Abstract
Herpes simplex virus 1 (HSV-1) enters neurons primarily by fusion of the viral envelope with the host cell plasma membrane, leading to the release of the capsid into the cytosol. The capsid travels via microtubule-mediated retrograde transport to the nuclear membrane, where the viral DNA is released for replication in the nucleus. In the present study, the composition and kinetics of incoming HSV-1 capsids during entry and retrograde transport in axons of human fetal and dissociated rat dorsal root ganglia (DRG) neurons were examined by wide-field deconvolution microscopy and transmission immunoelectron microscopy (TIEM). We show that HSV-1 tegument proteins, including VP16, VP22, most pUL37, and some pUL36, dissociated from the incoming virions. The inner tegument proteins, including pUL36 and some pUL37, remained associated with the capsid during virus entry and transit to the nucleus in the neuronal cell body. By TIEM, a progressive loss of tegument proteins, including VP16, VP22, most pUL37, and some pUL36, was observed, with most of the tegument dissociating at the plasma membrane of the axons and the neuronal cell body. Further dissociation occurred within the axons and the cytosol as the capsids moved to the nucleus, resulting in the release of free tegument proteins, especially VP16, VP22, pUL37, and some pUL36, into the cytosol. This study elucidates ultrastructurally the composition of HSV-1 capsids that encounter the microtubules in the core of human axons and the complement of free tegument proteins released into the cytosol during virus entry.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is a highly prevalent human pathogen, infecting 60 to 90% of the world's adult population (64). It is a member of the Alphaherpesvirinae subfamily of the Herpesviridae family of enveloped DNA-containing viruses. Like other alphaherpesviruses, HSV-1 has the ability to establish lifelong infections or latency in neurons within the peripheral nervous system of its human host. Although most clinical diseases caused by HSV-1 are mild, it can cause potentially fatal encephalitis in adults and disseminated infections in neonates (11, 60). All herpesviruses consist of an electron-dense core containing double-stranded DNA enclosed within an icosahedral capsid. The capsid is surrounded by a layer of proteins known as the tegument, which consists of approximately 23 proteins. The virus is then enclosed in a host cell-derived lipid envelope containing an estimated 16 membrane proteins (30, 37, 60).
HSV-1 enters the human body via breaks in the skin or intact mucosa, where it replicates in the epithelial cells (60). The virus then enters the nerve endings of dorsal root ganglion (DRG) neurons innervating the infected tissue and is transported retrogradely along the sensory axons to the neuronal cell body, where it can undergo a limited reproduction cycle or establish latency. Periodic reactivation from the latent state results in the virus being anterogradely transported to the nerve termini, where it either causes recurrent lesions or leads to asymptomatic viral shedding (60).
Entry of HSV-1 into neurons occurs by fusion of the viral envelope with the plasma membrane (1, 53, 63). Once inside the cell, the viral capsid must travel along the axon toward the nucleus in the cell body. The capsid docks at the nuclear pores to deposit the viral DNA into the nucleus, where viral transcription and replication occur (40, 54, 65, 69, 71).
The tegument composition of the incoming alphaherpesviruses, especially HSV-1 and porcine pseudorabies virus (PrV), has been the focus of recent studies in both neuronal and nonneuronal cells. It is believed that during entry of HSV-1 and PrV, most of the tegument proteins are lost, leaving the capsid along with some associated inner tegument proteins to be transported to the cell nucleus. Most studies so far have been done with either cultured epithelial cells (9, 18, 50) or sensory neurons of avian origin (3, 38). Cryo-electron tomography of adherent cell lines and synaptosomes has shown that most of the HSV-1 tegument proteins remain at the entry site, while the incoming capsids are largely devoid of tegument density (41). During entry of PrV, the inner tegument proteins VP1/2 (pUL36) and pUL37 have been shown to remain associated with the incoming capsids, while outer tegument proteins, such as VP13/14 (pUL47), VP16 (pUL48), and VP22 (pUL49), dissociate during PrV entry into nonneuronal cells (18) and chick sensory neurons (38). A recent study using time lapse microscopy to compare the postentry viral transport of HSV-1 and PrV in chick and mouse sensory neurons showed that the transport dynamics and composition of incoming HSV-1 particles was similar to that of PrV (3).
In the present study, the composition and kinetics of incoming HSV-1 capsids during entry and retrograde transport in axons of human fetal dorsal root ganglia (DRG) and dissociated rat DRG neurons were examined by serial fixation, fluorescence microscopy, and transmission immunoelectron microscopy (TIEM). Recombinant fluorescence-tagged viruses, belonging to different strains of HSV-1 (strains F and 17), in conjunction with antibody staining for specific viral proteins were used to determine the distribution and colocalization of HSV-1 capsid and tegument proteins at various times after virus entry. The composition and kinetics of incoming HSV-1 virions were then examined up to 4 h postinfection (hpi) using a Deltavision Core image restoration system and TIEM. We were able to show that HSV-1 tegument proteins, including VP16, VP22, most of pUL37, and some pUL36, dissociated from the incoming virions. Tegument proteins, including pUL36 and some pUL37, remained associated with the capsid during entry and transit toward the nucleus in the neuronal cell body. Our TIEM data suggested that although most of the tegument proteins were lost at the cell membrane, there was also a progressive loss of residual tegument proteins as capsids moved retrogradely toward the nucleus. Most of the tegument dissociated at the plasma membrane of the axons and neuronal cell body, with further dissociation taking place within the axons and cytosol of the cell body as the capsid moved toward the nucleus.
MATERIALS AND METHODS
Cells and viruses.
Three recombinant viruses were used in parallel in this study: vUL37-GFP (green fluorescent protein)-labeled HSV-1 (strain 17), mRFP1-VP26/pUL36-GFP (HSV F-GS2945) (strain F), and mRFP1-VP26/GFP-pUL37 (HSV F-GS3245) (strain F). These viruses have been described previously (3, 4, 48, 61). vUL37-GFP was kindly provided by Frazer Rixon (MRC Virology Unit, Institute of Virology, United Kingdom). Greg Smith (Northwestern University Medical School) kindly provided HSV F-GS2945 and HSV F-GS3245.
Preparation of virus stocks.
Viruses were grown and passaged in Vero cells. Briefly, Vero cells in 150-cm2 Falcon tissue culture flasks were infected at a multiplicity of infection (MOI) of 0.01 PFU per cell. Infected cells were frozen and thawed three times and subjected to sonication in a Branson sonicator (3 times for 20 s, 100% duty cycle). The cellular debris was removed by centrifugation at 15,800 × g for 10 min at 4°C in a Sorvall RC26 Plus ultracentrifuge. Virus was then pelleted by centrifugation through a 10% sucrose cushion at 64,000 × g for 2 h at 4°C in a Beckman Coulter Optima XL-100K ultracentrifuge. The pellet was resuspended in a small volume of Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 1% fetal calf serum (FCS) (vol/vol). Viral titers were measured by plaque assays performed on Vero cells.
Antibodies for TIEM.
Antibodies were kindly provided by the following investigators: rabbit antibody against VP5 (NC1) from Gary Cohen and Roselyn Eisenberg, University of Pennsylvania (8); rabbit antibody against VP22 from Peter O'Hare, Marie Curie Research Institute, Oxted, United Kingdom (15); mouse antibody against VP16 (LP1) from Tony Minson, University of Cambridge, United Kingdom (44); rabbit antibody against pUL37 (780 antiserum) from Frank Jenkins, Uniformed Services University of the Health Sciences, MD (62); and rabbit antibodies to pUL36 and pUL37 (35) from Thomas Mettenleiter, Freidrich Loeffler Institute, Insel Riems, Germany. The gold-conjugated antibodies were purchased from British Biocell International, United Kingdom.
Preparation of human fetal DRG explants.
DRG were prepared as described previously (61) from human fetal tissue obtained at therapeutic termination following the informed consent of the mother. Sydney West Area Health Services and the University of Sydney Human Research Ethics Committee approved these protocols. The DRG were dissected, cleansed of connective tissue, placed onto Matrigel (BD Biosciences)-coated coverslips, and cultured at 37°C with 5% CO2 in neurobasal medium supplemented with 4 mM l-glutamine (Invitrogen), 2% B-27 (Invitrogen), and 7S nerve growth factor (100 ng/ml) (Sigma) for 5 to 7 days for axon outgrowth prior to HSV-1 infection.
Preparation of dissociated rat neuronal cultures.
DRG neurons were prepared from 4-day-old Wistar rat neonates as previously described (49). Briefly, DRG were dissociated in Hanks calcium- and magnesium-free solution (Invitrogen) plus 0.25% trypsin (Sigma) and 0.05% collagenase (Worthington Biomedical Co.) for 30 min at 37°C, followed by DNase (10 mg/ml) (Sigma) for 5 min at 37°C, washed twice by centrifugation at 80 × g, and passed through 35% Percoll (Sigma). The cell pellet was resuspended in 500 μl of neurobasal medium (Invitrogen), plated on Matrigel-coated plastic coverslips, and cultured at 37°C with 5% CO2 in neurobasal medium supplemented with 4 mM l-glutamine (Invitrogen), 2% B-27 (Invitrogen), and 7S nerve growth factor (100 ng/ml) (Sigma) for 3 days prior to HSV-1 infection. Sydney West Area Health Services and the University of Sydney Animal Research Ethics Committees approved the use of rat neonates.
HSV-1 infection of DRG cultures.
For all time points, neuronal cultures in 24-well plates were infected with recombinant viruses at 2.0 × 105 PFU/coverslip. The cells were incubated at 4°C for 30 min to promote virus attachment. The cultures were then returned to 37°C with 5% CO2 and incubated for 30 min postinfection (mpi) or for 2, 4, or 24 h postinfection (hpi). The shift in temperature from 4°C to 37°C allowed the bound virus particles to enter the cells and give a synchronous infection. For incubation times longer than 2 h, the virus inoculum was removed at 2 hpi and the cells were washed with neurobasal medium. The neurons were always incubated at 37°C with 5% CO2. Mock-infected cultures were incubated in the same culture medium and fixed at the same time points. HSV F-GS2945 and HSV F-GS3245 were used for fluorescence microscopy studies, and vGFP-UL37 was used for TIEM studies.
Fluorescence microscopy and image analysis.
DRG cultures were processed for fluorescence studies as previously described (47). Briefly, neuronal cultures on coverslips were fixed in 3% formaldehyde for 30 min at room temperature. This was followed by six washes in phosphate-buffered saline (PBS) before the coverslips were mounted in Prolong Gold with 4′,6′-diamidino-2-phenylindole (DAPI) on glass slides. The slides were examined using a Deltavision Core image restoration system. Images were acquired using a Photometrics CoolSnap QE camera with sequential exposures to monomeric red fluorescent protein (mRFP) (600 ms) and GFP (1 s) and were deconvolved using Sedat-Agard algorithms (24) available through the Deltavision SoftWoRx software, version 3.0.0. Images were acquired as z-stack series and represented as volume projections (minimum number of stacks was 30). Background was subtracted after capture using SoftWoRx, through a single adjustment of the levels histogram. The images were then cropped using Adobe Photoshop CS5.
The proportion of mRFP1 capsids colocalizing with GFP during entry at 2 and 4 hpi were determined by manual counting of the total number of mRFP particles in the cytosol and at the nuclear rim and then counting the number of these particles colocalizing with GFP. The results were expressed as percentage of capsids that colocalized with GFP. For these counts, an average of 10 cells were used for each virus and time point. Both original (raw) and deconvolved images were compared to ensure no artifact (loss or addition of mRFP1 or GFP fluorescent puncta) during the counting process.
Imaging of extracellular virus particles.
To characterize the extracellular virus particles of fluorescence-tagged viruses (GS2945 and GS3245), supernatants from Vero cells infected with either virus were added to glass coverslips coated with Cell Tak adhesive (BD Pharmingen). The coverslips were spun at 1,200 × g for 30 min at 4°C in a benchtop centrifuge. The coverslips were rinsed once with PBS and fixed in 3% formaldehyde for 20 min at room temperature. After two rinses in PBS, the coverslips were mounted with Prolong gold and imaged using deconvolution microscopy as described above. The proportion of mRFP capsids emitting GFP fluorescence were manually counted, and results were expressed as averages from five separate counts.
TIEM.
DRG cultures were processed by modified freeze substitution as previously described (49, 61). Briefly, coverslips with DRG cultures in situ were fixed in 4% formaldehyde and 0.1% glutaraldehyde for 1 h and then washed in PBS. Coverslips were then trimmed, dipped in 10% gelatin, and placed in cryoprotectant (2.3 M sucrose) overnight. Preparations were then frozen by rapid plunging into liquid nitrogen followed by transfer to dry methanol at −90°C, freeze substituted from methanol, embedded in Lowicryl HM20, and polymerized with UV light at −45°C for 48 h in a Reichert AFS freeze substitution system (Leica Microsystems, Austria) as previously described (48).
Immunolabeling.
Tissue sections were collected on Formvar- and Pioloform-coated gilded nickel grids and immunolabeled as described previously (48). Briefly, tissue sections were incubated with 50 mM glycine for 15 min and in blocking buffer with 10% normal serum and acetylated bovine serum albumin (BSA) for 30 min. Primary antibodies were incubated overnight at 4°C, followed by incubation with secondary antibodies conjugated to 5- to 10-nm gold particles. Steps following primary antibody incubation, including washing steps and secondary-antibody incubation, were performed using a Leica EM IGL Immunostainer (Leica Microsystems, Austria). After immunolabeling, the sections were stained using 1% uranyl acetate (in 50% ethanol) followed by Reynolds lead citrate and examined with either a Philips CM10 or CM120 BioTWIN transmission electron microscope at 80 kV or 100 kV, respectively. Images were recorded on Kodak 4489 electron microscope film or collected using a SIS Morada digital camera.
RESULTS
The present study investigated the composition and kinetics of incoming HSV-1 virions in primary cultures of human and rat DRG neurons using TIEM and fluorescence microscopy. Fluorescently tagged recombinant viruses of two HSV-1 strains were used to ensure that results were not dependent on the strain. The first virus was vUL37-GFP, which carries a GFP tag on the C terminus of pUL37 in the HSV-1 strain 17 backbone (61). The second virus carries an mRFP1 tag at the N terminus of VP26 (UL35 gene) and a GFP tag at the N terminus of pUL37 (HSV F-GS3245) (strain F) (3). The third virus is a dually fluorescent recombinant carrying mRFP1-VP26 and a GFP tag at the C terminus of pUL36 (HSV F-GS2945) (strain F) (3). The recombinant viruses F-GS2945 and GS3245 were chosen to visualize capsids during entry and infection using a Deltavision Core image restoration system, while vUL37-GFP was used for electron microscopy studies.
Tegument composition of viral capsids after entry into neurons.
TIEM was used to examine the tegument composition of incoming HSV-1 virions in cultured human fetal explant and rat DRG neurons. Axons in DRG explant cultures are approximately 4 to 5 mm in length (61) and hence are optimal for visualizing viral particles during entry and retrograde transport in axons. As rat neonates are more readily available, cultures of dissociated rat DRG neurons, which yielded pure neuronal cultures, were chosen to examine virus particles in the neuronal cell body during virus entry.
Both rat and human DRG cultures were infected with vUL37-GFP, and the cultures were fixed at 30 mpi and 2, 4, and 24 hpi. Coverslips with DRG cultures in situ were fixed and processed for electron microscopy. Serial ultrathin sections were cut parallel to the growth plane (longitudinally) in order to examine proximal, middle, and distal regions of axons in situ and to ultrastructurally examine neuronal cell bodies from the cell surface to the nucleus without disrupting the arrangement of cells and axons.
Immunogold labeling (5- or 10-nm gold particles) with antibodies to the tegument proteins VP16, VP22, pUL36, and pUL37 and capsid VP5 was performed to examine the presence of these proteins on viral particles in the axons and cell body during the initial 4 hpi. The presence and intensity of immunolabeling of viral particles were classified semiquantitatively: the presence of 3 or fewer gold particles was considered moderate to weak labeling, and the presence of 4 or more gold particles was considered strong labeling. Using extracellular virions as controls, gold label was considered to be “on” a virus particle if it was located directly on the surface of the viral particle or within two gold particles of the target membrane (in the case of enveloped capsids or extracellular particles) or the edge of cytosolic capsids (with no envelope).
Extracellular virions and cytoplasmic enveloped virions were identified both on the basis of morphology, including the presence of an electron dense core or capsid, a well-developed tegument layer, and a distinct membrane representing viral envelope, and on the basis of size (170 to 220 nm in diameter) (20). Unenveloped capsids were distinguished from axonal vesicles on the basis of their size (approximately 125 nm in diameter) (74), thicker structure of the viral capsid compared to vesicle walls, and electron-dense DNA cores (26, 27, 48). In addition, viral capsids were further identified by immunolabeling with antibodies to the major capsid protein VP5. Electron micrographs presented here in support of the findings are representative of multiple observations.
Detection of unenveloped capsids in axons and in the cytoplasm of the cell body during virus entry.
Extracellular virions bound to the plasma membrane of axons and neuronal cell bodies, as well as unenveloped capsids within axons and cell body, were observed at all time points up to 4 hpi (see Fig. 1 to 5). However, the number of extracellular virions decreased progressively with time, and a reduction of almost 90% in the number of extracellular virions was seen from 30 mpi to 4 hpi (Tables 1 and 2), indicating efficient uptake of the virus by 4 hpi. While extracellular virions were readily observed, capsids within axons and cell bodies were considerably harder to find. Hence, few such capsids could be examined and counted. An average of 2,500 axonal processes and 200 cell body profiles were examined for each time point and each label (Tables 1 and 2). Unenveloped capsids could be detected within axons as early as 30 mpi and could also be seen in the cytosol of infected neurons at the same time point. By 4 hpi, capsids could be seen at the nuclear membrane and in the cytosol of the cell body. These unenveloped capsids were similar in morphology to those previously reported in axons of HSV-1-infected neurons (27, 47–49, 61) and were not present in uninfected cultures (data not shown). Furthermore, no enveloped capsids within vesicles or partially enveloped capsids were observed at any time between 30 mpi and 4 hpi (Tables 1 and 2).
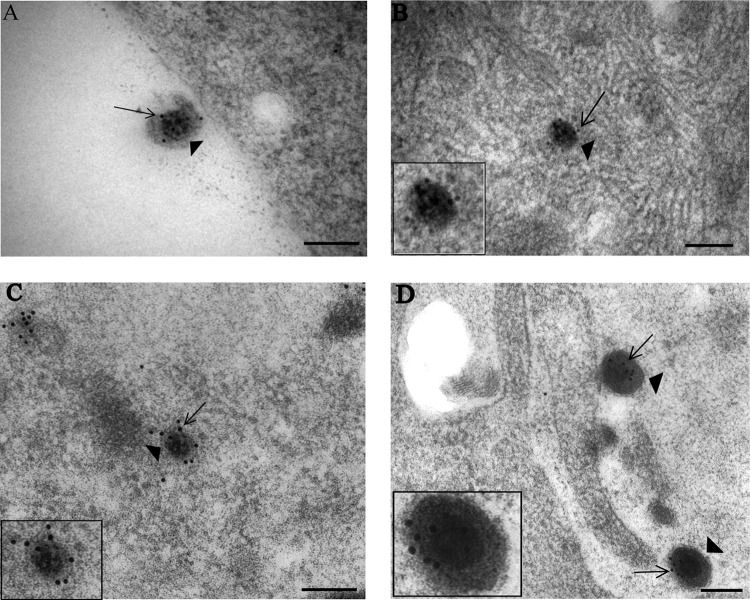
Fig 1.
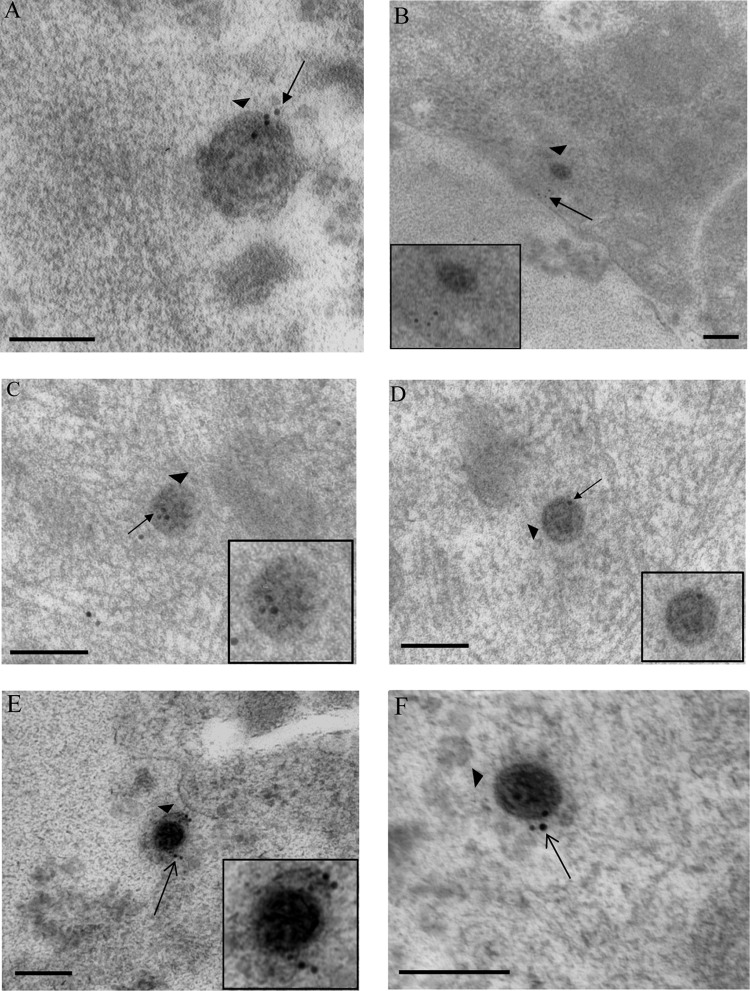
Immunogold labeling for capsid VP5 of HSV-1 particles in vUL37-GFP-infected human axons and rat DRG neurons. Coverslips with DRG cultures were processed with Lowicryl HM20, and immunogold labeling for VP5 of ultrathin sections was performed as described in Materials and Methods. (A) Extracellular virion (arrowhead) with label for VP5 (arrow) and lying close to a human DRG axon at 30 mpi. (B) Unenveloped capsid (arrowhead) within the cytosol of the cell body of a rat DRG neuron, labeled for VP5 (arrow), at 4 hpi. (C) Unenveloped capsid (arrowhead) within a neuronal cell body of a rat DRG neuron, labeled for VP5 (arrow), at 24 hpi. (D) Extracellular virions (arrowheads), labeled for VP5 (arrows) and lying close to a human axon at 24 hpi. (Insets) Enlargements of capsids in each panel. NM, nuclear membrane; PM, plasma membrane. Bars, 200 nm.
Fig 5.
Immunogold labeling for tegument protein pUL37 in human axons and rat DRG neurons infected with vUL37-GFP. (A) Extracellular virion (arrowhead), labeled for pUL37 (arrow), bound to the plasma membrane of a human DRG axon at 30 mpi. (B) Unenveloped capsid (arrowhead), labeled for pUL37 (long arrow), adjacent to the inner aspect of the plasma membrane of a human DRG axon at 30 mpi. Label for pUL37 is both on (long arrow) and off (short arrows) the capsid. (C) Unenveloped capsid (arrowhead) in the cytosol of cell body of a rat DRG neuron at 2 hpi, lying close to the plasma membrane and with label (arrow) off the capsid. (D) Unenveloped capsid (arrowhead) present deep in the cytosol of the cell body of a rat DRG neuron at 4 hpi and carrying no label for pUL37. (E and F) Extracellular virion (E) and cytoplasmic unenveloped capsid (F) (arrowheads) labeled for pUL37 (arrows) at 24 hpi. (Insets) Enlargements of virions in each panel. Bars, 200 nm.
Table 1.
Quantification of viral particles with or without label for outer tegument proteins VP16 and VP22 in infected human DRG axons and rat DRG neurons
| Particle type | Label intensityc | No. (%) of particles with label ina: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| VP16 |
VP22 |
||||||||
| Human axons |
Rat neuron cytoplasm |
Human axons |
Rat neuron cytoplasm |
||||||
| 30 mpib | 2 hpi | 4 hpi | 24 hpi | 30 mpib | 2 hpi | 4 hpi | 24 hpi | ||
| Extracellular virions | Total | 20 | 16 | 3 | 48 | 15 | 15 | 4 | 50 |
| Strong | 16 (80.0) | 12 (75.0) | 2 (66.6) | 40 (83.3) | 12 (80.0) | 11 (73.3) | 3 (75.0) | 40 (80.0) | |
| Moderate to weak | 4 (20.0) | 4 (25.0) | 1 (33.3) | 8 (16.6) | 3 (20.0) | 4 (17.6) | 1 (25.0) | 10 (20.0) | |
| None | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cytoplasmic unenveloped capsids | Total | 4 | 4 | 7 | 25 | 5 | 4 | 6 | 30 |
| Strong | 0 | 0 | 0 | 20 (80.0) | 0 | 0 | 0 | 0 | |
| Moderate to weak | 3 (75.0) | 0 | 0 | 4 (16.0) | 3 (60.0) | 1 (25.0) | 0 | 10 (33.0) | |
| None | 1 (25.0) | 4 (100) | 7 (100)d | 1 (4.0) | 2 (40.0) | 3 (75.0) | 6 (100)d | 20 (77.0) | |
| Cytoplasmic enveloped capsids | Total | 0 | 0 | 0 | 39 | 0 | 0 | 0 | 30 |
| Strong | 32 (82.0) | 24 (80.0) | |||||||
| Moderate to weak | 7 (18.0) | 5 (16.6) | |||||||
| None | 0 | 1 (3.3) | |||||||
On average, 2,500 axonal processes and 200 cell body profiles were examined for each time point and each label. Shading highlights the data for unenveloped capsids in the cytosol with no gold label after entry at 30 mpi and 2 and 4 hpi and during exit at 24 hpi.
Other time points, 2 and 4 hpi, were also examined. However, viral particles were counted only at 30 mpi.
Strong, ≥4 gold particles/virion; moderate to weak, ≤3 gold particles/virion.
The difference between the proportions of viral capsids with label for pUL36 (Table 2) and VP16 (P = 0.019) or VP22 (P = 0.028) at 4 hpi was significant (Fisher exact test).
Table 2.
Quantification of viral particles with or without label for inner tegument proteins pUL36 and pUL37 in infected human DRG axons and rat DRG neurons
| Particle type | Label intensityc | No. (%) of particles with label in: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| pUL36 |
pUL37 |
||||||||
| Human axonsa |
Rat neuron cytoplasma |
Human axonsa |
Rat neuron cytoplasma |
||||||
| 30 mpib | 2 hpi | 4 hpi | 24 hpi | 30 mpib | 2 hpi | 4 hpi | 24 hpi | ||
| Extracellular virions | Total | 38 | 15 | 4 | 71 | 25 | 17 | 3 | 60 |
| Strong | 3 (7.8) | 1 (6.6) | 0 | 6 (8.4) | 15 (60.0) | 10 (58.8) | 2 (66.6) | 41 (68.3) | |
| Moderate to weak | 25 (65.7)d | 10 (66.6)d | 3 (75.0)d | 49 (69.0)d | 6 (24.0) | 5 (29.4) | 1 (33.3) | 12 (20.0) | |
| None | 10 (26.3) | 4 (26.6) | 1 (25.0) | 16 (22.5) | 4 (16.0) | 2 (11.7) | 0 | 7 (11.6) | |
| Cytoplasmic unenveloped capsids | Total | 9 | 8 | 8 | 59 | 7 | 6 | 10 | 17 |
| Strong | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Moderate to weak | 5 (55.5)d | 4 (50.0)d | 5 (62.5)d | 40 (68.0)d | 4 (57.1) | 3 (50.0) | 2 (20.0) | 12 (70.5) | |
| None | 4 (44.5) | 4 (50.0) | 3 (37.5)e | 19 (32.0) | 3 (42.9) | 3 (50.0) | 8 (80.0) | 5 (29.4) | |
| Cytoplasmic enveloped capsids | Total | 0 | 0 | 0 | 43 | 0 | 0 | 0 | 44 |
| Strong | 6 (14.9) | 30 (68.1) | |||||||
| Moderate to weak | 28 (65.0) | 7 (16.0) | |||||||
| None | 9 (20.9) | 7 (16.0) | |||||||
On average, 2,500 axonal processes and 200 cell body profiles were examined for each time point and each label. Shaded areas highlight the data for unenveloped capsids in the cytosol with moderate to weak or no gold label after entry at 30 min, 2 and 4 hpi, and during exit at 24 hpi.
Other time points, 2 and 4 hpi, were also examined. However, viral particles were counted only at 30 mpi.
Strong, ≥4 gold particles/virion; moderate to weak, ≤3 gold particles/virion.
Viral unenveloped capsids at 30 mpi and 2 and 4 hpi carried weak label (on average, 1.0 gold particle) for pUL36, whereas unenveloped capsids at 24 hpi or extracellular virions (at any time point) had on average 3.0 gold particles for pUL36.
The difference between the proportions of viral capsids with label for pUL36 and VP16 (P = 0.019) or VP22 (P = 0.028) (Table 1) at 4 hpi was significant (Fisher exact test).
Fully enveloped and partially enveloped capsids enclosed within vesicles were, however, observed in varicosities and growth cones at 24 hpi, in accordance with our previous published findings (48, 61).
In addition to morphology, enveloped and unenveloped viral capsids in axons and neuronal cell bodies were identified by immunolabeling with antibodies to capsid protein VP5.
At early time points postinfection, strong label for capsid VP5 was present on extracellular virions that were either free or bound to the plasma membrane of axons and neuronal cell bodies (Fig. 1A). Label for VP5 could also be seen on unenveloped capsids within axons as previously published (47) and on unenveloped capsids in the cytosol at all time points postinfection, including 24 hpi (Fig. 1B and C). The density of label for VP5 (4 or 5 gold particles) on unenveloped capsids remained consistent and was similar to that on extracellular virions at early and late time points of infection (Fig. 1). Labeling for VP5 could not be performed for all experiments in order to allow labeling for other proteins. However, capsids could be readily identified by their size and morphology.
Dissociation of outer tegument VP16 from viral particles during entry.
At all time points examined during infection, about 75% of extracellular viral particles observed carried strong label for VP16 (Fig. 2A) (Table 1). These particles were usually found adjacent to cell bodies of infected neurons as well as to axonal processes. At 30 mpi, unenveloped capsids (75%; 3/4) (Table 1) carrying weak or no label for VP16 (Fig. 2B and C) were observed adjacent to the inner aspect of the axonal membrane. Label for VP16 was also present along the plasma membrane and in the cytosol of axons, in close proximity to the incoming capsids (Fig. 2B and C).
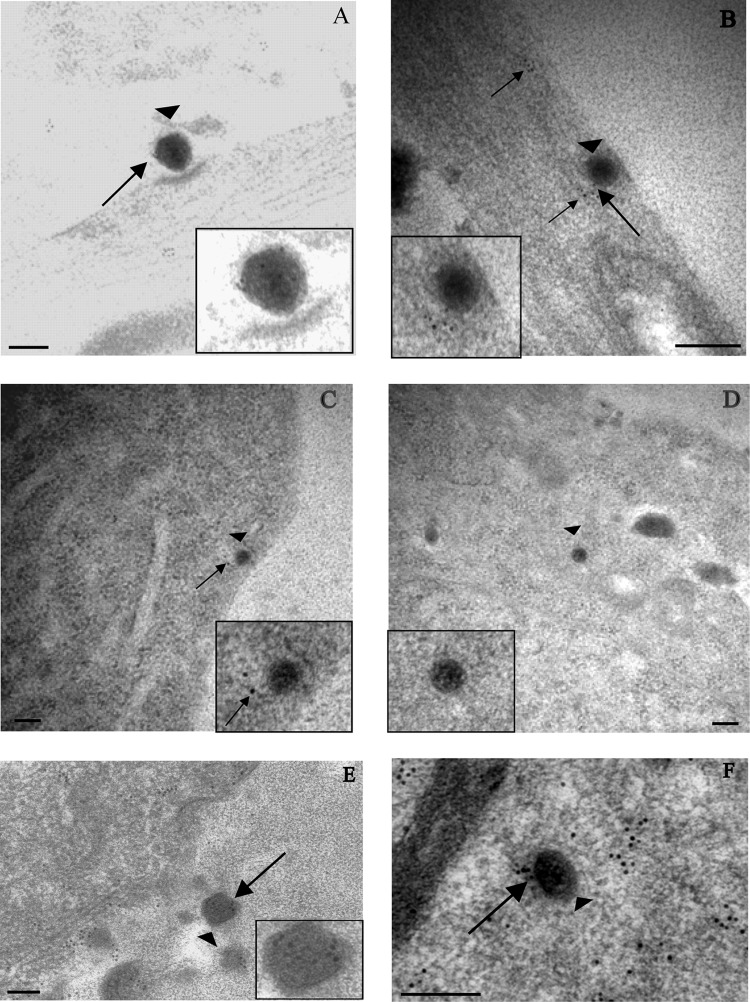
Fig 2.
Immunogold labeling for tegument protein VP16 in human axons and rat neurons infected with vUL37-GFP. (A) Extracellular virion (arrowhead) lying close to a human axon and carrying label for VP16 (arrow) at 30 mpi. (B and C) Unenveloped capsids (arrowheads) within human axons at 30 mpi. Label for VP16 (arrows) is present off the capsids and in panel C is on the plasma membrane. (D) Unenveloped capsid with no label for VP16, present in the cytosol of cell body of a rat DRG neuron at 2 hpi. Diffuse free label for VP16 is present in the cytosol (arrow). (E) Extracellular virion (arrowhead), labeled for VP16 (arrow), in close proximity to the plasma membrane of a DRG neuron at 24 hpi. (F) Unenveloped cytoplasmic capsids (arrowheads) inside the cytosol of the cell body of a rat DRG neuron at 24 hpi, carrying label for VP16 (arrows). (Insets) Enlargements of capsids in each panel. Gold particles were 5 nm (B and C) or 10 nm (other panels). Bars, 200 nm.
At 2 and 4 hpi, no label for VP16 could be seen on unenveloped capsids within axons. Similarly, no label for VP16 could be seen on capsids in the cytosol of infected neurons at these time points (4/4 and 7/7, respectively) (Table 1; Fig. 2D). Free label (not associated with capsid) for VP16 was, however, seen throughout the cytosols and nuclei of neuronal cell bodies (Fig. 2D). This labeling for VP16 was specific to infected neurons and was not detected in uninfected controls (data not shown).
Infected cultures fixed at 24 hpi served as a positive control for our experiments. At 24 hpi, strong label for VP16 was seen on extracellular virions (83.3%; 40/48) (Table 1; Fig. 2E) and unenveloped capsids (80%; 20/25) (Table 1; Fig. 2F) in the cytosol of the neuronal cell body. A direct comparison of VP16 label on unenveloped capsids during entry and egress revealed that while most capsids had 4 or more gold particles at 24 hpi (egress), only weak to moderate label (2 or 3 gold particles) for VP16 was on the capsids at 30 mpi (Table 1). By 4 hpi, capsids were devoid of any label for VP16 suggesting that there was then complete dissociation of the protein from incoming capsids (Table 1).
Dissociation of outer tegument protein VP22 from viral particles during entry.
As for VP16, strong label for VP22 was observed on almost 80% of extracellular virions at all time points postinfection (Table 1; Fig. 3A) and also on viral particles bound to the plasma membrane (Fig. 3B). At 30 mpi, weak or no label for VP22 was seen on unenveloped capsids (Table 1) lying close to the inner aspect of the axonal membrane (Fig. 3C). Label for VP22 was frequently detected beneath the axonal membrane in close proximity to incoming capsids (data not shown). This most likely represented residual dissociated protein from the incoming capsid. At 2 and 4 hpi, almost all the unenveloped capsids observed inside the cytosol of the neuronal cell body carried little or no label for VP22 (Table 1; Fig. 3D), suggesting that, like VP16, VP22 dissociated completely during transport of viral capsids toward the nucleus.
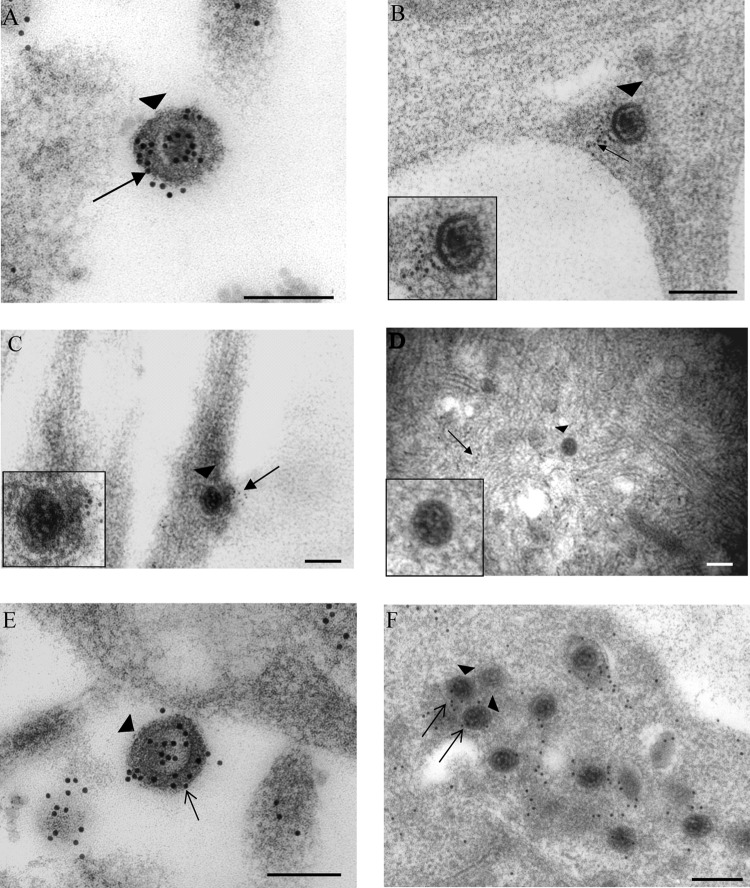
Fig 3.
Immunogold labeling for tegument protein VP22 in human axons and rat neurons infected with vUL37-GFP. Extracellular virions (arrowheads) bound to the plasma membrane of a rat DRG neuron (A) and human DRG axons (B) at 30 mpi. Label for VP22 is present on the virions (arrows). (C) Unenveloped capsid (arrowhead) within a human DRG axon at 30 mpi. Label for VP22 is off the capsid. (Inset) Enlargement of the capsid. (D) Unenveloped capsid (arrowhead) in the cytosol of cell body of a rat DRG neuron at 4 hpi with no label for VP22. (Inset) Enlargement of the capsid. (E) Extracellular virions (arrowheads) lining the cell surface at 24 hpi and labeled for VP22 (arrows). (Inset) Enlargement of a virion. (F) Unenveloped capsid (arrowhead) in the cytosol of the cell body of a rat DRG neuron carrying label for VP22 (arrow) at 24 hpi. (Inset) Enlargement of a capsid. Gold particles were 5 nm (C) or 10 nm (other panels). Bars, 200 nm.
At 24 hpi, label for VP22 was present on extracellular virions lying outside the cell membrane (80%; 40/50) (Table 1; Fig. 3E). In addition, strong specific label for VP22 was observed on unenveloped (33%; 10/30) and enveloped (80%; 24/30) capsids (Table 1) in the cytosol (Fig. 3F). As with VP16, there was a marked difference in the density of VP22 label between unenveloped capsids at 24 hpi and those at 30 mpi and at 2 and 4 hpi, suggesting that there was a gradual but complete loss of VP22 from the incoming capsids by 4 hpi.
Presence of inner tegument protein pUL36 on viral particles during entry.
In addition to VP16 and VP22, the presence of inner tegument proteins pUL36 and pUL37 on virus particles was also examined in axons and cell bodies of DRG neurons. Moderate label for pUL36 was seen on extracellular virions at all time points postinfection (Fig. 4A). The density of label for pUL36 on extracellular virions was consistently less than that for outer tegument proteins VP16 and VP22 (Fig. 2 and 3; Table 2). Both VP16 and VP22 are relatively abundant, being present at 1,000 to 2,000 copies per virion (22). In contrast, fewer than 500 copies of pUL36 and pUL37 are present in assembled virions (45, 62).
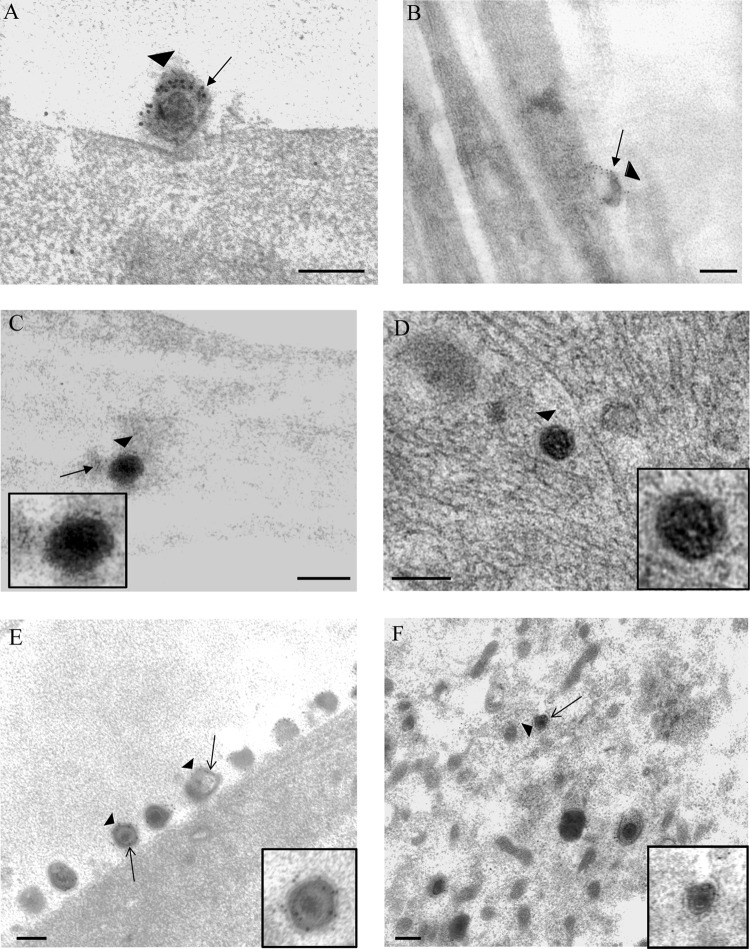
Fig 4.
Immunogold labeling for tegument pUL36 in human axons and rat DRG neurons infected with vUL37-GFP. (A) Extracellular virion (arrowhead), labeled for pUL36 (arrow), bound to the plasma membrane of the cell body of rat DRG neuron at 30 mpi. (B) Unenveloped capsid (arrowhead) in the cytosol of the cell body of a rat DRG neuron at 30 mpi. Label for pUL36 (arrow) is off the viral capsid. (C and D) Unenveloped capsids (arrowheads) in the cytosol of cell body of rat DRG neurons, carrying label for pUL36 (arrows) at 2 hpi and 4 hpi, respectively. (E and F) Extracellular virus (arrowhead) (E) and unenveloped capsid (arrowhead) (F) in the cytosol of cell body of a rat DRG neuron, carrying label for pUL36 (arrows), at 24 hpi. (Insets) Enlargements of viral particles in each panel. Bars, 200 nm.
Unenveloped capsids carrying weak to moderate label for pUL36 were seen within axonal processes at 30 mpi (55%; 5/9) (Table 2). By 2 and 4 hpi, unenveloped capsids, carrying weak to moderate residual label for pUL36, were detected in the cytosol of the infected neurons (4/8 and 5/8, respectively) (Table 2; Fig. 4C and D). The density of label on these unenveloped capsids was consistently less than that observed on extracellular adherent virions, suggesting a partial loss of the protein during virus entry. Residual dissociated label for pUL36 from incoming capsids was observed close to the plasma membrane (Fig. 4B) and in the cytosol of infected neurons. This label was specific only to infected neurons and was not detected in uninfected neurons.
At 24 hpi, extracellular virions (Fig. 4E) and cytoplasmic enveloped capsids carrying moderate to strong label for pUL36 were observed in infected controls (Table 2). In addition, unenveloped capsids, moderately labeled for pUL36 (68%; 40/59), could be observed throughout the neuronal cytosol (Fig. 4F). The density of pUL36 label on these unenveloped capsids was more than that observed on capsids at 4 hpi suggesting, that the reduced labeling for pUL36 at 4 hpi was not because of possible steric hindrance but due to a partial loss of pUL36 from the capsids upon entry.
Presence of inner tegument protein pUL37 on viral particles during entry.
At all time points examined during infection, majority of extracellular virions labeled densely for pUL37 (Table 2; Fig. 5A). At 30 mpi, approximately 57% of unenveloped capsids within axonal processes carried weak to moderate label for pUL37 (Fig. 5B; Table 2). As with pUL36, the density of pUL37 label on these unenveloped capsids was lower than that on extracellular virions, suggesting a partial loss of the protein upon entry (Table 2). At 2 hpi, half of incoming capsids, in close proximity to the plasma membrane, carried weak to moderate label for pUL37, albeit less than that present on extracellular virions (Table 2; Fig. 5C). However, by 4 hpi, most capsids deep within the cytosol had little or no label for pUL37 on them (Table 2; Fig. 5D), suggesting that there was a progressive loss of this protein as the capsids were translocated toward the nucleus. Free label for pUL37 was also seen in the cytosol of infected neurons but not in uninfected cells (data not shown).
At 24 hpi, extracellular virions surrounding the cell body carried strong label for pUL37 (Table 2; Fig. 5E). Unenveloped (Fig. 5F) and enveloped capsids labeled for pUL37 were also readily observed within the cytosol of infected neurons (Table 2). A comparison of pUL37 label on unenveloped capsids during early and late stages of infection revealed that, as with VP16 and VP22, there was a gradual loss of pUL37 from incoming capsids by 4 hpi.
Distribution of viral capsids and inner tegument proteins pUL36 and pUL37 during virus entry into rat DRG neurons.
To further investigate the association and/or dissociation of inner tegument proteins pUL36 and pUL37 from capsids during virus entry, their distribution and colocalization with viral capsids following virus entry into axons and cell body was examined using dually fluorescence-tagged viruses, HSV F-GS2945 (mRFP1-VP26 and pUL36-GFP) and GS3245 (mRFP1-VP26 and GFP-pUL37). Visualization of two different viral proteins was achieved by detection of endogenous mRFP1 (VP26) and GFP (pUL37 or pUL36) using a Deltavision Core image restoration system. Colocalization between capsids and pUL37 or pUL36 was quantified by manual counting of the total number of mRFP particles in the cytosol and at the nuclear rim and then counting the number of these particles colocalizing with GFP. The results were expressed as percentage of capsids that colocalized with GFP. For these counts, an average of 10 cells were used for each virus and time point.
Mock-infected neurons at the same time points postinfection were processed similarly to infected neurons and served as controls. Each experiment was repeated three times in replicate cultures.
Distribution of viral proteins at 30 mpi.
At 30 mpi, all the viral proteins examined, capsid VP26 and tegument proteins pUL36 and pUL37, were present as numerous distinct fluorescent puncta on the periphery of neuronal cell bodies and along axons from proximal to distal ends (Fig. 6 and 7). Colocalization of mRFP1 capsid VP26 with either pUL36-GFP or GFP-pUL37 was seen in puncta on the periphery of the cell body and along axons but most frequently in the proximal axons (Fig. 6A to D and Fig. 7A to D).
Fig 6.
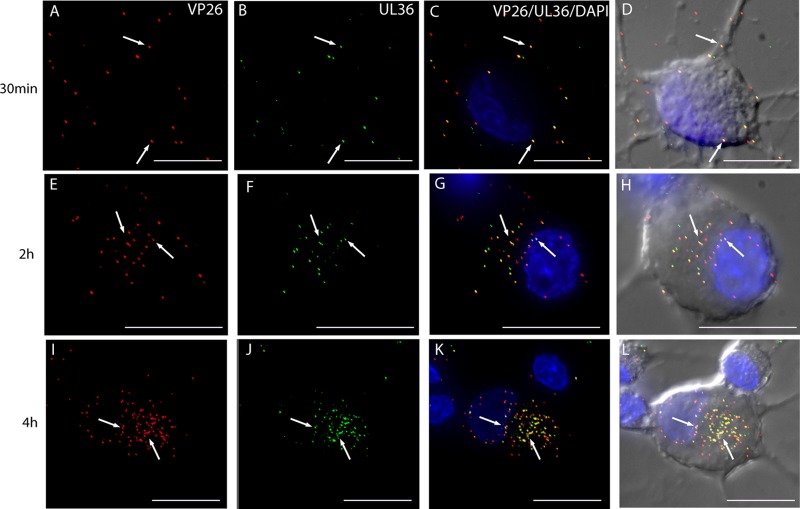
Visualization of mRFP1 capsids and pUL36-GFP in rat DRG neurons infected with HSV F-GS2945 from 30 mpi to 4 hpi by wide-field deconvolution microscopy. The images are three-dimensional (3D) reconstructed z series, and areas of colocalization are in yellow. Fluorescent puncta representing mRFP1 capsids (red) (A) and pUL36-GFP (green) (B) were detected at the cell periphery and along axons at 30 mpi (arrows). Most of the mRFP1 capsids colocalized with pUL36-GFP at the cell periphery and along axons at this time (C and D, arrows). By 2 hpi, fluorescent puncta for mRFP1 capsids (E) and for pUL36-GFP (F) were detected in the cytosol of the cell body and at the nuclear rim (arrows). The majority of mRFP1 capsids in the cytosol (78.9%; n = 71) and at the nuclear rim (71.2%; n = 59) colocalized with pUL36-GFP (G and H; arrows). Similarly, at 4 hpi, the majority of the fluorescent puncta for mRFP1 capsids (I) present in the cytosol (71.3%; n = 237) and at the nuclear rim (70.3%; n = 202) colocalized with fluorescent puncta (J) for pUL36-GFP (I to L; arrows). Bars, 10 μm.
Fig 7.
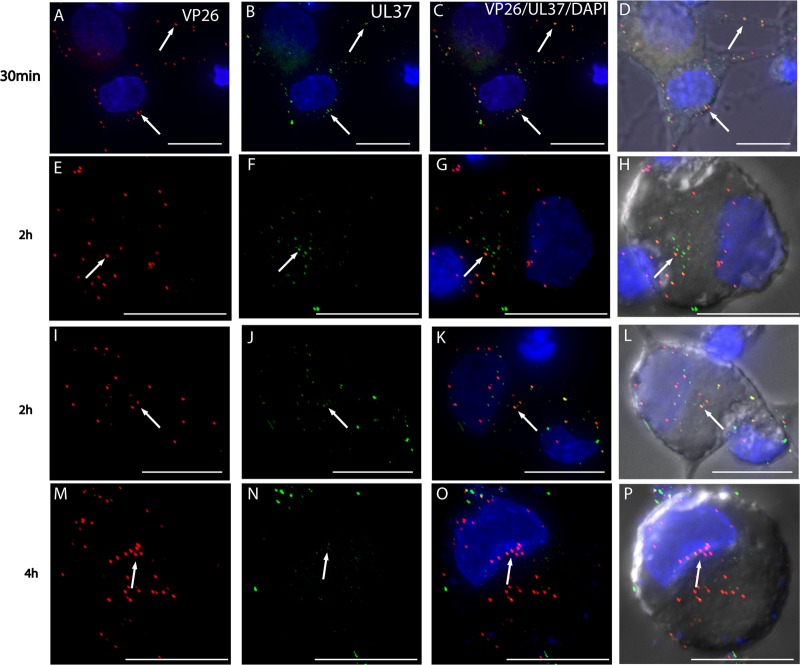
Visualization of mRFP1 capsids and GFP-pUL37 in rat DRG neurons infected with HSV F-GS3245 from 30 mpi to 4 hpi by wide-field deconvolution microscopy. These images are 3D reconstructed z series, and areas of colocalization appear in yellow. Fluorescent puncta representing mRFP1 capsids (red) (A) and GFP-pUL37 (green) (B) were detected at the cell periphery and along axons at 30 mpi (arrows). Most of the mRFP1 capsids colocalized with GFP-pUL37 at the cell periphery and along axons at this time (C and D; arrows). By 2 hpi, fluorescent puncta for mRFP1 capsids (E and I) and for GFP-pUL37 (F and J) were detected in the cytosol of the cell body and at the nuclear rim (arrows). About half of the mRFP1 capsids in the cytosol (55.2%; n = 67) and at the nuclear rim (51.3%; n = 76) colocalized with GFP-pUL37 (G, H, K, and L; arrows). By 4 hpi, less than half of fluorescent puncta for mRFP1 capsids (M) present in the cytosol (38.3%; n = 162) and at the nuclear rim (37.2%; n = 180) colocalized with fluorescent puncta (N) for GFP-pUL37 (O and P) (arrows). Bars, 10 μm.
The presence of capsid and tegument proteins on the periphery of the cell body, as well as axons, probably represents assembled input virions that had not yet been internalized. However, the virus inoculum could also include defective viral particles that remained bound to the plasma membrane of the axons and neuronal cell body. The viruses used for these experiments were subjected to high-speed centrifugation to remove membranous debris containing viral proteins. The virus preparation was further checked by negative staining and examined by electron microscopy (data not shown).
Distribution of viral proteins at 2 hpi.
At 2 hpi, fluorescent puncta representing mRFP1 capsids were detected in the cytosol of the cell body and at the nuclear rim (Fig. 6E to H and Fig. 7E to L). The majority of mRFP1 capsids (78.9% of a total of 71 capsids) in the cytosol contained pUL36-GFP whereas half of these mRFP1 capsids (55.2% of a total of 67 capsids) in the cytosol contained GFP-pUL37. By 2 hpi, mRFP1 capsids also reached the nuclear rim, and 71.2% (total, 59 capsids) of these contained pUL36-GFP and about half (51.3%; total, 76 capsids) contained GFP-pUL37.
GFP puncta for both pUL36 and pUL37 were also present in the cytosol and free of mRFP1 capsids, suggesting dissociation from incoming capsids.
Distribution of viral proteins at 4 hpi.
By 4 hpi, numerous fluorescent puncta for mRFP1 capsids could be seen in the cytosol and at the nuclear rim (Fig. 6I to L and Fig. 7M to P). In addition, a few discrete fluorescent puncta for mRFP1 capsids also remained on the cell periphery (Fig. 6 and 7). A proportion of fluorescent puncta for mRFP1 capsids (71.3%; total, 237 capsids) similar to that detected in the cytosol at 2 hpi were associated with pUL36-GFP, whereas 38.3% of fluorescent puncta for mRFP1 capsids (total, 162 capsids) in the cytosol at this time remained associated with GFP-pUL37.
The majority of mRFP1 capsids (70.3%; total, 202 capsids) at the nuclear rim contained pUL36-GFP, whereas only 37.2% of these mRFP1 capsids (total, 180 capsids) contained GFP-pUL37. In addition, GFP puncta for both pUL36 and pUL37 were also detected in the cytosol free (independent of mRFP1 capsids) as at 2 hpi. However, the number of GFP puncta for pUL37 markedly decreased at 4 hpi compared to 2 hpi, suggesting possible degradation of free pUL37.
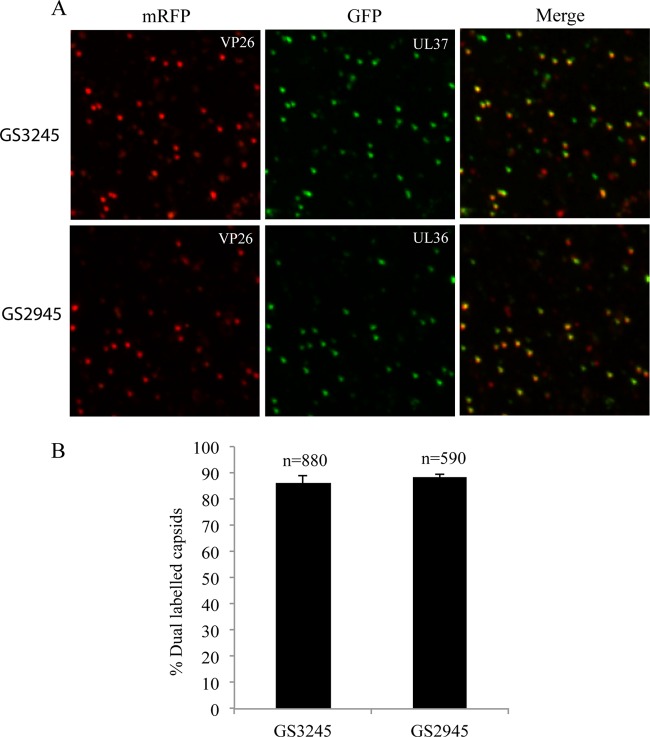
In order to confirm that the lack of colocalization of pUL37 with mRFP1 capsids during travel to the nucleus was due to dissociation and not to artifacts (from fixation and/or deconvolution processing), the virus inoculum was subjected to the fixation and detection processes performed with the infected neuronal cultures. For this, droplets of input virus suspensions were placed directly onto glass coverslips, fixed in 3% formaldehyde, and examined by deconvolution microscopy to determine the proportion of mRFP1 capsids expressing GFP. As shown in Fig. 8, 88.5% of mRFP-VP26 capsids were found to express pUL36-GFP (total, 590 capsids), while 86.2% of mRFP1 capsids expressed GFP-pUL37 (total, 880 capsids), showing no loss of GFP.
Fig 8.
Characterization of F-GS2945 and F-GS3245 extracellular virus particles. (A) Cell supernatants containing extracellular virus particles were spun down on Cell Tak-coated glass coverslips at 1,200 × g for 30 min at 4°C followed by fixation in 3% formaldehyde and imaged by deconvolution microscopy. F-GS3245 expresses mRFP1-VP26 and GFP-pUL37, while F-GS2945 expresses mRFP1-VP26 and pUL36-GFP. “Merge” panels show mRFP fluorescence superimposed on GFP fluorescence. (B) Percentage of mRFP capsids emitting GFP fluorescence from supernatants of F-GS3245 and F-GS2945 shown in panel A. Results are averages from five separate counts. Error bars represent standard errors of means. n, total number of capsids.
These results, obtained with dually fluorescence-tagged HSV-1, are consistent with our observations using TIEM in that there is some progressive loss of tegument pUL37 from capsids as capsids are transported from the cell periphery to the nuclear rim from 2 to 4 hpi.
DISCUSSION
In the present study, we sought to investigate the subcellular localization and kinetics of HSV-1 viral tegument and capsid proteins upon entry of HSV-1 into human and rat DRG neurons using serial fixation, wide-field deconvolution microscopy, and TIEM. In particular, TIEM was used to directly visualize the tegument composition of incoming HSV-1 capsids as they travel to the nucleus of the cell body.
In this study, TIEM was used to directly visualize the presence of the major tegument proteins VP16, VP22, pUL36, and pUL37 on incoming HSV-1 capsids during virus entry and subsequent transport toward the nucleus in rat and human DRG neurons. Human DRG explants were mainly used to study the viral transport in axons, while rat DRG neurons were used to study the virus transport within the cytosol. A progressive loss of tegument proteins, including VP16, VP22, and most of pUL37, during virus entry and transport in both types of neurons was observed. Most of the tegument proteins dissociated at the plasma membrane of both axons and cell body as the viral envelope fused with the cell membrane, and the capsid was released into the cytosol. Further dissociation of tegument proteins occurred within the cytosol during the transport of incoming unenveloped capsids toward the nucleus of the cell. Partial dissociation was also observed for the tegument protein pUL36 at the plasma membrane during virus entry. However, substantial residual label for pUL36 remained associated with the capsids in axons and cell bodies as they reached the nucleus.
Recombinant fluorescent-tagged viruses were used to determine the distribution and colocalization of inner tegument proteins (pUL36 and pUL37) and capsid (VP26) from 30 min up to 4 h after viral entry. Rat DRG neuronal cultures were infected with either HSV F-GS2945 (mRFP1-VP26 and pUL36-GFP) or HSV F-GS3245 (mRFP1-VP26 and GFP-pUL37). At 30 mpi, the majority of capsids were bound to the periphery of the neuronal cell body and along the axons. By 2 hpi, capsids had moved into the cytosol of the cell body and reached the nuclear rim. Capsids could be seen only in the proximal regions of axons at this time. By 4 hpi, capsids clustered at the nuclear rim. This was in contrast to kinetics studies carried out with epithelial cell lines, in which the virus began to accumulate at the nuclear membrane as early as 1 hpi (54, 65). pUL36 and pUL37 localized mainly to the periphery of the neuronal cell body and to axons at 30 mpi. Both pUL36 and pUL37 first appeared in the cytosol of the neuronal cell body at 2 hpi. At 2 and 4 hpi, the majority of capsids (70%) were transported to the nuclear rim in association with pUL36, while less than half of the capsids remained associated with pUL37 by 4 hpi. These findings are consistent with the TIEM observations, which revealed a similar proportion of capsids in the cytosol containing label for pUL36 and pUL37 after virus entry.
The proportion of capsids containing label for pUL37 at the nuclear rim as well as in the cytosol declined from 2 to 4 hpi. In addition, free (independent from capsids) pUL37 in the cytosol also markedly declined from 2 to 4 hpi, consistent with degradation of the protein. HSV pUL36 and pUL37 have been shown to directly interact (31, 70), and pUL37 would be expected to dissociate first, as pUL36 is the major determinant for the incorporation of pUL37 into the virion (32).
The recombinant singly fluorescence-tagged virus, vUL37-GFP, and the recombinant dually fluorescence-tagged viruses, HSV F-GS2945 and HSV F-GS3245, were constructed from different parental HSV1 strains. Strain 17 is the parental strain for vUL37-GFP, while strain F is the parental strain for HSV F-GS2945 and HSV F-GS3245 (3). HSV-1 strain 17 and strain F have been shown to differ in their genome sequences and pathogenicity (12, 42, 43, 68). Nevertheless, in this study, there appeared to be no difference in the kinetics of capsid entry and translocation to the nucleus of the neuronal cell body between these two strains.
As observed by TIEM, most of the label for outer tegument proteins VP16 and VP22 dissociated at the plasma membrane, as free label was found along the membranes of axons in close proximity to the incoming capsids. Further dissociation of these proteins occurred within the cytosol of axons and of cell body. It has been proposed that VP16 and VP22, along with tegument proteins pUL11, VP11/12, and VP13/14, are mainly present on the outer, distal part of the tegument (46), where they interact with the cytoplasmic tails of viral glycoproteins (7, 29, 67). In the case of PrV, it has been shown that VP22 interacts with the cytoplasmic tails of envelope proteins gM and gE (17), while HSV-1 VP22 interacts with gE (67), gD (7), and pUS9 (34). Similarly, HSV-1 VP16 interacts with the cytoplasmic tail of gH (19). Hence, it is likely that during the first stage of tegument disassembly, these proteins remain associated with the stripped envelope (41). Our findings are consistent with the existing model of HSV-1 entry in which the virus enters neuronal cells and certain nonneuronal cells by fusion of the viral envelope with the plasma membrane (39, 53, 65). The viral envelope is thus left behind as the capsid is released into the cell cytosol. A cryo-EM study of HSV-1 entry into rat brain synaptosomes and nonneuronal cell lines revealed the presence of glycoprotein spikes protruding from the cell membrane at the site of virus entry (41). The same study also showed that the glycoproteins were associated with retention of substantial tegument density at the cell membrane after the capsid was released into the cytosol. Little residual tegument could be observed on these capsids. Furthermore, treatment of virions in vitro with Triton X-100 similarly showed dissociation of most of the tegument, with “tufts” of proteins, most likely pUL36 and pUL37, remaining attached to the vertices of capsids (52).
Several tegument proteins play a variety of key roles during early stages in the infectious cycle of HSV-1 that include shutoff host cell protein synthesis and transactivation of viral immediate-early (IE) genes (6, 16, 21, 59, 66). Thus, release of soluble tegument proteins into the cytosol after infection requires the dissociation of much of the tegument during virus entry. VP16 plays an important role in stimulating transcription of the IE genes from the viral genome (6, 23, 59, 72). Hence, nuclear targeting of VP16 early in infection is a critical step in the virus replication cycle. Our data are also consistent with an earlier study by Yamauchi et al. that showed a similar nuclear localization pattern for VP16 at early time points in HSV-1-infected nonneuronal cells (73). The precise role of VP22 in the early stages of the HSV-1 replication cycle is yet to be defined, but it has been shown that phosphorylation of VP22 probably leads to its release from the incoming capsid (50, 51). Previous studies with HSV-1 and PrV have shown that both VP16 and VP22 dissociate from the incoming capsids in both epithelial cell lines and neuronal cells (3, 18, 38, 50). The same studies also reported that the inner tegument proteins pUL36 and pUL37 remain associated with the capsids.
Antinone et al. previously used combinations of dually fluorescence-tagged capsid and tegument proteins and real-time fluorescence microscopy in chick neurons to demonstrate the cotransport of pUL36 and pUL37 with the capsid and accumulation at the nuclear rim for both PrV and HSV-1 in DRG axons (3). They concluded that there is no loss of pUL36 and pUL37 from incoming capsids, and this appears at first glance to be contradictory to findings obtained by us using both fluorescence and TIEM. However, the study using chick neurons focused on the cotransport of HSV1 mRFP1-VP26 with pUL36 or pUL37 along axons and at the nuclear rim at 2 to 3 hpi by real-time fluorescence. The kinetics but not the composition of viral particles was compared with that of mouse DRG axons and human SK-N-SH neuronal cells. It did not focus on changes in pUL37 complement in the cytosol over 0.5 to 4 hpi nor the presence of free pUL37. The reasons for the differences between the studies are unclear but could be include differences in the techniques or cell types and/or anatomic and time-dependent differences. There may be a subset of capsids bearing pUL37 that arrive at the nuclear rim in chick neurons, and its presence may facilitate retrograde transport as for pseudorabies virus (33), but from our study it is not essential for most pUL37 to be on the capsid for transport to the nuclear rim. TIEM provided a more precise localization of protein in relation to viral particles and showed a partial dissociation of pUL36 and, to a greater extent, pUL37 as the capsids move from the plasma membrane to the nucleus. Thus, our findings are consistent with these previous studies in that there is cotransport of tegument pUL36 and some pUL37 with incoming capsids but provide new evidence for partial dissociation and eventual degradation of these inner tegument proteins from capsids as the capsids travel from the cell periphery to the nucleus.
A nonessential role for pUL37 during entry of both HSV-1 and PrV in nonneuronal cells has been described (9, 33, 58). In addition, a recent study demonstrated that pUL37 also has nonstructural functions and interacts with tumor necrosis factor receptor-associated factor 6 (TRAF6) with resulting activation of the NF-κB signaling pathway (36). Regulation of NF-κB activity most likely plays a role in transcriptional activity of IE genes such as that encoding ICP0 (2). Dissociation of pUL37 from incoming capsids and its subsequent localization to the cytosol early in infection may facilitate the activation of the NF-κB pathway in HSV-1-infected cells. Hence, it is possible that most pUL37, like VP16, is transported to the nucleus or to other cytoplasmic sites, independent of the capsids, to perform a key functional role early in infection.
pUL36, the largest tegument protein, is critical in early as well as late stages of the HSV-1 infectious cycle. Studies in both neuronal and nonneuronal cells have shown that pUL36 remains associated with capsids during entry and transit to the cell nucleus (3, 18, 38, 55). pUL36 is required for targeting capsids to the nuclear membrane and facilitates the docking of capsids by interacting with the nuclear pore complex (5, 9, 54). In addition, proteolytic cleavage of the N terminus of pUL36 is required for release of viral genomic DNA into the nucleus (10, 28). Consistent with the findings from other laboratories, we observed colocalization between pUL36 and VP26 at the nuclear rim at early time points of infection.
In contrast, the role of pUL37 in early stages of HSV-1 infection is less clear. Although studies with both PrV and HSV have reported that pUL37 remains associated with incoming capsids after virus entry (3, 9, 18, 38), pUL37 is not required for migration of capsids toward the nucleus (33, 58). Using an infectious-syncytium model in cultured fibroblasts, Roberts et al. showed that UL37 deletion mutants of HSV-1 are capable of transmitting HSV infection to all nuclei within a syncytium as efficiently as wild-type HSV-1 (58). In a study with PrV-infected cultured rabbit skin cells, Krautwald et al. used a UL37 deletion PrV mutant carrying a GFP tag on VP26 to show that the translocation of incoming virus particles was delayed but not abolished in the absence of pUL37 (33). In our study, a progressive and partial loss of pUL37 from capsids was observed over time (from 2 to 4 hpi) as the capsids moved from the cell periphery to the nucleus.
In summary, our wide-field deconvolution microscopy studies with recombinant fluorescence-tagged viruses in rat DRG neurons suggest that while there is partial dissociation of pUL37 from capsids, pUL36 and some pUL37 remain associated with HSV-1 capsids during virus entry and transit to the nucleus in the neuronal cell body. Immunoelectron microscopy revealed a progressive loss of tegument proteins, including VP16, VP22, and some pUL37, during HSV-1 entry in human and rat DRG axons and cell bodies of neurons. pUL36 was the main tegument protein demonstrated by TIEM to remain associated with incoming capsids despite undergoing partial loss at the cell surface. Colocalization of capsid VP26 and pUL37 was observed using wide-field deconvolution microscopy and is consistent with previous reports. Together, these observations suggest that lower levels of pUL37 than pUL36 may remain associated with capsids. Determining the tegument composition of capsids during their passage from the cell surface to the nuclear pore is a key step in understanding the mechanisms involved during virus entry and retrograde transport. Incoming capsids must engage molecular motors such as dynein for their transport along the cytoskeleton to the microtubule-organizing center (MTOC) and then probably kinesins for the journey to the nuclear pores (14, 25, 56, 57, 65). Thus, this study elucidates, at an ultrastructural level, the composition of HSV-1 capsids that encounter the microtubules in the core of human axons and also the complement of free tegument proteins released into the axons and cytosol of the cell body during virus entry. Identifying viral proteins that remain associated with translocating capsids would help in the elucidation of specific viral proteins that mediate capsid-motor interaction. These proteins are yet to be fully defined and are likely to be redundant (13, 14, 65). Further investigation into the fate, transport, and role of many of the tegument proteins in the virion released during virus entry will contribute to the better understanding of the mechanisms used by the virus for efficient replication and spread in the nervous systems.
ACKNOWLEDGMENTS
We thank Frazer Rixon, MRC Virology Unit, Institute of Virology, United Kingdom, for providing us with the vUL37-GFP used in this study. We also thank Greg Smith from Northwestern University Medical School for providing us with the HSV F-GS2945 and HSV F-GS3245. We also thank Carol Robinson, Levina Dear, and Gayle Versace-Avis, Electron Microscope Laboratory, ICPMR, Westmead Hospital, for assistance with electron microscopy. We thank the staff of the Westmead Department of Animal Care for their assistance with the maintenance and breeding of the Wistar rats. We also thank Hong Yu, Westmead Millennium Institute, for assistance with the Deltavision Core image restoration system.
This project was supported by project grant 570849 to A. L. Cunningham, R. J. Diefenbach, and M. Miranda-Saksena from the National Health and Medical Research Council of Australia.
Footnotes
Published ahead of print 28 March 2012
REFERENCES
- 1. Akhtar J, Shukla D. 2009. Viral entry mechanisms: cellular and viral mediators of herpes simplex virus entry. FEBS J. 276:7228–7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amici C, et al. 2006. Herpes simplex virus disrupts NF-κB regulation by blocking its recruitment on the IκBα promoter and directing the factor on viral genes. J. Biol. Chem. 281:7110–7117 [DOI] [PubMed] [Google Scholar]
- 3. Antinone SE, Smith GA. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J. Virol. 84:1504–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Antinone SE, Zaichick SV, Smith GA. 2010. Resolving the assembly state of herpes simplex virus during axon transport by live-cell imaging. J. Virol. 84:13019–13030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Batterson W, Furlong D, Roizman B. 1983. Molecular genetics of herpes simplex virus. VIII. further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Batterson W, Roizman B. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chi JH, Harley CA, Mukhopadhyay A, Wilson DW. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253–261 [DOI] [PubMed] [Google Scholar]
- 8. Cohen GH, et al. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Copeland AM, Newcomb WW, Brown JC. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83:1660–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delboy MG, Roller DG, Nicola AV. 2008. Cellular proteasome activity facilitates herpes simplex virus entry at a postpenetration step. J. Virol. 82:3381–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diefenbach RJ, Miranda-Saksena M, Douglas MW, Cunningham AL. 2008. Transport and egress of herpes simplex virus in neurons. Rev. Med. Virol. 18:35–51 [DOI] [PubMed] [Google Scholar]
- 12. Dix RD, McKendall RR, Baringer JR. 1983. Comparative neurovirulence of herpes simplex virus type 1 strains after peripheral or intracerebral inoculation of BALB/c mice. Infect. Immun. 40:103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dohner K, Radtke K, Schmidt S, Sodeik B. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 80:8211–8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dohner K, et al. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13:2795–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elliott G, Mouzakitis G, O'Hare P. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Everett RD. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761–770 [DOI] [PubMed] [Google Scholar]
- 17. Fuchs W, et al. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208–8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Granzow H, Klupp BG, Mettenleiter TC. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gross ST, Harley CA, Wilson DW. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1–12 [DOI] [PubMed] [Google Scholar]
- 20. Grunewald K, et al. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398 [DOI] [PubMed] [Google Scholar]
- 21. Hagglund R, Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heine JW, Honess RW, Cassai E, Roizman B. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14:640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herr W. 1998. The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation. Cold Spring Harbor Symp. Quant. Biol. 63:599–607 [DOI] [PubMed] [Google Scholar]
- 24. Hiraoka Y, Sedat JW, Agard DA. 1990. Determination of three-dimensional imaging properties of a light microscope system. Partial confocal behavior in epifluorescence microscopy. Biophys. J. 57:325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hirokawa N. 1998. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279:519–526 [DOI] [PubMed] [Google Scholar]
- 26. Holland DJ, Cunningham AL, Boadle RA. 1998. The axonal transmission of Herpes simplex virus to epidermal cells: a novel use of the freeze substitution technique applied to explant cultures retained on coverslips. J. Microsc. 192:69–72 [DOI] [PubMed] [Google Scholar]
- 27. Holland DJ, Miranda-Saksena M, Boadle RA, Armati P, Cunningham AL. 1999. Anterograde transport of herpes simplex virus proteins in axons of peripheral human fetal neurons: an immunoelectron microscopy study. J. Virol. 73:8503–8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jovasevic V, Liang L, Roizman B. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 82:3311–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kamen DE, Gross ST, Girvin ME, Wilson DW. 2005. Structural basis for the physiological temperature dependence of the association of VP16 with the cytoplasmic tail of herpes simplex virus glycoprotein H. J. Virol. 79:6134–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ. 2009. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 145:173–186 [DOI] [PubMed] [Google Scholar]
- 31. Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ko DH, Cunningham AL, Diefenbach RJ. 2010. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2). J. Virol. 84:1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krautwald M, Fuchs W, Klupp BG, Mettenleiter TC. 2009. Translocation of incoming pseudorabies virus capsids to the cell nucleus is delayed in the absence of tegument protein pUL37. J. Virol. 83:3389–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee JH, Vittone V, Diefenbach E, Cunningham AL, Diefenbach RJ. 2008. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378:347–354 [DOI] [PubMed] [Google Scholar]
- 35. Leege T, Granzow H, Fuchs W, Klupp BG, Mettenleiter TC. 2009. Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type 1. J. Gen. Virol. 90:1560–1568 [DOI] [PubMed] [Google Scholar]
- 36. Liu X, Fitzgerald K, Kurt-Jones E, Finberg R, Knipe DM. 2008. Herpesvirus tegument protein activates NF-κB signaling through the TRAF6 adaptor protein. Proc. Natl. Acad. Sci. U. S. A. 105:11335–11339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loret S, Guay G, Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luxton GW, et al. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lycke E, et al. 1988. Herpes simplex virus infection of the human sensory neuron. An electron microscopy study. Arch. Virol. 101:87–104 [DOI] [PubMed] [Google Scholar]
- 40. Lycke E, Kristensson K, Svennerholm B, Vahlne A, Ziegler R. 1984. Uptake and transport of herpes simplex virus in neurites of rat dorsal root ganglia cells in culture. J. Gen. Virol. 65(Pt. 1):55–64 [DOI] [PubMed] [Google Scholar]
- 41. Maurer UE, Sodeik B, Grunewald K. 2008. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc. Natl. Acad. Sci. U. S. A. 105:10559–10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McGeoch DJ, et al. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69(Pt. 7):1531–1574 [DOI] [PubMed] [Google Scholar]
- 43. McGeoch DJ, Dolan A, Donald S, Rixon FJ. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 181:1–13 [DOI] [PubMed] [Google Scholar]
- 44. McLean C, et al. 1982. Monoclonal antibodies to three non-glycosylated antigens of herpes simplex virus type 2. J. Gen. Virol. 63:297–305 [DOI] [PubMed] [Google Scholar]
- 45. McNabb DS, Courtney RJ. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221–232 [DOI] [PubMed] [Google Scholar]
- 46. Mettenleiter TC. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miranda-Saksena M, Armati P, Boadle RA, Holland DJ, Cunningham AL. 2000. Anterograde transport of herpes simplex virus type 1 in cultured, dissociated human and rat dorsal root ganglion neurons. J. Virol. 74:1827–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miranda-Saksena M, et al. 2009. Herpes simplex virus utilizes the large secretory vesicle pathway for anterograde transport of tegument and envelope proteins and for viral exocytosis from growth cones of human fetal axons. J. Virol. 83:3187–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miranda-Saksena M, Boadle RA, Armati P, Cunningham AL. 2002. In rat dorsal root ganglion neurons, herpes simplex virus type 1 tegument forms in the cytoplasm of the cell body. J. Virol. 76:9934–9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morrison EE, Stevenson AJ, Wang YF, Meredith DM. 1998. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J. Gen. Virol. 79(Pt. 10):2517–2528 [DOI] [PubMed] [Google Scholar]
- 51. Morrison EE, Wang YF, Meredith DM. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Newcomb WW, Brown JC. 2010. Structure and capsid association of the herpesvirus large tegument protein UL36. J. Virol. 84:9408–9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nicola AV, Hou J, Major EO, Straus SE. 2005. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J. Virol. 79:7609–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 20:4922–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83:6610–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Radtke K, Dohner K, Sodeik B. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol. 8:387–400 [DOI] [PubMed] [Google Scholar]
- 57. Radtke K, et al. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6:e1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Roberts AP, et al. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roizman B, Gu H, Mandel G. 2005. The first 30 minutes in the life of a virus: unREST in the nucleus. Cell Cycle. 4:1019–1021 [DOI] [PubMed] [Google Scholar]
- 60. Roizman B, Knipe DM, Whitley RJ. 2007. Herpes simplex viruses, p 2501–2601 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 61. Saksena MM, et al. 2006. Herpes simplex virus type 1 accumulation, envelopment, and exit in growth cones and varicosities in mid-distal regions of axons. J. Virol. 80:3592–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmitz JB, Albright AG, Kinchington PR, Jenkins FJ. 1995. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology 206:1055–1065 [DOI] [PubMed] [Google Scholar]
- 63. Simpson SA, et al. 2005. Nectin-1/HveC mediates herpes simplex virus type 1 entry into primary human sensory neurons and fibroblasts. J. Neurovirol. 11:208–218 [DOI] [PubMed] [Google Scholar]
- 64. Smith JS, Robinson NJ. 2002. Age-specific prevalence of infection with herpes simplex virus types 2 and 1: a global review. J. Infect. Dis. 186(Suppl. 1):S3–S28 [DOI] [PubMed] [Google Scholar]
- 65. Sodeik B, Ebersold MW, Helenius A. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stern S, Tanaka M, Herr W. 1989. The Oct-1 homeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature 341:624–630 [DOI] [PubMed] [Google Scholar]
- 67. Stylianou J, Maringer K, Cook R, Bernard E, Elliott G. 2009. Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 occurs via glycoprotein E-specific recruitment to the late secretory pathway. J. Virol. 83:5204–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Szpara ML, Parsons L, Enquist LW. 2010. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J. Virol. 84:5303–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Topp KS, Meade LB, LaVail JH. 1994. Microtubule polarity in the peripheral processes of trigeminal ganglion cells: relevance for the retrograde transport of herpes simplex virus. J. Neurosci. 14:318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vittone V, et al. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wolfstein A, et al. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227–237 [DOI] [PubMed] [Google Scholar]
- 72. Wysocka J, Herr W. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28:294–304 [DOI] [PubMed] [Google Scholar]
- 73. Yamauchi Y, et al. 2008. The UL14 tegument protein of herpes simplex virus type 1 is required for efficient nuclear transport of the alpha transinducing factor VP16 and viral capsids. J. Virol. 82:1094–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhou ZH, et al. 2000. Seeing the herpesvirus capsid at 8.5 A. Science 288:877–880 [DOI] [PubMed] [Google Scholar]