Abstract
Background:
Deoxyribonucleic acid (DNA) topology plays a critical role in maintaining the integrity of the genome and cellular functions. Although changes in DNA conformation and structural dynamics in the brain have been associated with various neurological disorders, its precise role in the pathogenesis is still unclear. Previous studies from our laboratory have shown that there is a conformational change in the genomic DNA of Parkinson's disease (PD) (B to altered B-DNA) and Alzheimer's disease brain (B to Z-DNA). However, there is limited information on the mechanism on DNA dynamics changes in brain.
Objective:
In the present study, we have investigated the DNA conformation and sequence specific binding ability of α-Synuclein and Tau with reference to B-DNA and Z-DNA using oligonucleotide (CGCGCGCG)2 as a novel model DNA system. This sequence is predominantly present in the promoter region of the genes of biological relevance.
Materials and Methods:
Natively, (CGCGCGCG)2 sequence exists in B-DNA conformation, but in the presence of high sodium concentration (4 M NaCl), the oligo converts into Z-DNA form. We used circular dichroism, melting temperature and fluorescence studies to understand protein-DNA interactions.
Results:
CD studies indicated that both α-Synuclein and Tau bind to B-DNA conformation of (CGCGCGCG)2 and induce altered B-form. Further, these proteins increased the melting temperature and decreased the number of EtBr molecules bound per base pair of DNA in B-form indicating that DNA stability is favored to alter B-DNA conformation, which could be an intermediate form favoring Z-DNA conformation. Moreover, both α-Synuclein and Tau also bound to disease-linked Z-DNA conformation of (CGCGCGCG)2 and further stabilized the Z-conformation.
Conclusions:
The present study provides vital mechanistic information on Synuclein and Tau binding to DNA in a conformation-specific manner causing conformational transition. Furthermore, both the proteins stabilize Z-DNA conformation. These have altered minor and major groove patterns and thus may have significant biological implications in relevance to gene expression pattern in neurodegeneration. We discuss the implications of α-Synuclein/Tau binding to DNA and stabilizing the altered conformations of DNA in neuronal cell dysfunction.
Keywords: α-Synuclein, Alzheimer's disease, oligonucleotide-protein interaction, Parkinson's disease, Tau, Z-DNA
DNA conformation and stability are critical for the normal cell functions. Deoxyribonucleic acid (DNA) is polymorphic in nature and adopts different conformations in the cell in different physiological conditions.[1] The altered conformation in DNA has been shown to cause altered gene expression in human diseases.[2] The role of DNA dynamics in brain disorders is not clearly understood.[3,4] Earlier studies from our laboratory have shown that genomic integrity is altered in Parkinson's disease (PD) and Alzheimer's Disease (AD).[5,6] The genomic DNA isolated from PD-affected human postmortem brain cells is altered from B-form to altered B conformation,[5] whereas in AD, a larger conformational transition from B-form to Z-form was observed.[6] The factors responsible for these alterations are not clear. We hypothesize that neuroproteins such as α-Synuclein, amyloid and Tau may be involved in the conformational and stability transitions in DNA.[3,4] In support of our hypothesis, there are evidences on the nuclear localization of α-Synuclein, amyloid and Tau,[7–9] but studies related to DNA binding abilities are limited[7–10] α-Synuclein (144 aa), a highly conserved, natively unfolded protein existing in random coil conformation is involved in the pathogenesis of PD.[9–13] The normal functions of α-Synuclein in the neuron is not clear.[13,14] However, the localization of α-Synuclein in the nucleus suggests that it may interact with chromatin.[15–17] Unlike α-Synuclein, Tau normal functions have been established. Tau's main function is to polymerize and stabilize the microtubules in the neuron.[18–20] Tau is responsible for the neurofibrillary tangles (NFTs) that are formed in AD.[18,19] Tau protein normally present in the cytoplasm is also localized in the nuclear region and may have the ability to bind to DNA.[8,20–22] But the mechanism of both α-Synuclein and Tau binding to DNA is not clear. In the present investigation, we investigate whether α-Synuclein and Tau bind to DNA in a conformation and sequence specific manner. We have selected oligonucleotide CGCGCGCG as a model DNA system that natively exists in B conformation and that, in the presence of 4 M NaCl, the oligo goes to Z conformation.[23] This sequence is present in the promoter region of the human genome and hence has biological implications.[24]
Materials and Methods
α-Synuclein and Tau were purchased from rPeptide, USA. CGCGCGCG oligonucleotide was custom synthesized from Sigma Aldrich, USA. Tris and HEPES were purchased from Sisco Research Lab, India. The self-complementary single stranded CGCGCGCG oligonucleotide was dissolved in triple distilled water and incubated in boiling water bath for 5 min and allowed to cool gradually at room temperature to generate annealed duplex structures.[25]
Circular dichroism studies
The Secondary conformation of oligonucleotide (CGCGCGCG)2 was analyzed using Circular dichroism (CD) spectroscopy. The (CGCGCGCG)2 oligonucleotide was diluted in Tris-Cl buffer (10 mM, pH 7.4) and CD spectrum was recorded using JASCO J700 spectropolarimeter at 25°C, with 2 mm cell length in the wavelength range between 200 and 320 nm. Each spectrum was the average of four scans. The (CGCGCGCG)2 oligonucleotide was converted from B-DNA to Z-DNA in the presence of 4 M NaCl (incubated for 12 h at 37°C) and later dialyzed to remove NaCl. The Z-form of the oligonucleotide was confirmed by CD. The oligonucleotide in B-form was incubated with α-Synuclein and Tau in the mass ratio of 1:2 (oligo/protein) in 10 mM Tris-HCl (pH 7.4) buffer for 12 hr at 37°C. The oligonucleotide alone (control) was also incubated in the 10 mM Tris-HCl (pH 7.4) buffer for 12 h at 37°C. The CD spectra of the B-DNA-α-Synuclein and B-DNA-Tau complexes were recorded. Similarly, the Z-form of the oligonucleotide was incubated with α-Synuclein and Tau in the mass ratio of 1:2 (oligo/protein) in 10 mM Tris-HCl (pH 7.4) buffer for 12 hr at 37°C. The CD spectra of Z-DNA-α-Synuclein and Z-DNA-Tau complexes were recorded. The background correction was done by deducting the spectral contribution from α-Synuclein and Tau alone at the concentrations used. The DNA conformations were assigned using the standard references.[26]
Thermal denaturation studies
The stability of B-DNA and Z-DNA forms of oligonucleotide and the oligo-protein complexes was studied using thermal denaturation studies and ethidium bromide binding studies. Thermal denaturation of oligonucleotide was carried out using UV-Visible Spectrophotometer equipped with thermostat control (Amarsham Bioscience, Hong Kong). The 2 μg of B-DNA and Z-DNA forms of (CGCGCGCG)2 was dissolved in 10 mM HEPES buffer (pH 7.4) and melting temperature (Tm) profile was recorded with 1°C raise of temperature per minute in the temperature range of 25–95°C. Both B-form and Z-form of the oligonucleotide were incubated with α-Synuclein and Tau proteins in the mass ratio of 1:2 (oligo/protein) in 10 mM HEPES (pH 7.4) buffer at 12 h at 37°C. The Tm (the temperature at which the absorbance is 50% between the minimum and maximum absorbance) was calculated using the graph plotted between temperature vs. absorbance, which is called the melting curve.
Ethidium bromide (EtBr) binding studies
The reaction mixtures for EtBr binding studies were prepared as described before for Tm studies, and the EtBr bound in moles per base pair of DNA was measured in Tris-Cl (10 mM, pH 7.4) using HITACHI F-2000 Fluorescence Spectrofluorimeter. The fluorescence was measured using a constant amount of scDNA (0.5 μg) with increasing amounts of EtBr. The fluorescence measurements were monitored with an excitation at 535 nm and emission at 600 nm using 10 mm path length.
The maximum amount of EtBr bound per base pair of DNA was calculated using Scatchard plots of ‘r’ vs. ‘r/Cf’. The concentration of bound EtBr in 1.0 ml dye-DNA mixture (Cb´) was calculated using the equation
Cb´ = Co´ [(F–Fo)/(V×Fo)], (1)
where
Co = EtBr (pmoles) present in the dye-DNA mixture,
F = observed fluorescence at any point of dye-DNA mixture,
Fo = observed fluorescence of EtBr with no DNA,
V = experimentally derived value, ratio of bound EtBr/free EtBr at saturation point.
The concentration of free dye (Cf΄) was then calculated by the relation
Cf´ = Co´ - Cb´, (2)
Where, Cf´, Co´, and Cb´ were expressed in pmoles. The amount of bound EtBr/base pair ‘r’ was calculated by
r = Cb´ (pmoles)/DNA concentration (pmoles of base pair). (3)
A plot with r vs. r/cf is plotted, point where the straight line intersects the X-axis was denoted as ‘n’. ‘n’ was the maximum amount of dye bound per base pair (n), where Cf = Cf´ × 1015 M.
The number of EtBr molecules bound per base pair DNA was calculated for B-DNA and Z-DNA in the presence and absence of Synuclein and Tau. The data was analyzed using Scatchard plots.[27,28]
Results
CD studies
The CD spectra of native (CGCGCGCG)2 showed a characteristic B-DNA conformation with a positive peak at 280 nm and a negative peak at 255 nm. Further, the CD spectrum of (CGCGCGCG)2 oligonucleotide in the presence of 4 M NaCl showed Z-DNA formation with characteristic 295 and 208 nm negative peak showing the conversion of B-form to Z-form [Figure 1a]. The CD spectrum of B-form of oligonucleotide in the presence of α-Synuclein complex showed a shift in the positive peak from 280 to 275 nm and a decrease in the intensity of the negative peaks at 235 and 210 nm compared to B-form alone [Figure 1b]. These changes were characteristic of altered B-DNA conformation.[26] Further, CD spectrum of B-DNA in the presence of Tau also showed similar changes [Figure 1c]. Thus it was inferred that both α-Synuclein and Tau induced altered B-DNA conformation in B-DNA. The CD spectra of Z-DNA in the presence of α-Synuclein and Tau did not alter any spectral changes of Z-form [Figures 1d and e]. Both the complexes showed characteristic 290 nm negative peaks indicating unaltered Z-DNA conformation.
Figure 1.
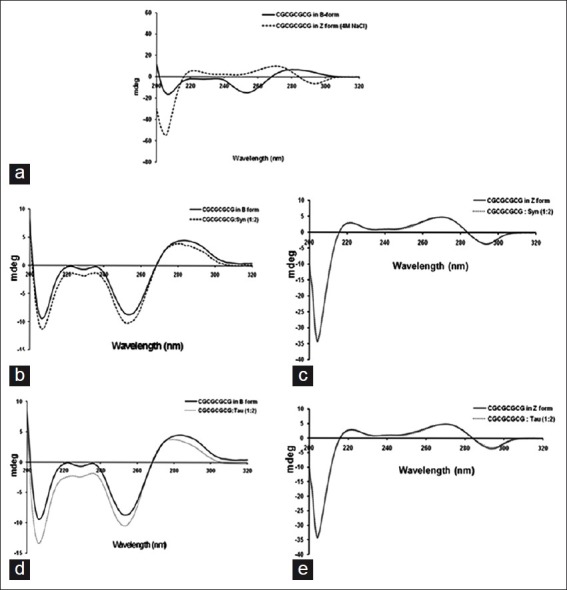
Circular dichroism spectroscopy of (CGCGCGCG)2 oligonucleotide complexes. (a) (CGCGCGCG)2 oligonucleotide in B-DNA conformation and Z-DNA conformation. (b) α-Synuclein induced altered B-conformation in B-DNA of (CGCGCGCG)2. (c) Tau induced altered B-conformation in B-DNA of (CGCGCGCG)2. (d) α-Synuclein - Z-DNA of (CGCGCGCG)2 complex in Z-DNA conformation. (e) Tau - Z-DNA of (CGCGCGCG)2 complex in Z-DNA conformation
Thermal denaturation studies
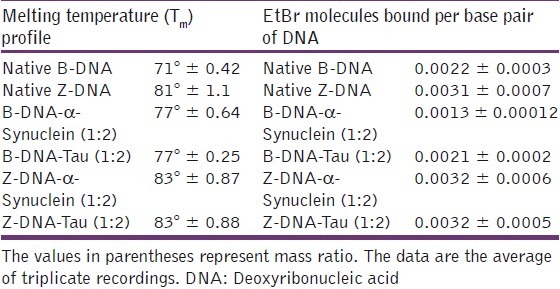
Melting temperature curves of oligonucleotides and their protein complexes were computed [Figure 2]. The melting temperature (Tm) of the B-form and Z-form of the oligonucleotide was 71 and 81°C, respectively. The Tm of the Z-form of the oligonucleotide was greater than the B-form, indicating that Z-form was more stable than B-form. The Tm of B-DNA in the presence of α-Synuclein and Tau was found to be similar (Tm = 77°C). The data indicated that both α-Synuclein and Tau stabilized B-DNA as evidenced by higher Tm. The Tm of Z-DNA in the presence of α-Synuclein and Tau was 83°C. The data indicate that both α-Synuclein and Tau further stabilized the Z-conformation of the oligonucleotide [Table 1]. The Tm values difference for Z-DNA was 2°C, but this difference was significant in light of the short length of oligonucleotides; for this reason, we concluded that both the proteins stabilized the Z-conformation.
Figure 2.
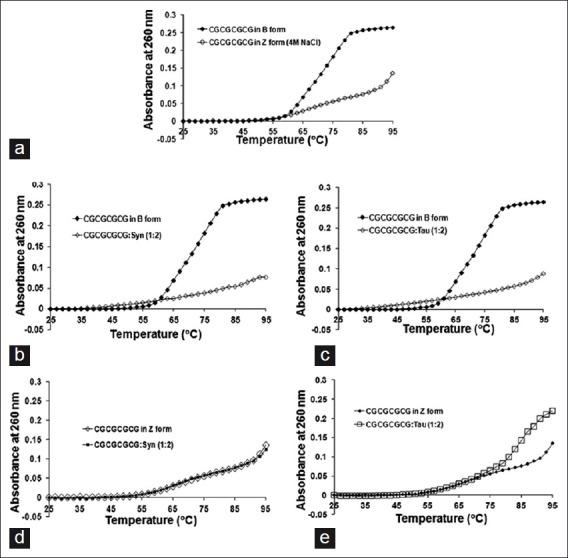
Melting temperature curves of (CGCGCGCG)2 oligonucleotide complexes. (a) Melting temperature (Tm) curve of (CGCGCGCG)2 oligonucleotide in B-DNA and Z-DNA conformations, respectively. (b) Melting temperature (Tm) curve of (CGCGCGCG)2 oligonucleotide in B-DNA and B-DNA-α-Synuclein complex (mass ratio 1:2). (c) Melting temperature (Tm) curve of (CGCGCGCG)2 oligonucleotide in B-DNA and B-DNA-Tau complex (mass ratio 1:2). (d) Melting temperature (Tm) curve of (CGCGCGCG)2 oligonucleotide in Z-DNA and Z-DNA- α-Synuclein complex (mass ratio 1:2). (e) Melting temperature (Tm) curve of (CGCGCGCG)2 oligonucleotide in Z-DNA and Z-DNA-Tau complex (mass ratio 1:2)
Table 1.
Melting temperature (Tm) and Ethidium bromide binding to (CGCGCGCG)2-α-Synuclein and (CGCGCGCG)2-Tau complexes both in B-form and Z-form
Ethidium bromide binding studies
The number of Ethidium bromide (EtBr) molecules bound per base pair DNA for B-form and Z-form of the oligonucleotide was 0.0022 and 0.0031, respectively. This indicates that Z-DNA has more EtBr binding ability. The B-DNA oligo was titrated with EtBr in the presence of α-Synuclein or Tau and the number EtBr molecules bound per base pair DNA was calculated to be 0.0013 and 0.0021, respectively. The results indicated that the number of EtBr molecules bound per base pair for complexes were less than the corresponding native B-form of oligonucleotide in the presence of α-Synuclein, indicating that α-Synuclein may be interfering with DNA intercalating properties of ETBR. On the other hand, Tau did not alter the DNA intercalating properties of ETBR. This may be attributed to differential Tau and Synuclein DNA binding properties,[3] although the mechanisms of DNA binding of these proteins are not clearly known. The number EtBr molecules bound per base pair DNA for Z-DNA in the presence of α-Synuclein and Tau was unaffected as expected [Table 1].
Discussion
DNA is polymorphic in nature existing in different conformations in cells under physiological conditions.[1] The B-DNA conformation is the normal conformation of DNA in cells and is attributed with a majority of biological functions.[29] The DNA also exists in other conformations like Z, A, C under experimental conditions.[30] These alternate conformations of DNA possess altered major and minor groove properties[30] and when present in genomes these non-B-conformations may alter gene expressions. Furthermore, as many transcription factors were minor grove binders,[31] the altered minor grove patterns in these alternative DNA conformations could affect their binding to DNA and thus impact gene expression. However, the biology of alternate DNA conformations in the regulation of gene functions is still unclear.[2] The conformation and physics of Z-DNA have been widely studied but the biological implications like transcription regulation, chromatin remodeling and nucleosomal positioning are still not completely understood.[32–35] The Z-DNA formation occurs in the stretches of alternative purine-pyrimidine residues.[36] And alternative purine-pyrimidine residues were widely distributed in the promoter regions of several genes.[24] A recent study showed that Z-DNA conformation was present in the promoter region of selected genes in the human genome.[36] Moreover, the study indicated that these alternative structures could be stabilized by specific proteins that promotealtered gene expression.[33] A similar kind of altered function in transcription activity by T7 RNA polymerase in relevance to Z-DNA was reported recently.[37] This suggests that alternate DNA conformations and their specific binding molecules might affect the normal genomic functions. We presume that similar mechanisms may be taking place in neurons in the process of neurodegeneration. One of the examples is the Vesicular Inhibitory Amino Acid Transporter (viaat) that transports GABA or glycine neurotransmitters into synaptic vesicles.[38] This promoter activity was dependent on the GC rich region that was adjacent to the SP and EGR transcription factor binding sites, and any modulation in GC rich region altered the neurobiology of GABA.[38] In the present study, we investigated the binding ability of neuroproteins like α-Synuclein and Tau to sequence and conformation specific oligonucleotide (CGCGCGCG)2. These sequences were involved in gene expression and the major component of promoter regions.[24,38] The present study thus provides insight into conformation specific DNA binding of α-Synuclein and Tau and has great relevance in understanding the genomic biology of neurodegeneration. The earlier studies from our lab have reported the presence of altered B-DNA and Z-DNA in PD and AD brains respectively.[5,6] Unlike the predominant B-DNA conformation in normal brain, in moderately affected AD brain, DNA was in B-Z intermediate form, while in severely affected AD brain, DNA was in Z-form.[6] We subsequently reported a strong DNA binding property of amyloid and synuclein modulating DNA topology.[3,10] Hegde et al.,[7] showed that Aβ is localized in the nuclear region of AD brain and proposed a possible mechanism for Aβ playing a role in DNA conformational change. Suram et al.,[6] reported a conformational change in genomic DNA from normal B-DNA to B-Z intermediate in moderate AD and B-DNA to Z-DNA in severe AD.
The present study provides vital mechanistic information on Synuclein and Tau binding to DNA in a conformation-specific manner causing conformational transition. Furthermore, both the proteins stabilize Z-DNA conformation. These have altered minor and major groove patterns and thus may have significant biological implications in relevance to gene expression pattern in neurodegeneration. We hypothesized the implication of α-Synuclein/Tau binding to DNA and stabilizing the altered conformations of DNA in neuronal cell dysfunction [Figure 3]. Both α-Synuclein/Tau may bind to CG rich region in the promoter region of the gene and alter the conformation from B-DNA to altered B-DNA. Altered conformation in the promoter region may allow the binding of transcription factors and alter gene expression. The altered regulation of the gene expression may result in neuronal cell dysfunction and finally lead to neuronal cell death. The negative supercoils generated during transcription may induce the formation of Z-DNA in CG rich regions. Normally, after transcription the negative supercoils are relaxed and the Z-DNA is again converted to B-DNA. Both α-Synuclein/Tau may bind to Z-DNA conformation that is already formed due to transcriptional stress, and further, these proteins may stabilize the Z-DNA conformation. The Z-DNA stabilization may result in the inhibition of transcription factors binding or may stop the movement of RNA polymerase. This may lead to the suppression of genes and finally to neuronal cell dysfunction.
Figure 3.
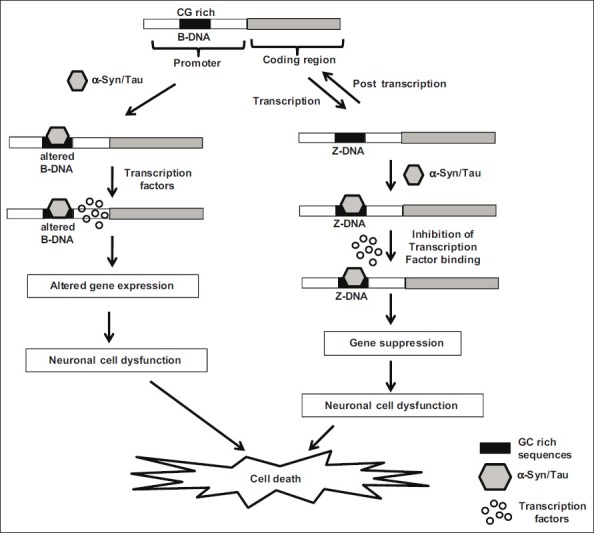
Our hypothesis on the potential implications of sequence specific binding of α-Synuclein/Tau in neuronal cell death. Both α-Synuclein/Tau may bind to CG rich regions in the promoters of various genes and alter the conformation from B-DNA to altered B-DNA. The altered conformation in the promoter region could alter the binding of transcription factors and alter gene expression. The altered regulation of the gene expression may result in neuronal cell dysfunction and finally lead to neuronal cell death. The negative supercoils generated during transcription may induce the formation of Z-DNA in CG rich regions. Normally, after transcription the negative supercoils are relaxed and the Z-DNA is again converted to B-DNA. Both α-Synuclein/Tau may bind to Z-DNA conformation formed due to transcriptional stress and stabilize the Z-DNA conformation. Z-DNA stabilization may result in the inhibition of transcription factors binding or may stop the movement of RNA polymerase. This may lead to the suppression of genes and finally to neuronal cell dysfunction
Conclusions
The binding of α-Synuclein and Tau changes DNA conformation to altered B and Z-form and thus alters minor and major groove patterns, which may have significant biological significance in relevance to gene expression pattern in neurodegeneration. We hypothesize that α-Synuclein/Tau binding to DNA and stabilizing the conformations of DNA plays a significant role in neuronal cell dysfunction.
Acknowledgement
The authors wish to thank Mr. Arturo Melo for supporting the study through Melo Brain Grant.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Rich A. DNA comes in many forms. Gene. 1993;135:99–109. doi: 10.1016/0378-1119(93)90054-7. [DOI] [PubMed] [Google Scholar]
- 2.Bacolla A, Wells RD. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol Carcinog. 2009;48:273–85. doi: 10.1002/mc.20507. [DOI] [PubMed] [Google Scholar]
- 3.Hegde ML, Vasudevaraju P, Rao KJ. DNA induced folding/fibrillation of alpha-synuclein: New insights in Parkinson's disease. Front Biosci. 2010;15:418–36. doi: 10.2741/3628. [DOI] [PubMed] [Google Scholar]
- 4.Vasudevaraju P, Bharathi, Garruto RM, Sambamurti K, Rao KS. Role of DNA dynamics in Alzheimer's disease. Brain Res Rev. 2008;58:136–48. doi: 10.1016/j.brainresrev.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Hegde ML, Gupta VB, Anitha M, Harikrishna T, Shankar SK, Muthane U, et al. Studies on genomic DNA topology and stability in brain regions of Parkinson's disease. Arch Biochem Biophys. 2006;449:143–56. doi: 10.1016/j.abb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Suram A, Rao KS, Latha KS, Viswamitra MA. First evidence to show the topological change of DNA from B-DNA to Z-DNA conformation in the hippocampus of Alzheimer's brain. Neuromolecular Med. 2002;2:289–97. doi: 10.1385/nmm:2:3:289. [DOI] [PubMed] [Google Scholar]
- 7.Hegde ML, Anitha S, Latha KS, Mustak MS, Stein R, Ravid R, et al. First evidence for helical transitions in supercoiled DNA by amyloid Beta Peptide (1-42) and aluminum: A new insight in understanding Alzheimer's disease. J Mol Neurosci. 2004;22:19–31. doi: 10.1385/jmn:22:1-2:19. [DOI] [PubMed] [Google Scholar]
- 8.Loomis PA, Howard TH, Castleberry RP, Binder LI. Identification of nuclear tau isoforms in human neuroblastoma cells. Proc Natl Acad Sci USA. 1990;87:8422–6. doi: 10.1073/pnas.87.21.8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maroteaux L, Campanelli JT, Scheller RH. Synuclein: A neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1998;8:2804–15. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegde ML, Rao KS. DNA induces folding in alpha-synuclein: Understanding the mechanism using chaperone property of osmolytes. Arch Biochem Biophys. 2007;464:57–69. doi: 10.1016/j.abb.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 11.Bisaglia M, Mammi S, Bubacco L. Structural insights on physiological functions and pathological effects of alpha-synuclein. FASEB J. 2009;23:329–40. doi: 10.1096/fj.08-119784. [DOI] [PubMed] [Google Scholar]
- 12.Broersen K, van den Brink D, Fraser G, Goedert M, Davletov B. Alpha-synuclein adopts an alpha-helical conformation in the presence of polyunsaturated fatty acids to hinder micelle formation. Biochemistry. 2006;45:15610–6. doi: 10.1021/bi061743l. [DOI] [PubMed] [Google Scholar]
- 13.Surguchov A. Molecular and cellular biology of synucleins. Int Rev Cell Mol Biol. 2008;270:225–317. doi: 10.1016/S1937-6448(08)01406-8. [DOI] [PubMed] [Google Scholar]
- 14.Waxman EA, Giasson BI. Molecular mechanisms of alpha-synuclein neurodegeneration. Biochim Biophys Acta. 2009;1792:616–24. doi: 10.1016/j.bbadis.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goers J, Manning-Bog AB, McCormack AL, Millett IS, Doniach S, Di Monte DA, et al. Nuclear localization of alpha-synuclein and its interaction with histones. Biochemistry. 2003;42:8465–71. doi: 10.1021/bi0341152. [DOI] [PubMed] [Google Scholar]
- 16.Lin WL, DeLucia MW, Dickson DW. Alpha-synuclein immunoreactivity in neuronal nuclear inclusions and neurites in multiple system atrophy. Neurosci Lett. 2004;354:99–102. doi: 10.1016/j.neulet.2003.09.075. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Zhang C, Zhu Y, Cai Q, Chan P, Uéda K, et al. Semi-quantitative analysis of alpha-synuclein in subcellular pools of rat brain neurons: An immunogold electron microscopic study using a C-terminal specific monoclonal antibody. Brain Res. 2008;1244:40–52. doi: 10.1016/j.brainres.2008.08.067. [DOI] [PubMed] [Google Scholar]
- 18.Brunden KR, Trojanowski JQ, Lee VM. Advances in tau-focused drug discovery for Alzheimer's disease and related tauopathies. Nat Rev Drug Discov. 2009;8:783–93. doi: 10.1038/nrd2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spires-Jones TL, Stoothoff WH, de Calignon A, Jones PB, Hyman BT. Tau pathophysiology in neurodegeneration: A tangled issue. Trends Neurosci. 2009;32:150–9. doi: 10.1016/j.tins.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proc Natl Acad Sci USA. 1975;72:1858–62. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis PK, Johnson GV. The microtubule binding of Tau and high molecular weight Tau in apoptotic PC12 cells is impaired because of altered phosphorylation. J Biol Chem. 1999;274:35686–92. doi: 10.1074/jbc.274.50.35686. [DOI] [PubMed] [Google Scholar]
- 22.Haque N, Tanaka T, Iqbal K, Grundke-Iqbal I. Regulation of expression, phosphorylation and biological activity of tau during differentiation in SY5Y cells. Brain Res. 1999;838:69–77. doi: 10.1016/s0006-8993(99)01622-4. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Thomas GA, Peticolas WL. Sequence dependent conformations of oligomeric DNA's in aqueous solutions and in crystals. J Biomol Struct Dyn. 1987;5:249–74. doi: 10.1080/07391102.1987.10506392. [DOI] [PubMed] [Google Scholar]
- 24.Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci. 2003;60:1647–58. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trewick SA, Dearden P. A rapid protocol for DNA extraction and primer annealing for PCR sequencing. Biotechniques. 1994;17:842–4. [PubMed] [Google Scholar]
- 26.Gray DM, Hung SH, Johnson KH. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 27.Chatterjee B, Rao GRK. Superhelical density of goat mitochondrial DNA: Fluorescent studies. Indian J Biochem Biophys. 1994;31:77–9. [PubMed] [Google Scholar]
- 28.Scatchard G. The attraction of proteins for small molecules and ions. Ann N Y Acad Sci (USA) 1949;51:660–72. [Google Scholar]
- 29.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–8. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh A, Bansal MA. Glossary of DNA structures from A to Z. Acta Crystallogr D Biol Crystallogr. 2003;59:620–6. doi: 10.1107/s0907444903003251. [DOI] [PubMed] [Google Scholar]
- 31.Hinoi E, Balcar VJ, Kuramoto N, Nakamichi N, Yoneda Y. Nuclear transcription factors in the hippocampus. Prog Neurobiol. 2002;68:145–65. doi: 10.1016/s0301-0082(02)00078-3. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Mulholland N, Fu H, Zhao K. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol Cell Biol. 2006;26:2550–9. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh DB, Kim YG, Rich A. Z-DNA-binding proteins can act as potent effectors of gene expression in vivo. Proc Natl Acad Sci USA. 2002;99:16666–71. doi: 10.1073/pnas.262672699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang AH, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel G, et al. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–6. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 35.Rich A, Zhang S. Timeline: Z-DNA: The long road to biological function. Nat Rev Genet. 2003;4:566–72. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Xiao J, Li J, Lu L, Feng S, Dröge P. Human genomic Z-DNA segments probed by the Z alpha domain of ADAR1. Nucleic Acids Res. 2009;37:2737–46. doi: 10.1093/nar/gkp124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dröge P, Pohl FM. The influence of an alternate template conformation on elongating phage T7 RNA polymerase. Nucleic Acids Res. 1991;19:5301–6. doi: 10.1093/nar/19.19.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh WJ, Noggle SA, Maddox DM, Condie BG. The mouse vesicular inhibitory amino acid transporter gene: Expression during embryogenesis, analysis of its core promoter in neural stem cells and a reconsideration of its alternate splicing. Gene. 2005;351:39–49. doi: 10.1016/j.gene.2005.01.009. [DOI] [PubMed] [Google Scholar]


