Background: KAP1 is important for epigenetic modifications and the DNA damage response.
Results: Distinct DNA damage signaling pathways regulate KAP1 Ser-473 phosphorylation (S473p). DNA damage-induced KAP1 S473p regulates KAP1-HP1 and KAP1-E2F1 interactions, gene transcription, DNA repair, and apoptosis.
Conclusion: KAP1 S473p is required for efficient DNA repair and cell survival under stress.
Significance: Our studies reveal functions for KAP1 S473p under stress.
Keywords: Apoptosis, Co-repressor Transcription, DNA Damage Response, E2F Transcription Factor, Protein Phosphorylation, Chk2, E2F1, HP1, KAP1, KRAB-ZNFs
Abstract
The Kruppel-associated box (KRAB)-associated co-repressor KAP1 is an essential nuclear co-repressor for the KRAB zinc finger protein superfamily of transcriptional factors. Ataxia telangiectasia mutated (ATM)-Chk2 and ATM- and Rad3-related (ATR)-Chk1 are two primary kinase signaling cascades activated in response to DNA damage. A growing body of evidence suggests that ATM and ATR phosphorylate KAP1 at Ser-824 in response to DNA damage and regulate KAP1-dependent chromatin condensation, DNA repair, and gene expression. Here, we show that, depending on the type of DNA damage that occurs, KAP1 Ser-473 can be phosphorylated by ATM-Chk2 or ATR-Chk1 kinases. Phosphorylation of KAP1 at Ser-473 attenuated its binding to the heterochromatin protein 1 family proteins and inhibited its transcriptional repression of KRAB-zinc finger protein (KRAB-ZFP) target genes. Moreover, KAP1 Ser-473 phosphorylation induced by DNA damage stimulated KAP1-E2F1 binding. Overexpression of heterochromatin protein 1 significantly inhibited E2F1-KAP1 binding. Elimination of KAP1 Ser-473 phosphorylation increased E2F1-targeted proapoptotic gene expression and E2F1-induced apoptosis in response to DNA damage. Furthermore, loss of phosphorylation of KAP1 Ser-473 led to less BRCA1 focus formation and slower kinetics of loss of γH2AX foci after DNA damage. KAP1 Ser-473 phosphorylation was required for efficient DNA repair and cell survival in response to DNA damage. Our studies reveal novel functions of KAP1 Ser-473 phosphorylation under stress.
Introduction
Kruppel-associated box (KRAB)3-associated protein 1 (KAP1; also named TRIM28, TIF1β, or KRIP1) was first identified as a universal transcriptional co-repressor for KRAB-ZFPs (1). It has essential roles in early embryonic development and cell differentiation (2, 3). Sequence analysis of KAP1 revealed several highly conserved motifs including an N-terminal RING finger B-box and coiled-coil (RBCC) domain, a C-terminal PHD finger, and bromodomain. The RBCC domain is necessary and sufficient for interaction with KRAB-ZFPs and for the formation of KAP1 oligomers (4). The C-terminal domains cooperatively function as a transcription repression module by recruiting the histone deacetylase complex and histone methyltransferase (5, 6). KAP1 also recruits heterochromatin protein 1 (HP1) to histones through a PXVXL motif in its centrally located HP1 binding domain (HP1BD). KAP1-HP1 interaction is crucial for dynamic regulation of histone modification and for the formation of facultative heterochromatin and is indispensable for KRAB-ZFP-mediated transcriptional repression (5, 7, 8). However, the molecular mechanism by which these interactions are regulated under stress remains poorly understood.
Our previous study revealed that KAP1 cooperates with mdm2 to inhibit p53 activity. KAP1 depletion increases γ-radiation-induced apoptosis (9). We also reported that KAP1 binds to E2F1 and regulates E2F1 transcriptional activity and that knockdown of KAP1 sensitizes cells to etoposide treatment (10). A decrease in KAP1 levels promotes p53-dependent p21 induction and inhibits cell proliferation in actinomycin D-treated cells (11). KAP1 mediates cell survival after DNA damage and may serve as a therapeutic target against cancer (11, 12). Several post-translational modifications are involved in regulating KAP1 activity. KAP1 is a strong substrate for sumoylation at sites clustered in the C-terminal portion of the protein, and sumoylation plays a major role in mediating KAP1 co-repressor activity (13, 14). KAP1 Ser-824 can be phosphorylated by ATM, contributes to global chromatin condensation following the formation of DNA double strand breaks (DSBs) (15), and participates in heterochromatic DNA double strand break repair (16–18). KAP1 Ser-824 phosphorylation (S824p) and sumoylation affect the transcription of a subset of genes involved in cell cycle control and apoptosis in response to genotoxic stresses (19). In addition, KAP1 Ser-473 has been suggested to be phosphorylated by PKCδ in early S phase under normal culture conditions, and KAP1-HP1β interaction is compromised by this modification (20). By quantitative mass spectrometry experiment-based proteomics, Bennetzen et al. (21) identified hundreds of ionizing radiation-induced phosphorylation sites and listed KAP1 Ser-473 as one potential phosphorylation site. While preparing our manuscript, Blasius et al. (22) also recently identified that phosphorylation of KAP1 Ser-473 is a novel DNA damage-induced Chk1 site by using a chemical genetics approach combined with high resolution mass spectrometry assay and that KAP1 Ser-473 phosphorylation (S473p) is a novel DNA damage-induced Chk1 site. However, the specific function of KAP1 Ser-473 phosphorylation in response to DNA damage was not found. The molecular mechanism of KAP1 Ser-473 phosphorylation in response to different DNA-damaging stimuli and the biochemical and biological differences between the two phosphorylations (S473p and S824p) are still unclear.
By sequence comparison of KAP1 orthologues, we found that KAP1 Ser-473 is highly evolutionarily conserved and fits the consensus sequence ((V/L)XRXXS) for a Chk2 substrate. Chk2 is an important regulator of diverse cellular responses to genotoxic stress and guards the integrity of the genome throughout eukaryotic evolution (23). Chk2 is activated in an ATM-dependent or -independent manner (24), and it phosphorylates proteins such as p53 (25), Cdc25c (26), Strap (27), and E2F1 (28) and plays crucial roles in mediating the cellular checkpoint response, DNA repair, and apoptosis. Results described below show that distinct DNA damage signaling pathways are required for KAP1 S473p in response to different DNA damage stimuli. KAP1 S473p regulates its transcriptional repression of KRAB-ZNF target genes. DNA damage-induced KAP1 S473p regulates its binding to HP1 and E2F1 and reduces its ability to repress the expression of E2F1-related proapoptotic genes and E2F1-induced apoptosis in response to DNA damage. KAP1 S824p and S473p participate in DNA repair in a spatial and temporal manner, and loss of KAP1 S473p leads to less BRCA1 focus formation and slower kinetics of γH2AX focus loss after DNA damage. KAP1 S473p is required for efficient DNA repair and cell survival in response to DNA damage under stress conditions.
EXPERIMENTAL PROCEDURES
Cell Lines, Plasmids, and Reagents
HeLa, HCT116, HCT15, HEK293T, and SaoS2 cells were maintained in DMEM, 10% fetal bovine serum. A rabbit antiserum against KAP1 phosphorylated at Ser-473 was raised by GL Biochem Ltd. (China) using the synthetic peptide (VKRSRS[p]GEGEVC) as antigen of which the second serine is phosphorylated and indicated as SCP. The antiserum was precleaned by affinity chromatography using the corresponding non-phosphorylated peptide (VKRSRSGEGEVC) coupled to SulfoLink Coupling Resin (Thermo Fisher Scientific) and then purified by affinity chromatography using phosphopeptide. Etoposide, Chk2 inhibitor II, and Chk1 inhibitor PD407824 were supplied by Sigma. Chk2 (1C12) and γH2AX (20E3) antibodies were supplied by Cell Signaling Technology. KAP1 S824p (ab70369), γH2AX (ab18311), and KAP1 ChIP grade (ab10483) antibodies were obtained from Abcam. TIF1β (H300), TIF1β (23), and ATR (N-19) were obtained from Santa Cruz Biotechnology. ATM (I1987), Chk1 (G282), PKCδ (G499), and MK2 (N266) were obtained from Bioworld. Calf intestine alkaline phosphatase (D2250) was obtained from Takara.
KAP1 mutant S473A, S473D, and S473E plasmids were generated with the QuikChange site-directed mutagenesis kit (Stratagene) using the following primers: S473A, AAACGGTCCCGCGCAGGTGAGGGCG; S473D, GTGAAACGGTCCCGCGATGGTGAGGGCGAGGTG; S473E, GTGAAACGGTCCCGCGAGGGTGAGGGCGAGGTG; S824A, CTGGTGCTGGCCTGAGTGCCCAGGAGC; S824D, GCCTGGTGCTGGCCTGAGTGACCAGGAGCTG; and S824E, TGGTGCTGGCCTGAGTGAGCAGGAGCTGTCTGGTG. GFP-tagged SUMO1 (GFP-SUMO1) plasmid was constructed by digesting the SUMO1 coding sequence from His-SUMO1 plasmid and cloning it into the XhoI site of pCMV-GFP vector. FLAG-tagged KAP1-SUMO1 fusion plasmid (F-KAP1-SUMO1) was made by inserting the SUMO1 coding sequence at the end of FLAG-KAP1 expression plasmid using the following primer pair: F, CGGATATCTCTGGACGACGATGACAAGGGATCTATG; and R, GACTCGAGATAAGATCTGGATCTTAAGGGGCC.
The sequence of the KAP1 knockdown double-stranded oligonucleotide (KAP1 target sequence underlined) was 5′-TGCGTCCTGGCACTAACTCATTCAAGAGATGAGTTAGTGCCAGGACGCTTTTTTC). The FLAG-KAP1–3K/R (K554R/K779R/K804R) sumoylation mutant was generated with the QuikChange site-directed mutagenesis kit (Stratagene) using the following primers: K554R, 5′-GCTGGCATGGCCATTGTCAGGGAGGAGGAG-3′; K779R, 5′-TTCAACAAGTTAACTGAGGACAGGGCAGACGTGCA-3′; and K804R, 5′-CCTTCGGTGACACCAGGTTCTCTGCTGTGCT-3′. To construct the KAP1 knockdown-rescue system, RNAi-resistant KAP1 mutants were generated using the kit described above with the following primer: CGAGGCCTTTGGCAAGATTGTGGCAGAACGGCCGGGAACGAATTCGACAGGCCCTGCAC (mutated nucleotides underlined). Lentivirus-based KAP1 knockdown was carried out using the LentiLox 3.7-DsRed/ZsGreen (pll3.7) lentiviral RNAi expression system. Lentivirus-based KAP1 and its mutant expression plasmids were constructed by using a pLVX-IRES-ZsGreen (Clontech) overexpression system. To establishing lentivirus-mediated knockdown of KAP1 and rescue of KAP1 knockdown, pll3.7-ZsGreen-shKAP1, pll3.7-DsRed-shKAP1, or lenti-FLAG-KAP1 plasmids were co-transfected with PMD2.G and psPAX2 plasmids into 293T cells for 2–3 days, and supernatants containing lentivirus were collected and concentrated. The packaged pll3.7-ZsGreen-shKAP1 was used for established KAP1 stable knockdown cell lines. The packaged pll3.7-DsRed-shKAP1 and lenti-ZsGreen-FLAG-KAP1 were used for established stable KAP1 rescue cell lines.
Real Time and Reverse Transcription-PCR Analysis
The levels of p53AIP1, Noxa, ZNF180, and RNA polymerase II (pol II) gene expression were determined by quantitative RT-PCR. Total RNA was extracted from 293T cells using TRIzol (Invitrogen) extraction reagent. Samples of 500 ng of total RNA were reverse transcribed by using Moloney murine leukemia virus reverse transcriptase (TaKaRa) and random primers. All reactions were performed on a Mx3005P real time PCR system (Stratagene) using the TaKaRa SYBR® Premix Ex TaqTM (Perfect Real Time) kit. The amplification was carried out as follows: initial enzyme activation at 95 °C for 30 s followed by 42 cycles of 95 °C for 5 s, 53 °C for 15 s, and 72 °C 15 s. A total of 60 ng of each diluted reverse transcription product was used for real time PCR in a final volume of 25 μl containing a 200 nm concentration of each specific primer. Each gene expression value was normalized to GAPDH. The following primers were used for real time PCR: p53AIP1: F, TCTTCCTCTGAGGCGAGC; R, AGGTGTGTGTGTCTGAGCCC; ZNF180: F, TGATGCACAATAAGTCGAGCA; R, TGCAGTCAATGTGGGAAGTC; Noxa: F, TGGAAGTCGAGTGTGCTACTCAAC; R, AGATTCAGAAGTTTCTGCCGGAA; GAPDH: F, CCTGTTCGACAGTCAGCCG; R, CGACCAAATCCGTTGACTCC; and pol II: F, GGTCGTTTTTGATGCTCGAT; R, TGGGAAGAGAGCAAATCCAA. E2F1 target gene expression levels were determined by semiquantitative RT-PCR. 500 ng of total RNA from each sample was subjected to PCR with reverse transcription using the same protocol as for real time PCR. PCR was carried out for 20–40 cycles with each cycle consisting of a denaturing step for 30 s at 95 °C, an annealing step for 30 s at 52 °C-55 °C, and a polymerization step for 30 s at 72 °C. The PCR product was separated on a 2.0% agarose gel containing ethidium bromide and photographed under ultraviolet light. The following primer sequences were used: ASK1: F, ACAGCAGATACTCTCAGCC; R, CATTGTCACCCTTTATGTCCC; Apaf-1: F, CACGTTCAAAGGTGGCTGAT; R, TGGTCAACTGCAAGGACCAT; Bim: F, GCTGTCTCGATCCTCCAGTG; R, GTTAAACTCGTCTCCAATACG; caspase 7: F, AGTGACAGGTATGGGCGTTCG; R, GCATCTATCCCCCCTAAAGTGG; p73: F, AACGCTGCCCCAACCACGA; R, GCCGGTTCATGCCCCCTACA; and actin: F, TCCTGTGGCATCCACGAA; R, TCGTCATACTCCTGCTTGC.
Immunoprecipitation Assay
Cells were lysed in lysis buffer (50 mm Tris-HCl, pH 8.0, 5 mm EDTA, 150 mm NaCl, 1% Triton X-100, 1 mm PMSF, 5 mm β-glycerophosphate, 0.1 mm Na3VO4, 5 mm NaF) and centrifuged for 10 min at 10,000 × g, and the insoluble debris was discarded. Cell lysate was immunoprecipitate with anti-FLAG M2-agarose or anti-Myc-agarose beads for 4–12 h at 4 °C.
Immunofluorescence Staining
Cells were seeded on coverslips in 6-well plates and treated with etoposide or transfected with plasmids if needed. Cells were washed in phosphate-buffered saline (PBS), fixed in 4% formaldehyde, immunostained for 2 h with primary antibodies, and washed three times with cold PBS buffer followed by a 1-h exposure to Alexa Fluor 488- or 568-conjugated secondary IgG. Immunofluorescence was visualized by conventional fluorescence microscopy or confocal microscopy.
In Vitro Phosphorylation Assay
FLAG-Chk2 plasmid was transfected into 293T cells for 30 h, and then FLAG-Chk2 protein was immunoprecipitated with anti-FLAG M2-agarose beads. GST-KAP1 protein was expressed in Escherichia coli BL21 (DE3) and purified using glutathione-Sepharose 4B resin (GE Healthcare). Kinase reactions contained Flag-Chk2 and GST-KAP1 substrates in 25 mm Tris-HCl, pH 7.5, 5 mm β-glycerophosphate, 2 mm DTT, 0.1 mm Na3VO4, 2 mm ATP, and 10 mm MgCl2 and were incubated for 30 min at 30 °C. The reaction was stopped by adding 20 μl of 4× SDS loading buffer. Phosphorylation of KAP1 Ser-473 was examined by Western blotting analysis using anti-KAP1 S473p antibody.
In Vivo Sumoylation Assay
The in vivo sumoylation assay was carried out by co-transfection of FLAG-KAP1 or its mutants and GFP-SUMO1 into 293T cells with calcium phosphate. Sumoylated KAP1 was detected by Western blotting analysis using anti-GFP antibody.
siRNA Transfection
To transiently knock down ATM, ATR, Chk1, Chk2, MK2, and PKCδ, short interfering RNA for ATM (5′-UGGUGCUAUUUACGGAGCUtt-3′), ATR (5′-CCUCCGUGAUGUUGCUUGAtt-3′), Chk1 (5′-GCAGUCGCAGUGAAGAUUGtt-3′), Chk2 (5′-GUGUCACUGAAGGAUCAGAUCtt-3′), MK2 (5′-UGACCAUCACCGAGUUUAUdTdT-3′), PKCδ (5′-GGCUGAGUUCUGGCUGGACtt-3′), and an irrelevant RNA oligo (5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized by Shanghai GenePharma Co. Double-stranded siRNA (40 pmol) was transiently transfected into 293T cells with Lipofectamine 2000 (Invitrogen).
WST-1 Cell Viability Assay
Cell survival was measured using the WST-1 assay. Briefly, 5 × 103 cells were cultured in a 96-well flat bottom plate in a final volume of 100 μl/well culture medium for 24 h followed by etoposide treatment. WST-1 reagent (Beyotime, China) was added to each well and then cultured for an additional 2 h. The absorbance of samples was measured at a wavelength of 450 nm using a microtiter plate reader.
Apoptosis Assay
SaoS2 cells in 10-cm plates were transfected with either HA-E2F1 and KAP1 wild type (WT) or KAP1 S473A expression plasmids for 20 h and then treated with etoposide (20 μm) for another 12 h. Cells were stained using the Annexin V-FITC/propidium iodide apoptosis detection kit and measured using a BD Biosciences FACSAria flow cytometer.
Luciferase Reporter Assay
KAP1 knockdown stable HeLa cells were plated in 24-well plates and transfected with a mixture of GAL4-TK-luciferase reporter and GAL4-KRAB or different amounts of RNAi-resistant FLAG-tagged KAP1 WT, S473A, S473D, or S473E plasmids for 36 h. For normalization of transfection efficiency, pRL-TK (Promega), which expressed luciferase from Renilla reniformis under the regulation of the herpes simplex virus TK promoter, was co-transfected. Luciferase was measured using the Dual-Luciferase assay kit (Promega).
RESULTS
KAP1 Is Phosphorylated on Ser-473 in Response to Different Types of DNA Damage
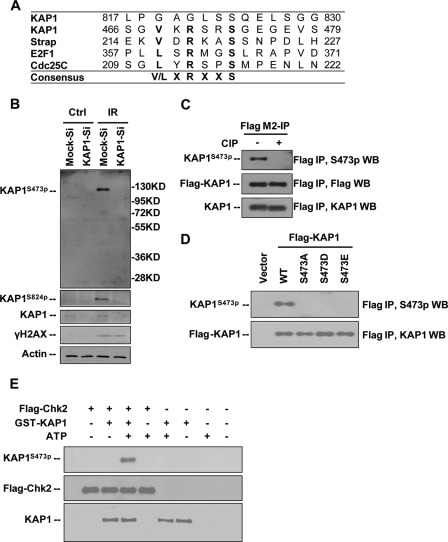
Sequence comparison of KAP1 orthologues revealed that the Ser-473 residue is highly evolutionarily conserved (supplemental Fig. S1), and this sequence fits the consensus sequence for a Chk2 substrate ((V/L)XRXXS) (Fig. 1A). In this study, we generated a KAP1 phospho-Ser-473-specific antibody (anti-S473p) as indicated under “Experimental Procedures,” and the specificity of this antibody was tested. We first characterized endogenous KAP1 phosphorylation in response to γ-irradiation in normal and KAP1 knockdown HeLa cells by Western blotting with anti-S473p antibody (Fig. 1B). A very low level of endogenous KAP1 S473p was detected in untreated cells, but higher levels of KAP1 S473p and S824p were detected in γ-irradiation (IR)-treated cells, and no phosphorylation signal was detected in IR-treated KAP1 knockdown cells. Next, ectopically expressed FLAG-KAP1 was purified from 293T cells with FLAG M2 beads followed by calf intestine alkaline phosphatase treatment and Western blotting with anti-S473p antibody. The representative band for S473p was diminished after calf intestine alkaline phosphatase treatment (Fig. 1C). These results suggest that this anti-S473p antibody can specifically recognize phosphorylated KAP1. To further verify the specificity of this antibody, we constructed plasmids encoding S473A mutant to eliminate phosphorylation and S473D and S473E mutants to mimic constitutive phosphorylation. Ectopically expressed FLAG-tagged KAP1 or mutants were immunoprecipitated from 293T cells with FLAG M2 beads and subjected to Western blotting (Fig. 1D). Unlike WT KAP1, KAP1 Ser-473 mutants were not detected by anti-S473p antibody. Collectively, these results demonstrated that this anti-S473p antibody can specifically recognize phosphorylated KAP1 at Ser-473. With aims to further check the specific of this antibody and to determine whether KAP1 is a direct Chk2 target, an in vitro kinase assay was performed using recombinant GST-KAP1 and 293T cells expressing FLAG-Chk2 kinase. Results showed that KAP1 Ser-473 was phosphorylated by Chk2 in vitro (Fig. 1E). This result further proved that this anti-S473p antibody can be used to detect KAP1 phosphorylation at Ser-473.
FIGURE 1.
Characterization of anti-KAP1 Ser-473 phosphorylation antibody. A, comparison of sequences of Chk2 peptide substrates of Strap, E2F1, and Cdc25C with KAP1 466–479 and KAP1 817–830. The consensus sequence is shown. B, HeLa cells stably infected with lentiviral vectors expressing control (Mock-Si) or KAP1 (KAP1-Si) shRNA were treated with 10 Gy of IR. Cell lysates were collected 5 h after radiation, and KAP1 phosphorylation at Ser-473 (KAP1S473p) and Ser-824 (KAP1S824p) and phosphorylation of H2AX (γH2AX) were analyzed by Western blotting (WB). C, FLAG M2-agarose-purified FLAG-KAP1 from 293T cells was treated with or without calf intestine alkaline phosphatase (CIP) (30 units) at 37 °C for 30 min, and KAP1 S473p and total KAP1 levels were analyzed by Western blotting. D, 293T cells were transfected with FLAG-tagged KAP1 WT or S473A, S473D, or S473E mutant plasmid. Cell lysates were immunoprecipitated with anti-FLAG M2-agarose beads and probed with anti-KAP1 S473p and anti-KAP1 antibodies. E, GST-KAP1 was expressed in E. coli BL21 and purified with GST beads, and FLAG-Chk2 was purified from FLAG-Chk2-transiently transfected 293T cells using anti-FLAG M2-agarose. In the presence or absence of ATP, GST-KAP1 was incubated with or without FLAG-Chk2. The reaction product was separated by SDS-PAGE and analyzed by Western blot analysis. Ctrl, control.
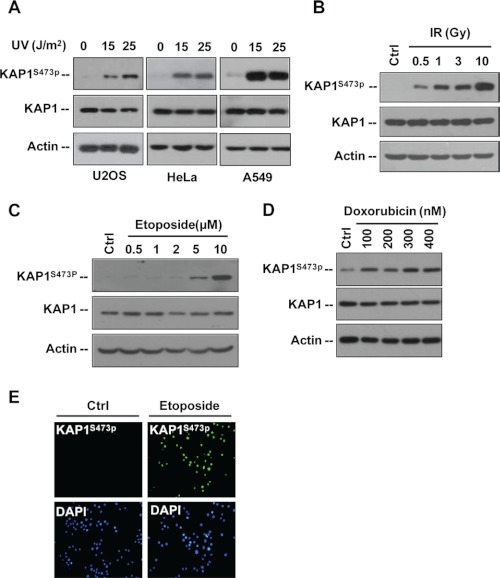
As the cellular DNA damage response is a very complicated signaling network that is activated by various genotoxic agents such as UV light, IR, and chemotherapeutic agents, we next examined the effects of different types of stresses on KAP1 S473p. Western blot analysis revealed that UV light, γ-radiation, etoposide, and doxorubicin could significantly induce KAP1 S473p in HeLa, A549, and U2OS cells in a dose-dependent manner (Fig. 2, A–D). Moreover, HeLa cells were treated with etoposide and subjected to immunofluorescence staining. Comparing with untreated cells, etoposide-treated cells exhibited a much stronger nuclear KAP1 S473p signal (Fig. 2E). These data suggest that KAP1 Ser-473 is a novel site phosphorylated in response to DNA damage.
FIGURE 2.
DNA damage induces KAP1 Ser-473 phosphorylation. A, U2OS, HeLa, and A549 cells were treated with 0, 15, or 25 J/m2 UV irradiation and harvested 12 h later. KAP1 S473p, total KAP1, and actin were analyzed by Western blot assay. B, HeLa cells were treated with different doses of IR as indicated and harvested 6 h later, and the total cell lysates were analyzed by immunoblotting with the indicated antibodies. C, HeLa cells were treated with different concentrations of etoposide for 8 h. KAP1 S473p, KAP1, and actin were analyzed by Western blot assay. D, HeLa cells were treated with different doses of doxorubicin as indicated for 12 h. KAP1 S473p, KAP1, and actin were analyzed by Western blot assay. E, HeLa cells were treated with 0 (Ctrl) or 30 μm etoposide for 8 h, and KAP1 S473p (green) was detected by immunofluorescence staining.
Distinct DNA Damage Signaling Pathways Are Required for KAP1 S473p in Response to Different DNA Damage Stimuli
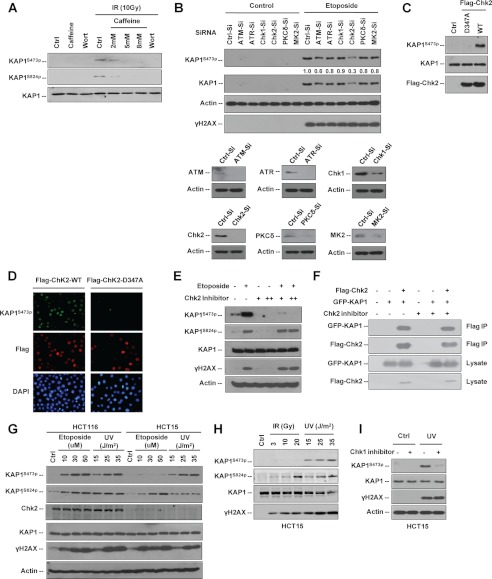
Different types of DNA damage can activate different molecular pathways. DNA damage signals immediately activate two related protein kinases, ATM and ATR, in mammalian cells (29). To determine how KAP1 Ser-473 is phosphorylated in response to DNA damage, we first tested whether KAP1 Ser-473 is a specific substrate for ATM or other PI3K-like serine/threonine protein kinase family members. HeLa cells were treated with two kinase inhibitors, wortmannin as a nonselective inhibitor for all PI3K-like serine/threonine protein kinase family members (30) and caffeine as an inhibitor of ATM and ATR, followed by IR. Both inhibitors decreased IR-induced KAP1 S473p and S824p levels, suggesting that ATM/ATR or their downstream kinases may phosphorylate KAP1 at Ser-473 in response to DNA damage (Fig. 3A).
FIGURE 3.
KAP1 Ser-473 is phosphorylated by ATM-Chk2 and ATR-Chk1 pathways in different cells under different types of DNA damage. A, HeLa cells were pretreated with different doses of wortmannin (Wort) or caffeine as indicated for 1 h and then treated with 10 Gy of γ-radiation. Whole cell lysates were collected 8 h after radiation, KAP1 S473p, KAP1 S824p, and total KAP1 were analyzed by Western blot assay. B, 293T cells were transfected with the indicated short interfering RNA (-Si) for ATM, ATR, Chk1, Chk2, PKCδ, or MK2 or irrelevant siRNA (Ctrl-Si) for 66 h and then treated with etoposide (30 μm) for another 6 h before harvesting. Cell lysates were subjected to Western blot analysis using the indicated antibodies. Relative KAP1 S473p levels were quantitated with NIH ImageJ software and are listed. C, 293T cells were transfected with vector (Ctrl), FLAG-Chk2 WT, or Chk2 kinase-dead mutant FLAG-Chk2 D347A (D347A) for 36 h. Cell lysates were analyzed by immunoblotting with the indicated antibodies. D, HeLa cells were transfected with FLAG-Chk2 WT or FLAG-Chk2 D347A for 24 h. FLAG-Chk2 WT (red) or D347A (red) and KAP1 S473p (green) were detected by immunofluorescence staining. E, HCT116 cells were treated with 0 (−), 5 (+), or 10 mm (++) Chk2 inhibitor II (Sigma-Aldrich, C3742) for 1 h followed by treatment of 30 μm etoposide for 6 h. Cell lysates were subjected to Western blotting as indicated. F, 293T cells were co-transfected with GFP-KAP1 and FLAG-Chk2 for 24 h, and then cells were either left untreated (−) or treated with 10 mm (+) Chk2 inhibitor II for another 2 h. KAP1-Chk2 binding was detected by FLAG M2-agarose bead IP and Western blot assay. G, HCT116 (Chk2 wild type) and HCT15 (Chk2-deficient) cells were treated with different doses of etoposide or UV irradiation as indicated for 6 h, and cell lysates were subjected to Western blot analysis using the indicated antibodies. H, HCT15 cells were treated with different doses of IR or UV irradiation as indicated, cells were harvested 6 h after treatment, and lysates were subjected to Western blotting as indicated. I, HCT15 cells were pretreated with 2 μm Chk1 inhibitor (PD407824, Sigma) for 1 h followed by 25 J/m2 UV treatment. Cells were harvested about 6 h after UV treatment, and cell lysates were then subjected to Western blot analysis as indicated.
As indicated in previous study, PKCδ may be involved in KAP1 S473p in early S phase under normal culture conditions (20). Therefore, to further identify which endogenous kinase(s) is involved in KAP1 S473p in response to DNA damage, PKC, ATM/ATR, and three checkpoint kinases, Chk1, Chk2, and Chk3 (31), were transiently knocked down by siRNA in 293T cells followed by etoposide treatment, and KAP1 S473p levels were examined (Fig. 3B). Results showed that Chk2 and ATM knockdown reduced etoposide-induced KAP1 S473p. Because KAP1 Ser-473 does not have the ATM phosphorylation SQ/TQ motifs (32), we suggest that Chk2 is the major endogenous kinase responsible for KAP1 S473p in response to etoposide treatment. This was confirmed in etoposide-treated Chk2 knockdown HeLa cells (supplemental Fig. S2). To further investigate the role of Chk2 in regulating KAP1 S473p, 293T cells were transiently transfected with FLAG-tagged Chk2 WT or dominant-negative Chk2 D347A plasmid and subjected to Western blot analysis (Fig. 3C). Only Chk2 WT, but not Chk2 D347A mutant, was able to induce KAP1 S473p. Next, HeLa cells were transiently transfected with Chk2 WT or Chk2 D347A plasmid and subjected to immunofluorescence double staining for FLAG-Chk2 and KAP1 S473p. Results showed that cells expressing wild-type Chk2, but not Chk2 D347A mutant, have visible elevated KAP1 S473p staining in the nucleus (Fig. 3D). To further determine whether endogenous Chk2 is required for etoposide-induced KAP1 S473p, Chk2 inhibitor II (33) was also used to abolish Chk2 activity in vivo. Western blot analysis showed that Chk2 inhibitor inhibited both basal and etoposide-induced KAP1 S473p levels in a dose-dependent manner in HCT116 cells, whereas the S824p level was not affected (Fig. 3E). Furthermore, Chk2 and KAP1 binding was detected in 293T cells overexpressing FLAG-Chk2 and GFP-tagged KAP1 (GFP-KAP1) (Fig. 3F), and Chk2 inhibitor showed no effect on KAP1-Chk2 binding.
To study the roles of Chk2 in regulating KAP1 S473p, we examined UV irradiation- and etoposide-induced KAP1 S473p and S824p in HCT15 (Chk2-deficient) cells (34) and HCT116 (Chk2 wild-type) cells. As expected, etoposide treatment could induce KAP1 S473p in HCT116 cells but not in HCT15 cells (Fig. 3G). IR did not significantly increase KAP1 S473p levels in HCT15 cells either (Fig. 3H). Interestingly, UV irradiation induced KAP1 S473p in HCT15 cells (Fig. 3, G and H). We suggest that this may be due to partially functional complementation of Chk1 in response to UV light.
Chk2 and Chk1 share a number of overlapping substrates (35). The ATM-Chk2 pathway is specifically activated by the generation of DNA double strand breaks that are induced by IR or radiomimetic drugs, whereas the ATR-Chk1 pathway by contrast is activated in response to stalled DNA replication forks that are induced by either UV light or drugs such as hydroxyurea (23). Therefore, we suggest that UV-induced KAP1 Ser-473 phosphorylation depends on the ATR-Chk1 pathway. This is further corroborated by the observation that Chk1 inhibitor (PD407824) specifically inhibited UV-induced KAP1 S473p level in HCT15 cells (Fig. 3I). Our data indicate that distinct DNA damage signaling pathways are required for KAP1 S473p in response to different DNA damage stimuli, and Chk2 is the major kinase responsible for KAP1 S473p in the etoposide- or IR-induced stress response, whereas Chk1 is required for KAP1 S473p in response to UV radiation.
Comparison of Two KAP1 Phosphorylation Sites in Response to DNA Damage
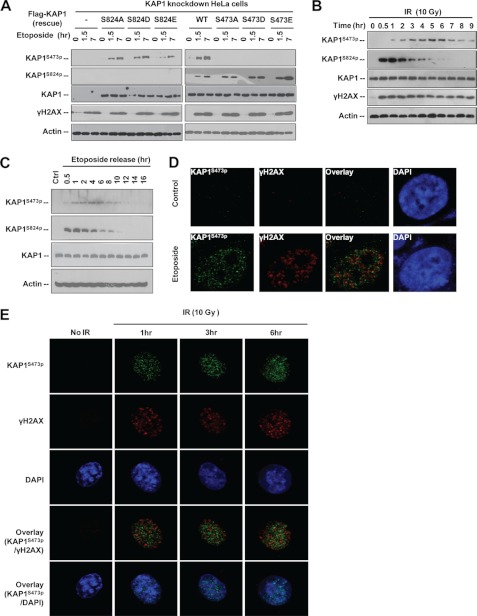
ATM-induced KAP1 S824p is involved in chromatin relaxation, heterochromatic DSB repair, and KAP1-mediated gene silencing (13, 15, 16, 36, 37). Identification of KAP1 S473p modified by Chk2/Chk1 adds additional complexity to analyze the function of KAP1 in regulating the DNA damage response and may reveal a new step be important for DNA repair and cell survival under stress. However, DNA damage-induced KAP1 S473p function remains unknown. To address whether different phosphorylations may have specific roles in the DNA damage response, we first examined whether there is cross-talk between these two modifications. To minimize the interference of endogenous KAP1, a KAP1 knockdown HeLa stable cell line (∼90% reduction) was selected and transfected with siRNA-resistant FLAG-KAP1 WT or S473A, S473D, or S473E mutant followed by etoposide treatment. KAP1 S473p and S824p levels were analyzed by Western blotting (Fig. 4A). Results showed that these two phosphorylations occurred independently. We next compared the kinetics of S824p and S473p in response to γ-radiation. Interestingly, KAP1 S824p was strongly induced by IR and reached the maximal level in 0.5–1 h after IR and then faded rapidly in 2–4 h after IR. In contrast to S824p, KAP1 S473p reached the maximal levels at 5–6 h after IR (Fig. 4B). This is consistent with different kinases being involved in these two modifications. We also examined the putative dephosphorylation rate of these two sites after etoposide withdrawal and found that S824p decreased faster than S473p, whereas the total KAP1 protein level was constant (Fig. 4C), suggesting that different phosphatases may be involved in KAP1 S473p and S824p dephosphorylation. Moreover, KAP1 S824p was reported to repress its sumoylation (36, 37), but we found that phosphomimetic mutations of KAP1 Ser-473 had no effect on its sumoylation (supplemental Fig. S3A). KAP1 sumoylation was reported to inhibit Ser-824 phosphorylation (19), but KAP1 sumoylation was not able to influence Ser-473 phosphorylation (supplemental Fig. S3B). These data indicated that KAP1 S473p is not involved in cross-talk with KAP1 sumoylation. As the Ser-473 residue is adjacent to KAP1 RBCC domains, which are required for KAP1-KRAB interaction and KAP1 self-oligomerization (4), we examined the effect of S473p on KAP1 oligomerization, but S473p did not affect KAP1 trimerization either (supplemental Fig. S3, C and D).
FIGURE 4.
Comparison of KAP1 S824p and S473p in response to DNA damage. A, KAP1 knockdown stable HeLa cells were transfected with vector (−) or siRNA-resistant FLAG-KAP1 WT or Ser-473 or Ser-824 site mutants for 24 h, and then treated with etoposide (30 μm) for 1.5 and 7 h. Cell lysates were subjected to Western blot analysis using the indicated antibodies. B, HeLa cells were treated with IR and harvested at different time points. Cell lysates were subjected to Western blot analysis with antibodies as indicated. C, HeLa cells were pretreated with 30 μm etoposide for 1 h and then transferred to drug-free medium. Cells were harvested at different time point as indicated, and cell lysates were subjected to Western blotting. D, HeLa cells were treated with 0 (control) or 30 μm etoposide for 8 h, fixed, and stained for KAP1 S473p (green) and γH2AX (red). DNA (blue) is stained with DAPI. The images were analyzed by confocal microscopy. E, HeLa cells were irradiated with 10 Gy of IR, harvested at different time points, fixed, and stained for KAP1 S473p (green) and γH2AX (red). DNA (blue) is stained with DAPI. Ctrl, control.
In response to DNA damage, histone H2AX is phosphorylated at Ser-139 (γH2AX) by ATM/ATR and forms foci at DNA lesions. Several DNA repair factors such as 53BP1, TopBP1, and BRCA1 co-localize with γH2AX and are involved in the DNA repair process (15, 16, 38). Ser-824-phoshorylated KAP1 co-localizes with γH2AX, 53BP1, TopBP1, and BRCA1 after ionizing radiation (39). To determine whether Ser-473-phosphorylated KAP1 is recruited to the DNA damage sites, etoposide-treated (Fig. 4D) and IR-treated HeLa cells (Fig. 4E) were subjected to immunofluorescence staining using anti-S473p and anti-γH2AX antibodies. Interestingly, KAP1 S473p showed a nuclear punctate pattern in both etoposide- and IR-treated cells, but only a small portion of KAP1 S473p punctate staining (less than 5%) co-localized with γH2AX, and this is very different from KAP1 S824p distribution (15, 16). Collectively, these data suggest that KAP1 S473p and S824p have different functions.
DNA Damage-induced KAP1 Ser-473 Phosphorylation Attenuates Its Binding to HP1 Family Proteins
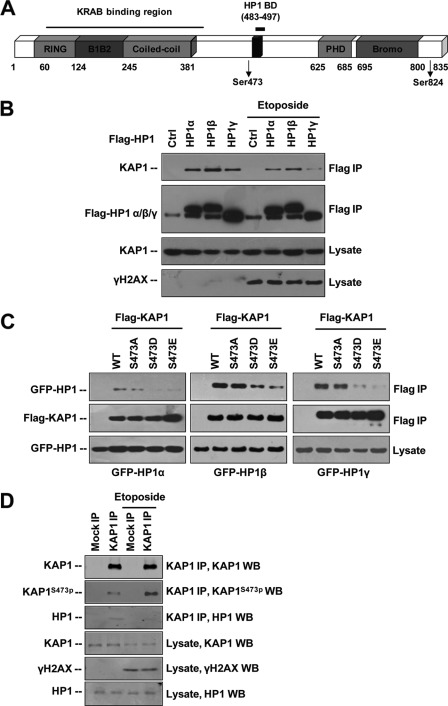
The KAP1 Ser-473 residue is located close to the HP1-interacting PXVXL motif (HP1BD; amino acids 486–490) (7) (Fig. 5A). Previous study has shown that the interaction between KAP1 S473A mutant and HP1β is stronger than that of KAP1 S473E mutant and HP1β (20). As KAP1-HP1 interaction is crucial for chromatin dynamics, maintenance, and gene regulation (8), we examined whether DNA damage-induced KAP1 S473p can regulate its binding to HP1 family proteins (HP1 α, β, and γ). HeLa cells were transfected with FLAG-tagged HP1 α, β, or γ plasmids and treated with etoposide. Western blotting of immunoprecipitated FLAG-HP1 complexes using anti-KAP1 antibody revealed a decreased level of co-immunoprecipitated KAP1 after etoposide treatment, indicating the etoposide treatment could reduce KAP1-HP1 binding (Fig. 5B). In a reverse IP, 293T cells were co-transfected with FLAG-tagged KAP1 WT or Ser-473 mutants (S473A, S473D, or S473E) and GFP-tagged HP1 (α, β, or γ) plasmids. Ser-473 constitutive phosphorylation-mimicking mutants (S473D and S473E) inhibited the KAP-HP1 interaction (Fig. 5C). The binding between endogenous KAP1 and HP1 was also inhibited by etoposide treatment (Fig. 5D). These results indicate that DNA damage promotes phosphorylation of KAP1 on Ser-473, thereby attenuating the binding of KAP1 to HP1 family proteins. To further delineate how KAP1 S473p regulates its binding affinity to HP1, we used the known amino acid sequence of KAP1 WT and S473E mutant to derive a predicted protein structure of KAP1 amino acids 423–584 based on multiple threading alignments by LOMETS and iterative TASSER simulations using the I-TASSER server (40). The HP1 binding domain (amino acids 483–497) of KAP1 S473E is significantly altered compared with the wild-type model (supplemental Fig. S4), suggesting that S473p might change the structure of KAP1 HP1BD and attenuate the KAP1-HP1 interaction.
FIGURE 5.
DNA damage-induced KAP1 S473p attenuates KAP1 binding affinity to HP1 family proteins by changing structure at HP1 binding domain. A, schematic representation of the domain organization of KAP1. Bromo, bromodomain. Phosphorylated residues Ser-473 and Ser-824 are indicated. B, HeLa cells were transfected with vector (ctrl) or FLAG-HP1α, β, or γ for 24 h followed by 30 μm etoposide treatment for 12 h. KAP1-HP1 binding was detected by anti-FLAG M2-agarose bead immunoprecipitation and Western blot assay. C, FLAG-KAP1 WT or Ser-473 site mutant plasmids were co-transfected with GFP-HP1 α-, β-, or γ-overexpressing plasmids as indicated into 293T cells for 24 h. KAP1-HP1 binding was detected by anti-FLAG M2-agarose bead IP and Western blot assay. D, HeLa cells were treated with etoposide for 6 h, endogenous KAP1 was precipitated using KAP1 antibody (ab10483), and co-precipitation of endogenous HP1 was detected by Western blot.
KAP1 Ser-473 Phosphorylation Resulted in Loss of KRAB Domain-mediated Transcription Repression and Inhibited Its Transcriptional Repression Activity to KRAB-ZNFs
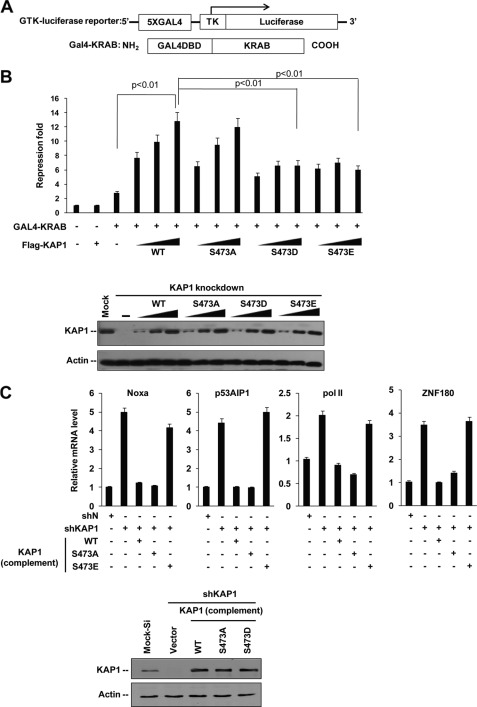
Because HP1 facilitates KRAB-KAP1-mediated transcriptional repression (5, 7, 8), we examined whether KAP1 S473p regulates KRAB-KAP1-mediated repression by a luciferase reporter assay using a GAL4-TK-luciferase reporter and a GAL4-KRAB repressor (Fig. 6A). Results showed that both S473D and S473E mutants can reduce KRAB-KAP1 transcriptional activity, suggesting that DNA damage-induced S473p plays a role in KRAB-KAP1-mediated repression (Fig. 6B).
FIGURE 6.
Phosphorylation of KAP1 Ser-473 regulates its transcription co-repression activity. A, schematic illustration of GAL4-TK-luciferase reporter (GTK-luciferase reporter) and GAL4-KRAB repressor protein. B, KAP1 knockdown stable HeLa cells were co-transfected with GAL4-TK-luciferase reporter and Renilla luciferase plasmid or different amounts of RNAi-resistant FLAG-tagged KAP1 WT, S473A, S473D, or S473E plasmid in the presence or absence of GAL4-KRAB plasmid. Relative luciferase repression (upper panel) was calculated by comparing firefly luciferase activity with Renilla luciferase activity. Error bars represent mean ± S.D. (n = 4) in each case. Phosphorylation-mimicking mutants (S473D and S473E) increased (p < 0.01) KAP1 transcriptional repression activity compared with WT KAP1, and there is no significant difference between wild-type KAP1 and S473A mutant. KAP1 levels were confirmed by Western blot (lower panel). C, HeLa cells were infected with pll3.7-DsRed (shN) or pll3.7-DsRed-shKAP1 (shKAP1) lentiviral vectors, and siRNA-resistant lenti-ZsGreen-FLAG-KAP1 (WT, S473A, S473D, or S473E) plasmids for 60 h, green and red positive cells were collected by FACS, a quantitative RT-PCR assay was performed to analyze KAP1-ZNF target gene expression as indicated, and KAP1 expression was detected by Western blots.
Moreover, KAP1 was found to cooperate with KRAB-ZFP Apak (ZNF420) to repress p53 activity, and knockdown of KAP1 increased transcription of p53AIP1 and Noxa (41). KAP1 also binds to KRAB-ZFP ZNF274 and is recruited to KRAB-ZFP ZNF180 promoter (42), cooperating with KRAB-ZFP KS1 (ZNF382) to repress RNA pol II transcription (43). Therefore, we examined whether KAP1 S473p regulates its transcription repression of p53AIP1, Noxa, ZNF180, and RNA pol II genes by quantitative RT-PCR assay (Fig. 6C). To minimize the interference of endogenous KAP1, KAP1 knockdown and rescue cell lines were used. Results showed that KAP1 knockdown increased the expression of these genes. The KAP1 wild type and S473A mutant complements significantly repressed these genes, whereas phosphomimetic KAP1 S473E mutant rescue did not have such effects. Thus, KAP1 S473p inhibits its transcriptional repression of KRAB-ZNFs.
Roles of KAP1 Ser-473 Phosphorylation in Regulating Apoptosis and Cell Survival in Response to DNA Damage
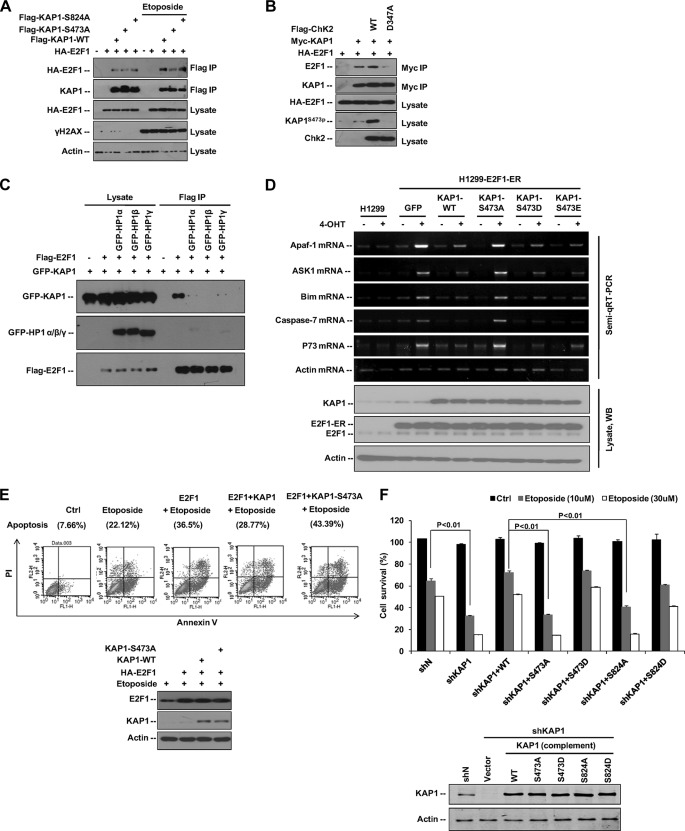
Our previous study found that KAP1 inhibits E2F1-induced apoptosis in response to DNA damage (10). However, the mechanism by which KAP1 regulates E2F1-induced apoptosis in response to DNA damage remains to be elucidated. Therefore, we first examined the interaction between E2F1 and KAP1 under DNA damage stress. 293T cells transfected with HA-E2F1 and FLAG-KAP1 WT, S473A, or S824A plasmids were treated with etoposide, and the E2F1-KAP1 binding was analyzed (Fig. 7A). Etoposide treatment increased the E2F1-KAP1 WT and E2F1-KAP1 S824A interactions but not E2F1-KAP1 S473A binding. Thus, KAP1 S473p may increase KAP1-E2F1 interaction in response to DNA damage. Next, we examined whether Chk2 regulates KAP1-E2F1 interaction and found that KAP1-E2F1 binding increased in the presence of wild-type Chk2 but not Chk2 D473A mutant (Fig. 7B). Taken together, these data indicate that DNA damage-induced KAP1 phosphorylation at Ser-473 increases its ability to bind E2F1.
FIGURE 7.
KAP1 Ser-473 phosphorylation regulates E2F1 and etoposide-induced cell apoptosis. A, HA-tagged E2F1 was co-transfected with FLAG-KAP1 WT, S473A, or S824A expression plasmid into 293T cells for 24 h followed by treatment with etoposide (30 μm) for another 10 h, and KAP1-E2F1 binding was detected by anti-FLAG M2-agarose bead IP and Western blot assay. B, 293T cells were co-transfected with HA-E2F1, Myc-KAP1, FLAG-Chk2 WT, or FLAG-Chk2 D347A plasmid as indicated for 36 h. KAP1-E2F1 binding was detected by anti-Myc-agarose IP and Western blot assay. C, 293T cells were transfected with FLAG-tagged E2F1, GFP-tagged KAP1, and HP1 expression plasmids as indicated. Cells were harvested 36 h after transfection, and E2F1-KAP1 binding was detected by anti-FLAG M2-agarose bead IP and Western blot assay. D, an H1299 cell line stably expressing the E2F1-ER construct was transfected with KAP1 wild type or Ser-473 mutant plasmid as indicated for 24 h, and then cells were either left untreated (−) or treated with 100 nm 4-hydroxytamoxifen (4-OHT) (+) for another 12 h. The mRNA levels of E2F1-mediated apoptosis genes (Apaf-1, ASK1, Bim, p73, and caspase 7) were determined by semiquantitative RT (Semi-qRT)-PCR, and KAP1 protein expression levels were analyzed by Western blot assay. E, SaoS2 cells were co-transfected with either vector (Ctrl) or HA-E2F1 and KAP1 WT or KAP1 S473A expression plasmids for 20 h, and then cells were treated with etoposide (20 μm) for another 12 h. Cells were stained with Annexin V-FITC and propidium iodide (PI), and analyzed by FACS (upper panel). KAP1, E2F1, and actin protein levels were examined by Western blot (WB) (lower panel). F, Mock control (shN) or KAP1 knockdown (shKAP1) HeLa cells were infected with siRNA-resistant lenti-ZsGreen-FLAG-KAP1 (WT, S473A, S473D, S824A, or S824D) for 36 h followed by treatment with different doses of etoposide for another 24 h. Cell survival was analyzed by WST-1 assay. Error bars represent mean ± S.D. (n = 4) in each case.
As indicated above, the Ser-473-phosphorylated KAP1 showed lower binding affinity to HP1 family proteins. To address how KAP1 Ser-473 phosphorylation could decrease KAP1-HP1 binding but increase KAP1-E2F1 binding, we examined whether HP1 competes with E2F1 for binding to KAP1. Results showed that HP1 overexpression dramatically inhibited E2F1 binding to KAP1 (Fig. 7C). These results indicate that DNA damage-induced dissociation of HP1 from KAP1 may lead to an increase in KAP1-E2F1 binding and also suggest that DNA damage-induced KAP1 S473p may constitute a molecular switch that regulates specific gene expression in response to DNA damage.
E2F1 has well documented target genes that are proapoptotic such as Apaf-1; p73; ASK1; caspases 3, 7, and 9; and Bim (44–47). We next examined whether phosphorylation of KAP1 Ser-473 may regulate E2F1-mediated proapoptotic gene expression in response to DNA damage. An H1299 cell line stably expressing an E2F1-ER construct was transfected with plasmids for KAP1 WT or S473A, S473D, or S473E mutant and then treated with 4-hydroxytamoxifen to activate the E2F1 fusion protein (9, 48). The ASK1, Apaf-1, Bim, p73, and caspase 7 mRNA levels were measured by semiquantitative RT-PCR (Fig. 7D). Results showed that E2F1 activity in 4-hydroxytamoxifen-induced ER-E2F1 cells was effectively repressed by overexpression of wild-type or S473D/E mutant KAP1 but not by S473A mutant, suggesting that KAP1 S473p may increase KAP1 repression of E2F1-induced proapoptotic gene expression.
E2F1 is critical for etoposide-induced apoptosis in the absence of p53 (49). To determine whether KAP1 S473p is required for KAP1 to repress E2F1-induced apoptosis, Saos2 (p53-, mdm2-, and Rb-null) cells co-transfected with E2F1 and KAP1 WT or S473A expression plasmids were treated with etoposide and then analyzed for apoptosis by an Annexin V apoptosis assay (Fig. 7E). Results showed that wild-type KAP1 inhibited E2F1-induced apoptosis, whereas S473A did not, suggesting that DNA damage-induced KAP1 S473p may increase KAP1-E2F1 interaction and further inhibit E2F1-induced apoptosis. Finally, we used molecular replacement experiments using a knockdown-rescue approach to show that KAP1 S473p increased cell survival after etoposide treatment, whereas KAP1 S473A mutant significantly increased etoposide-induced cell death in HeLa cells (Fig. 7F). Loss of KAP1 Ser-824 phosphorylation (S824A) also increased cellular sensitivity to DNA damage, which is consistent with a previous report (15). These results suggest that KAP1 S824p and S473p have a synergistic impact on cell survival in response to DNA damage and that KAP1 S473p can inhibit E2F1-induced apoptosis by enhancing its binding to E2F1.
KAP1 Ser-473 Mutant Is Associated with Defective DNA Repair
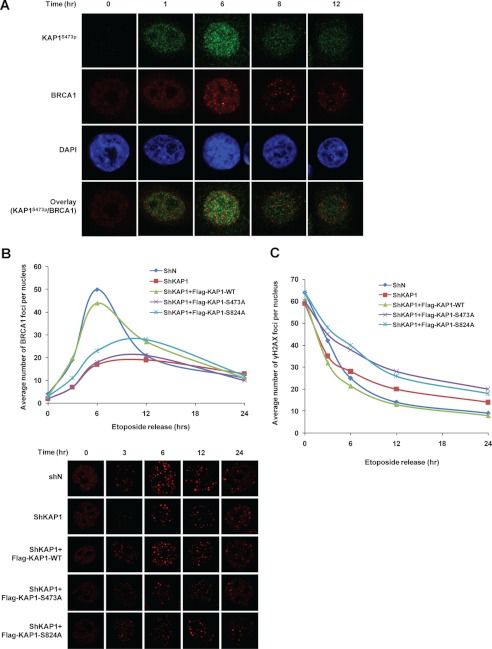
Growing evidence suggests that KAP1 Ser-824 phosphorylation is associated with regulation of chromatin relaxation (15) and heterochromatic DNA DSB repair (17) and that Ser-824-phosphorylated KAP1 co-localizes with various damage-responsive proteins including H2AX, 53BP1, TopBP1, and BRCA1 at DNA lesions with differing kinetics (39). Because DNA damage also induces KAP1 Ser-473 phosphorylation and S473p could negatively regulate DNA damage-induced cell death, we thus examined the potential role of KAP1 S473p in DNA repair. A previous study also reported that Ser-824-phosphorylated KAP1 co-localizes with 53BP1 and γH2AX within 3 h after 9 Gy of ionizing radiation, whereas it co-localizes with BRCA1 about 6–8 h after radiation (39). Interestingly, in this study, we observed that KAP1 S473p reached its peak level at 5–6 h after 10 Gy of ionizing radiation (Fig. 4B). Thus, we first determined the co-localization of BRCA1 and Ser-473-phosphorylated KAP1 in response to DNA damage. HeLa cells were pretreated for 1 h with etoposide and then released, and the distributions of BRCA1 and Ser-473-phosphorylated KAP1 at different time points were observed by confocal microscopy. However, no obvious co-localization of BRCA1 and Ser-473-phosphorylated KAP1 was observed (Fig. 8A).
FIGURE 8.
A, HeLa cells were treated with 20 μm etoposide for 1 h and washed with PBS three times. Cells were collected at different time points after etoposide withdrawal, and KAP1 S473p (green), BRCA1 (red), and DAPI (blue) were detected by immunofluorescence staining. B, Mock control or KAP1 knockdown (shKAP1) stable HeLa cells were co-transfected with GFP and KAP1 siRNA-resistant FLAG-tagged KAP1 wild type, S473A, or S824A plasmid for 24 h, and then cells were treated the same as in A. Cells were stained for BRCA1 (red) (lower panels) and analyzed by confocal microscopy. The average number of BRCA1 foci per nucleus of GFP-positive cells was quantified (upper panel). A minimum of 30 cells per sample were counted in triplicate. C, the average number of γH2AX foci per nucleus was determined using the same assay as in B.
We next examined whether KAP1 or KAP1 S473p could influence the dynamics of BRCA1 focus formation after DNA damage. KAP1 stable knockdown and transiently rescued cell lines were pretreated for 1 h with etoposide and then released, and BRCA1 foci were enumerated (Fig. 8B). Interestingly, knockdown of KAP1 decreased BRCA1 focus formation after DNA damage, and recomplementation of wild-type KAP1 significantly increased BRCA1 focus formation, whereas recomplementation of S473A or S824A mutant showed less BRCA1 focus formation. These data indicate that DNA damage-induced KAP1 S473p participates in BRCA1 focus formation. The kinetics of γH2AX focus disappearance closely resembles the kinetics of DSB repair (50–52). We next studied the function of KAP1 S473p in response to DNA damage. The DNA repair dynamics was characterized by counting the loss of γH2AX foci in etoposide-treated KAP1 stable knockdown and rescue cell lines (Fig. 8C). Results showed that KAP1-deficient cells or recomplementation of KAP1 S473A or S824A mutant has slower kinetics of loss of γH2AX foci, indicating a reduced ability to carry out DNA DSB repair. Taken together, these results indicate that KAP1 S473p participates in BRCA1 focus formation and is required for efficient DNA repair in response to DNA damage.
DISCUSSION
In this study, we showed that, depending on the type of DNA damage that occurs, KAP1 Ser-473 is phosphorylated by different DNA damage signaling pathways. Chk2 is the major kinase responsible for KAP1 S473p in the etoposide- or IR-induced stress response, whereas Chk1 is required for KAP1 S473p in response to UV radiation. Consistent with our results, the most recent report of Blasius et al. (22) also identified human Chk1 and Chk2 as two novel KAP1 Ser-473 kinases. However, they have not carried out any functional studies of these phosphorylation changes. In this study, we found that KAP1 S473p proceeds in a slower kinetic manner compared with S824p and also showed that DNA damage-induced KAP1 S473p attenuates its binding to HP1 family proteins, which may lead to an increased binding of KAP1-E2F1 and reduce the ability of E2F1 to activate the expression of a subset of proapoptotic genes and apoptosis. Intriguingly, we found that loss of phosphorylation on KAP1 Ser-473 leads to less BRCA1 focus formation, slower kinetics of γ-H2AX focus loss, and more cell death after DNA damage. These findings represent the first report of the functions of KAP1 Ser-473 phosphorylation in DNA repair and cell survival under DNA damage stress.
KAP1 may serve as a universal co-repressor for the over 200 zinc finger-containing site-specific transcriptional repressors. The functional model for KAP1-mediated KRAB-ZFPs transcriptional regulation is that KRAB-ZFPs bind to their corresponding DNA sequence, triggering the recruitment of KAP1 and subsequently forming a scaffold complex containing HP1, SETDB1, and histone deacetylases to silence gene expression by forming facultative heterochromatin at the targeted locus (53). The intact PHD finger and bromodomain of KAP1 and the KAP1-HP1 interaction are both required for KAP1 transcriptional repression. Sumoylation at the PHD finger and bromodomain of KAP1 was reported to be required for KAP1-mediated gene silencing by directly recruiting the SETDB1 histone methyltransferase and the CHD3/Mi2 component of the Mi-2/nucleosome remodeling and deacetylase complex (13). ATM was found to selectively stimulate KAP1 Ser-824 phosphorylation in the PHD domain, leading to repressed KAP1 sumoylation and attenuating its transcriptional repression activity. These studies suggest that ATM-regulated KAP1 transcriptional repression activity depends mainly on interfering with KAP1 sumoylation (14, 15, 19). The KAP1-HP1 interaction is essential for KAP1 co-repressor activity, and disruption of the interaction between KAP1 and HP1 cripples the co-repressor activity of KAP1. In this study, we show that in response to DNA damage Chk2 and Chk1 specifically phosphorylate KAP1 at residue Ser-473 located near the HP1-interacting PXVXL motif. KAP1 S473p may change its protein structure at HP1BD. Phosphomimetic mutants of KAP1, S473D and S473E, reduced KRAB domain-mediated transcription repression and failed to repress KRAB-ZNF downstream gene transcription. Therefore, DNA damage-activated ATM-Chk2 and ATR-Chk1 kinase cascades may sequentially phosphorylate KAP1 PHD (Ser-824) and HP1BD domain (Ser-473), and S824p and S473p may corporately regulate KAP1 transcriptional repression activity in response to DNA damage.
Genomic analysis of KAP1 binding has identified about 7000 KAP1 binding sites throughout the human genome (54). Interestingly, the identified KAP1 targets are highly enriched for C2H2 ZNFs, particularly those containing KRAB domains, suggesting that KAP1 may also be involved in mediating transcription of numerous KRAB-ZFPs (42, 54). Our data clearly show that phosphomimetic mutants S473D and S473E decrease KAP1 transcriptional repression of p53AIP1, Noxa, RNA pol II, and KRAB-ZNF protein ZNF180 genes, suggesting that Chk2/Chk1 and KAP1 may coordinate to regulate the expression of some KRAB-ZNFs or their target genes. By mediating KAP1 phosphorylation, ATM-Chk2 and ATR-Chk1 kinase cascades may be involved in the finely tuned regulation of numerous Kruppel-type zinc finger proteins, which are supposed to be the largest class of transcription factors in the human genome.
In response to DNA damage, ATM-dependent Ser-824-phosphorylated KAP1 co-localizes well with several DNA damage response proteins including γH2AX, TopBP1, and BRCA1 at DNA lesions and is required with ATM for DSB repair. By comparing the kinetics of phosphorylation of KAP1 Ser-824 and Ser-473, we identified that in response to DNA damage like IR KAP1 S824p is induced earlier than S473p, and S824p fades faster than S473p. Although no obvious co-localization of Ser-473-phosphorylated KAP1, γH2AX, and BRCA1 was found, we observed that loss of KAP1 or restoration of KAP1 S473A mutant in KAP1 knockdown cells led to a BRCA1 focus formation defect and slower kinetics of γH2AX focus loss after DNA damage. These results suggest that phosphorylation of Ser-824 and Ser-473 of KAP1 participates in DNA repair in a spatial and temporal manner, and these sequential modifications may act synergistically to repair damaged DNA and contribute to cell survival under stress conditions.
In this study, we also revealed that DNA damage-induced phosphorylation of KAP1 Ser-473 increases its binding to E2F1 and reduces the ability of E2F1 to activate the expression of a subset of proapoptotic genes and apoptosis. Members of the E2F family, particularly E2F1, have important roles in regulating cell proliferation and apoptosis (44, 55). DNA damage is known to activate the ATM-Chk2-E2F1 apoptosis pathway (28, 56), which makes E2F1 a versatile target to develop anticancer chemotherapy drugs such as etoposide and doxorubicin. However, DNA damage always triggers negative regulatory networks to inhibit E2F1-induced apoptosis, which makes the tumor cells insensitive to chemotherapy drugs targeting E2F1 activation. For example, our previous study established that, to antagonize etoposide-induced E2F1 activity, an NAD+-dependent class III deacetylase SirT1 is transcriptionally activated by E2F1, and SirT1 interacts with E2F1 and inhibits E2F1-induced apoptosis (48). We also reported that KAP1 inhibits E2F1 activity by recruiting histone deacetylase 1 to deacetylate E2F1, and knockdown KAP1 increases etoposide-induced cell death in pRb-deficient Saos2 tumor cells (10). In this study, we extend our research and demonstrate that KAP1 S473p increases E2F1-KAP1 binding, and KAP1 S473A mutant overexpression increases etoposide-induced cell death. Our data suggest that Chk2-actived KAP1 phosphorylation may counter Chk2-induced E2F1 activity in response to DNA damage. Combination chemotherapy using etoposide and appropriate peptides that block KAP1 S473p may be useful to induce tumor cell death.
Supplementary Material
This work was supported by the National Natural Science Foundation of China (Grants 31071248 and 31171361), the National Basic Research Program of China (Grant 2009CB918401), the program for New Century Excellent Talents in University (Grant NCET-08-0194), and the Science and Technology Commission of Shanghai Municipality (Grant 11DZ2260300) (to C. W.).

This article contains supplemental Figs. S1 and S2.
- KRAB
- Kruppel-associated box
- S473p
- Ser-473 phosphorylation
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM- and Rad3-related
- ZNF
- zinc finger
- KAP1
- KRAB-associated protein 1
- ZFP
- zinc finger protein
- PHD
- plant homeodomain
- HP1
- heterochromatin protein 1
- HP1BD
- HP1 binding domain
- DSB
- double strand break
- S824p
- Ser-824 phosphorylation
- pol
- polymerase
- F
- forward
- R
- reverse
- TK
- thymidine kinase
- IR
- γ-irradiation
- IP
- immunoprecipitation
- ER
- estrogen receptor
- Gy
- grays.
REFERENCES
- 1. Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., Neilson E. G., Rauscher F. J., 3rd (1996) KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10, 2067–2078 [DOI] [PubMed] [Google Scholar]
- 2. Cammas F., Mark M., Dollé P., Dierich A., Chambon P., Losson R. (2000) Mice lacking the transcriptional corepressor TIF1β are defective in early postimplantation development. Development 127, 2955–2963 [DOI] [PubMed] [Google Scholar]
- 3. Weber P., Cammas F., Gerard C., Metzger D., Chambon P., Losson R., Mark M. (2002) Germ cell expression of the transcriptional co-repressor TIF1β is required for the maintenance of spermatogenesis in the mouse. Development 129, 2329–2337 [DOI] [PubMed] [Google Scholar]
- 4. Peng H., Begg G. E., Schultz D. C., Friedman J. R., Jensen D. E., Speicher D. W., Rauscher F. J., 3rd (2000) Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J. Mol. Biol. 295, 1139–1162 [DOI] [PubMed] [Google Scholar]
- 5. Lechner M. S., Begg G. E., Speicher D. W., Rauscher F. J., 3rd (2000) Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol. Cell. Biol. 20, 6449–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schultz D. C., Friedman J. R., Rauscher F. J., 3rd (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 15, 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ryan R. F., Schultz D. C., Ayyanathan K., Singh P. B., Friedman J. R., Fredericks W. J., Rauscher F. J., 3rd (1999) KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 19, 4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sripathy S. P., Stevens J., Schultz D. C. (2006) The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 26, 8623–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang C., Hou X., Mohapatra S., Ma Y., Cress W. D., Pledger W. J., Chen J. (2005) Activation of p27Kip1 expression by E2F1. A negative feedback mechanism. J. Biol. Chem. 280, 12339–12343 [DOI] [PubMed] [Google Scholar]
- 10. Wang C., Rauscher F. J., 3rd, Cress W. D., Chen J. (2007) Regulation of E2F1 function by the nuclear corepressor KAP1. J. Biol. Chem. 282, 29902–29909 [DOI] [PubMed] [Google Scholar]
- 11. Okamoto K., Kitabayashi I., Taya Y. (2006) KAP1 dictates p53 response induced by chemotherapeutic agents via Mdm2 interaction. Biochem. Biophys. Res. Commun. 351, 216–222 [DOI] [PubMed] [Google Scholar]
- 12. Wang C., Ivanov A., Chen L., Fredericks W. J., Seto E., Rauscher F. J., 3rd, Chen J. (2005) MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 24, 3279–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ivanov A. V., Peng H., Yurchenko V., Yap K. L., Negorev D. G., Schultz D. C., Psulkowski E., Fredericks W. J., White D. E., Maul G. G., Sadofsky M. J., Zhou M. M., Rauscher F. J., 3rd (2007) PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mascle X. H., Germain-Desprez D., Huynh P., Estephan P., Aubry M. (2007) Sumoylation of the transcriptional intermediary factor 1β (TIF1β), the co-repressor of the KRAB Multifinger proteins, is required for its transcriptional activity and is modulated by the KRAB domain. J. Biol. Chem. 282, 10190–10202 [DOI] [PubMed] [Google Scholar]
- 15. Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D. C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. (2006) Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8, 870–876 [DOI] [PubMed] [Google Scholar]
- 16. Noon A. T., Shibata A., Rief N., Löbrich M., Stewart G. S., Jeggo P. A., Goodarzi A. A. (2010) 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat. Cell Biol. 12, 177–184 [DOI] [PubMed] [Google Scholar]
- 17. Goodarzi A. A., Noon A. T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., Jeggo P. A. (2008) ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31, 167–177 [DOI] [PubMed] [Google Scholar]
- 18. Goodarzi A. A., Kurka T., Jeggo P. A. (2011) KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 18, 831–839 [DOI] [PubMed] [Google Scholar]
- 19. Li X., Lee Y. K., Jeng J. C., Yen Y., Schultz D. C., Shih H. M., Ann D. K. (2007) Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J. Biol. Chem. 282, 36177–36189 [DOI] [PubMed] [Google Scholar]
- 20. Chang C. W., Chou H. Y., Lin Y. S., Huang K. H., Chang C. J., Hsu T. C., Lee S. C. (2008) Phosphorylation at Ser473 regulates heterochromatin protein 1 binding and corepressor function of TIF1β/KAP1. BMC Mol. Biol. 9, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bennetzen M. V., Larsen D. H., Bunkenborg J., Bartek J., Lukas J., Andersen J. S. (2010) Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol. Cell. Proteomics 9, 1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blasius M., Forment J. V., Thakkar N., Wagner S. A., Choudhary C., Jackson S. P. (2011) A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 12, R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bartek J., Falck J., Lukas J. (2001) CHK2 kinase—a busy messenger. Nat. Rev. Mol. Cell. Biol. 2, 877–886 [DOI] [PubMed] [Google Scholar]
- 24. Hirao A., Cheung A., Duncan G., Girard P. M., Elia A. J., Wakeham A., Okada H., Sarkissian T., Wong J. A., Sakai T., De Stanchina E., Bristow R. G., Suda T., Lowe S. W., Jeggo P. A., Elledge S. J., Mak T. W. (2002) Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol. Cell. Biol. 22, 6521–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shieh S. Y., Ahn J., Tamai K., Taya Y., Prives C. (2000) The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14, 289–300 [PMC free article] [PubMed] [Google Scholar]
- 26. Ahn J., Prives C. (2002) Checkpoint kinase 2 (Chk2) monomers or dimers phosphorylate Cdc25C after DNA damage regardless of threonine 68 phosphorylation. J. Biol. Chem. 277, 48418–48426 [DOI] [PubMed] [Google Scholar]
- 27. Adams C. J., Graham A. L., Jansson M., Coutts A. S., Edelmann M., Smith L., Kessler B., La Thangue N. B. (2008) ATM and Chk2 kinase target the p53 cofactor Strap. EMBO Rep. 9, 1222–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stevens C., Smith L., La Thangue N. B. (2003) Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 5, 401–409 [DOI] [PubMed] [Google Scholar]
- 29. Durocher D., Jackson S. P. (2001) DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell Biol. 13, 225–231 [DOI] [PubMed] [Google Scholar]
- 30. Hall T. J., Jeker H., Schaueblin M. (1995) Wortmannin, a potent inhibitor of phosphatidylinositol 3-kinase, inhibits osteoclastic bone resorption in vitro. Calcif. Tissue Int. 56, 336–338 [DOI] [PubMed] [Google Scholar]
- 31. Manke I. A., Nguyen A., Lim D., Stewart M. Q., Elia A. E., Yaffe M. B. (2005) MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol. Cell 17, 37–48 [DOI] [PubMed] [Google Scholar]
- 32. Kim S. T., Lim D. S., Canman C. E., Kastan M. B. (1999) Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274, 37538–37543 [DOI] [PubMed] [Google Scholar]
- 33. Arienti K. L., Brunmark A., Axe F. U., McClure K., Lee A., Blevitt J., Neff D. K., Huang L., Crawford S., Pandit C. R., Karlsson L., Breitenbucher J. G. (2005) Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimidazoles. J Med. Chem. 48, 1873–1885 [DOI] [PubMed] [Google Scholar]
- 34. Wu X., Webster S. R., Chen J. (2001) Characterization of tumor-associated Chk2 mutations. J. Biol. Chem. 276, 2971–2974 [DOI] [PubMed] [Google Scholar]
- 35. Antoni L., Sodha N., Collins I., Garrett M. D. (2007) CHK2 kinase: cancer susceptibility and cancer therapy—two sides of the same coin? Nat. Rev. Cancer 7, 925–936 [DOI] [PubMed] [Google Scholar]
- 36. Lee Y. K., Thomas S. N., Yang A. J., Ann D. K. (2007) Doxorubicin down-regulates Kruppel-associated box domain-associated protein 1 sumoylation that relieves its transcription repression on p21WAF1/CIP1 in breast cancer MCF-7 cells. J. Biol. Chem. 282, 1595–1606 [DOI] [PubMed] [Google Scholar]
- 37. Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krum S. A., la Rosa Dalugdugan E., Miranda-Carboni G. A., Lane T. F. (2010) BRCA1 forms a functional complex with γ-H2AX as a late response to genotoxic stress. J. Nucleic Acids 2010, pii: 801594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. White D. E., Negorev D., Peng H., Ivanov A. V., Maul G. G., Rauscher F. J., 3rd (2006) KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 66, 11594–11599 [DOI] [PubMed] [Google Scholar]
- 40. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tian C., Xing G., Xie P., Lu K., Nie J., Wang J., Li L., Gao M., Zhang L., He F. (2009) KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat. Cell Biol. 11, 580–591 [DOI] [PubMed] [Google Scholar]
- 42. Frietze S., O'Geen H., Blahnik K. R., Jin V. X., Farnham P. J. (2010) ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS One 5, e15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gebelein B., Urrutia R. (2001) Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Kruppel-associated box protein. Mol. Cell. Biol. 21, 928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nahle Z., Polakoff J., Davuluri R. V., McCurrach M. E., Jacobson M. D., Narita M., Zhang M. Q., Lazebnik Y., Bar-Sagi D., Lowe S. W. (2002) Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 4, 859–864 [DOI] [PubMed] [Google Scholar]
- 45. Kherrouche Z., Blais A., Ferreira E., De Launoit Y., Monté D. (2006) ASK-1 (apoptosis signal-regulating kinase 1) is a direct E2F target gene. Biochem. J. 396, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stiewe T., Pützer B. M. (2000) Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat. Genet. 26, 464–469 [DOI] [PubMed] [Google Scholar]
- 47. Zhao Y., Tan J., Zhuang L., Jiang X., Liu E. T., Yu Q. (2005) Inhibitors of histone deacetylases target the Rb-E2F1 pathway for apoptosis induction through activation of proapoptotic protein Bim. Proc. Natl. Acad. Sci. U.S.A. 102, 16090–16095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang C., Chen L., Hou X., Li Z., Kabra N., Ma Y., Nemoto S., Finkel T., Gu W., Cress W. D., Chen J. (2006) Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat. Cell Biol. 8, 1025–1031 [DOI] [PubMed] [Google Scholar]
- 49. Pediconi N., Ianari A., Costanzo A., Belloni L., Gallo R., Cimino L., Porcellini A., Screpanti I., Balsano C., Alesse E., Gulino A., Levrero M. (2003) Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell Biol. 5, 552–558 [DOI] [PubMed] [Google Scholar]
- 50. Rothkamm K., Krüger I., Thompson L. H., Löbrich M. (2003) Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 23, 5706–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Löbrich M., Shibata A., Beucher A., Fisher A., Ensminger M., Goodarzi A. A., Barton O., Jeggo P. A. (2010) γH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle 9, 662–669 [DOI] [PubMed] [Google Scholar]
- 52. Banáth J. P., Macphail S. H., Olive P. L. (2004) Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 64, 7144–7149 [DOI] [PubMed] [Google Scholar]
- 53. Iyengar S., Farnham P. J. (2011) KAP1 protein: an enigmatic master regulator of the genome. J. Biol. Chem. 286, 26267–26276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. O'Geen H., Squazzo S. L., Iyengar S., Blahnik K., Rinn J. L., Chang H. Y., Green R., Farnham P. J. (2007) Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 3, e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu X., Levine A. J. (1994) p53 and E2F-1 cooperate to mediate apoptosis. Proc. Natl. Acad. Sci. U.S.A. 91, 3602–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin W. C., Lin F. T., Nevins J. R. (2001) Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 15, 1833–1844 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.