Abstract
Colorectal cancer (CRC) remains a major cause of cancer mortality worldwide. Murine models have yielded critical insights into CRC pathogenesis, but they often fail to recapitulate advanced-disease phenotypes, notably metastasis and chromosomal instability (CIN). New models are thus needed to understand disease progression and to develop therapies. We sought to model advanced CRC by inactivating two tumor suppressors that are mutated in human CRCs, the Fbw7 ubiquitin ligase and p53. Here we report that Fbw7 deletion alters differentiation and proliferation in the gut epithelium and stabilizes oncogenic Fbw7 substrates, such as cyclin E and Myc. However, Fbw7 deletion does not cause tumorigenesis in the gut. In contrast, codeletion of both Fbw7 and p53 causes highly penetrant, aggressive, and metastatic adenocarcinomas, and allografts derived from these tumors form highly malignant adenocarcinomas. In vitro evidence indicates that Fbw7 ablation promotes genetic instability that is suppressed by p53, and we show that most Fbw7−/−; p53−/− carcinomas exhibit a CIN+ phenotype. We conclude that Fbw7 and p53 synergistically suppress adenocarcinomas that mimic advanced human CRC with respect to histopathology, metastasis, and CIN. This model thus represents a novel tool for studies of advanced CRC as well as carcinogenesis associated with ubiquitin pathway mutations.
INTRODUCTION
Colorectal cancers (CRCs) progress through well-characterized histological stages as they develop from benign adenomas to metastatic adenocarcinomas (8). Mutations in early adenomas target genes such as adenomatous polyposis coli (Apc) and KRAS, whereas later mutations affect pathways involving p53, transforming growth factor β (TGF-β), and phosphatidylinositol-3′-kinase, among others (9, 41). CRCs contain ∼80 gene mutations per tumor, although only ∼15 of these are thought to be causal or driver mutations (41). CRCs frequently exhibit genomic instability, which may facilitate the acquisition of mutations that drive progression. Three forms of genomic or epigenomic instability define CRC subclasses: impaired DNA mismatch repair (MMR), aberrant DNA CpG methylation, and most commonly, chromosomal instability (CIN) (16, 31).
CRC has been extensively studied in murine models, and Apc mutations are often utilized to initiate tumorigenesis. ApcMin is the most common allele used and typically causes numerous adenomas in the small intestine and fewer adenomas in the colon (34). Disease incidence and latency in Apc models can be modified by additional oncogenic mutations, but lesions rarely progress beyond adenomas, in part because mice die from an overwhelming benign tumor burden (15). MMR gene mutations in mice also produce intestinal tumors, including adenocarcinomas, but most Apc and MMR models rarely exhibit metastasis or CIN (34). Although more-recent approaches reproduce some of these phenotypes, robust models of invasive, metastatic, and CIN+ CRCs are needed (10, 12, 37).
Fbw7 is the substrate receptor of a ubiquitin ligase that degrades more than a dozen proteins, including products of proto-oncogenes that are activated when Fbw7 is disabled (e.g., Myc, Notch, cyclin E, Jun) (40). Accordingly, Fbw7 is a tumor suppressor that is inactivated by deletions and point mutations in tumors (1). Fbw7 was identified as the fourth most commonly mutated gene in CRCs and is involved in 8 to 10% of cases (14, 28, 41). Fbw7 pathway mutations cause genomic instability and induce p53 activity. This homeostatic p53 response limits the consequences of Fbw7 loss, including genomic instability, and may underlie the findings that combined Fbw7 and TP53 mutations accelerate leukemogenesis (17, 20, 23, 25, 27, 28, 33). Conditional approaches are used to study Fbw7 null adult tissues because constitutive Fbw7 loss is embryonic lethal (35, 38). Fbw7 loss impacts cell division and differentiation in stem cell compartments, and Notch, Myc, and Jun have been implicated in these phenotypes (11, 20, 30, 36). Fbw7 deletion in hematopoietic cells is sufficient to cause blood cancers, but this has not been seen in other organs. Indeed, although Fbw7 mutations are found in early human adenomas, gut-specific Fbw7 deletion does not cause intestinal tumors in mice, although it does alter adenoma latency and incidence in ApcMin mice (2, 30).
Because the p53 pathway suppresses the effects of Fbw7 loss (and both genes are mutated in human CRCs), we sought to develop a model of advanced CRC based upon deletions of these two tumor suppressors. Remarkably, p53 loss fundamentally altered the impact of Fbw7 deletion in the gut, and deletion of both genes caused adenocarcinomas that frequently metastasized to lymph nodes and liver and often exhibited CIN. Notably, these tumors mimic critical phenotypes of advanced human CRC.
MATERIALS AND METHODS
Mice.
The floxed Fbw7 (Fbxw7tm1Kei) strain was obtained from A. Balmain (27). The floxed p53 (Trp53tm1Brn/J) and Villin-Cre [Tg(Vil-cre)997Gum] strains were purchased from Jackson Laboratory (13, 19). All strains were backcrossed onto C57BL/6 at least 5 generations prior to transfer to our laboratory, and experimental mice were segregated for the C57BL/6J and S129 genomes at an approximate 90:10 ratio. Animal studies and all animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) and were carried out at the Fred Hutchinson Cancer Research Center (FHCRC). To analyze growth of FPV cell lines in vivo, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (originally obtained from Jackson Laboratory and bred at FHCRC) were treated with low-dose radiation (275 rads) prior to injection of 5 million tumor cells subcutaneously (n ≥ 2 per cell line) (FPV1, FPV2, and FPV3) according to standard protocols. Animals with subcutaneous tumors were sacrificed when masses were palpable, generally by 4 weeks postinjection.
Pathology.
Mice were sacrificed via carbon dioxide inhalation, and full necropsies were performed according to standard techniques. In all animals, the gastrointestinal tract (from stomach to anus) and the cecum were harvested, flushed with ice-cold phosphate-buffered saline (PBS), opened longitudinally, divided into 4 sections (proximal small intestine, mid-small intestine, distal small intestine, and colon) and fixed flat on bibulous paper in 10% formalin for at least 24 h at room temperature. After fixation, intestines were rolled and stored in 70% ethanol until submitted for pathological evaluation. Individual tumors were sometimes dissected and fixed separately. A full gross inspection of all animals, including lymph nodes, breast tissue, thymus, lungs, heart, stomach, liver, spleen, pancreas, kidneys, and reproductive organs, was performed. In a subset of representative animals (>10 per genotype), all tissues were fixed in 10% formalin for 7 days and then submitted for pathological evaluation. In subsequent necropsies, all tissues were examined grossly and any abnormal tissues were submitted for pathology. After fixation, samples were submitted to the Experimental Histopathology Core Facility at the FHCRC, where tissues were processed, embedded in paraffin, cut onto slides, and stained according to standard techniques. The stains used included hematoxylin/eosin (H&E), periodic acid-Schiff (PAS), and Alcian blue (AB). In some cases, tumors were harvested and embedded in OCT compound (Tissue-Tek) and frozen at −80°C. Slides were cut from these blocks both for hematoxylin/eosin staining and for laser capture microdissection with an Arcturus Veritas laser capture microdissection system to enrich for tumor tissue. All tissue slides were reviewed by a board-certified veterinary pathologist (S. E. Knoblaugh).
Immunohistochemistry.
Immunohistochemistry was performed by the Experimental Histopathology Core Facility at the FHCRC. Four-micrometer sections were cut, deparaffinized, and rehydrated in Dako wash buffer (Carpinteria, CA). Steam antigen retrieval was performed in a Black and Decker steamer. Ki-67 slides were steamed for 40 min and β-catenin slides were steamed for 20 min in preheated target retrieval solution (pH 6; Dako) and cooled for 20 min. Lysozyme slides were treated with proteinase K (Dako) for 5 min. Slides were rinsed 3 times in wash buffer, and all subsequent staining steps were performed at room temperature using the Dako Autostainer. Endogenous peroxide activity was blocked using 3% H2O2 for 8 min, followed by protein blocking with 0.25% casein and 0.1% Tween 20 in Tris-buffered saline (TBS) for 10 min. In addition to protein blocking, the Ki-67 slides were blocked with biotin block (Dako) for 10 min. Ki-67 was detected using a rat monoclonal antibody from Dako (Tec-3 M7249) at a dilution of 1:25 for 30 min, followed by biotinylated goat anti-rat antibody at 1:200 for 30 min (112-065-167; Jackson ImmunoResearch) and horseradish peroxidase (HRP)-labeled streptavidin (016-030-084; Jackson ImmunoResearch) at 1:2,000 for 30 min. Lysozyme was detected using rabbit polyclonal antibody from Dako (A0099) at 1:200 for 30 min, followed by Mach 2 anti-rabbit HRP-labeled polymer (Biocare Medical, Walnut Creek, CA) for 30 min. β-Catenin staining was detected using a mouse monoclonal antibody from BD Biosciences (610154; San Jose, CA) at a dilution of 1:100, conjugated to a rabbit anti-mouse Fab fragment (Jackson ImmunoResearch; 315-007-003), followed by Mach 2 anti-rabbit HRP-labeled polymer for 30 min. The staining for all slides was visualized with 3,3′-diaminobenzidine (DAB) (Dako) for 8 min, and the sections were counterstained with hematoxylin (Dako) for 2 min. Concentration-matched isotype control slides were run for each tissue sample (Jackson ImmunoResearch Laboratories).
Digital imaging.
All images were obtained using a Nikon Eclipse 50i microscope and a Digital Sight DS-L1 camera. Images were imported into Photoshop (Adobe Systems), and the white balance was automatically adjusted using the levels tool.
Quantitation of proliferation and differentiation in normal intestines.
To evaluate proliferation and differentiation in normal tissues, intestinal rolls from mice of each genotype were stained for lysozyme (to stain Paneth cells) (n = 2 per genotype), PAS/AB (to stain goblet cells) (n = 3 per genotype), or Ki-67 (to stain proliferating cells) (n = 3 per genotype) as outlined above. To quantitate Paneth cells, stained sections were microscopically examined using the 10× objective. The positive crypts per field were counted for at least 5 fields. Goblet cells were quantitated by counting 30 villi per mouse, and Ki-67-positive cells were counted in 25 crypts per mouse, both using the 20× objective.
Immunoblots.
Small intestines were harvested from mice, flushed extensively with PBS, and opened longitudinally. To enrich for crypt cells, we modified a published protocol (22). Briefly, intestines were gently scraped with a microscope slide. They were then cut into ∼1-cm pieces, incubated in Hanks' balanced salt solution (HBSS) and 8 mM EDTA for 5 min at room temperature, and then shaken vigorously. The intact tissue pieces were allowed to settle, and the supernatant was discarded. These initial steps serve to remove the majority of the villi from the intestine pieces. Next, the tissue pieces were resuspended in 10 ml of ice-cold HBSS and 8 mM EDTA in 15-ml conical tubes and then incubated at 4 degrees with constant rotation for a total of 45 min. Every 15 min, the tubes were shaken vigorously for 2 min. At the end of this incubation, the intact tissue fragments were removed with forceps and the remaining material was pelleted in a refrigerated centrifuge for 5 min at 1,000 × g. The resulting pellet (∼500 μl of volume) was highly enriched for crypt cells. It was resuspended in PBS and equally aliquoted to 4 microcentrifuge tubes, followed by a second spin at 1,000 × g at 4 degrees. The supernatant was removed, and pellets were stored at −80°C.
For immunoblotting, cell pellets were lysed in TENT buffer (50 mM Tris-Cl [pH 8.0], 2 mM EDTA, 150 mM NaCl, 1% Triton X-100) and analyzed using standard techniques. The following primary antibodies were used: c-Myc (a mixture of monoclonal antibodies 143 and 274, both provided by N. Ikegaki, University of Illinois, Chicago, IL) (6), Myc-pT58 (ab28842; Abcam), cyclin E (26), c-Jun (H-79; Santa Cruz Biotechnology), transforming growth factor interacting factor (TGIF) (H-172; Santa Cruz Biotechnology), p53 (1C12; Cell Signaling Technology), bax (1063-1; Epitomics), and gamma tubulin (C-20; Santa Cruz Biotechnology). Cyclin E-associated kinase assays were performed as previously described using histone H1 as the substrate (5).
qRT-PCR.
Crypt fractions were isolated as described above, and total RNA was purified from cell pellets using TRIzol (Invitrogen). cDNA was synthesized with a high-capacity cDNA reverse transcription kit (Applied Biosystems), and quantitative reverse transcription-PCR (qRT-PCR) was performed using Platinum SYBR green qPCR SuperMix UDG (Invitrogen) with an ABI 7900HT real-time PCR system. Data were analyzed using SDS 2.3 software (Applied Biosystems). Results were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression and compared across genotypes. The results are expressed as fold change relative to the control genotype (FP). The following primers were used (forward primer listed first in each case): Atoh1 (TATCCCGTCCTTCAACAACGA and TGGTCATTTTTGCAGGAAGCT), Myc (TCCCTGAATTGGAAAACAACG and TGCTCGTCTGCTTGAATGGA), Fbxw7 (GAGACTTCATCTCCTTGCTTCCTAAA and CGCTTGCAGCAGGTCTTTG), Gapdh (GCAAAGTGGAGATTGTTGCCA and ATTTGCCGTGAGTGGAGTCAT), Hes1 (TCAACACGACACCGGACAAA and TTATTCTTGCCCTTCGCCTC), Hes5 (GGTACAGTTCCTGACCCTGCA and CCGCTGGAAGTGGTAAAGCA), Insm1 (CAGGTGATCCTCCTTCAGGT and CTCTTTGTGGGTCTCCGAGT), Lgr5 (CCAATGGAATAAAGACGACGGCAACA and GGGCCTTCAGGTCTTCCTCAAAGTCA), Neurod1 (CCTGCGCTCAGGCAAAA and GCTGGGACAAACCTTTGCA), ngn3 (CCGGATGACGCCAAACTTA and GAGTCAGTGCCCAGATGTAGTTGT), and Trp53 (GCAACTATGGCTTCCACCTG and CTCCGTCATGTGCTGTGACT).
Flow cytometry.
All analyses were performed in the flow cytometry resource at the University of Washington. For formalin-fixed paraffin-embedded (FFPE) tumors, four 60-μm sections were cut, dewaxed, rehydrated, and digested with 1% pepsin for 1 h at 37°C. Samples were washed in bovine serum albumin saline-Tris buffer (ph 7.8), filtered though nylon mesh, and resuspended in isotonic (pH 7.4) buffered solution with 0.1% Nonidet P-40 detergent (NP-40) and 10 μg/ml diamidino-2-phenylindole (DAPI). Flow cytometric analysis was performed on a Cytopeia InFlux cytometer using UV excitation. A known diploid cell line and normal cells contained within each tumor were used to determine the position of the diploid peak. A total of 50,000 cells were analyzed, if available, and in all cases, acceptable histograms contained at least 10,000 cells and a coefficient of variation (CV) below 6.0%. DNA content and cell cycles were analyzed using MultiCycle software (Phoenix Flow Systems, San Diego, CA). For FPV cell lines, cell pellets of approximately 500,000 cells were resuspended in 1,000 μl of an isotonic (pH 7.4) buffered solution with 0.1% NP-40 detergent and 10 μg/ml DAPI and analyzed as described above. Normal mouse kidney cells were used to determine the position of the diploid peak.
Derivation of FPV cell lines and tumor allografting.
Tumors were rinsed 5 times with Dulbecco's modified Eagle medium (DMEM) containing 10% serum as well as penicillin-streptomycin (pen-strep) and gentamicin. After washing, tumors were minced with a razor blade, scraped into 10 ml of 0.5% trypsin-EDTA, and incubated at 37 degrees for 1 h. At the end of this incubation, tissue fragments were put through a 25-gauge needle several times to further break up cell clumps. Cells were pelleted for 5 min at 800 × g, resuspended in DMEM with 20% serum and the antibiotics listed above, and plated into tissue culture dishes. These cells were considered passage 0 (p0). Cells were allowed to grow to confluence and then were harvested via trypsinization and replated into new dishes. After initial passage, cells were grown in DMEM with 10% serum and pen-strep. Cells were frozen back at various passages (p5, p10, and p15). After p20, cells were considered to be stable cell lines.
Array CGH.
A total of 50 ng of DNA was whole-genome amplified (WGA) using the GenomePlex WGA kit (Sigma). A total of 500 ng each of the amplified control and the tumor DNA was fluorescently labeled with either Cy3 or Cy5 using the genomic DNA ULS labeling kit (Agilent Technologies). The labeled control and tumor DNAs were combined, and mouse Cot-1, hybridization buffer, and blocking agent were added. The hybridization mixture was heated to 95°C for 3 min, followed by incubation at 37°C for 30 min. An Agilent comparative genomic hybridization (CGH) block was then added, and the entire mixture was hybridized to Agilent SurePrint G3 mouse CGH 4×180K microarrays at 65°C for 24 h with rotation. Hybridization was followed by washes using Agilent array CGH (aCGH) commercial wash solutions according to Agilent's aCGH procedures. Arrays were subsequently scanned using the Agilent microarray scanner system, and feature intensity data were extracted from the images using the Agilent feature extraction software.
RESULTS
Conditional deletion of p53 and Fbw7 causes metastatic intestinal adenocarcinomas.
Fbw7flox/flox mice were crossed with a Villin-Cre strain to produce Fbw7flox/flox; Villin-Cre mice (termed FV), resulting in widespread Fbw7 deletion in the intestinal epithelium which was confirmed by genotyping and mRNA expression (19, 27) (data not shown and see Fig. 2F). Homozygous FV mice followed for 18 months exhibited previously described abnormal epithelial phenotypes associated with Fbw7 loss (see Fig. 2) but did not develop intestinal neoplasms (2, 30).
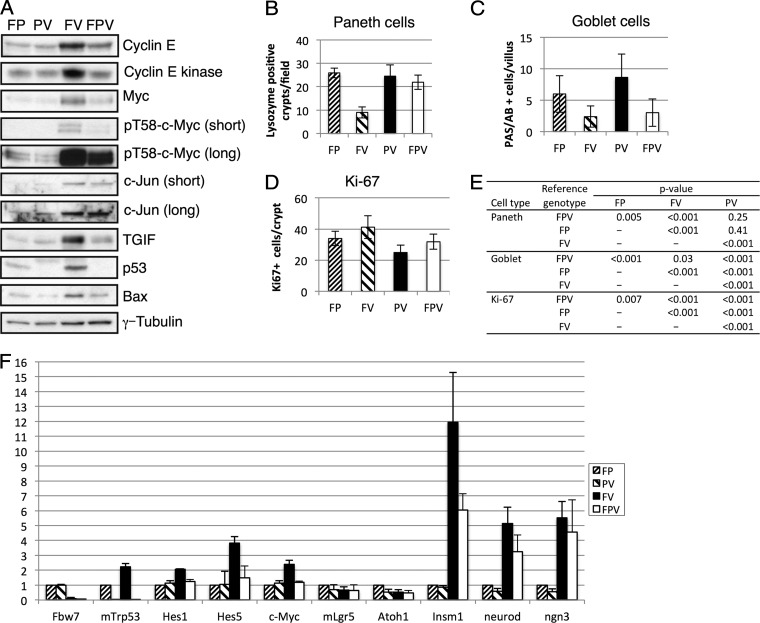
Fig 2.
p53 status impacts Fbw7 substrate regulation, proliferation, and cell fate anomalies in the Fbw7 null gut. (A) Western blot analyses of the indicated proteins in lysates obtained from intestinal epithelial cells of the indicated genotypes. (B to E) Quantitated immunostaining of intestines is shown for Paneth cells (lysozyme) (B), goblet cells (combined periodic acid-Schiff and Alcian blue) (C), and proliferating cells (Ki-67) (D). The statistical significance of these findings is shown in panel E. (F) Real-time PCR analyses of the indicated genes in gut epithelial cells from all four genotypes. Triplicate runs of three biologic replicates were performed for each genotype, and data were normalized with GAPDH and expressed relative to the data for the control genotype (FP).
We interbred mice containing a floxed p53 allele (p53flox/flox) with Fbw7flox/flox and Villin-Cre mice to delete p53 (p53flox/flox; Villin-Cre [termed PV]) or both Fbw7 and p53 (Fbw7flox/flox; p53flox/flox; Villin-Cre [termed FPV]) from the gut (13). Compared with control mice (Fbw7flox/flox; p53 flox/flox [termed FP]) or mice containing only an Fbw7 (FV) or p53 (PV) deletion, FPV animals showed highly significantly decreased survival and a dramatic intestinal cancer phenotype (Fig. 1). Most of the evaluable animals in a cohort of 51 mice (28/51, 55%) developed invasive and highly malignant adenocarcinomas (Fig. 1A). Tumors were detected at between 43 and 101 weeks of age, when mice were sacrificed after becoming premorbid (median, 70 weeks). Necropsies usually demonstrated a single intestinal tumor without additional neoplasms or polyposis, although a few older mice (greater than 80 weeks) had multiple tumors.
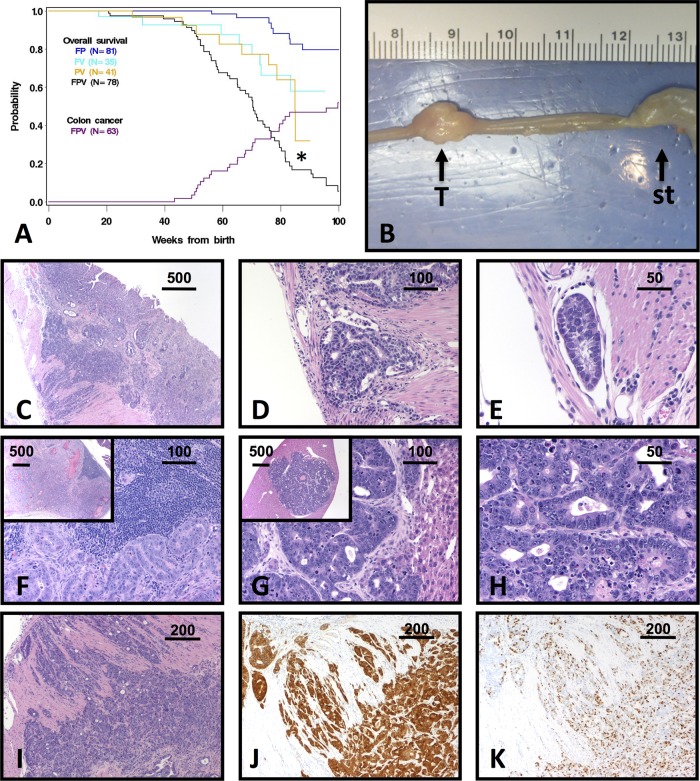
Fig 1.
Tumorigenesis in FPV mice. (A) Kaplan-Meier overall survival curves for the indicated genotypes, with the cumulative incidence of colon cancer superimposed (purple curve). *, P < 0.01 for FPV compared to all other groups by a log rank test (FPV versus FP, P < 0.001; FPV versus FV, P = 0.006; FPV versus PV, P = 0.005; FP versus FV, P < 0.001; FP versus PV, P = 0.001). (B) Gross appearance of an FPV tumor (T). st, stomach. The ruler shown is in cm. (C to I) Representative FPV tumor histopathology is shown. All sections are stained with hematoxylin and eosin except those in panels J and K. (C) Low-power image of an infiltrative adenocarcinoma. (D and E) High-power image of the section from panel C, showing penetration of neoplastic glands into the muscularis (D) and lymphatics (E). (F) Lymph node metastasis. The architecture is effaced by neoplastic glands (inset, low-power view). (G and H) Low (G, inset)-, medium (G)-, and high-power (H) views of liver metastasis. (I to K) Representative FPV invasive adenocarcinoma, stained with hematoxylin and eosin (I), anti-β-catenin antibody (J), and anti-Ki-67 antibody (K). Diffuse cytoplasmic and nuclear accumulation of β-catenin is indicative of aberrant Wnt activity, whereas Ki-67 staining shows the high proliferative rate of this tumor. Scale bars are in micrometers.
FPV adenocarcinomas occurred in the small intestine (22/28), cecum (4/28), proximal colon (1/28), and rectum (1/28) and were usually grossly visible (Fig. 1B). Most tumors were classified as adenocarcinomas with transmural effacement by neoplastic glands that penetrated through the muscularis mucosa and extended into the serosa and lymphatics (Fig. 1C to E) (4). Adenocarcinomas with large lakes of tumor-produced mucin were classified as mucinous adenocarcinomas, whereas others had less glandular differentiation or were poorly differentiated (not shown). Almost all tumors exhibited uniformly positive Ki-67 immunostaining, indicating a high proliferative index (Fig. 1K). Concurrent adenomas were found in some FPV mice with adenocarcinomas (7/28, 25%) and one animal without a carcinoma (1/23, 4%). In contrast, gross or microscopic intestinal lesions were not found in any controls (FP, FV, and PV), which were sacrificed when obviously ill or at 100 weeks. FPV mice also exhibited renal cysts (5/51, 10%) and polycystic kidneys with hydronephrosis (11/51, 22%) that were not seen in any controls, likely reflecting the known expression of Villin-Cre in the renal epithelium (21). Other causes of death in FPV mice (also observed in control mice) included extraintestinal carcinomas (2/51, 4%), sarcomas (3/51, 6%), lymphomas (2/51, 4%), and unknown causes (4/51, 8%).
Human CRCs typically metastasize first to regional lymph nodes, followed by distant sites (most commonly the liver). Likewise, many FPV mice with adenocarcinomas exhibited gross and/or microscopic metastases in regional lymph nodes (10/28, 36%) and the liver (4/28, 14%) (Fig. 1F to H). All liver lesions were found in mice with concurrent lymph node metastases, suggesting that FPV tumors phenocopy patterns of human CRC metastasis. Human and mouse CRCs share activated oncogenic pathways, and we examined Wnt and TGF-β signaling in FPV tumors. Wnt pathway activation occurs in most CRCs, but some mouse CRCs are Wnt independent (37). Most FPV adenocarcinomas contained areas of intense β-catenin immunostaining, consistent with Wnt pathway activation (Fig. 1J). Furthermore, examination of 10 tumors revealed low or absent phospho-Smad2 expression, suggesting that TGF-β pathway activation is not a prominent feature of these tumors (data not shown).
Impact of Fbw7 and p53 deletions on Fbw7 substrate abundance and intestinal cell differentiation.
We isolated epithelial cells from normal intestines from age- and sex-matched animals of all four genotypes (FP, PV, FV, and FPV) and examined the abundance of Fbw7 substrates with proposed roles in colon cancer. Cyclin E, Myc, Jun, and transforming growth factor interacting factor (TGIF) were all elevated in FV mice (Fig. 2A). We also observed elevated cyclin E-associated kinase activity and increased phospho-T58 Myc abundance, both of which measure the pools of these substrates that are directly regulated by Fbw7 in FV mice (Fig. 2A). Although we could not reliably detect Notch proteins in these preparations, Notch activity was increased in FV animals, as shown by increased mRNA expression of Notch target genes (Hes1, Hes5, and Myc genes) (Fig. 2F).
Because Fbw7 depletion activates p53 in cell culture models, we examined p53 abundance in FV mice. p53 protein and mRNA expression was elevated compared to that of FP mice, indicating that Fbw7 loss in primary intestinal epithelial cells in vivo also induces p53 expression (Fig. 2A and F). However, in contrast to what is seen when cyclin E is overexpressed in mouse and human cells, p53 ser15 phosphorylation was not elevated in FV intestinal epithelia (data not shown) and p53 induction appears to result largely from increased p53 gene transcription (17, 25). We examined two p53 response genes in FV epithelial cells and found that Bax expression was elevated (Fig. 2A) whereas p21 was undetectable (data not shown). Because our previous work found a synergistic increase in cyclin E activity when both p53 and Fbw7 are inactivated in primary fibroblasts, we anticipated that p53 loss would similarly exacerbate substrate deregulation in the intestines of the FPV mice (17, 23–25). However, this was not the case. In fact, concordant p53 deletion actually reverted cyclin E and TGIF expression to near-normal levels and partly reverted Myc expression while having little impact on Jun abundance (Fig. 2A).
We next examined cell fate decisions in the crypt/villus axis in order to understand how p53 loss modifies the impact of Fbw7 loss from the gut (2, 30). FV mice exhibited markedly reduced numbers of both Paneth and goblet cells, which are specialized cells that comprise the stem cell niche and secrete mucin, respectively (Fig. 2B, C, and E). In addition to these histochemical studies, we also used quantitative PCR to study the expression of genes associated with gut differentiation. Consistent with previous reports, markers of intestinal stem cells and/or early progenitor cells were unaffected by Fbw7 status (lgr5, atoh1), whereas markers of the enteroendocrine lineage (insm1, neuroD, and ngn3) were all overexpressed in FV animals (Fig. 2F). p53 loss had a mixed impact on these Fbw7-associated differentiation phenotypes and reverted the Paneth cell and enteroendocrine anomalies in FV mice but not the reduced number of goblet cells. We also observed modest hyperproliferation in Fbw7 null crypts, as evidenced by Ki-67 immunostaining (Fig. 2D and E), and these data are similar to those of cell proliferation studies in Fbw7 null murine intestines that examined bromodeoxyuridine incorporation (30). In contrast, p53 deletion, either alone or in combination with Fbw7 loss, decreased Ki-67 staining, indicating that crypt cell proliferation per se does not correlate with tumorigenesis in FPV mice. The decreased proliferation in FPV crypts likely contributes to the decreased cyclin E expression seen in these animals, since cyclin E is expressed only in proliferating cells in the mouse (17). In sum, aspects of both the abnormal Fbw7 substrate regulation and the aberrant cellular differentiation seen in FV animals appear to be attenuated in FPV mice (see Discussion).
Establishing tumorigenic cell lines from FPV tumors.
In vitro cultures of advanced intestinal cancers from genetically defined models have great potential for studies of metastasis and therapy but are difficult to establish. To test whether FPV tumors can grow as cell lines in culture, we processed tumor samples as described in Materials and Methods. Early-passage cells derived from these tumors contained fibroblasts and other cell types in addition to tumor cells, but cytokeratin immunostaining revealed that cultures consisted largely of adenocarcinoma cells by passage 5 (data not shown). Remarkably, 3/3 FPV adenocarcinomas readily established cell lines that were p53 and Fbw7 null and that formed invasive cancers that recapitulated the histology of primary FPV adenocarcinomas when injected subcutaneously into immunocompromised mice (Fig. 3A to F).
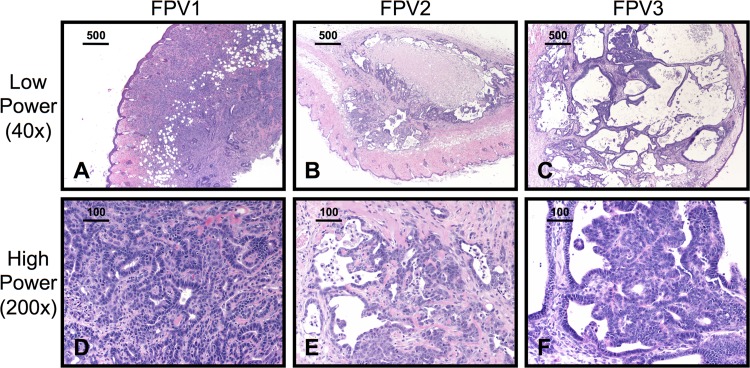
Fig 3.
FPV cell lines grow as malignant tumors in NOD/scid/gamma (NSG) mice. Three cell lines derived from FPV tumors, termed FPV1, FPV2, and FPV3, were implanted subcutaneously in NSG mice (n ≥ 2 per cell line; representative sections are shown). H&E-stained sections of subcutaneous tumors are shown at low power (A, B, and C) and high power (D, E, and F). Bars indicate the scale in micrometers.
FPV adenocarcinomas exhibit CIN.
Combined Fbw7 and p53 pathway mutations cause genomic instability in vitro, and we hypothesized that FPV tumors might also demonstrate CIN. Flow cytometry and metaphase chromosome counts revealed that each FPV cell line is aneuploid and that the number of chromosomes in each line correlated with the DNA content determined by flow cytometry (Fig. 4A to C). FPV1 and FPV3 have largely stable karyotypes (with most cells containing 72 and 52 chromosomes, respectively), whereas FPV2 exhibited greater variability. We also demonstrated ongoing genomic instability in each FPV line by scoring micronucleation, which occurred at rates similar to those found when Fbw7 and p53 are codeleted in primary human fibroblasts and other Fbw7 null cell models (Fig. 4A) (23, 28).
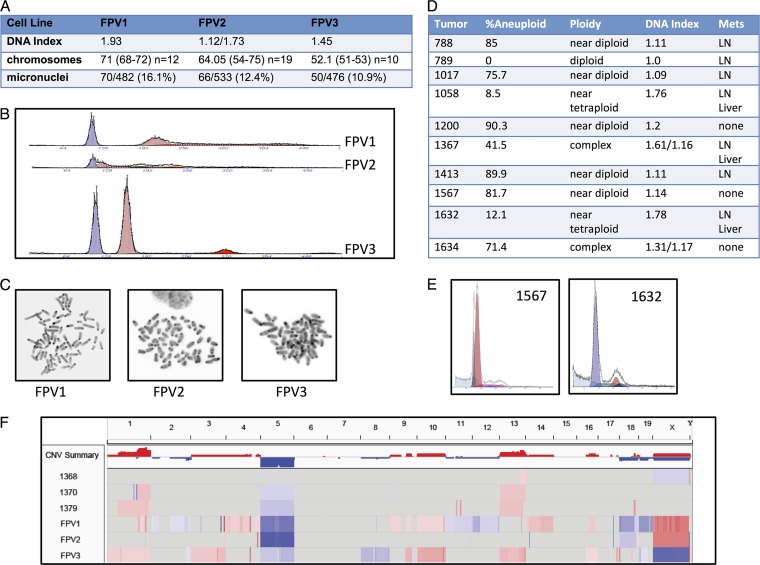
Fig 4.
Aneuploidy in FPV cell lines and tumors. (A) Table summarizing the features of the three FPV cell lines described below. The DNA index is calculated as aneuploid G1 DNA content/diploid G1 DNA content. Note that FPV2 has two separate aneuploid peaks. Mean chromosome numbers are listed, with the ranges indicated in parentheses. The number of metaphase spreads counted is indicated for each cell line. Micronucleation is expressed as a ratio of the cells with micronuclei to the total cell number. (B) Flow cytometry profiles of FPV1, FPV2, and FPV3. Each sample contained normal diploid nucleus kidney cells as controls (blue peaks) in addition to the established tumor cells (red peaks). (C) Representative FPV cell line metaphase spreads. (D) Table summarizing the ploidy of 10 FPV carcinomas determined by flow cytometry. Nuclei were isolated from fixed and embedded tumor samples and processed for flow cytometry. The percentages of total cells that were aneuploid and the DNA indexes as well as the general phenotypes of the aneuploid cells are indicated. The presence of metastases (Mets) in each animal is indicated. LN, lymph node. Note that tumors 1367 and 1634 have two separate aneuploid peaks. (E) Representative profiles of a near-diploid tumor (left) and a near-tetraploid tumor (right). Aneuploid peaks are shown in red, and normal diploid cells are shown in dark blue. (F) Array comparative genomic hybridization of three FPV tumors (after laser capture microdissection) and three FPV cell lines. DNA was labeled after whole-genome amplification and hybridized to Agilent CGH arrays. Normal tissues were used as control DNAs, and the cell lines were compared with sex-mismatched normal spleen DNA.
Because the genomic instability in FPV cell lines can reflect in vitro selection rather than primary tumor characteristics, we used flow cytometry to analyze 10 FPV adenocarcinomas and found that almost all of these primary tumors contained discrete aneuploid populations (Fig. 4D and E) (3, 39). These samples were isolated from fixed and embedded samples of grossly dissected tumors, and each contained normal diploid cells (in addition to tumor cells) that provided internal standards. Most investigators define the DNA index (DI) of CRCs determined by flow cytometry to be indicative of CIN+ disease if cells with a DI of >1.1 to 1.2 are detected (39). Thus, 5/10 FPV tumors met the strictest criteria for CIN (DNA index = 1.2 or greater) and most (8/10) fell within the more commonly used DI criterion of >1.1. Interestingly, all three tumors with DIs of >1.6 occurred in mice with gross liver metastases, suggesting that greater aneuploidy correlates with more-aggressive disease, a finding which has also been suggested for human CRC (3, 39).
Finally, we used array comparative genomic hybridization (aCGH) to examine chromosomal losses and gains in three primary tumors (after laser capture microdissection) and the three FPV cell lines. The aCGH confirmed the CIN+ phenotype observed by flow cytometry (Fig. 4F). Although aCGH does not adequately reveal large copy number variations (CNVs) in highly aneuploid cells (e.g., near-tetraploid cells) due to normalization methods, each of the samples demonstrated shared chromosomal gains and losses affecting chromosomes 1, 5, and 13 as well as unique and focal copy number alterations. However, the resolution of these arrays does not allow conclusions about specific genes that may be targeted by any of the CNVs, and these conclusions will require extensive genome-wide analyses of many additional tumors.
DISCUSSION
Mechanisms of disease progression in FPV mice.
Why did the combined deletion of Fbw7 and p53 produced advanced CRCs that fundamentally differed from the benign adenomas seen when Fbw7 was deleted in ApcMin mice (30)? This may reflect, in part, p53's role in suppressing the consequences of Fbw7 mutations. However, because most FPV mice develop a single primary tumor, tumorigenesis is rare and requires genetic events in addition to the p53 and Fbw7 deletions. It remains uncertain whether CIN is a cause or consequence of CRC progression (29, 32). Because Fbw7 and p53 mutations synergistically induce CIN in cultured cells, our similar findings in FPV adenocarcinomas may also result from the direct consequences of Fbw7 and p53 loss, and we speculate that CIN contributes to advanced disease in FPV mice. Cyclin E deregulation drives chromosomal instability in cultured cells that overexpress cyclin E or underexpress Fbw7, and it is tempting to suggest that cyclin E also contributes to CIN+ disease in FPV mice. However, the abnormally high cyclin E activity found in Fbw7 null intestinal cells was largely reverted when both p53 and Fbw7 were deleted. Thus, while cyclin E may play a role in CIN+ disease in FPV mice, its activity in the normal gut epithelium does not correlate with tumorigenesis in FPV and FV mice. It is possible, however, that abnormal cyclin E activity promotes CIN in a subset of target cells that are difficult to detect by immunoblotting and/or kinase assays.
p53 activation is a homeostatic response that protects cells against deregulated oncogenes, including the cyclin E and Myc genes, and p53 is activated by Fbw7 pathway mutations (17, 18, 25). Accordingly, p53 expression is elevated in gut epithelial cells in FV mice (Fig. 2A and F), and we speculate that p53 loss allows unrestrained oncogene activation in FPV mice. However, our finding that concordant p53 loss reverts differentiation and Fbw7 substrate abnormalities in FV mice seems paradoxical in the setting of increased tumorigenesis in FPV animals. One possibility is that p53 loss reduces populations of proliferating gut epithelial cells that overexpress Fbw7 substrates in FPV mice but allows Fbw7 to drive tumorigenesis and/or genetic instability in a population of rare, yet critical, CRC target cells. However, the mechanisms through which Fbw7 and p53 synergistically suppress gut carcinogenesis require further investigation, such as by using mouse models in which the associated genes are deleted in defined intestinal stem cells.
The extent of aneuploidy varies among the FPV tumors, and some were near diploid. These findings are reminiscent of a study of human CRCs that found Fbw7 mutations in both CIN+ and CIN− tumors (14). Thus, while Fbw7 and p53 loss leads to CIN+ CRCs in the mouse, other mechanisms almost certainly contribute to carcinogenesis in FPV mice. For example, p53 loss mitigates many of the differentiation abnormalities seen in the Fbw7 null gut epithelium, and it may also impact tumor cell phenotypes, such as metastasis. Fbw7 mutations are thought to contribute to carcinogenesis by allowing oncogenic substrates, several of which (Notch1, c-Jun, and c-Myc) have been implicated in intestinal neoplasia, to accumulate. We found that expression of cyclin E, Myc, Jun, and TGIF was elevated in FV animals and was differentially impacted by concordant p53 loss. It thus seems likely that these, and perhaps other oncogenic Fbw7 substrates, contribute to FPV carcinogenesis. However, in light of the multiple broadly acting oncogenes impacted by Fbw7 loss in the gut epithelium, we cannot presently identify specific roles played by individual substrates in FPV disease progression.
Murine models of advanced CRC.
Mouse models of advanced CRC are needed for mechanistic studies and therapy development, and recent approaches have made advances in this area. For example, intracolonic delivery of Cre recombinase into mice harboring Apc and KrasG12D mutations caused colonic neoplasms (including adenocarcinomas) and liver metastases (12). The combination of a TGF-β receptor deletion with oncogenic KrasG12D also resulted in adenocarcinomas and metastases (37). Mice in which a Cdx2 Cre driver was used to delete Apc also developed colonic tumors, including locally invasive (but not metastatic) adenocarcinomas, and primary cell cultures derived from these tumors were aneuploid, implicating the CIN pathway (10). Finally, a recent study found that animals with combined deletions of both p53 and casein kinase Iα develop microinvasive intestinal carcinomas (7). FPV mice represent a new mouse model of advanced disease that mimics a unique combination of advanced human CRC phenotypes and should prove to be an important tool for future studies of the pathogenesis and treatment of metastatic and chromosomally unstable CRC.
ACKNOWLEDGMENTS
We thank Peter Rabinovitch for expert advice and interpretation of the flow cytometry data.
This work was supported by NIH grants R01CA084069 and R01CA102742 (to B.E.C.) and K08 CA109124 (to J.E.G.).
Footnotes
Published ahead of print 2 April 2012
REFERENCES
- 1. Akhoondi S, et al. 2007. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 67:9006–9012 [DOI] [PubMed] [Google Scholar]
- 2. Babaei-Jadidi R, et al. 2011. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J. Exp. Med. 208:295–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bauer KD, et al. 1993. Consensus review of the clinical utility of DNA flow cytometry in colorectal cancer. Cytometry 14:486–491 [DOI] [PubMed] [Google Scholar]
- 4. Boivin GP, et al. 2003. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology 124:762–777 [DOI] [PubMed] [Google Scholar]
- 5. Clurman BE, Sheaff RJ, Thress K, Groudine M, Roberts JM. 1996. Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes Dev. 10:1979–1990 [DOI] [PubMed] [Google Scholar]
- 6. Conacci-Sorrell M, Ngouenet C, Eisenman RN. 2010. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell 142:480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elyada E, et al. 2011. CKIα ablation highlights a critical role for p53 in invasiveness control. Nature 470:409–413 [DOI] [PubMed] [Google Scholar]
- 8. Fearon ER, Vogelstein B. 1990. A genetic model for colorectal tumorigenesis. Cell 61:759–767 [DOI] [PubMed] [Google Scholar]
- 9. Grady WM, Markowitz SD. 2002. Genetic and epigenetic alterations in colon cancer. Annu. Rev. Genomics Hum. Genet. 3:101–128 [DOI] [PubMed] [Google Scholar]
- 10. Hinoi T, et al. 2007. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 67:9721–9730 [DOI] [PubMed] [Google Scholar]
- 11. Hoeck JD, et al. 2010. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat. Neurosci. 13:1365–1372 [DOI] [PubMed] [Google Scholar]
- 12. Hung KE, et al. 2010. Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc. Natl. Acad. Sci. U. S. A. 107:1565–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jonkers J, et al. 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29:418–425 [DOI] [PubMed] [Google Scholar]
- 14. Kemp Z, et al. 2005. CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer Res. 65:11361–11366 [DOI] [PubMed] [Google Scholar]
- 15. Kwong LN, Dove WF. 2009. APC and its modifiers in colon cancer. Adv. Exp. Med. Biol. 656:85–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lengauer C, Kinzler KW, Vogelstein B. 1998. Genetic instabilities in human cancers. Nature 396:643–649 [DOI] [PubMed] [Google Scholar]
- 17. Loeb KR, et al. 2005. A mouse model for cyclin E-dependent genetic instability and tumorigenesis. Cancer Cell 8:35–47 [DOI] [PubMed] [Google Scholar]
- 18. Lowe SW, Sherr CJ. 2003. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 13:77–83 [DOI] [PubMed] [Google Scholar]
- 19. Madison BB, et al. 2002. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 277:33275–33283 [DOI] [PubMed] [Google Scholar]
- 20. Matsuoka S, et al. 2008. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 22:986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maunoury R, et al. 1992. Developmental regulation of villin gene expression in the epithelial cell lineages of mouse digestive and urogenital tracts. Development 115:717–728 [DOI] [PubMed] [Google Scholar]
- 22. Merlos-Suarez A, et al. 2011. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8:511–524 [DOI] [PubMed] [Google Scholar]
- 23. Minella AC, Grim JE, Welcker M, Clurman BE. 2007. p53 and SCFFbw7 cooperatively restrain cyclin E-associated genome instability. Oncogene 26:6948–6953 [DOI] [PubMed] [Google Scholar]
- 24. Minella AC, et al. 2008. Cyclin E phosphorylation regulates cell proliferation in hematopoietic and epithelial lineages in vivo. Genes Dev. 22:1677–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Minella AC, et al. 2002. p53 and p21 form an inducible barrier that protects cells against cyclin E-cdk2 deregulation. Curr. Biol. 12:1817–1827 [DOI] [PubMed] [Google Scholar]
- 26. Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. 1995. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 15:2612–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Onoyama I, et al. 2007. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J. Exp. Med. 204:2875–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rajagopalan H, et al. 2004. Inactivation of hCDC4 can cause chromosomal instability. Nature 428:77–81 [DOI] [PubMed] [Google Scholar]
- 29. Rusan NM, Peifer M. 2008. Original CIN: reviewing roles for APC in chromosome instability. J. Cell Biol. 181:719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sancho R, et al. 2010. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology 139:929–941 [DOI] [PubMed] [Google Scholar]
- 31. Shen L, et al. 2007. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc. Natl. Acad. Sci. U. S. A. 104:18654–18659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sieber OM, Heinimann K, Tomlinson IP. 2003. Genomic instability—the engine of tumorigenesis? Nat. Rev. Cancer 3:701–708 [DOI] [PubMed] [Google Scholar]
- 33. Spruck CH, Won KA, Reed SI. 1999. Deregulated cyclin E induces chromosome instability. Nature 401:297–300 [DOI] [PubMed] [Google Scholar]
- 34. Taketo MM, Edelmann W. 2009. Mouse models of colon cancer. Gastroenterology 136:780–798 [DOI] [PubMed] [Google Scholar]
- 35. Tetzlaff MT, et al. 2004. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. U. S. A. 101:3338–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson BJ, et al. 2008. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J. Exp. Med. 205:1395–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trobridge P, et al. 2009. TGF-β receptor inactivation and mutant Kras induce intestinal neoplasms in mice via a β-catenin-independent pathway. Gastroenterology 136:1680–1688.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsunematsu R, et al. 2004. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279:9417–9423 [DOI] [PubMed] [Google Scholar]
- 39. Walther A, Houlston R, Tomlinson I. 2008. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut 57:941–950 [DOI] [PubMed] [Google Scholar]
- 40. Welcker M, Clurman BE. 2008. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8:83–93 [DOI] [PubMed] [Google Scholar]
- 41. Wood LD, et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science 318:1108–1113 [DOI] [PubMed] [Google Scholar]