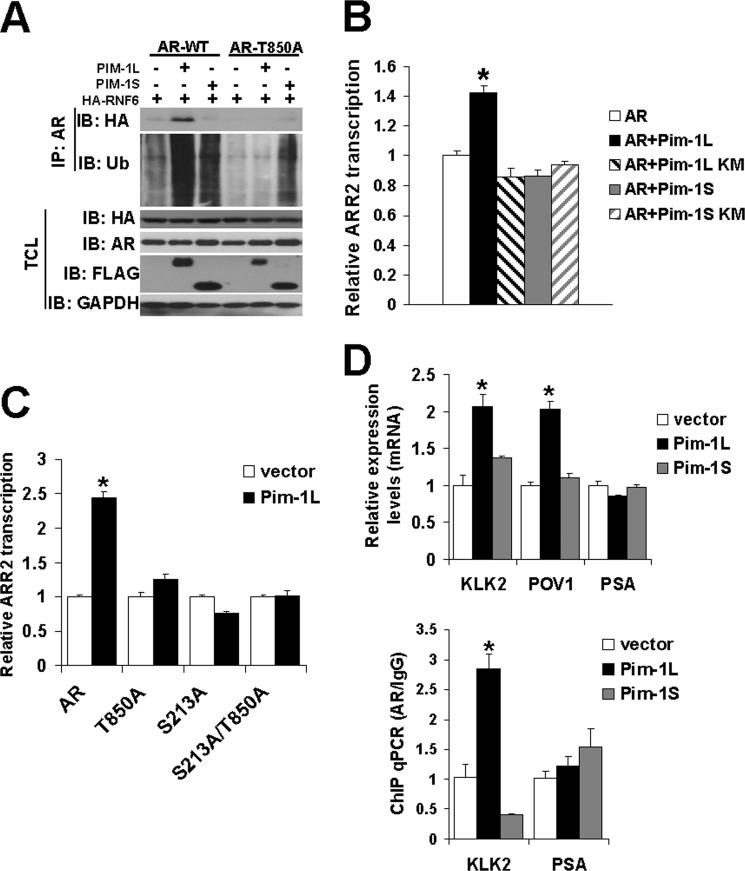
Meanwhile, PIM-1L had little effect on the interaction between AR and Mdm2 and Ser-213 mutation did not compromise PIM-1L-induced AR polyubiquitination. This raises a possibility that Thr-850 phosphorylation may antagonize Ser-213 phosphorylation and recruit another ubiquitin E3 ligase. We previously showed that RNF6 interacts with AR and induces AR ubiquitination in noncanonical manner that did not cause AR destabilization (52). To test whether PIM-1L could enhance the interaction between AR and RNF6, we performed a co-IP experiment with overexpressed RNF6, AR, and PIM-1 kinases in COS-1 cells. As predicted, PIM-1L was able to enhance the association of RNF6 and AR, while PIM-1S had no effect (Fig. 4A). This supported our earlier observation that PIM-1L could increase AR ubiquitination, but did not reduce AR stability. Thr-850 phosphorylation appeared to be responsible for PIM-1L promoted interaction of AR and RNF6 as T850A mutation diminished its interaction with RNF6. In support of this observation, PIM-1L failed to induce ubiquitination of T850A. Meanwhile, S213A mutation enhanced RNF6-induced AR ubiquitination (supplemental Fig. S3), suggesting that Ser-213 phosphorylation may suppress RNF6-induced ubiquitination. Taken together, these experiments supported the notion that PIM-1 kinases induce AR ubiquitination through different E3 ligases in a phosphorylation-dependent manner.
FIGURE 4.
PIM-1L enhances AR transcriptional activity via recruitment of RNF6. A, COS-1 cells were cotransfected with FLAG-tagged PIM-1 kinases, HA-tagged RNF6, and AR constructs. Lysates were subjected to denaturing conditions before an IP using an AR antibody. Immunoprecipitates were immunoblotted with HA and Ub antibodies. TCL: total cell lysates. B, effects of PIM-1 kinases on AR regulated ARR2 reporter. AR and FLAG-tagged PIM-1 kinases were transfected into COS-1 cells with the ARR2-LUC reporter. Approximately 24 h post-transfection, cells were maintained in medium containing 0.1% FBS overnight and the effects of PIM-1 kinases on AR transcriptional activity were determined by luciferase assays. Data is shown relative to AR transfected alone whose value was set at 1. *p < 0.01. C, wild type AR and AR mutants were cotransfected into COS-1 cells with FLAG-tagged PIM-1L and luciferase activity of ARR2 reporter measured as in B. Data are shown relative to AR transfected alone whose value was set at 1. *, p < 0.01. D. LNCaP cells were infected with lentiviruses encoding FLAG-tagged PIM-1 kinases or vector control. Approximately 24 h postinfection, cells were maintained in media containing 0.1% FBS and total RNAs were collected 18 h later. The level of AR target genes KLK2, POV1, and PSA were detected by real time RT-PCR (left panel). Data are shown relative to vector control infection whose value was set at 1. *, p < 0.01. The recruitment of AR to the KLK2 and PSA promoter regions was examined using ChIP assays (right panel). The value was normalized with IgG and input level. Data were shown relative to vector control infection whose value was set at 1. *, p < 0.01.
To examine whether PIM-1-induced AR phosphorylation plays a role in regulating its activity, we measured the effect of PIM-1 kinases on a probasin promoter based ARR2-Luciferase reporter, which we have shown provides reliable readout for AR-mediated transcriptional activation (17). The luciferase assay revealed that only kinase active PIM-1L was capable of enhancing ARR2 transcription in kinase-dependent manner under low serum conditions (Fig. 4B). Mutation of either Ser-213 or Thr-850 diminished PIM-1L induced AR transcriptional activity (Fig. 4C). These results suggest that PIM-1L can enhance AR transcriptional activity through phosphorylation of both Ser-213 and Thr-850. We further examined the effects of PIM-1L and PIM-1S on expression of endogenous AR target genes. As shown in Fig. 4D (top panel), PIM-1L, but not PIM-1S, was able to induce expression of KLK2 and POV1. Interestingly, PIM-1L did not increase expression of PSA, which belongs to the same Kallikrein family as KLK2. This suggests that PIM-1L mediated effects on AR signaling may depend on promoter context. Our ChIP assays further validated that PIM-1L promoted a significant increase in AR recruitment to the KLK2 promoter, but not the PSA promoter (Fig. 4D, bottom panel). These results suggest a novel mechanism by which PIM-1L can selectively activate AR target gene transcription under specific circumstances.
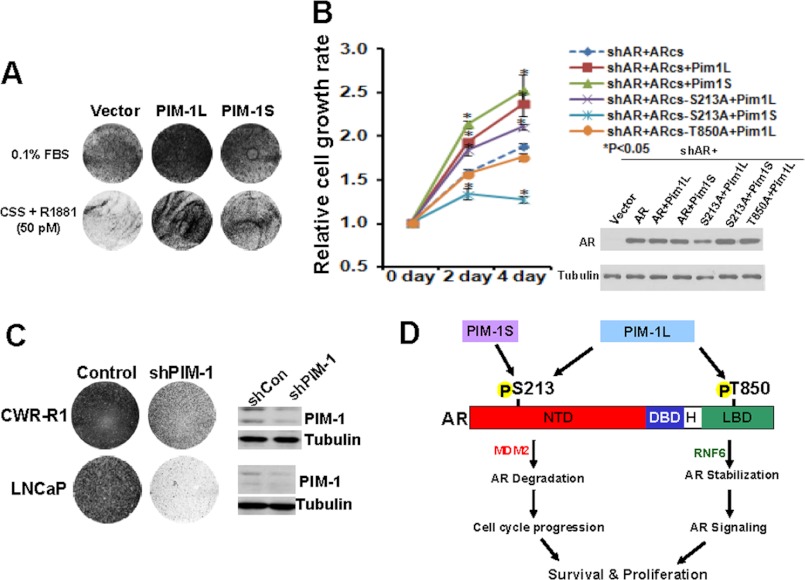
In addition, the effects of overexpression of PIM-1 kinases on growth were examined in LNCaP cells under low androgen conditions. Fig. 5A shows that both PIM-1 kinases greatly increased LNCaP cell growth under these conditions. Replacement of endogenous AR with the codon-switched AR S213A or T850A mutant significantly compromised cell proliferation promoted by PIM-1S and PIM-1L respectively (Fig. 5B). On the other hand, specific knock down of endogenous PIM-1 kinases in CWR-R1 and LNCaP cells significantly reduced cell growth (Fig. 5C). These results suggested that PIM-1 kinases could sensitize prostate cancer cells to low levels of androgens, possibly through phosphorylating different sites.
FIGURE 5.
PIM-1 kinases promote prostate cancer growth. A, LNCaP cells were infected with lentiviruses encoding PIM-1 kinases and maintained in medium containing 0.1% FBS (top panel) or 5% charcoal-stripped serum supplemented with 50 pm R1881 (bottom panel) for 6 days. Cell cultures were then fixed and stained using Coomassie Blue dye. B, LNCaP cells were infected with the indicated lenti-virus. At 24 h postinfection cells were cultured in CS medium containing 0.1 nm DHT. Cell proliferation was measured using the CCK8 assays. The experiments were repeated three times and the representative data were shown. C, LNCaP cells were infected with lentiviruses encoding control shRNA or target specific for PIM-1. Cells were grown for 6 days in complete media and stained with Coomassie Blue dye. D, proposed model of the effects of PIM-1 kinases on modulating AR function. PIM-1S induces phosphorylation at serine 213, a residue, which can also be targeted by PIM-1L. Phosphorylation of Ser-213 can promote AR turnover mediated by Mdm2 and cell cycle progression. PIM-1L is able to induce phosphorylation at an addition residue Thr-850 in the ligand binding domain. Phosphorylation of Thr-850 can stabilize AR protein and enhance its binding to RNF6 to promote its transcriptional activity in regulating expression of a subset target genes.
DISCUSSION
Although the majority of research on PIM-1 kinase has been carried out in the context of hematologic cancers, increasing evidence suggest that PIM-1 kinases may also play a significant role in solid tumors such as prostate cancer. In addition, most PIM-1 literature is centered on studies of the 33 kDa PIM-1S isoform while very little is known about the specific role of the 44 kDa PIM-1L. Our results suggest that PIM-1 kinase isoforms may play distinct roles in prostate cancer progression by modulating AR protein turnover and transcriptional activity. Both PIM-1 isoforms could associate with AR via the LXXLL motifs in their kinase domains. We also demonstrated, for the first time, that both PIM-1 isoforms could induce AR phosphorylation at Ser-213 while only PIM-1L was capable of phosphorylating AR at Thr-850 in vivo. The underlying mechanisms for this differential phosphorylation have yet to be investigated. It is possible that these two phosphorylation events could occur separately in different subcellular compartments depending on the accessibility of these two phosphorylation sites to PIM-1 kinases because PIM-1L is mainly localized to the plasma membrane and cytosol while PIM-1S is largely in the nucleus (51 and supplemental Fig. S4). Alternatively, the antagonizing phosphotases may differ for these sites and several of which are inhibited by PIM-1 kinase activity (34, 71, 72). Despite the fact that both isoforms could phosphorylate Ser-213, only PIM-1S was capable of promoting AR degradation. Since T850D and S213A mutants were more stable than wild-type AR in the presence of PIM-1S, Ser-213 phosphorylation might promote AR degradation while Thr-850 phosphorylation might stabilize AR.
The differential effects on AR induced by PIM-1 kinases may likely result from a phosphorylation-dependent recruitment of different ubiquitin E3 ligases. We showed that both PIM-1 isoforms promoted AR ubiquitination. However, PIM-1S could enhance AR association with Mdm2 while PIM-1L promoted AR complexing with RNF6. The interaction between AR and Mdm2 has previously been shown to promote proteasomal degradation of AR (73). Although Ser-213 was originally identified as an AKT phosphorylation site, PIM-1 kinases appear to be a stronger Ser-213 kinase at least in our system. Our observation is consistent with an independent study from the Logan laboratory in which they also showed that purified recombinant PIM-1 can directly phosphorylate AR at Ser-213.4 Increased PIM-1S expression and activity coincided with decreased AR expression in M-phase in LNCaP cells. The increased Ser-213 phosphorylation of endogenous AR at the M-phase was correlated with an increase in PIM-1S expression while little change in AKT expression or activity was detected under these conditions. In addition, PIM-1S failed to promote degradation of AR-S213A mutant which was stabilized during M-phase. Taken together, these data suggest that PIM-1S may play a role in modulating AR degradation during mitosis, which is required for cell cycle progression and proliferation in AR-positive prostate cancer cells (29).
Although PIM-1L can also target Ser-213, PIM-1L-induced Thr-850 phosphorylation appears to serve a protective role in AR stability. AR stabilization has previously been described as promoting AR activity in advanced prostate cancer (74). PIM-1L promoted recruitment of RNF6 and subsequent ubiquitination and stabilization of AR, leading to increased AR transcriptional activity (52). Mutation of threonine 850 of AR to alanine abrogated its ability to associate with and be ubiquitinated by RNF6. Furthermore, PIM-1L was capable of promoting AR transcription of an androgen responsive promoter ARR2 reporter through its kinase activity. Given that PIM-1S does not enhance AR activity through its phosphorylation at Ser-213 under androgen-depleted conditions, it is interesting that S213A mutation alone diminished PIM-1L-promoted AR transcription activity. These data suggested that phosphorylation of both Ser-213 and Thr-850 is required for PIM-1L induced AR activity. This is consistent with our previous report that PIM-1S alone fails to increase AR transcription while it can synergize with Etk to activate AR reporter constructs (75). It is possible that ETK could complex with and activate endogenous PIM-1L as we showed previously (51) and PIM-1L could in turn phosphorylate AR at Thr-850. Fig. 5C summarizes our proposed model for how PIM-1 kinases modulate AR during prostate cancer progression. Future work will be needed to delineate the dynamics of single versus dual phosphorylation of AR induced by PIM-1 kinases.
While both PIM-1 kinases were able to promote androgen-sensitive LNCaP cell growth under low androgen conditions, only PIM-1L could enhance a subset of AR target genes. The effects are more apparent under low androgen conditions, which is more relevant to castration resistance. This finding was particularly intriguing given that PIM-1L increased KLK2 expression yet had little effect on PSA expression despite both genes belonging to the Kallikrein family. PIM-1L likely regulates distinct AR targets under specific conditions, a concept that should be further explored. A ChIP assay with an anti-AR antibody supported differential recruitment of AR to the KLK2 and PSA promoters. A recent study showed that AR regulates a distinct transcription program in androgen-independent prostate cancer with enrichment in M-phase-related genes (41). It would be interesting to test in the future whether PIM-1 induced AR phosphorylation is involved in modulating AR transcription specificity. These studies will provide new insights into the mechanisms by which AR activity is regulated by low levels of androgens and growth factors and their role in castration resistance.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health (CA106504) (to Y. Q.) and pre-doctoral fellowships (W81XWH-08-1-0068) (to D. L.) and (W81XWH-10-1-0492) (to H. S.).

This article contains supplemental Figs. S1–S4.
Personal communication.
- AR
- androgen receptor
- ARE
- androgen response element
- Co-IP
- coimmunoprecipitation
- KM
- kinase-dead mutant.
REFERENCES
- 1. Chen Y., Sawyers C. L., Scher H. I. (2008) Targeting the androgen receptor pathway in prostate cancer. Curr. Opin. Pharmacol. 8, 440–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Debes J. D., Tindall D. J. (2004) Mechanisms of androgen-refractory prostate cancer. N. Engl. J. Med. 351, 1488–1490 [DOI] [PubMed] [Google Scholar]
- 3. McPhaul M. J. (2003) Factors that mediate and modulate androgen action. J. Investig. Dermatol. Symp. Proc. 8, 1–5 [DOI] [PubMed] [Google Scholar]
- 4. Culig Z., Hobisch A., Bartsch G., Klocker H. (2000) Androgen receptor–an update of mechanisms of action in prostate cancer. Urol. Res. 28, 211–219 [DOI] [PubMed] [Google Scholar]
- 5. Marcelli M., Cunningham G. R. (1999) Hormonal signaling in prostatic hyperplasia and neoplasia. J. Clin. Endocrinol. Metab. 84, 3463–3468 [DOI] [PubMed] [Google Scholar]
- 6. Chen C. D., Welsbie D. S., Tran C., Baek S. H., Chen R., Vessella R., Rosenfeld M. G., Sawyers C. L. (2004) Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 10, 33–39 [DOI] [PubMed] [Google Scholar]
- 7. Hong H., Kao C., Jeng M. H., Eble J. N., Koch M. O., Gardner T. A., Zhang S., Li L., Pan C. X., Hu Z., MacLennan G. T., Cheng L. (2004) Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer 101, 83–89 [DOI] [PubMed] [Google Scholar]
- 8. Zegarra-Moro O. L., Schmidt L. J., Huang H., Tindall D. J. (2002) Disruption of androgen receptor function inhibits proliferation of androgen-refractory prostate cancer cells. Cancer Res. 62, 1008–1013 [PubMed] [Google Scholar]
- 9. Culig Z., Klocker H., Bartsch G., Hobisch A. (2002) Androgen receptors in prostate cancer. Endocr. Relat. Cancer 9, 155–170 [DOI] [PubMed] [Google Scholar]
- 10. Gelmann E. P. (2002) Molecular biology of the androgen receptor. J. Clin. Oncol. 20, 3001–3015 [DOI] [PubMed] [Google Scholar]
- 11. Feldman B. J., Feldman D. (2001) The development of androgen-independent prostate cancer. Nat. Rev. Cancer 1, 34–45 [DOI] [PubMed] [Google Scholar]
- 12. Guo Z., Yang X., Sun F., Jiang R., Linn D. E., Chen H., Chen H., Kong X., Melamed J., Tepper C. G., Kung H. J., Brodie A. M., Edwards J., Qiu Y. (2009) A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 69, 2305–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang X., Guo Z., Sun F., Li W., Alfano A., Shimelis H., Chen M., Brodie A. M., Chen H., Xiao Z., Veenstra T. D., Qiu Y. (2011) Novel membrane-associated androgen receptor splice variant potentiates proliferative and survival responses in prostate cancer cells. J. Biol. Chem. 286, 36152–36160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo Z., Qiu Y. (2011) A new trick of an old molecule: androgen receptor splice variants taking the stage?! Int. J. Biol. Sci. 7, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gioeli D., Ficarro S. B., Kwiek J. J., Aaronson D., Hancock M., Catling A. D., White F. M., Christian R. E., Settlage R. E., Shabanowitz J., Hunt D. F., Weber M. J. (2002) Androgen receptor phosphorylation. Regulation and identification of the phosphorylation sites. J. Biol. Chem. 277, 29304–29314 [DOI] [PubMed] [Google Scholar]
- 16. Gioeli D., Paschal B. M. (2012) Post-translational modification of the androgen receptor. Mol. Cell. Endocrinol. 352, 70–78 [DOI] [PubMed] [Google Scholar]
- 17. Guo Z., Dai B., Jiang T., Xu K., Xie Y., Kim O., Nesheiwat I., Kong X., Melamed J., Handratta V. D., Njar V. C., Brodie A. M., Yu L. R., Veenstra T. D., Chen H., Qiu Y. (2006) Regulation of androgen receptor activity by tyrosine phosphorylation. Cancer Cell 10, 309–319 [DOI] [PubMed] [Google Scholar]
- 18. Kraus S., Gioeli D., Vomastek T., Gordon V., Weber M. J. (2006) Receptor for activated C kinase 1 (RACK1) and Src regulate the tyrosine phosphorylation and function of the androgen receptor. Cancer Res. 66, 11047–11054 [DOI] [PubMed] [Google Scholar]
- 19. Mahajan N. P., Liu Y., Majumder S., Warren M. R., Parker C. E., Mohler J. L., Earp H. S., Whang Y. E. (2007) Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 104, 8438–8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dai B., Chen H., Guo S., Yang X., Linn D. E., Sun F., Li W., Guo Z., Xu K., Kim O., Kong X., Melamed J., Qiu S., Qiu Y. (2010) Compensatory upregulation of tyrosine kinase Etk/BMX in response to androgen deprivation promotes castration-resistant growth of prostate cancer cells. Cancer Res. 70, 5587–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin H. K., Yeh S., Kang H. Y., Chang C. (2001) Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proc. Natl. Acad. Sci. U.S.A. 98, 7200–7205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yeh S., Lin H. K., Kang H. Y., Thin T. H., Lin M. F., Chang C. (1999) From HER2/Neu signal cascade to androgen receptor and its coactivators: a novel pathway by induction of androgen target genes through MAP kinase in prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 96, 5458–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gregory C. W., Fei X., Ponguta L. A., He B., Bill H. M., French F. S., Wilson E. M. (2004) Epidermal growth factor increases coactivation of the androgen receptor in recurrent prostate cancer. J. Biol. Chem. 279, 7119–7130 [DOI] [PubMed] [Google Scholar]
- 24. Ueda T., Mawji N. R., Bruchovsky N., Sadar M. D. (2002) Ligand-independent activation of the androgen receptor by interleukin-6 and the role of steroid receptor coactivator-1 in prostate cancer cells. J. Biol. Chem. 277, 38087–38094 [DOI] [PubMed] [Google Scholar]
- 25. Gordon V., Bhadel S., Wunderlich W., Zhang J., Ficarro S. B., Mollah S. A., Shabanowitz J., Hunt D. F., Xenarios I., Hahn W. C., Conaway M., Carey M. F., Gioeli D. (2010) CDK9 regulates AR promoter selectivity and cell growth through serine 81 phosphorylation. Mol. Endocrinol. 24, 2267–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen S., Xu Y., Yuan X., Bubley G. J., Balk S. P. (2006) Androgen receptor phosphorylation and stabilization in prostate cancer by cyclin-dependent kinase 1. Proc. Natl. Acad. Sci. U.S.A. 103, 15969–15974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cinar B., De Benedetti A., Freeman M. R. (2005) Post-transcriptional regulation of the androgen receptor by Mammalian target of rapamycin. Cancer Res. 65, 2547–2553 [DOI] [PubMed] [Google Scholar]
- 28. Litvinov I. V., Vander Griend D. J., Antony L., Dalrymple S., De Marzo A. M., Drake C. G., Isaacs J. T. (2006) Androgen receptor as a licensing factor for DNA replication in androgen-sensitive prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 103, 15085–15090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vander Griend D. J., Litvinov I. V., Isaacs J. T. (2007) Stabilizing androgen receptor in mitosis inhibits prostate cancer proliferation. Cell Cycle 6, 647–651 [DOI] [PubMed] [Google Scholar]
- 30. Cuypers H. T., Selten G., Quint W., Zijlstra M., Maandag E. R., Boelens W., van Wezenbeek P., Melief C., Berns A. (1984) Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell 37, 141–150 [DOI] [PubMed] [Google Scholar]
- 31. Wang Z., Bhattacharya N., Mixter P. F., Wei W., Sedivy J., Magnuson N. S. (2002) Phosphorylation of the cell cycle inhibitor p21Cip1/WAF1 by Pim-1 kinase. Biochim. Biophys. Acta 1593, 45–55 [DOI] [PubMed] [Google Scholar]
- 32. Leverson J. D., Koskinen P. J., Orrico F. C., Rainio E. M., Jalkanen K. J., Dash A. B., Eisenman R. N., Ness S. A. (1998) Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol. Cell 2, 417–425 [DOI] [PubMed] [Google Scholar]
- 33. Mochizuki T., Kitanaka C., Noguchi K., Muramatsu T., Asai A., Kuchino Y. (1999) Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J. Biol. Chem. 274, 18659–18666 [DOI] [PubMed] [Google Scholar]
- 34. Bachmann M., Kosan C., Xing P. X., Montenarh M., Hoffmann I., Möröy T. (2006) The oncogenic serine/threonine kinase Pim-1 directly phosphorylates and activates the G2/M specific phosphatase Cdc25C. Int. J. Biochem. Cell Biol. 38, 430–443 [DOI] [PubMed] [Google Scholar]
- 35. Koike N., Maita H., Taira T., Ariga H., Iguchi-Ariga S. M. (2000) Identification of heterochromatin protein 1 (HP1) as a phosphorylation target by Pim-1 kinase and the effect of phosphorylation on the transcriptional repression function of HP1(1). FEBS Lett. 467, 17–21 [DOI] [PubMed] [Google Scholar]
- 36. Ellwood-Yen K., Graeber T. G., Wongvipat J., Iruela-Arispe M. L., Zhang J., Matusik R., Thomas G. V., Sawyers C. L. (2003) Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4, 223–238 [DOI] [PubMed] [Google Scholar]
- 37. Wang J., Kim J., Roh M., Franco O. E., Hayward S. W., Wills M. L., Abdulkadir S. A. (2010) Pim1 kinase synergizes with c-MYC to induce advanced prostate carcinoma. Oncogene 29, 2477–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Verbeek S., van Lohuizen M., van der Valk M., Domen J., Kraal G., Berns A. (1991) Mice bearing the E mu-myc and E mu-pim-1 transgenes develop pre-B-cell leukemia prenatally. Mol. Cell Biol. 11, 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y., Wang Z., Li X., Magnuson N. S. (2008) Pim kinase-dependent inhibition of c-Myc degradation. Oncogene 27, 4809–4819 [DOI] [PubMed] [Google Scholar]
- 40. Zippo A., De Robertis A., Serafini R., Oliviero S. (2007) PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat. Cell Biol. 9, 932–944 [DOI] [PubMed] [Google Scholar]
- 41. Wang Q., Li W., Zhang Y., Yuan X., Xu K., Yu J., Chen Z., Beroukhim R., Wang H., Lupien M., Wu T., Regan M. M., Meyer C. A., Carroll J. S., Manrai A. K., Jänne O. A., Balk S. P., Mehra R., Han B., Chinnaiyan A. M., Rubin M. A., True L., Fiorentino M., Fiore C., Loda M., Kantoff P. W., Liu X. S., Brown M. (2009) Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ashcroft M., Ludwig R. L., Woods D. B., Copeland T. D., Weber H. O., MacRae E. J., Vousden K. H. (2002) Phosphorylation of HDM2 by Akt. Oncogene 21, 1955–1962 [DOI] [PubMed] [Google Scholar]
- 43. Aho T. L., Sandholm J., Peltola K. J., Mankonen H. P., Lilly M., Koskinen P. J. (2004) Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 571, 43–49 [DOI] [PubMed] [Google Scholar]
- 44. Wood N. T., Meek D. W., Mackintosh C. (2009) 14–3-3 Binding to Pim-phosphorylated Ser166 and Ser186 of human Mdm2–Potential interplay with the PKB/Akt pathway and p14(ARF). FEBS Lett. 583, 615–620 [DOI] [PubMed] [Google Scholar]
- 45. Friedmann M., Nissen M. S., Hoover D. S., Reeves R., Magnuson N. S. (1992) Characterization of the proto-oncogene pim-1: kinase activity and substrate recognition sequence. Arch. Biochem. Biophys. 298, 594–601 [DOI] [PubMed] [Google Scholar]
- 46. Wang S., Gao J., Lei Q., Rozengurt N., Pritchard C., Jiao J., Thomas G. V., Li G., Roy-Burman P., Nelson P. S., Liu X., Wu H. (2003) Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 4, 209–221 [DOI] [PubMed] [Google Scholar]
- 47. Valdman A., Fang X., Pang S. T., Ekman P., Egevad L. (2004) Pim-1 expression in prostatic intraepithelial neoplasia and human prostate cancer. Prostate 60, 367–371 [DOI] [PubMed] [Google Scholar]
- 48. Cibull T. L., Jones T. D., Li L., Eble J. N., Ann Baldridge L., Malott S. R., Luo Y., Cheng L. (2006) Overexpression of Pim-1 during progression of prostatic adenocarcinoma. J. Clin. Pathol. 59, 285–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dhanasekaran S. M., Barrette T. R., Ghosh D., Shah R., Varambally S., Kurachi K., Pienta K. J., Rubin M. A., Chinnaiyan A. M. (2001) Delineation of prognostic biomarkers in prostate cancer. Nature 412, 822–826 [DOI] [PubMed] [Google Scholar]
- 50. Saris C. J., Domen J., Berns A. (1991) The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 10, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xie Y., Xu K., Dai B., Guo Z., Jiang T., Chen H., Qiu Y. (2006) The 44 kDa Pim-1 kinase directly interacts with tyrosine kinase Etk/BMX and protects human prostate cancer cells from apoptosis induced by chemotherapeutic drugs. Oncogene 25, 70–78 [DOI] [PubMed] [Google Scholar]
- 52. Xu K., Shimelis H., Linn D. E., Jiang R., Yang X., Sun F., Guo Z., Chen H., Li W., Chen H., Kong X., Melamed J., Fang S., Xiao Z., Veenstra T. D., Qiu Y. (2009) Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell 15, 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xie Y., Xu K., Linn D. E., Yang X., Guo Z., Shimelis H., Nakanishi T., Ross D. D., Chen H., Fazli L., Gleave M. E., Qiu Y. (2008) The 44-kDa Pim-1 kinase phosphorylates BCRP/ABCG2 and thereby promotes its multimerization and drug-resistant activity in human prostate cancer cells. J. Biol. Chem. 283, 3349–3356 [DOI] [PubMed] [Google Scholar]
- 54. Jiang T., Guo Z., Dai B., Kang M., Ann D. K., Kung H. J., Qiu Y. (2004) Bi-directional regulation between tyrosine kinase Etk/BMX and tumor suppressor p53 in response to DNA damage. J. Biol. Chem. 279, 50181–50189 [DOI] [PubMed] [Google Scholar]
- 55. Blom N., Gammeltoft S., Brunak S. (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 56. Bullock A. N., Debreczeni J., Amos A. L., Knapp S., Turk B. E. (2005) Structure and substrate specificity of the Pim-1 kinase. J. Biol. Chem. 280, 41675–41682 [DOI] [PubMed] [Google Scholar]
- 57. Qian K. C., Wang L., Hickey E. R., Studts J., Barringer K., Peng C., Kronkaitis A., Li J., White A., Mische S., Farmer B. (2005) Structural basis of constitutive activity and a unique nucleotide binding mode of human Pim-1 kinase. J. Biol. Chem. 280, 6130–6137 [DOI] [PubMed] [Google Scholar]
- 58. McInerney E. M., Rose D. W., Flynn S. E., Westin S., Mullen T. M., Krones A., Inostroza J., Torchia J., Nolte R. T., Assa-Munt N., Milburn M. V., Glass C. K., Rosenfeld M. G. (1998) Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12, 3357–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Taneja S. S., Ha S., Swenson N. K., Huang H. Y., Lee P., Melamed J., Shapiro E., Garabedian M. J., Logan S. K. (2005) Cell-specific regulation of androgen receptor phosphorylation in vivo. J. Biol. Chem. 280, 40916–40924 [DOI] [PubMed] [Google Scholar]
- 60. Lin H. K., Wang L., Hu Y. C., Altuwaijri S., Chang C. (2002) Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 21, 4037–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liang H., Hittelman W., Nagarajan L. (1996) Ubiquitous expression and cell cycle regulation of the protein kinase PIM-1. Arch. Biochem. Biophys. 330, 259–265 [DOI] [PubMed] [Google Scholar]
- 62. Morishita D., Katayama R., Sekimizu K., Tsuruo T., Fujita N. (2008) Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 68, 5076–5085 [DOI] [PubMed] [Google Scholar]
- 63. Bhatt A. S., Erdjument-Bromage H., Tempst P., Craik C. S., Moasser M. M. (2005) Adhesion signaling by a novel mitotic substrate of src kinases. Oncogene 24, 5333–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Song G., Ouyang G., Bao S. (2005) The activation of Akt/PKB signaling pathway and cell survival. J. Cell Mol. Med. 9, 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Alessi D. R., Cohen P. (1998) Mechanism of activation and function of protein kinase B. Curr. Opin. Genet. Dev. 8, 55–62 [DOI] [PubMed] [Google Scholar]
- 66. Gaughan L., Logan I. R., Neal D. E., Robson C. N. (2005) Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 33, 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Feng J., Tamaskovic R., Yang Z., Brazil D. P., Merlo A., Hess D., Hemmings B. A. (2004) Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J. Biol. Chem. 279, 35510–35517 [DOI] [PubMed] [Google Scholar]
- 68. Milne D., Kampanis P., Nicol S., Dias S., Campbell D. G., Fuller-Pace F., Meek D. (2004) A novel site of AKT-mediated phosphorylation in the human MDM2 onco-protein. FEBS Lett. 577, 270–276 [DOI] [PubMed] [Google Scholar]
- 69. Gottlieb T. M., Leal J. F., Seger R., Taya Y., Oren M. (2002) Cross-talk between Akt, p53 and Mdm2: possible implications for the regulation of apoptosis. Oncogene 21, 1299–1303 [DOI] [PubMed] [Google Scholar]
- 70. Hogan C., Hutchison C., Marcar L., Milne D., Saville M., Goodlad J., Kernohan N., Meek D. (2008) Elevated levels of oncogenic protein kinase Pim-1 induce the p53 pathway in cultured cells and correlate with increased Mdm2 in mantle cell lymphoma. J. Biol. Chem. 283, 18012–18023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen W. W., Chan D. C., Donald C., Lilly M. B., Kraft A. S. (2005) Pim family kinases enhance tumor growth of prostate cancer cells. Mol. Cancer Res. 3, 443–451 [DOI] [PubMed] [Google Scholar]
- 72. Wang Z., Bhattacharya N., Meyer M. K., Seimiya H., Tsuruo T., Tonani J. A., Magnuson N. S. (2001) Pim-1 negatively regulates the activity of PTP-U2S phosphatase and influences terminal differentiation and apoptosis of monoblastoid leukemia cells. Arch. Biochem. Biophys. 390, 9–18 [DOI] [PubMed] [Google Scholar]
- 73. Lin H. K., Altuwaijri S., Lin W. J., Kan P. Y., Collins L. L., Chang C. (2002) Proteasome activity is required for androgen receptor transcriptional activity via regulation of androgen receptor nuclear translocation and interaction with coregulators in prostate cancer cells. J. Biol. Chem. 277, 36570–36576 [DOI] [PubMed] [Google Scholar]
- 74. Gregory C. W., Johnson R. T., Jr., Mohler J. L., French F. S., Wilson E. M. (2001) Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Res. 61, 2892–2898 [PubMed] [Google Scholar]
- 75. Kim O., Jiang T., Xie Y., Guo Z., Chen H., Qiu Y. (2004) Synergism of cytoplasmic kinases in IL6-induced ligand-independent activation of androgen receptor in prostate cancer cells. Oncogene 23, 1838–1844 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.