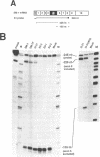
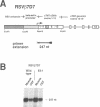
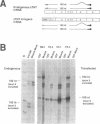
Abstract
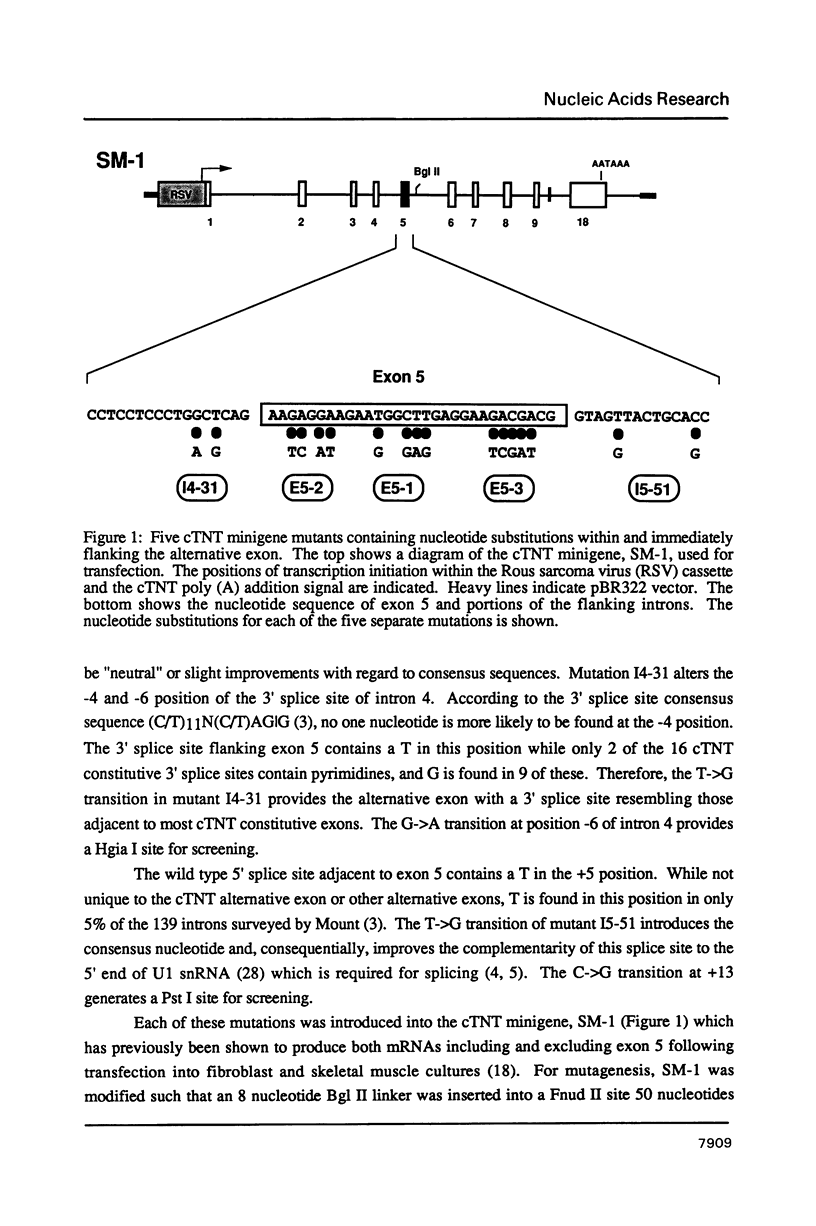
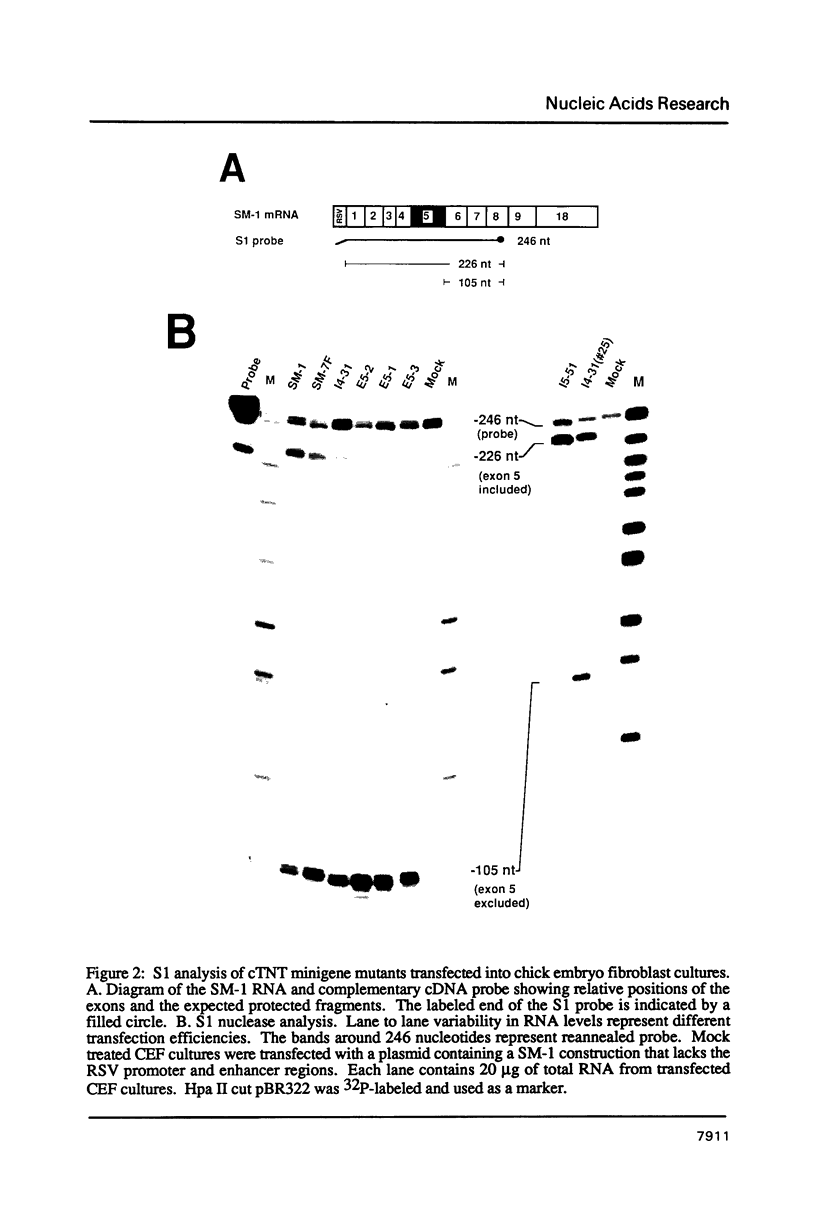
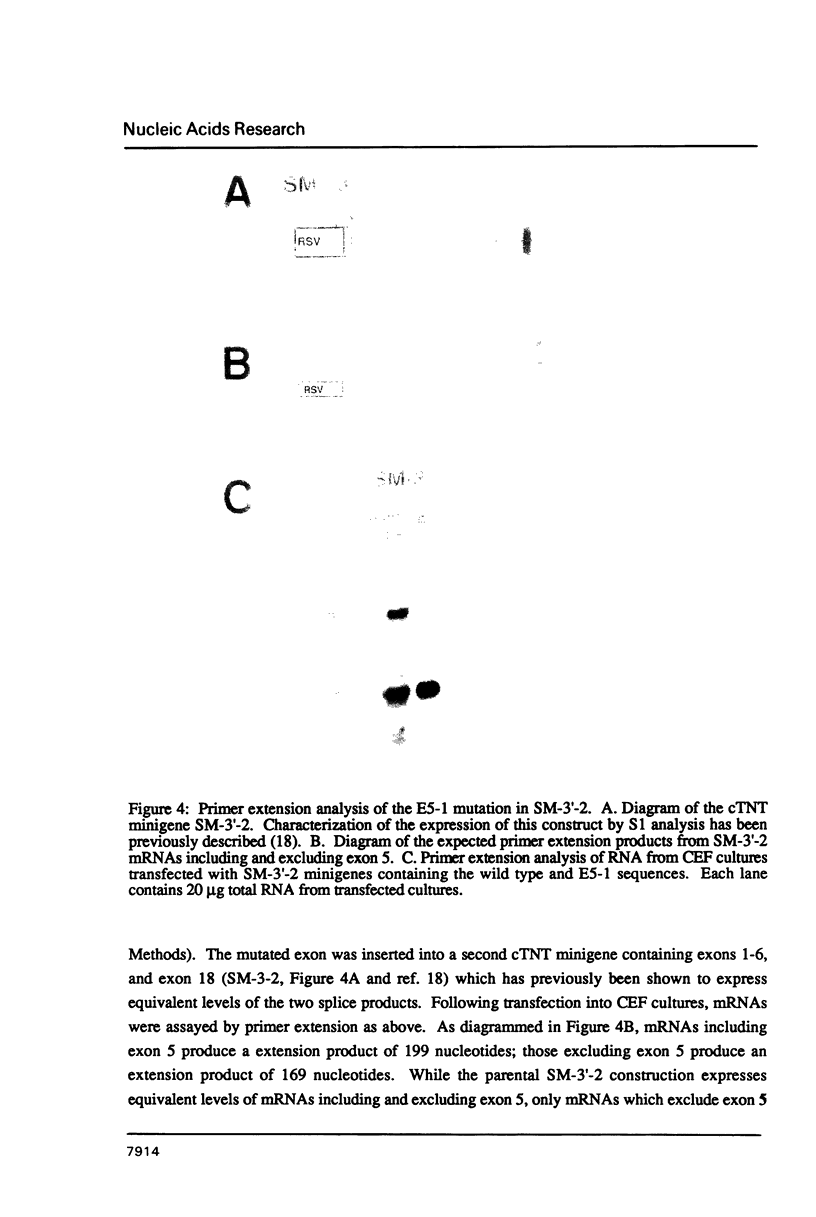
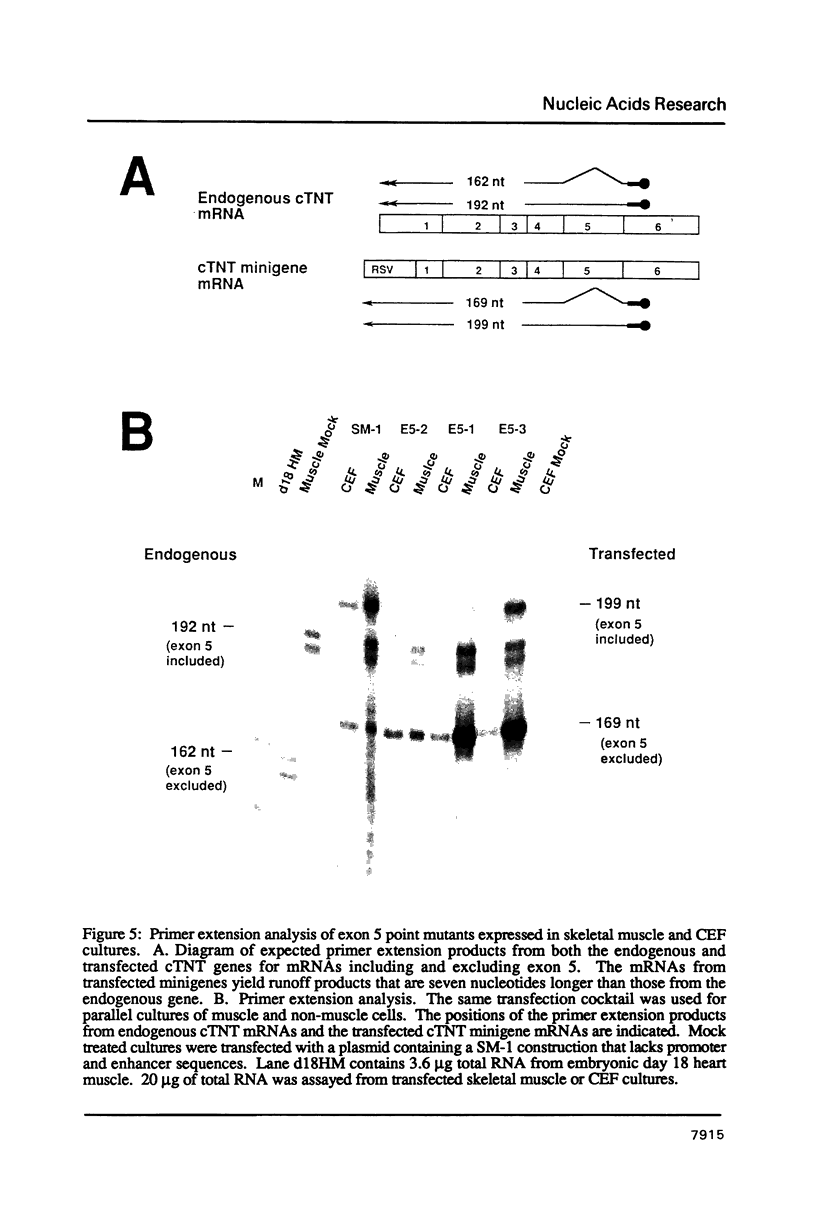
The cardiac troponin T (cTNT) pre-mRNA contains a single alternative exon (exon 5) which is either included or excluded from the processed mRNA. Using transient transfection of cTNT minigenes, we have previously localized pre-mRNA cis elements required for exon 5 alternative splicing to three small regions of the pre-mRNA which include exons 4, 5, and 6. In the present study, nucleotide substitutions were introduced into the region containing exon 5 to begin to define specific nucleotides required for exon 5 alternative splicing. A mutation within the 5' splice site flanking the cTNT alternative exon that increases its homology to the consensus sequence improves splicing efficiency and leads to increased levels of mRNAs that include the alternative exon. Surprisingly, substitution of as few as four nucleotides within the alternative exon disrupts cTNT pre-mRNA alternative splicing and prevents recognition of exon 5 as a bona fide exon. These results establish that the cTNT alternative exon contains information in cis that is required for its recognition by the splicing machinery.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Baldwin A. S., Jr, Kittler E. L., Emerson C. P., Jr Structure, evolution, and regulation of a fast skeletal muscle troponin I gene. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8080–8084. doi: 10.1073/pnas.82.23.8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat B. M., Wold W. S. A small deletion distant from a splice or polyadenylation site dramatically alters pre-mRNA processing in region E3 of adenovirus. J Virol. 1987 Dec;61(12):3938–3945. doi: 10.1128/jvi.61.12.3938-3945.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindereif A., Green M. R. Ribonucleoprotein complex formation during pre-mRNA splicing in vitro. Mol Cell Biol. 1986 Jul;6(7):2582–2592. doi: 10.1128/mcb.6.7.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Botstein D., Shortle D. Strategies and applications of in vitro mutagenesis. Science. 1985 Sep 20;229(4719):1193–1201. doi: 10.1126/science.2994214. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Nadal-Ginard B. Developmentally induced, muscle-specific trans factors control the differential splicing of alternative and constitutive troponin T exons. Cell. 1987 Jun 19;49(6):793–803. doi: 10.1016/0092-8674(87)90617-9. [DOI] [PubMed] [Google Scholar]
- Burtis K. C., Baker B. S. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989 Mar 24;56(6):997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Chabot B., Black D. L., LeMaster D. M., Steitz J. A. The 3' splice site of pre-messenger RNA is recognized by a small nuclear ribonucleoprotein. Science. 1985 Dec 20;230(4732):1344–1349. doi: 10.1126/science.2933810. [DOI] [PubMed] [Google Scholar]
- Chabot B., Steitz J. A. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol. 1987 Jan;7(1):281–293. doi: 10.1128/mcb.7.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. A., Cardone M. H., Ordahl C. P. Cis requirements for alternative splicing of the cardiac troponin T pre-mRNA. Nucleic Acids Res. 1988 Sep 12;16(17):8443–8465. doi: 10.1093/nar/16.17.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. A single cardiac troponin T gene generates embryonic and adult isoforms via developmentally regulated alternate splicing. J Biol Chem. 1985 Sep 15;260(20):11140–11148. [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. A single troponin T gene regulated by different programs in cardiac and skeletal muscle development. Science. 1984 Nov 23;226(4677):979–982. doi: 10.1126/science.6095446. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Estibeiro J. P., Eperon I. C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986 Nov 20;324(6094):280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- Fu X. Y., Colgan J. D., Manley J. L. Multiple cis-acting sequence elements are required for efficient splicing of simian virus 40 small-t antigen pre-mRNA. Mol Cell Biol. 1988 Sep;8(9):3582–3590. doi: 10.1128/mcb.8.9.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdon P. J., Kole R. The length of the downstream exon and the substitution of specific sequences affect pre-mRNA splicing in vitro. Mol Cell Biol. 1988 Feb;8(2):860–866. doi: 10.1128/mcb.8.2.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel L. I., Davidson N. Developmentally regulated expression of a truncated myosin light-chain 1F/3F gene. Mol Cell Biol. 1987 Oct;7(10):3826–3829. doi: 10.1128/mcb.7.10.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V., Steitz J. A. A protein associated with small nuclear ribonucleoprotein particles recognizes the 3' splice site of premessenger RNA. Cell. 1986 Dec 26;47(6):973–984. doi: 10.1016/0092-8674(86)90812-3. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Merlino G. T., Willingham M. C., Pastan I., Howard B. H. The Rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6777–6781. doi: 10.1073/pnas.79.22.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hwang L. S., Park J., Gilboa E. Role of intron-contained sequences in formation of moloney murine leukemia virus env mRNA. Mol Cell Biol. 1984 Nov;4(11):2289–2297. doi: 10.1128/mcb.4.11.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes D. H., Steitz J. A. Accurate 5' splice-site selection in mouse kappa immunoglobulin light chain premessenger RNAs is not cell-type-specific. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7928–7932. doi: 10.1073/pnas.84.22.7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P., Dhar R., Lai C. J. Processing and expression of early SV40 mRNA: a role for RNA conformation in splicing. Cell. 1979 Sep;18(1):85–92. doi: 10.1016/0092-8674(79)90356-8. [DOI] [PubMed] [Google Scholar]
- Kost T. A., Theodorakis N., Hughes S. H. The nucleotide sequence of the chick cytoplasmic beta-actin gene. Nucleic Acids Res. 1983 Dec 10;11(23):8287–8301. doi: 10.1093/nar/11.23.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühne T., Wieringa B., Reiser J., Weissmann C. Evidence against a scanning model of RNA splicing. EMBO J. 1983;2(5):727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Mar J. H., Antin P. B., Cooper T. A., Ordahl C. P. Analysis of the upstream regions governing expression of the chicken cardiac troponin T gene in embryonic cardiac and skeletal muscle cells. J Cell Biol. 1988 Aug;107(2):573–585. doi: 10.1083/jcb.107.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Urlaub G., Chasin L. Spontaneous splicing mutations at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1986 Jun;6(6):1926–1935. doi: 10.1128/mcb.6.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovits W., Jr, Kuncio G., Ordahl C. P. The chicken fast skeletal troponin I gene: exon organization and sequence. Nucleic Acids Res. 1986 Apr 25;14(8):3377–3390. doi: 10.1093/nar/14.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton P. A., Hynes R. O. Alternative splicing of chicken fibronectin in embryos and in normal and transformed cells. Mol Cell Biol. 1987 Dec;7(12):4297–4307. doi: 10.1128/mcb.7.12.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Parent A., Zeitlin S., Efstratiadis A. Minimal exon sequence requirements for efficient in vitro splicing of mono-intronic nuclear pre-mRNA. J Biol Chem. 1987 Aug 15;262(23):11284–11291. [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol Cell Biol. 1989 Feb;9(2):726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R. Compilation of small RNA sequences. Nucleic Acids Res. 1988;16 (Suppl):r71–r85. doi: 10.1093/nar/16.suppl.r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Ricketts M. H., Simons M. J., Parma J., Mercken L., Dong Q., Vassart G. A nonsense mutation causes hereditary goitre in the Afrikander cattle and unmasks alternative splicing of thyroglobulin transcripts. Proc Natl Acad Sci U S A. 1987 May;84(10):3181–3184. doi: 10.1073/pnas.84.10.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Specific and stable intron-factor interactions are established early during in vitro pre-mRNA splicing. Cell. 1985 Nov;43(1):131–142. doi: 10.1016/0092-8674(85)90018-2. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985 Aug 12;13(15):5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli M., Saito H. Regulation of tissue-specific alternative splicing: exon-specific cis-elements govern the splicing of leukocyte common antigen pre-mRNA. EMBO J. 1989 Mar;8(3):787–796. doi: 10.1002/j.1460-2075.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. S., Dreyfuss G. RNA binding specificity of hnRNP proteins: a subset bind to the 3' end of introns. EMBO J. 1988 Nov;7(11):3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Alibert C., Temsamani J., Reveillaud I., Cathala G., Brunel C., Jeanteur P. A protein that specifically recognizes the 3' splice site of mammalian pre-mRNA introns is associated with a small nuclear ribonucleoprotein. Cell. 1986 Dec 5;47(5):755–766. doi: 10.1016/0092-8674(86)90518-0. [DOI] [PubMed] [Google Scholar]
- Turnbull-Ross A. D., Else A. J., Eperon I. C. The dependence of splicing efficiency on the length of 3' exon. Nucleic Acids Res. 1988 Jan 25;16(2):395–411. doi: 10.1093/nar/16.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulfendahl P. J., Pettersson U., Akusjärvi G. Splicing of the adenovirus-2 E1A 13S mRNA requires a minimal intron length and specific intron signals. Nucleic Acids Res. 1985 Sep 11;13(17):6299–6315. doi: 10.1093/nar/13.17.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibe-Pedersen K., Kornblihtt A. R., Baralle F. E. Expression of a human alpha-globin/fibronectin gene hybrid generates two mRNAs by alternative splicing. EMBO J. 1984 Nov;3(11):2511–2516. doi: 10.1002/j.1460-2075.1984.tb02165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaud M., Gattoni R., Stevenin J., Vidaud D., Amselem S., Chibani J., Rosa J., Goossens M. A 5' splice-region G----C mutation in exon 1 of the human beta-globin gene inhibits pre-mRNA splicing: a mechanism for beta+-thalassemia. Proc Natl Acad Sci U S A. 1989 Feb;86(3):1041–1045. doi: 10.1073/pnas.86.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Leung H., Weiner A. M. The natural 5' splice site of simian virus 40 large T antigen can be improved by increasing the base complementarity to U1 RNA. Mol Cell Biol. 1987 Aug;7(8):3018–3020. doi: 10.1128/mcb.7.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]