Background: Phenol-soluble modulins (PSMs) are small peptides of Staphylococcus aureus with immunosuppressive and antimicrobial properties.
Results: Imaging mass spectrometry (IMS) identified PSM derivatives with properties different from those of the parent forms.
Conclusion: S. aureus generates truncated PSMs with altered antimicrobial and immunostimulatory properties and aureolysin may contribute to processing of some PSMs.
Significance: Observations using the technology of IMS expand our understanding of S. aureus PSMs.
Keywords: Bacteria, Bacterial Infections, Bacterial Toxins, Mass Spectrometry (MS), Staphylococcus aureus, Virulence Factors, Imaging Mass Spectrometry, Phenol-soluble Modulins (PSMs)
Abstract
Staphylococcus aureus causes a wide range of human disease ranging from localized skin and soft tissue infections to potentially lethal systemic infections. S. aureus has the biosynthetic ability to generate numerous virulence factors that assist in circumventing the innate immune system during disease pathogenesis. Recent studies have uncovered a set of extracellular peptides produced by community-associated methicillin-resistant S. aureus (CA-MRSA) with homology to the phenol-soluble modulins (PSMs) from Staphylococcus epidermidis. CA-MRSA PSMs contribute to skin infection and recruit and lyse neutrophils, and truncated versions of these peptides possess antimicrobial activity. In this study, novel CA-MRSA PSM derivatives were discovered by the use of microbial imaging mass spectrometry. The novel PSM derivatives are compared with their parent full-length peptides for changes in hemolytic, cytolytic, and neutrophil-stimulating activity. A potential contribution of the major S. aureus secreted protease aureolysin in processing PSMs is demonstrated. Finally, we show that PSM processing occurs in multiple CA-MRSA strains by structural confirmation of additional novel derivatives. This work demonstrates that IMS can serve as a useful tool to go beyond genome predictions and expand our understanding of the important family of small peptide virulence factors.
Introduction
Systemic infection by a bacterial pathogen is the result of a stepwise process achieved by circumventing the host immune response. The ability to evade innate immunity, in part, is defined by the metabolic output of the bacterium. Specific components of a pathogen's metabolic output, experimentally proven to contribute to infection, are defined as virulence determinants. Among the virulence determinants used by bacterial pathogens to establish themselves on or within a host are molecular entities possessing adherence, invasion, inflammatory, cytotoxic, or molecular mimicry functions (1–3). One of the most prominent human pathogens, the Gram-positive coccus Staphylococcus aureus, has evolved to synthesize and express a large number of virulence determinants. This abundance of molecular weaponry contributes to the significant morbidity and mortality associated with S. aureus infections.
A challenge to public health, S. aureus is increasingly resistant to first-line antibiotics, as exemplified by methicillin-resistant strains (MRSA).4 Hospital-associated MRSA is a longstanding concern within hospitals and healthcare centers, primarily due to transmission between healthcare workers, patients, and surgical instrumentation (4). However, over a decade ago, the epidemiology of MRSA infection shifted, as infections also spread to the community (CA-MRSA), frequently arising in healthy individuals with no predisposed risks (5). CA-MRSA strains typically present in localized skin or soft tissue infections but also can spread systemically to cause potentially life-threatening diseases, including sepsis and endocarditis (6). Soon after the emergence of CA-MRSA, the pore-forming cytotoxin Panton-Valentine leukocidin, present in many strains, was suggested to be the critical underlying determinant of infection (7). Controversy has arisen surrounding this proposal since murine models of infection using isogenic Panton-Valentine leukocidin-deficient CA-MRSA strains failed to show conclusive virulence contributions (8), and the role Panton-Valentine leukocidin plays in the infectious cycle of CA-MRSA continues to be examined in differential infection models with differing results (9).
In 2007, Wang et al. (10) characterized a set of ribosomally encoded peptides related to the phenol-soluble modulins (PSMs) found in Staphylococcus epidermidis and demonstrated their contribution to CA-MRSA virulence in the murine model. Candidate S. aureus PSMs are present within the genome sequences of all of the available sequence strains as short open reading frames, which many gene identification computer algorithms do not easily recognize. Using analytical methods, it was shown under in vitro culture conditions that CA-MRSA strains produced higher concentrations of PSMs compared with hospital-associated MRSA, prompting the hypothesis that these peptides contribute to the unusual degree of virulence among CA-MRSA strains. Targeted mutagenesis studies showed a significant reduction in the ability of PSM knock-out strains to cause infection in murine abscess and bacteremia models. More recently, the same investigators determined the existence of truncated forms of PSMs with antimicrobial activity against other Gram-positive bacteria. It was proposed that the truncated PSMs contribute to CA-MRSA niche establishment by providing a competitive advantage over other microflora, thereby allowing skin colonization, which predisposes to subsequent soft tissue infection (11).
Liquid chromatography and mass spectrometry was previously used to identify PSMs among the S. epidermidis proteins differentially regulated by quorum sensing during growth in liquid culture (12). In this paper, the emerging technique of microbial imaging mass spectrometry (IMS) was applied to a representative strain of the epidemic CA-MRSA USA300 clone to gain insight into the metabolic fingerprint displayed by this organism when grown as colonies on agar. Spatial distribution patterns of the observed ions showed differential arrangements between neighboring ion masses, particularly within the 1000–3000 Da range. Investigations into the identity of the ions by tandem mass spectrometry demonstrated that they belonged to the PSMα family. Ions with spatial distributions localized to the CA-MRSA colony were determined to be the previously characterized full-length PSMα1 to -4 and the pore forming cytolysin δ-toxin or PSMγ (13). The ions with distributions extending beyond the colony were identified as novel truncated PSMα derivatives, herein annotated as dPSMα1 and dPSMα4. In this work, we present a functional analysis of the antimicrobial, cytotoxic, and immunostimulatory properties of these novel truncated PSMs, study the potential contribution of the highly conserved S. aureus exoprotease aureolysin in their processing, and demonstrate how PSM truncation occurs in multiple MRSA strains by discovery and structural confirmation of other derivatives.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
S. aureus USA300 strains TCH1516 (14), LAC (15), and UAMS1182 (provided by G. Somerville at the University of Nebraska, Lincoln, NE), and USA1000 strain S. aureus ST59 (16) were used in the study. Additionally, the strains S. epidermidis ATCC35984 (17) and group A Streptococcus serotype M3 strain 4041-05 (Centers for Disease Control) were studied. All strains were grown in Todd Hewitt broth or Todd Hewitt agar (THA) except S. epidermidis, which was grown in tryptic soy broth. All strains were propagated at 37 °C unless indicated in each specific method.
Preparation of CA-MRSA Samples for Imaging Mass Spectrometry
THA was prepared and 10 ml of autoclaved agar was poured into Petri dishes under sterile conditions. Colony growth was initiated by spotting 3 μl of an overnight CA-MRSA TCH1516 culture on the prepared THA plates. The bacterial colony was allowed to grow for 72 h at 30 °C, excised, and transferred to a Bruker MSP 96 MALDI anchor plate. Thereafter, the target plate was uniformly covered with Sigma universal matrix (α-cyano-4-hydroxycinnamic acid and 2,5-dihydroxybenzoic acid) by the use of a 50-μm sieve. Once the sample was uniformly covered with matrix, it was placed in a 37 °C oven for 3–4 h or until dried.
Imaging Mass Spectrometry
The Bruker MSP 96 anchor plate containing the sample was inserted into a Microflex Bruker Daltonics mass spectrometer outfitted with the Compass version 1.2 software suite (which consists of FlexImaging version 2.0, FlexControl version 3.0, and FlexAnalysis version 3.0). The sample was run in positive mode, with 600-μm laser intervals in xy and 60% laser power. A photomicrograph of the colony undergoing IMS was loaded onto the FlexImaging command window. Three teach points were selected in order to align the background image with the sample target plate. After the target plate calibration was complete, the AutoXecute command was used to analyze the samples. The method setting under the FlexControl panel was ImaingRPpepmix, which consisted of the following settings. Laser: Fuzzy Control: On, Weight 1.00. Laser power varied between 60% and 70%; Matrix Blaster 0. Evaluation: Peak Selection Masses from m/z 400 to 4000, Mass Control List: Off. Peak Exclusion: Off. Peak Evaluation Processing Method Default, Smoothing: Off, Base-line Subtraction: On, Peak: Resolution higher than 100. Accumulation: Parent Mode: On, Sum up to 20 satisfactory shots in 20 shots, Dynamic Termination: Off. Movement: Random Walk: Two shots at raster spot. Quit sample after: two subsequent failed attempts. Processing: Flex analysis. Method: none, Boito's MS method: none. Sample Carrier: nothing. Spectrometer: On, Ion Source 1–19.00 mV, Ion Source 2: 16.40 mV, Lens: 9.45 mV, Reflector 20.00, Pulsed Ion Extraction: 190 ns, Polarity: Positive. Matrix Suppression: Deflection, Suppress up to: m/z 400. Detector Gain: Reflector 4.1. Sample Rate: 2.00 GS/s, Mode low range, Electronic Gain: Enhanced, 100 mV. Real-time Smooth: off. Spectrometer, Size: 81040, Delay 42968. Processing Method: Factory method RP_2465. Setup: Mass Range: Low. Laser Frequency: 20 Hz, Autoteaching: Off. Instrument-specific Settings: Digitizer Trigger Level: 2000 mV, Digital off Linear: 127 cant, Digital off Reflector: 127 cant. Detector Gain Voltage Offset, Linear: 1300 V, Reflector: 1400 V. Laser Attenuator, Offset: 12%, Range: 30%, Electronic Gain Button Definitions, Regular: 100 me (offset line) 100 mV (offset ref) 200 mV/full scale. End: 51 mV (offset line), 51 mV (offset ref) 100 mV/full scale. Highest: 25 mV (offsetting) 25 mV (offset ref) 50 mV/full scale. Calibration: Calibration was accomplished using a Bruker Daltonics Pepmix 4 as an external standard. Zoom Range 1.0% Peak Assignment Tolerance-User Defined-500 pap. After acquisition, the data were analyzed using the FlexImaging software. The resulting mass spectrum was filtered manually in 0.5–3.0-absorbance unit increments with individual colors assigned to the specific masses.
Identification of PSM Derivatives
To prepare samples for MALDI-TOF-TOF analysis, 50 ml of Todd Hewitt broth was inoculated separately with a cell stock of CA-MRSA strain TCH1516, LAC, UAMS1182, or ST59 and grown overnight at 37 °C with shaking. The saturated medium was then split into two 50-ml conical tubes and extracted with 25 ml of either 1-butanol or ethyl acetate. Extractions were then concentrated to ∼500 μl by the use of a SpeedVac and subjected to gel filtration chromatography using methanol as the mobile phase. Fractions were collected at 15, 20, 25, 40, and 50 ml. Samples were then dried by a SpeedVac and resuspended in a 1:1 mixture of methanol/water in preparation for high pressure liquid chromatography (HPLC). Samples were further separated and purified by the use of an Agilent Technologies 1200 series high pressure liquid chromatograph using a linear gradient from 5% acetonitrile to 95% in 40 min. Fractions resulting from the HPLC run were brought to complete dryness by a SpeedVac and then resuspended in 10 μl of the appropriate acetonitrile/water ratio, depending on the HPLC elution time, to ensure maximum solubility. One μl of each collected fraction was then mixed 1:1 with Sigma-Aldrich universal MALDI matrix resuspended in 78% acetonitrile and 0.1% trifluoric acid. One μl of the mixture was plated on an Allied Biosystems (ABI) MALDI target plate for analysis by ABI 4800 MALDI-TOF-TOF. Masses that corresponded to potential dPSMs based on intact mass were isolated and fragmented. Ions that were amenable to tandem mass spectrometry were manually annotated by de novo sequencing. Sequences identified to be dPSMα1 and dPSMα4 were purchased as synthetic peptides through American Peptide Corporation.
IMS of Bacterial-Bacterial Interactions
CA-MRSA TCH1516, S. epidermidis, and group A Streptococcus (GAS) were grown to A600 = 0.4 and plated as adjacent spots on either THA or tryptic soy broth agar. The co-cultures were allowed to grow for 48 h at 37 °C. After incubation, the agar surrounding the colonies was excised and transferred to a MALDI target plate in preparation for IMS as previously described.
Antimicrobial Assays
Minimum inhibitory concentration (MIC) assays were performed by broth microdilution. Vancomycin and gentamicin (Hospira, Lake Forest, IL) were used as control antibiotics. MIC was determined as the lowest concentration of antibiotic that inhibited bacterial growth by A600. In agar inhibition assays, PSMα1 (20 μg), dPSMα1 (20 μg), PSMα4 (20 μg), and dPSMα4 (20 μg) were resuspended in 70% ethanol, and 20 μg of compound in a 10-μl volume were spotted in triplicate onto prewarmed agar plates and allowed to dry. GAS (2 × 105 cfu) was pipetted onto agar at the location of the compounds, and plates were incubated at 37 °C for 2 h. Spots were then excised and placed into 2-ml screw cap tubes containing 1 mm silica/zirconia beads in 1 ml of PBS. The samples were homogenized in a mini-BeadBeater-8 for 1 min at full speed twice, and the tubes were placed on ice in between. Homogenized samples were serially diluted in sterile PBS and plated on THA plates for enumeration. Inhibition of bacterial growth by purified compounds was calculated as a percentage of initial inoculum.
Assay for GAS Inhibition on Murine Skin
Permission to undertake animal experiments was obtained from the University of California, San Diego Institutional Animal Care and Use Committee. Flanks of 8–10-week-old female CD1 mice were shaved. The following day, mice were anesthetized with ketamine/xylazine and placed on heating pads to maintain body temperature. Full-length PSMs and dPSMs were resuspended in 70% ethanol alone, and 20 μg of compound in 10 μl were pipetted onto premarked spots on the mouse flank and allowed to dry. Five spots per mouse were prepared as follows: negative control (70% ethanol), PSMα1 (20 μg), dPSMα1 (20 μg), PSMα4, and dPSMα4. GAS was grown to midlogarithmic phase (A600 = 0.4), washed with sterile PBS, and diluted to 2 × 107 cfu/ml. Ten μl (∼2 × 105 cfu) of bacteria were spotted onto agar plates in triplicate and allowed to dry. Once the spots had dried, agar discs were excised using a 6-mm biopsy punch. One agar disc containing 2 × 105 cfu of GAS M3 was placed onto each spot (5 discs/mouse) and affixed to the mouse with a Tegaderm transparent wound dressing. After 2 h, both the skin and the agar disc were placed into 2-ml screw cap tubes containing 1-mm silica/zirconia beads in 1 ml of PBS. The samples were homogenized in a mini-BeadBeater-8 for 1 min at full speed twice, placing the tubes on ice in between. Homogenized samples were serially diluted in sterile PBS and plated on THA plates for enumeration of CFU. Inhibition of bacterial growth by purified compounds was calculated as a percentage of initial inoculum. Statistics analyses for the acquired data sets were performed by one-way analysis of variance.
Neutrophil Lysis Assays
Blood was drawn from healthy donors after informed consent, and neutrophils were isolated using the PolyMorphPrep kit as per the manufacturer's instructions (Axis-Shield, Rodeløkka, Norway). The Promega Cyto Tox 96 non-radioactive cytotoxicity assay kit was used to monitor lactate dehydrogenase release from neutrophils upon incubation with PSMs or dPSMs. Briefly, a round-bottom 96-well plate was used to mix the following reaction mixtures: (i) no cells or medium only, (ii) untreated neutrophils, (iii) lysis control, (iv) DMSO, (v) PSM1 (100 and 10 μg/ml), (vi) dPSM1 (100 and 10 μg/ml), (vii) PSM4 (100 and 10 μg/ml), (viii) dPSM4 (100 and 10 μg/ml). Each well contained 2.5 × 106 neutrophil cells/ml in RPMI plus 2% heat-inactivated FBS. After mixing, PSM peptides as well as all controls were incubated at 37 °C and 5% CO2. The lactate dehydrogenase release assay was measured at 60 and 120 min by reading A490.
Neutrophil Extracellular Trap Assays
Neutrophils at 2 × 105 cells/well were pipetted into a 48-well tissue culture plate in RPMI. Twenty-five nm phorbol 12-myristate 13-acetate stimulation was used as a positive control to monitor extracellular trap formation. Neutrophils incubated in DMSO were used as a background control. PSM1 and PSM4, including their corresponding truncated derivatives, were incubated for 180 min at concentrations of 100 and 10 μg/ml. Thereafter, 500 milliunits of micrococcal nuclease in a volume of 50 μl/well were added for 10 min. The reaction was then halted with 5 mm EDTA. The plate was then centrifuged at 200 × g for 8 min. After centrifugation, 100 μl of the supernatant were transferred to 96-flat bottom well plates in preparation for the Quant-iT Picogreen (Invitrogen) assay. The Quant-iT Picogreen assay was performed as described in the provided protocol. Briefly, Picogreen reagent was diluted 1:200 and was mixed 1:1 with samples. The mixture was allowed to incubate for 5 min at room temperature in dark conditions. After incubation, the excitation 480 nm, emission A520 nm (fluorescein) for each well was recorded.
Fluorescence Microscopy of Neutrophil Extracellular Traps (NETs)
For visualization of extracellular traps, neutrophils were placed on poly-l-lysine-coated glass slides and treated with PSMs or vehicle controls. NETs were visualized using a rabbit antimyeloperoxidase antibody followed by a secondary goat anti-rabbit Alexa 488 antibody; samples were embedded in 4′,6-diamidino-2-phenylindole (DAPI) to counterstain DNA in blue. Mounted samples were examined using an inverted confocal laser-scanning two-photon microscope, Olympus Fluoview FV1000, with FluoviewTM spectral scanning technology (Olympus) and a ×20/0.75 Olympus objective.
Hemolysis Assays
Defribinated sheep or human whole blood was washed in PBS and diluted to a final concentration of 1:25 (v/v) in PBS. Whole blood (100 μl) was then placed into individual wells of a flat-bottom 96-well microtiter plate (Costar, Corning). PSM1, dPSM1, PSM4, and dPSM4 at a concentration of 100 and 10 μg/ml were added directly to the wells, and the mixture was incubated for 60 min at 37 °C. After incubation, plates were centrifuged at 500 × g for 10 min, and an aliquot of supernatant (100 μl) was placed in a separate microtiter plate to measure hemoglobin absorbance at 450 nm on a microplate reader. One μl of the erythrocyte and PSM mixtures was aliquoted into separate Eppendorf tubes and examined by a Riveal contrast microscope (Quorum Technologies) to confirm erythrocyte lysis.
Generation of CA-MRSA Aureolysin Mutant
The wild type strain LAC is an isolate of the MRSA USA300 clone. A markerless aur deletion was created in each strain by transduction of the pKOR1Δaur plasmid (18) by 80α phage and utilization of the pKOR1 knock-out strategy, as described previously (19). To construct the aureolysin complementation plasmid, pDB59 (20) was digested with BamHI and EcoRI to remove the P3 promoter driving YFP. Oligonucleotides CLM525 (5′-gatccatcgatcgatgctagcattcgatcatg-3′) and CLM526 (5′-aattcatgatcgaatgctagcatcgatcgatg-3′) were annealed and ligated into pDB59 digested by BamHI and EcoRI in order to introduce a NheI site. This intermediate plasmid was designated pCM46. The aur gene was amplified from the USA300 LAC genome by PCR using primers CLM529 (5′-gttgttggatccgttatctcacatatttcaagcattg-3′) and CLM522 (5′-gttgttgctagcggtataccccatatataagctg-3′). Digestion of the PCR product by BamHI and NheI and ligation with pCM46 digested by the same enzymes yielded complementation plasmid pAur. The aureolysin mutant was subsequently transformed with pAur by electroporation.
Dried Droplet MALDI-MS of LAC Strains
Wild type LAC, LAC Δaur, and LAC Δaur + pAur were prepared and analyzed with MALDI-MS as described previously. Repeated experiments were performed to generate a mass list of the ions observed within a 400–5000 Da range for each strain. Ultimately, each mass list for a particular strain was compiled and used to identify potential dPSMs based on intact mass.
Recombinant Aureolysin Cleavage Assays
Recombinant aureolysin was purchased from Biocentrum and resuspended as recommended in the factory protocol. Thereafter, activity was assessed by incubating 2 μg of aureolysin with 4 μg of either PSM1 or PSM4 in 20 mm Tris-HCl, pH 7.8, containing 5 mm CaCl2. Control samples were included to further verify aureolysin activity on PSMs. Observation of PSM cleavage was performed by dried droplet MALDI-MS.
RESULTS
Imaging Mass Spectrometry of CA-MRSA
The technique of thin layer agar IMS is an effective tool for characterizing microbial metabolic output within a defined mass range (21, 22). The strategy lends a molecular view, in particular spatial distributions, of the different small molecules and peptides an organism produces when grown on solid agar media and exposed to abiotic or biotic factors (23). Our goal was to apply IMS to study the metabolic behavior of the notable human pathogen, CA-MRSA. To this end, a representative strain (TCH1516) of the epidemic CA-MRSA USA300 clone was grown on agar medium and then subjected to IMS in order to gain a molecular fingerprint of the bacterial pathogen within a 400–3500 Da mass range (supplemental Fig. S1). The resulting IMS data revealed several small molecules (<1000 Da) and a set of molecular entities falling in the range of peptides (1000–3000 Da). Many of the observed molecular species had spatial distribution patterns, indicating them to be physically bound to, or contained within, the CA-MRSA colony, whereas others had spatial distribution patterns revealing the ions to be released beyond the colony into its environment. The identities of the molecular entities observed in the mass spectral fingerprint were pursued, with a particular interest in the peptides that displayed a spatial distribution consistent with release.
Identification of PSM Derivatives
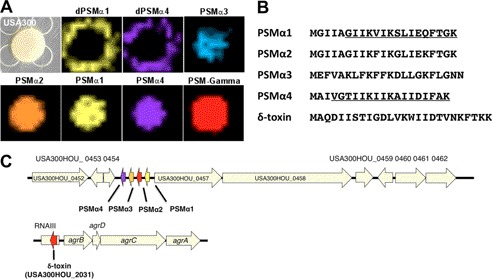
To identify the released peptides observed by IMS, CA-MRSA was grown in liquid broth medium overnight and extracted with various solvent-solvent systems as a preparatory step. Samples were further processed by HPLC and then analyzed by dried droplet MALDI-MS/MS. Ions within the 1000–3000 Da range were targeted for fragmentation to gain structural information in the form of a peptide sequence tag (24). Data were collected for those ions that were amenable to collision-induced fragmentation (supplemental Figs. S2–S8). BLAST analysis was performed on the acquired peptide sequence tags searched against the CA-MRSA USA300 TCH1516 genome. The initial BLAST analysis identified two of the acquired sequence tags related to the previously characterized PSMα1 (10). One of the fragmented peptides was identified as full-length PSMα1, based on the BLAST analysis and the corresponding observed intact mass at an error of 170 ppm. The released ion detected by IMS at mass 1773 Da corresponded to a five-amino acid truncated form of PSMα1, with an observed mass error relative to the theoretical value of 112 ppm (Fig. 1A and supplemental Fig. S2).
FIGURE 1.
Visualization of dPSM by imaging mass spectrometry. A, imaging mass spectrometry of S. aureus TCH1516 USA300, a representative CA-MRSA strain. Full-length PSMs are displayed through various false coloring, indicating a colony-bound spatial distribution. Truncated peptides dPSM1 and dPSM4 display a spatial distribution extending beyond the colony, indicating their release into the agar medium. B, amino acid sequences of PSMs identified by MALDI-TOF/TOF (spectra provided in the supplemental material). Characterized derivative peptide regions are underlined. C, location of PSMs in CA-MRSA TCH1516 genome. PSMα1 to -4 are adjacent, small ribosomally encoded peptides. PSMγ (also known as delta-toxin) is located within the agr gene cluster embedded within the rnaIII gene.
BLAST analysis of the remaining peptide sequence tags identified PSMα2 and PSMγ, whereas a number of others remained unidentified and were not annotated in the CA-MRSA USA300 TCH1516 genome (Fig. 1C and supplemental Table S1). Therefore, a six-frame translation of the genome was performed, creating a database with protein sequences for every potential open reading frame (ORF). Using the database, the unidentified peptide sequence tags were manually searched by peptidogenomics (25), and PSMα3 and PSMα4 were identified (Fig. 1B). Like our observation with the truncated form of PSMα1, another derivative was identified originating from PSMα4 (supplemental Fig. S3), again with an IMS spatial distribution extending beyond the bacterial colony (Fig. 1A). With this structural evidence in hand, two PSMα derivatives (dPSMα) at masses 1773 and 1855 Da were commercially synthesized and designated dPSMα1 and dPSMα4, respectively, for functional characterization studies.
Antimicrobial Activity of PSM Derivatives
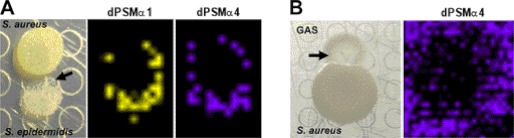
Otto and colleagues (11) suggested that PSM derivatives might serve antimicrobial functions relevant for CA-MRSA niche establishment. To this end, we set up S. aureus bacterial-bacterial interactions, first with the common skin microbe S. epidermidis and second with a leading human skin pathogen, GAS. The CA-MRSA colony achieved similar size and morphology when grown alone or in the co-culture experiment. However, both S. epidermidis and GAS showed inhibition of growth at the bacterial interface presumably caused by the metabolic output of the CA-MRSA colony (Fig. 2, A and B). When the interactions between CA-MRSA and S. epidermidis or GAS were analyzed by IMS, dPSMα1 and dPSMα4 were both localized to the inhibition zones, suggesting that the peptides could contribute to the growth-suppressive phenotype.
FIGURE 2.
Antimicrobial activity exhibited by PSMs and identified derivatives. A, bacterial-bacterial interaction between CA-MRSA and S. epidermidis. The optical image of the interaction shows a perturbed morophology of the S. epidermidis colony (black arrow) presumably caused by the metabolic output of the adjacent CA-MRSA colony. IMS demonstrated dPSMα peptides localized within the zone of growth inhibition. B, IMS of the interaction between CA-MRSA and GAS. The optical image shows that CA-MRSA has the ability to inhibit the growth (black arrow) of the competing GAS colony. IMS showed that dPSMα is released and therefore potentially contributes to growth inhibition. Shown is the spatial distribution of dPSMα4 (dPSMα1 is omitted).
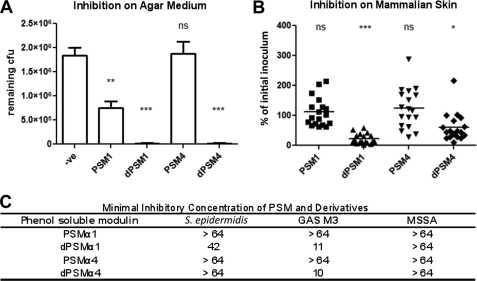
Suppression of GAS growth on solid agar was further analyzed using synthetic dPSMα1 and dPSMα4, which showed inhibitory activity greater than their parent peptides (Fig. 3A). In MIC testing, peptide dPSMα1 had activity against GAS at 11 μm and S. epidermidis at 42 μm but no activity at the highest concentration tested (64 μm) against a methicillin-susceptible S. aureus strain (MSSA in Fig. 3C). The parent peptide PSMα1 showed no activity against any of the strains at the upper concentration limit. Peptide dPSMα4 showed activity against GAS at 10 μm (but not S. epidermidis), whereas the parent peptide PSMα4 showed no activity. We next tested if bioactivity could be maintained in an in vivo setting. Flanks of CD1 mice were shaved and individually treated with PSMα1, PSMα4, dPSMα1, or dPSMα4 and subsequently inoculated with GAS. Both dPSMα1 and dPSMα4 restricted growth of GAS on mammalian skin at 4 h because a significant decrease in cfu count was produced by both derivatives compared with the parent peptides (Fig. 3B).
FIGURE 3.
Antimicrobial activity exhibited by PSMs and derivatives. A, inhibition of GAS growth on solid agar. GAS was grown on agar plates in the presence of synthetic PSMs. **, p < 0.01; ***, p < 0.001; ns, no significant difference. B, in vivo growth inhibition of GAS on mammalian skin in the presence of PSMs. Inhibition of bacterial growth was calculated as a percentage of the initial inoculum. *, p < 0.05; ***, p < 0.001; ns, no significant difference. C, MIC of the synthetic peptides dPSMα1 and dPSM4. Derivative peptides displayed antimicrobial activity in contrast to their respective parent forms. Numerical values displayed in the table indicate the concentrations tested (μm).
Mammalian Cell Lysis Caused by PSM Derivatives
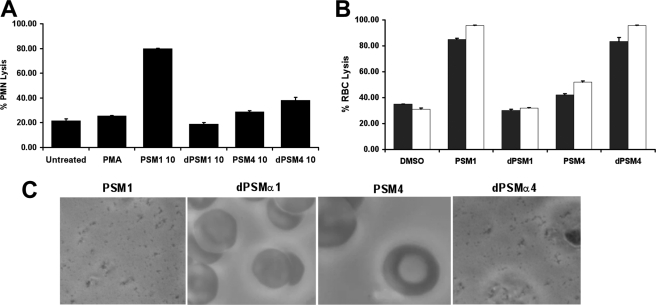
An essential first line of defense in the mammalian innate immune response is the ability of phagocytic cells, such as neutrophils, to accumulate at the site of infection and intercept invading foreign pathogens (26). One strategy bacterial pathogens have evolved to circumvent neutrophil killing is to secrete enzymes and peptides cytotoxic to the phagocyte. To test if dPSMα1 and dPSMα4 exhibited cytolytic activity, neutrophils were isolated from human whole blood and incubated with the peptides at different concentrations and time points. At 10 μg/ml through a 90-min exposure, dPSMα1 showed no cytolytic activity, whereas full-length PSMα1 parent peptide caused 59% neutrophil lysis. In parallel experiments, the truncated peptide dPSMα4 caused 13% and its parent peptide PSMα4 induced 5% neutrophil lysis (Fig. 4A). No neutrophil cytolytic activity of any of the full-length or truncated peptides was detected at a lower concentration of 1 μg/ml.
FIGURE 4.
Neutrophil and erythrocyte lysis caused by PSMs. A, neutrophils were incubated with 10 μg/ml synthetic PSMs and their respective derivatives for 180 min. Lysis was assessed by monitoring the release of lactate dehydrogenase by colorimetric spectroscopy. Absorbances were normalized by the use of a lysis reagent. B, human (white bars) and 5% sheep (black bars) blood was incubated with 10 μg/ml synthetic PSMs and their respective derivatives for 60 min. Lysis was then assessed by monitoring the release of heme spectroscopically at A450. C, human blood samples incubated with PSMs were further assessed with a Riveal contrast microscope to confirm erythrocyte lysis.
To determine if dPSMα1 and dPSMα4 or their parent peptides had hemolytic activity against human erythrocytes (27, 28), whole blood was washed and incubated with 10 μg/ml peptide for 60 min, and the A450 was measured to monitor the release of hemoglobin. Peptide dPSMα1 showed no hemolytic activity, whereas the parent peptide PSMα1 released 58% hemoglobin compared with the Triton X-100 lysis control. In contrast, the truncated dPSMα4 had a stronger hemolytic phenotype (52% lysis) than its parent peptide (19% lysis) (Fig. 4B). Similar hemolytic activities were observed using 5% difibrinated sheep blood (Fig. 4B) and validated by microscopy to visualize erythrocyte lysis from each sample (Fig. 4C).
Immunostimulation of Neutrophil Extracellular Traps
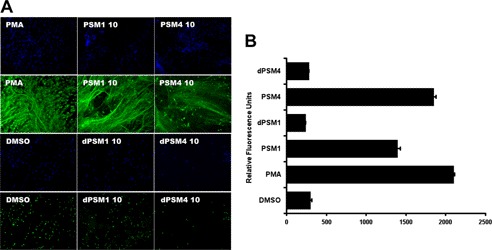
Now recognized as a key component of their innate immune function, neutrophils release DNA-based NETs to immobilize bacteria at the tissue site of infection, exposing them to embedded antimicrobial peptides and proteases. Because PSMs and their respective derivatives are released extracellularly in high concentrations by CA-MRSA isolates, we explored their influence on NET formation. As a positive control, neutrophils were incubated with phorbol 12-myristate 13-acetate, and a robust formation of NETs was visualized by microscopy using an anti-myeloperoxidase antibody and DAPI nuclear staining. The parent peptides PSMα1 and PSMα4 at 10 μg/ml both stimulated NET formation to a level comparable with phorbol 12-myristate 13-acetate as observed by microscopy. In contrast, both derivative peptides dPSMα1 and dPSMα4 did not demonstrate the ability to stimulate NETs (Fig. 5A). The differential effects on NET stimulation were corroborated quantitatively using Picogreen dye to measure extracellular DNA (Fig. 5B).
FIGURE 5.
Immunostimulation of NETs. A, for visualization of NETs, neutrophils were placed on poly-l-lysine-coated glass slides and treated with PSMs or vehicle controls. NETs were visualized using a rabbit antimyeloperoxidase antibody followed by a secondary goat anti-rabbit Alexa 488 antibody; samples were embedded in DAPI to counterstain DNA in blue. B, in parallel with microscopy, the Quant-iT Picogreen assay was used to quantify the extracellular DNA upon stimulation by PSMs and the controls by colorimetric spectroscopy. Error bars, S.D.
MALDI-MS of Multiple MRSA Strain Extracts Identifies Additional PSM Derivatives
Next we set out to determine if other MRSA strains biosynthesized an array of dPSMs similar to those we observed with CA-MRSA USA300 strain TCH1516. To this end, CA-MRSA strains ST59 (USA1000), UAMS1182 (USA300), and CA-MRSA LAC (USA300) were grown, and supernatants were extracted as described previously for MALDI-TOF/TOF analysis. In each strain, several ions were observed, corresponding to dPSMα candidates based on intact mass (supplemental Table S2). All ion candidates were subjected to tandem mass spectrometry in order gain structural information in the form of peptide sequence tags, and the generated tags were studied by either BLAST analysis or manually using six-frame translation databases as described above. We found that the majority of the ions were not amenable to tandem mass spectrometry either because the ions were of low concentration or because they were not easily fragmented by collision-induced dissociation. Nevertheless, two additional novel dPSMs, originating from PSMα2 and dPSMα4, were structurally verified, as was one novel derivative of PSMλ supplemental Figs. S9–S12).
Aureolysin Activity Influences Processing of PSMs
Finally, we performed initial investigations of contributing mechanisms for the proteolytic cleavage of native full-length PSMs into their truncated dPSM form. First, the gene cluster containing the PSMs was examined in various CA-MRSA USA300 genomes to assess adjacent or neighboring genes (supplemental Table S1), recognizing that because PSMs are short peptides, they are commonly missed by sequencing algorithms and often not annotated. We found that PSMα1 to -4 are located near the cobalamin biosynthesis protein and adjacent to small hypothetical proteins (47 and 42 amino acids) of unknown function. Downstream to these PSMs are proteins annotated as NADH dehydrogenase, PAP2 superfamily phosphatase, carboxyesterase, or hypothetical proteins. The gene encoding PSMγ is distant from the PSMα1 gene, contained within the agr gene cluster, and embedded within rnaIII (29). Because no gene immediately adjacent to the PSMs showed clear homology to known proteinases, we explored the potential contribution of the well studied zinc-dependent S. aureus metalloprotease aureolysin. Previous studies have shown that aureolysin is promiscuous in its enzymatic activity, with the ability to cleave its own extracellular cell wall proteins, the human-derived antimicrobial peptide LL-37, and several other substrates (30, 31).
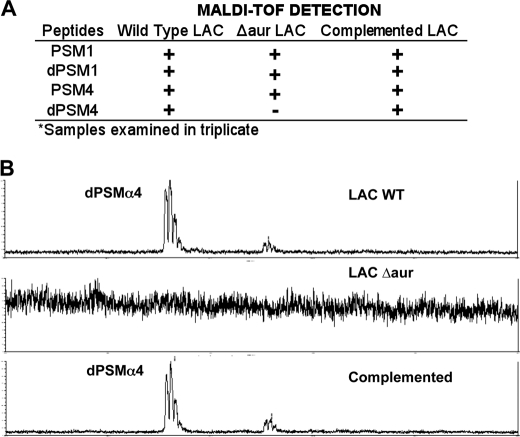
To investigate if aureolysin contributes to processing PSMs, we compared the mass spectrometry profile of PSMα1 and PSMα4 peptides produced by WT CA-MRSA LAC, its isogenic Δaur mutant, and a complemented strain in which the Δaur mutant is transformed with an expression vector expressing the aur gene (Fig. 6A). The WT CA-MRSA LAC produced detectable ions at 1855 and 1774 Da as expected; however, the isogenic Δaur strain showed only the ability to produce the ion at 1774 Da but not the 1855 Da ion. The presence of the 1855-Da ion was restored in the complemented mutant strain (Fig. 6B). Aureolysin's activity on PSMs was further validated when recombinant protein was incubated with PSMα4 and PSMα1. In both cases, peptide truncated products were observed, which were absent in the control samples (supplemental Figs. S13–S15).
FIGURE 6.
Aureolysin PSM processing. A, dried droplet MALDI-TOF analysis was used to examine processed extracts originating from CA-MRSA LAC, LAC Δaur, and LAC Δaur + pAur for the presence and absence of dPSM1 and dPSM4. B, MALDI-TOF data showing the detection of dPSM4 in wild type CA-MRSA LAC, the loss of the dPSM4 in the LAC Δaur background, and its restoration in the complemented strain.
DISCUSSION
We have demonstrated that IMS can be used as a tool to discover previously undetermined biological phenomena by providing a molecular view of the vast number of small molecules and peptides a bacterial colony biosynthesizes during growth. CA-MRSA pathogenesis is the integrated outcome of multiple interactions of bacterial and host factors with different components acting alone or in synergy. The cytolytic peptides known as PSMs were shown to contribute to CA-MRSA pathogenesis, and more recently, the same set of peptides, once proteolytically cleaved, demonstrated a gain of antimicrobial function. Using the strategy of IMS, we were able to uncover previously unidentified PSM derivatives produced by prototypical CA-MRSA strains. The peptides were initially targeted based on the spatial distribution patterns observed by IMS, which showed that the dPSMs were released into the solid agar medium during growth, whereas the parent PSMs remained colony-bound. Once the structural identity of dPSMα1 and dPSMα4 was confirmed, we verified that dPSMs have selective antimicrobial activity, a characteristic not displayed by the PSM until proteolytic cleavage of the peptide's N terminus. The potential role that dPSM modifications played in influencing host cells was studied by monitoring neutrophil and erythrocyte lysis, followed by stimulation of NETs. PSMα1 demonstrated lytic activity against human neutrophils and erythrocytes, but these activities diminished in the truncation to the dPSMα1 form. Conversely, the modest lytic activities of the dPSMα4 were significantly greater than those of its parent peptide, PSMα4. Stimulation of NET production by human neutrophils was readily observed in response to either of the parent PSMs, whereas no detectable stimulation was observed with either dPSM. Thus, the processing of PSM peptides not only influences their ability to diffuse further away from the parent cell but influences their antimicrobial, cytolytic, and proinflammatory capacities. In the two studied PSM peptides, processing was associated with increased antimicrobial activity and decreased NET-stimulating capacity, whereas cytolytic activity was affected differentially in the two instances.
The mechanism for processing PSMs into their truncated form has not previously been investigated, and a candidate coordinately regulated protease gene does not appear in the gene cluster containing the PSMs. Our studies using isogenic aureolysin-producing and -deficient strains suggest that expression of the broad spectrum metalloprotease was associated with the appearance of dPSMα4. Additionally, recombinant aureolysin cleaves PSMα4 into smaller peptides, although we were not successful in capturing the ion corresponding to dPSM4 at the mass 1855 Da. In contrast, dPSM1 was present in both the WT and aureolysin mutant CA-MRSA strains. We conclude that aureolysin may influence processing of a subset of PSMs. Given that the genome of CA-MRSA encodes several other known proteases, alternative activities probably work together to generate a multitude of dPSM derivatives. The hypothesis is consistent with our analysis of three additional CA-MRSA strains in which a large number of potential dPSMs were identified based on mass alone but not structurally verified, given the peptides were either of low concentration or were not amenable to collision-induced dissociation leading to a peptide sequence tag. Ultimately, the work herein demonstrates that IMS can serve as a useful tool to go beyond genome predictions and expands our understanding of the important family of small peptide virulence factors, such as PSMs in S. aureus, S. epidermidis, and other important human pathogens. Recognition of differences in spatial distribution and biological activities of derivative peptides will aid in elucidating their influence on the establishment of infection and their broader impacts on the pathogen-host interaction.
Supplementary Material
Acknowledgment
We appreciate the support of the Bruker Therapeutic Discovery Mass Spectrometry Center at the University of California at San Diego.
This work was supported, in whole or in part, by National Institutes of Health grants GM086283 and GM094802 (to P. C. D.) and AI077780 and AR05278 (to V. N.).

This article contains supplemental Tables S1 and S2 and Figs. S1–S15.
- MRSA
- methicillin-resistant S. aureus
- CA-MRSA
- community-associated methicillin-resistant S. aureus
- PSM
- phenol-soluble modulin
- IMS
- imaging mass spectrometry
- THA
- Todd Hewitt agar
- MIC
- minimum inhibitory concentration
- NET
- neutrophil extracellular traps.
REFERENCES
- 1. Pizarro-Cerdá J., Cossart P. (2006) Bacterial adhesion and entry into host cells. Cell 124, 715–727 [DOI] [PubMed] [Google Scholar]
- 2. Kobayashi S. D., DeLeo F. R. (2009) An update on community-associated MRSA virulence. Curr. Opin. Pharmacol. 9, 545–551 [DOI] [PubMed] [Google Scholar]
- 3. Molloy E. M., Cotter P. D., Hill C., Mitchell D. A., Ross R. P. (2011) Streptolysin S-like virulence factors. The continuing sagA. Nat. Rev. Microbiol. 9, 670–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maree C. L., Daum R. S., Boyle-Vavra S., Matayoshi K., Miller L. G. (2007) Community-associated methicillin-resistant Staphylococcus aureus isolates causing healthcare-associated infections. Emerg. Infect. Dis. 13, 236–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gorak E. J., Yamada S. M., Brown J. D. (1999) Clin. Infect. Dis. 29, 29797–29800 [DOI] [PubMed] [Google Scholar]
- 6. King M. D., Humphrey B. J., Wang Y. F., Kourbatova E. V., Ray S. M., Blumberg H. M. (2006) Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft tissue infections. Ann. Intern. Med. 144, 309–317 [DOI] [PubMed] [Google Scholar]
- 7. Labandeira-Rey M., Couzon F., Boisset S., Brown E. L., Bes M. (2007) Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science 315, 1130–1133 [DOI] [PubMed] [Google Scholar]
- 8. Bubeck Wardenburg J., Palazzolo-Ballance A. M., Otto M., Schneewind O., DeLeo F. R. (2008) Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J. Infect. Dis. 198, 1166–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lipinska U., Hermans K., Meulemans L., Dumitrescu O., Badiou C., Duchateau L., Haesebrouck F., Etienne J., Lina G. (2011) Panton-Valentine leukocidin does play a role in the early stage of Staphylococcus aureus skin infections. A rabbit model. PLoS One 6, e22864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang R., Braughton K. R., Kretschmer D., Bach T. H., Queck S. Y., Li M., Kennedy A. D., Dorward D. W., Klebanoff S. J., Peschel A., DeLeo F. R., Otto M. (2007) Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13, 1510–1514 [DOI] [PubMed] [Google Scholar]
- 11. Joo H. S., Cheung G. Y., Otto M. (2011) Antimicrobial activity of community-associated methicillin-resistant Staphylococcus aureus is caused by phenol-soluble modulin derivatives. J. Biol. Chem. 286, 8933–8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li M., Villaruz A. E., Vadyvaloo V., Sturdevant D. E., Otto M. (2008) AI-2-dependent gene regulation in Staphylococcus epidermidis. BMC Microbiol. 8, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kerr I. D., Doak D. G., Sankararamakrishnan R., Breed J., Sansom M. S. (1996) Molecular modeling of staphylococcal δ-toxin ion channels by restrained molecular dynamics. Protein Eng. 9, 161–171 [DOI] [PubMed] [Google Scholar]
- 14. Highlander S. K., Hultén K. G., Qin X., Jiang H., Yerrapragada S., Mason E. O., Jr., Shang Y., Williams T. M., Fortunov R. M., Liu Y., Igboeli O., Petrosino J., Tirumalai M., Uzman A., Fox G. E., Cardenas A. M., Muzny D. M., Hemphill L., Ding Y., Dugan S., Blyth P. R., Buhay C. J., Dinh H. H., Hawes A. C., Holder M., Kovar C. L., Lee S. L., Liu W., Nazareth L. V., Wang Q., Zhou J., Kaplan S. L., Weinstock G. M. (2007) Subtle genetic changes enhance virulence of methicillin-resistant and sensitive Staphylococcus aureus. BMC Microbiol. 7, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kennedy A. D., Otto M., Braughton K. R., Whitney A. R., Chen L., Mathema B., Mediavilla J. R., Byrne K. A., Parkins L. D., Tenover F. C., Kreiswirth B. N., Musser J. M., DeLeo F. R. (2008) Epidemic community-associated methicillin-resistant Staphylococcus aureus. Recent clonal expansion and diversification. Proc. Natl. Acad. Sci. U.S.A. 105, 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang Y. C., Ho C. F., Chen C. J., Su L. H., Lin T. Y. (2008) Clin. Microbiol. Infect. 14, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 17. Liu W. T., Yang Y. L., Xu Y., Lamsa A., Haste N. M., Yang J. Y., Ng J., Gonzalez D., Ellermeier C. D., Straight P. D., Pevzner P. A., Pogliano J., Nizet V., Pogliano K., Dorrestein P. C. (2010) Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 107, 16286–16290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kavanaugh J. S., Thoendel M., Horswill A. R. (2007) A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol. Microbiol. 65, 780–798 [DOI] [PubMed] [Google Scholar]
- 19. Bae T., Schneewind O. (2006) Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63 [DOI] [PubMed] [Google Scholar]
- 20. Yarwood J. M., Bartels D. J., Volper E. M., Greenberg E. P. (2004) Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang Y. L., Xu Y., Straight P., Dorrestein P. C. (2009) Translating metabolic exchange with imaging mass spectrometry. Nat. Chem. Biol. 5, 885–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Watrous J. D., Dorrestein P. C. (2011) Imaging mass spectrometry in microbiology. Nat. Rev. Microbiol. 9, 683–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gonzalez D. J., Haste N. M., Hollands A., Fleming T. C., Hamby M., Pogliano K., Nizet V., Dorrestein P. C. (2011) Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology 157, 2485–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Steen H., Mann M. (2004) The ABC's (and XYZ's) of peptide sequencing. Nat. Rev. Mol. Cell Biol. 5, 699–711 [DOI] [PubMed] [Google Scholar]
- 25. Kersten R. D., Yang Y. L., Xu Y., Cimermancic P., Nam S. J., Fenical W., Fischbach M. A., Moore B. S., Dorrestein P. C. (2011) A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 7, 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nathan C. (2006) Neutrophils and immunity. Challenges and opportunities. Nat. Rev. Immunol. 6, 173–182 [DOI] [PubMed] [Google Scholar]
- 27. Datta V., Myskowski S. M., Kwinn L. A., Chiem D. N., Varki N., Kansal R. G., Kotb M., Nizet V. (2005) Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol. Microbiol. 56, 681–695 [DOI] [PubMed] [Google Scholar]
- 28. Doran K. S., Liu G. Y., Nizet V. (2003) Group B streptococcal β-hemolysin/cytolysin activates neutrophil signaling pathways in brain endothelium and contributes to development of meningitis. J. Clin. Invest. 112, 736–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reynolds J., Wigneshweraraj S. (2011) Molecular insights into the control of transcription initiation at the Staphylococcus aureus agr operon. J. Mol. Biol. 412, 862–881 [DOI] [PubMed] [Google Scholar]
- 30. Beaufort N., Wojciechowski P., Sommerhoff C. P., Szmyd G., Dubin G., Eick S., Kellermann J., Schmitt M., Potempa J., Magdolen V. (2008) The human fibrinolytic system is a target for the staphylococcal metalloprotease aureolysin. Biochem. J. 410, 157–165 [DOI] [PubMed] [Google Scholar]
- 31. Sieprawska-Lupa M., Mydel P., Krawczyk K., Wójcik K., Puklo M., Lupa B., Suder P., Silberring J., Reed M., Pohl J., Shafer W., McAleese F., Foster T., Travis J., Potempa J. (2004) Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 48, 4673–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.