Abstract
Although the morphological and physiological changes involved in pregnancy in live-bearing reptiles are well studied, the genetic mechanisms that underlie these changes are not known. We used the viviparous African Ocellated Skink, Chalcides ocellatus, as a model to identify a near complete gene expression profile associated with pregnancy using RNA-Seq analyses of uterine transcriptomes. Pregnancy in C. ocellatus is associated with upregulation of uterine genes involved with metabolism, cell proliferation and death, and cellular transport. Moreover, there are clear parallels between the genetic processes associated with pregnancy in mammals and Chalcides in expression of genes related to tissue remodeling, angiogenesis, immune system regulation, and nutrient provisioning to the embryo. In particular, the pregnant uterine transcriptome is dominated by expression of proteolytic enzymes that we speculate are involved both with remodeling the chorioallantoic placenta and histotrophy in the omphaloplacenta. Elements of the maternal innate immune system are downregulated in the pregnant uterus, indicating a potential mechanism to avoid rejection of the embryo. We found a downregulation of major histocompatability complex loci and estrogen and progesterone receptors in the pregnant uterus. This pattern is similar to mammals but cannot be explained by the mammalian model. The latter finding provides evidence that pregnancy is controlled by different endocrinological mechanisms in mammals and reptiles. Finally, 88% of the identified genes are expressed in both the pregnant and the nonpregnant uterus, and thus, morphological and physiological changes associated with C. ocellatus pregnancy are likely a result of regulation of genes continually expressed in the uterus rather than the initiation of expression of unique genes.
Keywords: placenta, pregnancy, RNA-Seq, transcriptome, uterus, viviparity
Introduction
Reproductive mode is subject to intense selective pressure and any modification will directly affect an organism's fitness. Thus, the transition between oviparity (egg-laying) and viviparity (live-bearing) is one of the most dramatic changes in the evolution of vertebrate animals. As a result of constraints on reproductive function, most vertebrate clades show a high phylogenetic conservatism in reproductive mode. For example, therian mammals represent only a single (albeit the best-known) origin of viviparity, whereas birds, alligators, and turtles exhibit only oviparity. By contrast, the evolutionary history of squamate reptiles (lizards and snakes) includes over 100 transitions from oviparity to viviparity, more than all other lineages of vertebrates combined (Blackburn 2006), with an extraordinary diversity of placental morphology and physiology (Blackburn et al. 1984; Blackburn 1993; Stewart 1993; Thompson and Speake 2006). Squamates are therefore an ideal model system with which to explore the processes that shape the evolution of viviparity and placentation in vertebrates.
Viviparity and placentation in squamate reptiles have been the subject of extensive morphological and physiological studies (e.g., Weekes 1935; Blackburn 1993; Thompson et al. 2000; Stewart and Thompson 2000; Thompson and Speake 2006). These studies have described a continuum of placental complexity, ranging from simple placentae formed by the apposition of uterine and embryonic tissue (e.g., most Eulamprus quoyii group skinks; Murphy et al. 2012) to highly complex placentae characterized by extensive folding of the uterus associated with blood vessels and containing enlarged uterine and chorionic epithelial cells (e.g., some Mabuya group skinks; Jerez and Ramírez-Pinilla 2001; Ramírez-Pinilla et al. 2006; Vieira et al. 2007; Blackburn and Flemming 2011). For the purposes of this discussion, we refer to these ends of the morphological complexity spectrum as “simple” and “complex,” although we note these categories are an oversimplification of the true underlying diversity (see Blackburn 1993). Subsequent studies have quantified nutrient exchange across these different placental types and have concluded that very little maternal to embryonic transfer occurs via simple placentae, but substantial transfer occurs in species with more complex placentae. In one extreme, the embryos of the South American skink, Mabuya bistrata experience a 47,400% increase in dry mass (Vitt and Blackburn 1991) compared with an average of a 25% decrease in dry mass of oviparous species (Thompson et al. 2000).
Although much is known about the morphological and physiological changes involved in reptilian pregnancy, there has been little examination of the genetic architecture that underlies them (for review, see Murphy and Thompson 2011). Multiple studies have used a targeted gene approach to assess the expression of genes involved with remodeling of embryonic and maternal tissue (Biazik et al. 2007, 2008; Wu et al. 2011), increasing uterine blood supply to facilitate nutrient and gas exchange with the embryo (Murphy et al. 2009), modifying the maternal immune system to prevent rejection of embryos (Paulesu et al. 1995; Romagnoli et al. 2003), and supplying nutrients to the embryo in addition to those provided in the yolk (Biazik et al. 2009).
Although these studies provided important information into genes critical to maintaining squamate reptile pregnancy, the candidate gene approach used in those studies is not adequate to profile the potentially thousands of genes that are regulated during pregnancy in squamate reptiles. Indeed, until recently, it has not been technically feasible to investigate the genetic controls of viviparity and placental development in nonmammalian vertebrates because the necessary baseline genomic data have not been available. However, recent technological advances (RNA-Seq; Marioni et al. 2008) provide the first opportunity to both identify and quantify almost all of the genes expressed in the reptilian uterus. The integration of transcriptomic data with existing morphological and physiological data now allows us to elucidate the genetic mechanisms that underlie reptilian pregnancy, which are subject to drastic change during evolutionary transitions between reproductive modes.
Here, we conduct the first comprehensive examination of gene regulation in a pregnant and nonpregnant viviparous lizard uterus using RNA-Seq. Using the viviparous Ocellated Skink, Chalcides ocellatus as a model system, we provide a near complete gene expression profile associated with maintaining pregnancy (supplementary information 1, Supplementary Material online). Chalcides ocellatus is an ideal subject for this study because it possesses a relatively complex placenta with endometrial folds and histotrophic activity (Corso et al. 1988, 2000; it is roughly in the middle of the continuum of simple to very complex placenta) and because partial gene expression data exist on Chalcides chalcides, a congener used in previous candidate gene analyses (Paulesu et al. 1995, 2001; Romagnoli et al. 2003). We focus particularly on suites of genes associated with growth and remodeling of the uterus, the maternal immune system, transport across the placenta, and uterine metabolism because they are important functions of maintaining pregnancy and have also been the subject of mammalian, and to a lesser extent, squamate reptile gene expression analyses. We compare the gene expression profiles of the uterus of C. ocellatus with mammalian models to determine whether similar genetic mechanisms have evolved to maintain pregnancy mammals and reptiles. Similarities between reptiles and mammals in uterine gene expression would suggest convergent evolution of genetic mechanisms in response to similar natural selection pressures. On the other hand, significant differences in gene expression would be evidence that there are potentially many genetic pathways that permit the transition to viviparity (i.e., “many means to an end”). Both possibilities are inherently interesting because they allow us to interpret the general processes under which pregnancy has evolved in vertebrate animals.
Materials and Methods
Adult C. ocellatus individuals were purchased from a commercial dealer. Phylogenetic comparison of a 396-nt fragment of cytochrome b mitochondrial DNA (mtDNA) from these individuals with a comprehensive Chalcides mtDNA data set (Carranza et al. 2008) revealed them to be from Egyptian populations of C. ocellatus ocellatus (results not shown). The lizards were housed at Yale University in ∼40 l glass aquaria with a 12 h light/dark photoperiod, a 24–37 °C thermal gradient (during the day), free access to water, and fed crickets three times a week. From a euthanized pregnant female, we dissected out a uterine egg chamber, removed the embryonic tissues attached to the uterus, and stored the uterus in RNA-Later (Ambion). The developmental state of the embryo was ∼37 according to the normal development table of Defaure and Hubert (1961). For a sample of nonpregnant uterus, we obtained an additional uterus from a female that was isolated for 6 weeks postpartum. There is no record of sperm retention in C. ocellatus, and the uterus showed no signs of pregnancy.
Total RNA was extracted from the uterus using an RNeasy Mini Kit (Qiagen). cDNA library construction and sequencing was performed by the Yale Center for Genome Analysis. The Illumina Genome Analyzer was used to sequence 72-bp pair-end fragments of cDNA libraries from both the pregnant and the nonpregnant samples.
There is no sequenced complete genome for C. ocellatus; indeed, only one complete genome of any nonavian reptile is currently sequenced (Anolis carolinensis), and it shares a common ancestor with C. ocellatus approximately 175 Ma (Brandley et al. 2008, 2011). To facilitate identification of sequenced transcripts, we therefore used ABySS 1.2.4 to assemble individual raw Illumina reads into larger contigs that are more easily identified. We used the subtractive multiple-k method of Surget-Groba and Montoya-Burgos (2010) and constructed multiple assemblies using a range of k-mer lengths (25, 29, 33, 37, 41, 45, 49, 53, 57, 61, 65, and 69). We then used CD-HIT-EST (Li and Godzik 2006) to remove redundant contigs and then removed contigs less than 100 bp (table 1).
Table 1.
Characteristics of Contigs Assembled by ABySS Using the Subtractive Multiple-k Method (Surget-Groba and Montoya-Burgos 2010)
| Number of Contigs | 300,967 |
| Maximum contig length | 15,179 |
| Mean contig length | 530 |
| Median contig length | 165 |
| N50 | 1,454 |
To identify the contigs, we used BlastX with an e value of 10−5 to compare them with a database including 38 complete sequenced genomes of vertebrate animals (Ensembl build 61). We note that this methodology will not identify recently diverged paralogs and may therefore underestimate the true diversity of genes expressed in the uterus but that this is a potential problem for all transcriptomic studies and does not alter our broad conclusions. The raw Illumina data were filtered to remove any reads less than 30 bp. We then used the short-read aligner Bowtie 0.12.7 (Langmead et al. 2009) to align these filtered reads from both the nonpregnant and the pregnant uterine samples to a database composed of the identified contigs, allowing two nucleotide mismatches. Because of differing sequence quality between the forward and the reverse pair-end reads, we chose to only use the forward reads for alignment. Because both reads were sequenced from the same fragment, this should introduce little or no bias in the gene counts.
We assessed differential gene expression of normalized numbers of transcripts between the nonpregnant and the pregnant samples using the DEGseq (Wang et al. 2010) R package. Preliminary analysis revealed that some genes are massively upregulated in the pregnant uterus (e.g., 497,141 reads of cathepsin L1 in the pregnant uterus compared with 917 reads in the nonpregnant; see Results and Discussion). Because the number of cDNA fragments sequenced is finite, we were concerned that several highly expressed genes were sequenced at the expense of others, thereby artificially lowering the apparent expression of other genes (Robinson and Oshlack 2010). To try to mitigate this sequencing artifact, we used the trimmed mean of M values (TMM) method of Robinson and Oshlack (2010) in the edgeR R package (Robinson et al. 2010) to normalize the data and create an effective library size for both samples. We scaled both the pregnant and the nonpregnant libraries to “counts per 107 reads” using the following formula:
where the TMM estimated normalization factor is 0.698231 for the pregnant library and 1.0 for the nonpregnant library and the original library sizes of 20,060,751 and 23,120,916, respectively. We then estimated differential expression of each gene between the nonpregnant and the pregnant samples using the MA plot–based method with random sampling model (MARS; Wang et al. 2010) in the DEGseq using a P value cutoff of 0.001 and a false discovery rate of 0.1%.
We used Gene Ontology (GO) analyses to broadly infer the biological function of the genes significantly regulated (as identified in the MARS differential expression analysis) in the uterine transcriptomes. We imported gene symbol lists of the significantly regulated genes in both the pregnant and the nonpregnant uteri into the Database for Annotation, Visualization, and Integrated Discovery (DAVID) functional GO annotation program and calculated GO terms corresponding to biological processes (BP-FAT). To more easily interpret the hundreds of individual GO biological functions inferred by DAVID, we aggregated similar processes to the following broad functional groups:
Catabolism—processes that breakdown complex molecules. This group contains any GO biological processes that were explicitly catabolic or proteolytic.
Cell death and differentiation—processes that regulate cellular proliferation, mitosis, and death.
Cell morphogenesis and development—processes involved with cell organization, including localization, cytoskeleton maintenance, tissue development, and motility.
Gene expression—processes that regulate transcription, translation, and posttranscriptional modification.
Homeostasis and response to stimuli—processes involved with maintaining homeostasis or reaction to stimuli.
Metabolism and energy—processes involved with the metabolism of macromolecules such as amino acids, lipids, nucleic acids, and proteins for the production of energy that are not explicitly catabolic (see above). This group also contains those mitochondrial processes involved in ATP production.
Molecule biosynthesis—processes that result in the production of macromolecules.
Signaling—processes involved with intracellular communication.
Transport—processes that transport both organic and inorganic molecules both intracellularly and intercellularly.
Other—those processes that do not naturally fit into any of the above categories.
We grouped individual GO biological processes into the above categories provided they were represented by at least 50 unique genes. We note that these categories are not necessarily mutually exclusive as, for example, those processes involved in catabolism are also metabolic. However, we chose to identify separately explicit catabolic functions given the substantial upregulation of proteolytic and hydrolytic enzymes in the pregnant uterus (see Results and Discussion).
Results and Discussion
Our study is the first to survey the almost complete gene expression profile associated with maintaining pregnancy in a live-bearing reptile. Indeed, it is one of few studies that have used RNA-Seq to assess gene expression in any reptile (Gracheva et al. 2010; Schwartz et al. 2010; Wall et al. 2011). Our transcript identification analysis positively identified 12,070,543 of 20,060,751 (60.2%) transcripts in the pregnant and 12,330,621 of 23,120,916 (53.8%) transcripts in the nonpregnant C. ocellatus uterine transcriptome (supplementary information 1, Supplementary Material online). If we assume a minimum cutoff of ≥2 transcripts per million reads as our conservative criterion for calling a gene expressed, we identified 8,846 unique genes expressed in the pregnant and 8,881 unique genes expressed in the nonpregnant uteri, 8,296 (88%) of which are expressed in both the nonpregnant and the pregnant uterus. Our differential expression analysis using the MARS method in DEGseq identified a total of 3,903 upregulated and 2,709 downregulated genes in the pregnant C. ocellatus uterus, respectively. The 50 most upregulated and downregulated genes (as assessed by z-score) in the uterus of the pregnant C. ocellatus uterus, as well as specific genes discussed in the remainder of the paper, are provided in tables 2 and 3.
Table 2.
A Selection of Significantly Upregulated Genes in the Pregnant Chalcides ocellatus Uterus as Inferred by the MARS Differential Gene Expression Analysis of TMM Transformed Transcript Counts
| Gene Symbol | Gene Name | Pregnant Transcript Count (Raw) | Nonpregnant Transcript Count (Raw) | Pregnant Transcript Count (Transformed) | Nonpregnant Transcript Count (Transformed) | Log2-Fold Change | Z-Score | Expression Rank |
| ACTB | Actin, beta | 95,506 | 39,786 | 133,776 | 55,728 | 1.3 | 154.2 | 16 |
| AGTRAP | Angiotensin II receptor–associated protein | 1,177 | 657 | 840 | 284 | 1.6 | 16.9 | 1,058 |
| ALDOA | Aldolase A | 10,459 | 9,017 | 7,467 | 3,900 | 0.9 | 33.7 | 384 |
| ALDOB | Aldolase B | 7,194 | 291 | 5,136 | 126 | 5.3 | 74.4 | 94 |
| ALDOC | Aldolase C | 275 | 123 | 196 | 53 | 1.9 | 9.3 | 1,991 |
| ANG | Angiogenin | 14,189 | 4,337 | 10,130 | 1,876 | 2.4 | 78.7 | 80 |
| ANGPT4 | Angiopoietin 4 | 451 | 199 | 322 | 86 | 1.9 | 12.0 | 1,575 |
| ANXA4 | Annexin A4 | 12,674 | 3,524 | 9,048 | 1,524 | 2.6 | 76.8 | 87 |
| APOA1 | Apolipoprotein A-I | 102 | 2 | 73 | 1 | 6.2 | 8.9 | 2,090 |
| APOA2 | Apolipoprotein A-II | 1,123 | 0 | 802 | 0 | 10.6 | 23.4 | 720 |
| APOA4 | Apolipoprotein A-IV | 331 | 109 | 236 | 47 | 2.3 | 11.7 | 1,617 |
| APOE | Apolipoprotein E | 8,568 | 2,879 | 6,117 | 1,245 | 2.3 | 59.1 | 145 |
| APOM | Apolipoprotein M | 34 | 2 | 24 | 1 | 4.6 | 5.0 | 3,188 |
| AQPP11 | Aquaporin 11 | 123 | 28 | 88 | 12 | 2.9 | 8.0 | 2,279 |
| AQPP3 | Aquaporin 3 | 859 | 837 | 613 | 362 | 0.8 | 8.1 | 2,266 |
| AQPP5 | Aquaporin 5 | 2,391 | 18 | 1,707 | 8 | 7.7 | 41.0 | 279 |
| ARG2 | Amino acid acetyltransferase | 16,563 | 81 | 23,200 | 113 | 7.7 | 128.0 | 22 |
| ATP1A1 | ATPase, Na+/K+ transporting, alpha 1 | 54,438 | 10,240 | 38,865 | 4,429 | 3.1 | 176.0 | 11 |
| BPI2 | Bactericidal/permeability-increasing protein 2 | 17,327 | 97 | 12,370 | 42 | 8.2 | 107.9 | 40 |
| BTG1 | B-cell translocation gene 1 | 16,577 | 1,535 | 11,835 | 664 | 4.2 | 107.9 | 41 |
| CA2 | Carbonic anhydrase II | 7,873 | 749 | 5,621 | 324 | 4.1 | 74.1 | 95 |
| CAT | Catalase | 41,302 | 604 | 57,852 | 846 | 6.1 | 211.2 | 8 |
| CDH1 | Cadherin 1 | 12,711 | 3,753 | 9,075 | 1,623 | 2.5 | 75.4 | 91 |
| CDH11 | Cadherin 11 | 20,807 | 634 | 14,855 | 274 | 5.8 | 126.8 | 24 |
| CDH15 | Cadherin 15 | 1,735 | 1,214 | 1,239 | 525 | 1.2 | 17.2 | 1,037 |
| CDH2 | Cadherin 2 | 685 | 717 | 489 | 310 | 0.7 | 6.4 | 2,732 |
| CDH24 | Protocadherin 24 | 99 | 60 | 71 | 26 | 1.4 | 4.7 | 3,305 |
| CDH4 | Cadherin 4 | 4,170 | 21 | 2,977 | 9 | 8.4 | 52.5 | 185 |
| CHY | Chymosin | 12,199 | 12 | 8,709 | 5 | 10.8 | 76.2 | 88 |
| CKB | Creatine kinase | 20,847 | 3,963 | 14,883 | 1,714 | 3.1 | 108.6 | 38 |
| CLDN1 | Claudin 1 | 9,577 | 601 | 6,837 | 260 | 4.7 | 84.4 | 67 |
| CLDN12 | Claudin 12 | 214 | 222 | 153 | 96 | 0.7 | 3.6 | 3,732 |
| CLDN23 | Claudin 23 | 92 | 60 | 66 | 26 | 1.3 | 4.2 | 3,478 |
| CLDN3 | Claudin 3 | 2,806 | 2,719 | 2,003 | 1,176 | 0.8 | 14.8 | 1,245 |
| CLDN4 | Claudin 4 | 8,586 | 4,014 | 6,130 | 1,736 | 1.8 | 51.0 | 197 |
| CLDN5 | Claudin 5 | 557 | 587 | 398 | 254 | 0.6 | 5.7 | 2,963 |
| CLDN6 | Claudin 6 | 3,666 | 2,451 | 2,617 | 1,060 | 1.3 | 26.1 | 613 |
| CLDN7 | Claudin 7 | 3,596 | 1,716 | 2,567 | 742 | 1.8 | 32.6 | 417 |
| CLDN8 | Claudin 8 | 5,015 | 111 | 3,580 | 48 | 6.2 | 62.1 | 135 |
| CO3 | Cytochrome c oxidase subunit 3 | 37,325 | 14,477 | 52,281 | 20,278 | 1.4 | 102.2 | 48 |
| COL6A3 | Collagen, type VI, alpha 3 | 14,847 | 1,170 | 20,796 | 1,639 | 3.7 | 116.0 | 28 |
| CPA2 | Carboxypeptidase A2 | 18,052 | 4,020 | 25,286 | 5,631 | 2.2 | 98.1 | 50 |
| CSTC | Cystatin C | 18,365 | 1,827 | 13,111 | 790 | 4.1 | 112.7 | 35 |
| CSTM | Cystatin 6/M/E | 30,367 | 95 | 21,680 | 41 | 9.0 | 136.2 | 20 |
| CTSA | Cathepsin A | 6,942 | 1,022 | 4,956 | 442 | 3.5 | 65.8 | 119 |
| CTSB | Cathepsin B | 17,222 | 5,309 | 12,295 | 2,296 | 2.4 | 86.5 | 64 |
| CTSD | Cathepsin D | 10,474 | 7,033 | 7,478 | 3,042 | 1.3 | 43.9 | 253 |
| CTSE | Cathepsin E | 67 | 7 | 48 | 3 | 4.0 | 6.8 | 2,630 |
| CTSF | Cathepsin F | 1,269 | 1,105 | 906 | 478 | 0.9 | 11.6 | 1,633 |
| CTSL | Cathepsin L1 | 497,141 | 918 | 354,922 | 397 | 9.8 | 523.9 | 1 |
| CTSV | Cathepsin V (cathepsin L2) | 106,855 | 129 | 76,287 | 56 | 10.4 | 232.0 | 6 |
| CTSZ | Cathepsin Z | 65,117 | 999 | 46,489 | 432 | 6.7 | 221.5 | 7 |
| CYB | Cytochrome b | 39,111 | 14,696 | 54,783 | 20,585 | 1.4 | 107.3 | 44 |
| DDIT4 | DNA damage–inducible transcript 4 | 15,069 | 569 | 10,758 | 246 | 5.5 | 107.8 | 42 |
| DSG2 | Desmoglein 2 | 2,002 | 946 | 1,429 | 409 | 1.8 | 24.5 | 676 |
| ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 | 13,373 | 60 | 9,547 | 26 | 8.5 | 93.2 | 56 |
| EPAS1 | Endothelial PAS domain protein 1 | 41,345 | 2,946 | 29,517 | 1,274 | 4.5 | 174.1 | 12 |
| FABP1 | Fatty acid–binding protein 1 | 14,898 | 0 | 10,636 | 0 | 14.4 | 60.2 | 140 |
| FABP2 | Fatty acid–binding protein 2 | 97 | 2 | 69 | 1 | 6.1 | 8.6 | 2,147 |
| FABP3 | Fatty acid–binding protein 3 | 2,247 | 395 | 1,604 | 171 | 3.2 | 36.2 | 340 |
| FABP4 | Fatty acid–binding protein 4 | 3,051 | 523 | 2,178 | 226 | 3.3 | 42.4 | 268 |
| FABP5 | Fatty acid–binding protein 5 | 1,198 | 388 | 855 | 168 | 2.3 | 22.4 | 765 |
| FABP7 | Fatty acid–binding protein 7 | 12,174 | 2 | 8,691 | 1 | 13.1 | 61.9 | 136 |
| FN1 | Fibronectin protein | 15,382 | 2,046 | 21,546 | 2,866 | 2.9 | 106.9 | 45 |
| FTL | Ferritin, light polypeptide | 36,692 | 10,208 | 51,395 | 14,298 | 1.8 | 126.0 | 25 |
| FUCA1 | Fucosidase, alpha-L1 | 145,697 | 220 | 104,017 | 95 | 10.1 | 277.5 | 5 |
| FUCA2 | Fucosidase, alpha-L2 | 129,789 | 765 | 92,660 | 331 | 8.1 | 296.6 | 4 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 29,268 | 15,408 | 20,895 | 6,664 | 1.6 | 87.8 | 62 |
| GGH | Gamma-glutamyl hydrolase | 12,615 | 85 | 17,670 | 119 | 7.2 | 113.8 | 33 |
| GLA | Galactosidase, alpha | 16,970 | 486 | 12,115 | 210 | 5.9 | 114.5 | 32 |
| GLB1 | Galactosidase, beta 1 | 14,934 | 513 | 10,662 | 222 | 5.6 | 107.4 | 43 |
| GLUL | Glutamate–ammonia ligase (glutamine synthetase) | 50,207 | 3,729 | 35,844 | 1613 | 4.5 | 191.3 | 10 |
| GRN | Granulin | 10,060 | 1,385 | 7,182 | 599 | 3.6 | 80.0 | 74 |
| GSN | Gelsolin | 19,199 | 5,489 | 13,707 | 2,374 | 2.5 | 93.7 | 54 |
| GPX1 | Glutathione peroxidase 1 | 3,871 | 1,307 | 5,422 | 1,831 | 1.6 | 36.4 | 337 |
| GPX3 | Glutathione peroxidase 3 | 2,969 | 23 | 4,159 | 32 | 7.0 | 55.6 | 161 |
| GPX4 | Glutathione peroxidase 4 | 2,996 | 957 | 4,197 | 1,340 | 1.6 | 33.2 | 396 |
| GPX5 | Glutathione peroxidase 5 | 1,484 | 22 | 2,079 | 31 | 6.1 | 40.0 | 287 |
| HEXA | Hexosaminidase A | 196,426 | 183 | 140,234 | 79 | 10.8 | 305.3 | 3 |
| HEXB | Hexosaminidase B | 15,870 | 192 | 11,330 | 83 | 7.1 | 108.3 | 39 |
| HMGCS2 | 3-Hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | 9,362 | 55 | 6,684 | 24 | 8.1 | 79.7 | 76 |
| HYAL3 | Hyaluronoglucosaminidase 1 | 17,923 | 46 | 12,796 | 20 | 9.3 | 102.8 | 47 |
| HYAL4 | Hyaluronoglucosaminidase 3 | 12,479 | 0 | 8,909 | 0 | 14.1 | 56.5 | 157 |
| IFI30 | Interferon gamma–inducible protein 30/GILT | 11,020 | 138 | 15,436 | 193 | 6.3 | 108.8 | 37 |
| IL20RB | Interleukin 20 receptor beta | 8,497 | 261 | 6,066 | 113 | 5.7 | 81.0 | 69 |
| KRT18 | Keratin 18 | 19,527 | 3,222 | 27,352 | 4,513 | 2.6 | 113.5 | 34 |
| KRT8 | Keratin 8 | 20,650 | 3,619 | 28,925 | 5,069 | 2.5 | 114.5 | 31 |
| LDHA | Lactate dehydrogenase A | 10,376 | 1,447 | 7,408 | 626 | 3.6 | 81.1 | 68 |
| LGMN | Legumain | 57,922 | 430 | 41,352 | 186 | 7.8 | 201.3 | 9 |
| MDH1 | Malate dehydrogenase 1 | 8,071 | 3,725 | 5,762 | 1,611 | 1.8 | 49.7 | 207 |
| MDH2 | Malate dehydrogenase 2 | 5,076 | 3,831 | 3,624 | 1,657 | 1.1 | 27.4 | 572 |
| NAGA | Alpha-N-acetylgalactosaminidase | 25,652 | 354 | 18,314 | 153 | 6.9 | 138.4 | 19 |
| NDRG1 | N-myc downstream–regulated 1 | 26,922 | 1,595 | 19,220 | 690 | 4.8 | 142.0 | 18 |
| NEU1 | Sialidase 1 | 29,234 | 76 | 20,871 | 33 | 9.3 | 131.4 | 21 |
| NFAT5 | Nuclear factor of activated T-cells 5 | 10,795 | 785 | 15,121 | 1,100 | 3.8 | 100.0 | 49 |
| OVGP1 | Oviductal glycoprotein 1, 120 kDa | 29,231 | 217 | 40,944 | 304 | 7.1 | 174.0 | 13 |
| PCK2 | Phosphoenolpyruvate carboxykinase 2 | 8,634 | 511 | 6,164 | 221 | 4.8 | 80.4 | 73 |
| PGAM1 | Phosphoglycerate mutase 1 | 11,543 | 2,573 | 8,241 | 1,113 | 2.9 | 77.9 | 83 |
| PKM2 | Pyruvate kinase, isozymes M1/M2 | 25,858 | 6,134 | 18,461 | 2,653 | 2.8 | 114.8 | 30 |
| PLA2G1B | Phospholipase A2, group IB | 22,787 | 187 | 16,268 | 81 | 7.6 | 127.0 | 23 |
| PPIB | Peptidyl-prolyl cis-trans-isomerase B | 16,423 | 2,048 | 23,004 | 2,869 | 3.0 | 112.1 | 36 |
| PRCP | Prolylcarboxypeptidase (angiotensinase C) | 2,944 | 173 | 2,102 | 75 | 4.8 | 47.0 | 229 |
| PRSS1 | Protease, serine, 1 | 2,681 | 0 | 1,914 | 0 | 11.9 | 32.4 | 420 |
| PRSS12 | Protease, serine, 12 | 1,290 | 49 | 921 | 21 | 5.5 | 31.5 | 443 |
| PRSS16 | Protease, serine, 16 | 90,516 | 9 | 64,622 | 4 | 14.0 | 154.5 | 15 |
| PRSS2 | Protease, serine, 2 | 148,046 | 2 | 105,694 | 1 | 16.7 | 147.5 | 17 |
| PRSS27 | Protease, serine, 27 | 19,852 | 1,096 | 14,173 | 474 | 4.9 | 122.3 | 26 |
| PRSS3 | Protease, serine, 3 | 161,271 | 5 | 115,136 | 2 | 15.8 | 169.8 | 14 |
| PRSS33 | Protease, serine, 33 | 994 | 60 | 710 | 26 | 4.8 | 27.3 | 578 |
| PRSS36 | Protease, serine, 36 | 466 | 12 | 333 | 5 | 6.1 | 19.0 | 935 |
| PRSS8 | Protease, serine, 8 | 11,814 | 400 | 8,434 | 173 | 5.6 | 95.5 | 52 |
| REN | Renin | 15,355 | 32 | 10,962 | 14 | 9.6 | 93.3 | 55 |
| SLC15A1 | Solute carrier family 15, member 1 | 12,659 | 22 | 17,732 | 31 | 9.2 | 103.2 | 46 |
| SPINT1 | Serine peptidase inhibitor, Kunitz type 1 | 10,400 | 1,020 | 7,425 | 441 | 4.1 | 84.9 | 66 |
| SPINT2 | Serine protease inhibitor, Kunitz type 2 | 14,494 | 4,573 | 10,348 | 1,978 | 2.4 | 78.7 | 81 |
| TFPI2 | Tissue factor pathway inhibitor 2 | 17,908 | 393 | 12,785 | 170 | 6.2 | 117.3 | 27 |
| TPP1 | Polynucleotide 3′-phosphatase | 162,913 | 2,062 | 116,308 | 892 | 7.0 | 347.8 | 2 |
| VEGFA | Vascular endothelial growth factor A | 3,348 | 1,727 | 2,390 | 747 | 1.7 | 30.1 | 495 |
| WSB1 | WD repeat and SOCS box–containing 1 | 14,527 | 1,088 | 20,348 | 1,524 | 3.7 | 115.6 | 29 |
NOTE.—Only the top 50 most upregulated genes (as assessed by z-score), and those genes discussed in the text, are shown. The expression rank represents the order of expression in the entire upregulated transcriptome from highest to lowest (e.g., a rank of 1 is the highest change in expression). Complete results for the MARS analysis are provided in supplementary information 1 (Supplementary Material online).
Table 3.
A Selection of Significantly Downregulated Genes in the Pregnant Chalcides ocellatus Uterus as Inferred by the MARS Differential Gene Expression Analysis of TMM Transformed Transcript Counts
| Gene Symbol | Gene Name | Pregnant Transcript Count (Raw) | Nonpregnant Transcript Count (Raw) | Pregnant Transcript Count (Transformed) | Nonpregnant Transcript Count (Transformed) | Log2-Fold Change | Z-Score | Expression Rank |
| ACTA2 | Actin, alpha 2 | 1,340 | 7,007 | 1,877 | 16,201 | −2.4 | −64.8 | 19 |
| AHNAK | AHNAK nucleoprotein | 6,465 | 15,345 | 9,056 | 35,479 | −1.2 | −61.0 | 23 |
| BTF3 | Basic transcription factor 3 | 3,393 | 9,969 | 4,753 | 23,049 | −1.6 | −58.1 | 28 |
| C3 | Complement component 3 | 2,256 | 10,334 | 3,160 | 23,893 | −2.2 | −74.8 | 9 |
| CALD1 | Caldesmon 1 | 9,97 | 5,162 | 1,397 | 11,935 | −2.4 | −55.4 | 31 |
| CD74 | CD74 molecule | 503 | 4,806 | 705 | 11,112 | −3.3 | −62.9 | 21 |
| CDH13 | Cadherin 13 | 48 | 105 | 67 | 243 | −1.1 | −4.7 | 2164 |
| CDH5 | Cadherin 5 | 40 | 246 | 56 | 569 | −2.6 | −12.8 | 686 |
| CNN1 | Calponin 1 | 612 | 4,810 | 857 | 11,121 | −3.0 | −60.4 | 25 |
| COL12A1 | Collagen, type XII, alpha 1 | 306 | 6,613 | 429 | 15,290 | −4.4 | −82.0 | 6 |
| CXCL14 | Chemokine (C-X-C motif) ligand 14 | 56 | 3,551 | 78 | 8,210 | −6.0 | −62.0 | 22 |
| DCN | Decorin | 361 | 3,191 | 506 | 7,378 | −3.1 | −50.5 | 41 |
| DDX50 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 | 8,260 | 23,789 | 11,570 | 55,002 | −1.5 | −88.6 | 4 |
| DES | Desmin | 1,883 | 9,420 | 2,638 | 21,780 | −2.3 | −73.9 | 11 |
| EFEMP1 | EGF-containing fibulin-like extracellular matrix protein 1 | 448 | 3,065 | 628 | 7,087 | −2.8 | −46.5 | 48 |
| ESR1 | Estrogen receptor 1 | 499 | 7,745 | 699 | 17,907 | −4.0 | −86.0 | 5 |
| GAS1 | 1,3-Beta-glucanosyltransferase GAS1 | 1304 | 4,980 | 1,827 | 11,514 | −1.9 | −47.8 | 42 |
| GPI | Glucose-6-phosphate isomerase | 6,118 | 14,803 | 8,570 | 34,226 | −1.3 | −61.0 | 24 |
| GPR124 | G protein–coupled receptor 124 | 228 | 4,267 | 319 | 9,866 | −4.2 | −65.1 | 18 |
| HMGB1 | High-mobility group 1 | 856 | 4,490 | 1,199 | 10,381 | −2.4 | −51.9 | 39 |
| HSP90AB1 | Heat shock protein 90 kDa alpha, class B member 1 | 6,564 | 15,181 | 9,194 | 35,100 | −1.2 | −59.3 | 27 |
| IFITM3 | Interferon-induced transmembrane protein 3 | 568 | 3,435 | 796 | 7,942 | −2.6 | −47.6 | 44 |
| IGFBP5 | Insulin-like growth factor–binding protein 5 | 289 | 5,002 | 405 | 11,565 | −4.1 | −69.9 | 12 |
| LGR5 | Leucine-rich repeat-containing G protein–coupled receptor 5 | 436 | 8,664 | 611 | 20,032 | −4.3 | −93.2 | 1 |
| MEIS1 | Meis homeobox 1 | 316 | 2,680 | 443 | 6,196 | −3.1 | −45.9 | 49 |
| MHCIA | MHC Class IA | 1,651 | 11,834 | 2,313 | 27,361 | −2.8 | −92.6 | 2 |
| MHCIIA | MHC Class IIA | 637 | 5,550 | 892 | 12,832 | −3.1 | −66.4 | 17 |
| MHCIIB | MHC Class IIB | 835 | 4,703 | 1,170 | 10,874 | −2.5 | −54.4 | 34 |
| MYH11 | Myosin, heavy chain 11 | 1,940 | 9,598 | 2,717 | 22,191 | −2.3 | −74.3 | 10 |
| PCLP1 | Podocalyxin-like protein 1 | 965 | 6,741 | 1,352 | 15,586 | −2.8 | −69.4 | 13 |
| PDLIM4 | PDZ and LIM domain 4 | 364 | 6,474 | 510 | 14,968 | −4.2 | −79.8 | 8 |
| PGR | Progesterone receptor | 283 | 2,345 | 396 | 5,422 | −3.1 | −42.7 | 61 |
| PTGIS | Prostaglandin I2 (prostacyclin) synthase | 131 | 1,580 | 183 | 3,653 | −3.6 | −37.6 | 93 |
| RPL10A | 60S ribosomal protein L10A | 5,856 | 17,979 | 8,203 | 41,569 | −1.6 | −80.4 | 7 |
| RPL14 | 50S ribosomal protein L14 | 3,239 | 8,857 | 4,537 | 20,478 | −1.5 | −52.1 | 38 |
| RPL17 | 50S ribosomal protein L17 | 6,268 | 13,974 | 8,780 | 32,309 | −1.2 | −54.9 | 32 |
| RPL18A | 50S ribosomal protein L18A | 2,656 | 10,063 | 3,720 | 23,267 | −1.9 | −67.7 | 16 |
| RPL19 | 50S ribosomal protein L19 | 4,645 | 10,270 | 6,506 | 23,745 | −1.1 | −46.6 | 47 |
| RPL27A | 50S ribosomal protein L27A | 2,685 | 8,070 | 3,761 | 18,659 | −1.6 | −53.1 | 35 |
| RPL32 | 60S ribosomal protein L32 | 3,513 | 8,440 | 4,921 | 19,514 | −1.3 | −45.7 | 50 |
| RPL37 | 60S ribosomal protein L37A | 2,163 | 8,028 | 3,030 | 18,561 | −1.9 | −59.9 | 26 |
| RPL4 | 60S ribosomal protein L4 | 8,509 | 19,769 | 11,919 | 45,708 | −1.2 | −67.9 | 15 |
| RPLP2 | 60S ribosomal protein LP2 | 2,023 | 6,650 | 2,834 | 15,375 | −1.7 | −51.0 | 40 |
| RPS13 | 40S ribosomal protein S13 | 3,213 | 9,151 | 4,500 | 21,158 | −1.5 | −54.5 | 33 |
| RPS2 | 40S ribosomal protein S2 | 12,945 | 22,719 | 18,132 | 52,528 | −0.8 | −52.1 | 37 |
| RPS23 | 40S ribosomal protein S23 | 2,612 | 7,930 | 3,659 | 18,335 | −1.6 | −53.0 | 36 |
| RPS7 | 40S ribosomal protein S7 | 2,504 | 13,326 | 3,507 | 30,811 | −2.4 | −89.9 | 3 |
| RPS8 | 40S ribosomal protein S8 | 3,464 | 11,054 | 4,852 | 25,558 | −1.7 | −64.6 | 20 |
| RPS9 | 40S ribosomal protein S9 | 5,272 | 11,266 | 7,385 | 26,048 | −1.1 | −47.2 | 45 |
| SLC6A6 | Solute carrier family 6, member A6 | 845 | 4,998 | 1,184 | 11,556 | −2.6 | −57.0 | 29 |
| SLIT2 | Slit homolog 2 | 276 | 2,727 | 387 | 6,305 | −3.3 | −47.7 | 43 |
| SOX7 | SRY (sex-determining region Y)-box 7 | 318 | 2,784 | 445 | 6,437 | −3.1 | −47.1 | 46 |
| TNS3 | Tensin 3 | 675 | 6,013 | 945 | 13,903 | −3.2 | −69.4 | 14 |
| TPBG | Trophoblast glycoprotein | 271 | 3,464 | 380 | 8,009 | −3.7 | −56.1 | 30 |
NOTE.—Only the top 50 most downregulated genes (as assessed by z-score), and those genes discussed in the text, are shown. The expression rank represents the order of expression in the entire nonpregnant uterine transcriptome from highest to lowest (e.g., a rank of 1 is the highest change in expression). Complete results for the MARS analysis are provided in supplementary information 1 (Supplementary Material online).
It is notable that many of the genes downregulated in the pregnant C. ocellatus uterus are genes involved with basic cellular housekeeping processes (e.g., ribosomal subunit proteins, desmin, myosin; table 3), which could indicate that the TMM normalization was inadequate and that these genes were undersampled. However, if these data were improperly normalized, then properly normalizing them would only increase the relative number of highly expressed transcripts in the pregnant uterus (table 2) and therefore does not severely impact our conclusions. We also emphasize that, given the lack of comparative genomic resources, we were unable to identify almost half of the sequenced transcripts and we have likely undersampled particularly quickly evolving genes—a challenge inherent in any RNA-Seq study of an organism with no closely related sequenced genome.
Most of the genes expressed in the pregnant uterus are also expressed in the nonpregnant uterus, which suggests that the morphological and physiological processes that maintain reptilian pregnancy are mostly a function of differential regulation of genes continually expressed in the uterus rather than the new expression of unique genes. If a similar pattern exists in the expression profiles of other oviparous and viviparous squamate reptiles, it would suggest that the evolutionary transition to viviparity is relatively “easy” given that the necessary genes are already expressed in the uterus when not gravid/pregnant and only become modulated in their expression level rather reprogrammed.
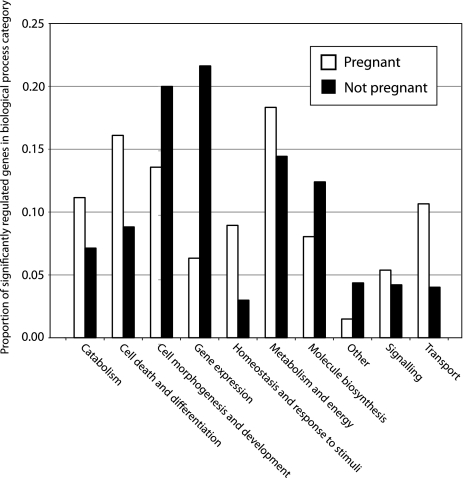
The GO biological function analysis revealed that C. ocellatus pregnancy is associated with upregulation of genes involved with catabolism, cell death and differentiation, homeostasis and response to stimuli, cellular signaling, and transport are upregulated in the pregnant uterus. By contrast, the nonpregnant uterus is dominated by genes that regulate cell morphogenesis and development, gene expression, and other processes.
Given the impracticality of examining the expression and function of each gene, we limit our discussion primarily to suites of genes that are associated with the biological functions that are apparently critical to maintaining pregnancy, including growth and remodeling of the uterus, nutrient transport across the placenta, and uterine metabolism and histotrophy. We also focus on the regulation of the maternal immune system and estrogen and progesterone activity, given their importance in maintaining mammalian pregnancy. We particularly focus on the most highly upregulated and downregulated genes identified by the MARS analysis and genes that have been assessed previously in squamate reptiles (table 4). Unless otherwise stated, we limit our discussion to only genes that are significantly upregulated or downregulated (P ≤ 0.001) according to our MARS differential expression analysis. The entire results of the differential expression analysis for all genes are provided in the supplementary information 1 (Supplementary Material online).
Table 4.
Genes Whose Regulation Has Been Assessed Previously in Squamate Reptiles and Comparison with Chalcides ocellatus
| Gene | Species | Regulatory Activity during Pregnancy | Reference |
| Desmoglein 2 (DSG2) | Chalcides ocellatus | Increase | This study |
| Lerista bougainvilliia | No change | Biazik et al. (2010) | |
| Pseudemoia entrecasteauxii | Decrease | Biazik et al. (2010) | |
| Pseudemoia spenceri | Decrease | Biazik et al. (2010) | |
| Saiphos equalisa | No change | Biazik et al. (2010) | |
| Occludin (OCLN) | C. ocellatus | No change | This study |
| Eulamprus tympanum | Absent | Biazik et al. (2007) | |
| P. entrecasteauxii | Absent | Biazik et al. (2007) | |
| P. spenceri | Increase | Biazik et al. (2007) | |
| S. equalisa | Increase | Biazik et al. (2007) | |
| Claudin 5 (CLDN5) | C. ocellatus | Increase | This study |
| E. tympanum | Present | Biazik et al. (2008) | |
| L. bougainvilliia | Present | Biazik et al. (2008) | |
| P. entrecasteauxii | Present | Biazik et al. (2008) | |
| P. spenceri | Present | Biazik et al. (2008) | |
| S. equalisa | Present | Biazik et al. (2008) | |
| Cadherin (CDH)b | C. ocellatus | Varied, but mostly increase | This study |
| L. bougainvilliia | Decrease | Wu et al. (2011) | |
| Niveoscincus metallicus | Decrease | Wu et al. (2011) | |
| Niveoscincus ocellatus | Decrease | Wu et al. (2011) | |
| Vascular endothelial growth factor A (VEGFA) | C. ocellatus | Increase | This study |
| E. tympanum | Present | Murphy, Belov, et al. (2010) | |
| N. metallicus | Present | Murphy, Belov, et al. (2010) | |
| P. entrecasteauxii | Present | Murphy, Belov, et al. (2010) | |
| S. equalisa | Increase | Murphy, Belov, et al. (2010) | |
| Interleukin 1A (IL1A) | C. ocellatus | Absent | This study |
| Chalcides chalcides | Present | Paulesu et al. (1995) | |
| Zootoca (Lacerta) viviparaa | Present | Paulesu et al. (2005) | |
| Interleukin 1B (IL1B) | C. ocellatus | Absent | This study |
| C. chalcides | Present | Paulesu et al. (1995) | |
| Z. viviparab | Present | Paulesu et al. (2005) |
NOTE.—Gene expression is identified as “present” if expression was inferred, but levels were not quantified between pregnant and nonpregnant individuals. Gene expression is identified as “no change” if expression was quantified, but there is no change in expression between pregnant and nonpregnant individuals.
Reproductively bimodal species. Only data for the viviparous individuals are presented.
Nonspecific “pan-cadherin” antibody used.
Growth and Remodeling of the Uterus
The increasing demands of the growing embryo, including gas exchange, nutrition, and uterine space, require both fundamental destruction and growth of uterine tissue to both increase the size of the uterus and maximize the surface area of the maternal–embryonic interface. Genes likely involved in these remodeling process are some of the most highly upregulated in the pregnant C. ocellatus uterus (table 2) and include cytoskeletal elements (e.g., β-actin and keratins) and gelsolin (a powerful actin regulator), genes that promote (e.g., N-myc downstream regulated 1, Granulin) and inhibit (e.g., B-cell translocation gene 1, DNA damage–inducible transcript four) cell proliferation, and hyaluronoglucosaminidases 3 and 4, that break down the extracellular matrix.
Although there are potentially hundreds or thousands of genes involved in the uterine remodeling process, we focus on the proteolytic enzymes because they are massively upregulated and have been the subjects of multiple studies of mammalian pregnancy. We also evaluate expression of the highly upregulated cell adhesion and angiogenic genes because they have been subject to previous studies in squamate reptiles (Biazik et al. 2007, 2008, 2010; Murphy, Belov, et al. 2010; Wu et al. 2011) and therefore provide a rare opportunity to assess the diversity of molecular mechanisms involved in reptilian pregnancy.
Proteases
Because the close apposition of maternal and fetal tissue facilitates gas and nutrient exchange, uterine and placental tissues are remodeled to minimize this distance, regardless of whether the placental type is epitheliochorial, endothelialchorial, or hemochorial (Enders and Carter 2004). The destruction of this tissue is achieved, in part, via the actions of both cysteine and serine proteases (Salamonsen and Nie 2002). Perhaps the most striking gene expression upregulation in the pregnant C. ocellatus uterine transcriptome is the enormous upregulation of cysteine and serine proteases. Cathepsins (CTS) are lysosomal cysteine proteases that degrade extracellular matrix and catabolize intracellular proteins (Kirschke et al. 1997). Cathepsins make up over 8% of the entire upregulated transcriptome, with CTSL alone representing 5% of the transcriptome. The serine proteases PRSS2 (trypsin) and PRSS3 (mesotrypsin) have the highest log2-fold changes of all the upregulated genes (16.7 and 15.5, respectively; table 2). Taken together with other highly upregulated proteases such as legumain (LGMN) and tripeptidyl peptidase I (TPP1), these results reveal that the breakdown of tissue is one of the most active physiological processes in the C. ocellatus pregnant uterus.
Although the roles of cathepsins, especially CTSB and CTSL, in uterine remodeling has been elucidated in several mammal species (e.g., Afonso et al. 1997; Divya et al. 2002; Song et al. 2006, 2010; Varanou et al. 2006), comparison of cathepsin expression between C. ocellatus lizards and pigs is most instructive because both possess an epitheliochorial placenta. In pigs, CTSB and CTSL are expressed in both uterine and chorionic epithelia, and this expression increases as pregnancy progresses (Song et al. 2010). Both CTSB and CTSL are upregulated in the pregnant C. ocellatus uterus, CTSL being the most abundant transcript in the transcriptome. Although we cannot localize expression of these genes to a specific region of the uterus, we nonetheless speculate that, as in mammals, these genes may have a primary role in degrading uterine tissue during the remodeling process in the C. ocellatus uterus.
Placental remodeling is an interplay between proteases and their inhibitors (Afonso et al. 1997; Salamonsen and Nie 2002; Song et al. 2006, 2007, 2010), especially the reciprocal actions of cathepsins and their inhibitors, cystatins (CST). Cystatin C (CSTC) is a powerful inhibitor of cathepsins L and B and is highly upregulated in C. ocellatus. CTSL and two other lysosomal cysteine proteases, CTSV (also called CTSL2) and LGMN, are also inhibited by cystatin M/E (also called cystatin 6), a very highly upregulated gene in the pregnant C. ocellatus transcriptome. Moreover, multiple serine protease inhibitors, including tissue factor pathway inhibitor 2 (TFPI2), serine peptidase inhibitor, Kunitz types 1 and 2 (SPINT1 and 2), are significantly upregulated, thereby suggesting active regulation of serine protease activity. Thus, there is a clear pattern of protease expression and concomitant regulation by their inhibitors. Moreover, the genes likely responsible for these processes are identical to those used by mammals.
Cell Adhesion
Remodeling of the uterus throughout pregnancy requires the growth and movement of the closely apposed maternal and embryonic surface epithelial cells and presumably the molecules that permit adhesion of these cells to one another. These cellular adhesion proteins, including those critical to the development of desmosomes, tight junctions, and adherens junctions, are some of the few proteins extensively studied in squamate reptiles (Australian skinks; Biazik et al. 2007, 2008, 2010; Wu et al. 2011) and therefore provide a rare opportunity to assess gene expression differences amongst multiple species of viviparous reptiles (table 4).
Desmoglelin 2 (DSG2) is a protein involved in the formation of desmosomes and its expression has been evaluated in the uteri of four scincid lizards representing the extremes of placental development (Biazik et al. 2010). DSG2 expression is not different in pregnant and nonpregnant females in two species with simple placentae (Lerista bougainvillii and Saiphos equalis) but is lower during pregnancy in two species with complex placentae (Pseudemoia entrecasteauxii and Pseudemoia spenceri), which suggests that a decrease in desmosome number may allow for easier deformation and remodeling of the uterus. In contrast, our transcriptome results demonstrate a statistically significant increase in DSG2 expression in the uterus of pregnant C. ocellatus and in the absence of corroborating morphological data, presumably an increasing number of desmosomes.
In addition to simple cell adhesion, tight junctions regulate paracellular transport by controlling the permeability of the interstitial space between adjacent cells to molecules of specific sizes. Occludin (OCLN) and claudins (CLDN) are the dominant proteins in tight junctions. OCLN expression increases as pregnancy progresses in the uteri of two viviparous species of skinks with complex placentation (P. entrecasteauxii and P. spenceri), suggesting decreased paracellular permeability with compensation by transcellular transport (Biazik et al. 2007). OCLN expression was not detected in two species with simple placentae (L. bougainvillii and S. equalis; Biazik et al. 2007). Similarly, our transcriptomic analysis finds no significant difference in OCLN expression between nonpregnant and pregnant C. ocellatus females (P = 0.187), although this species has a more complex placenta. CLDN5 is expressed over the entire surface of the uterine epithelial cell in early pregnancy in four viviparous skink species but becomes localized at the tight junctions, suggesting an increased role of the tight junctions in limiting paracellular transport (Biazik et al. 2008). As no study has examined tight junction distribution in C. ocellatus, we cannot directly compare our results with Biazik et al. (2008), but we do find significant upregulation of nine claudin loci, including CLDN1, 3–8, 12, and 23, suggesting that they have an important role in maintaining pregnancy in C. ocellatus.
Cadherins (CDH) are a family of proteins that are components of adherens junctions between adjacent cells. Cadherin expression is high near ovulation, as revealed using an antibody that labels all cadherins but significantly decreases as pregnancy progresses and is absent in late pregnancy in three species of Australian skinks (Wu et al. 2011). Thus, decreasing cadherin expression, and therefore few adherens junctions, is speculated to allow the uterus to expand to accommodate the growing embryo (Wu et al. 2011). In C. ocellatus, we detected a far more complicated expression profile of cadherins with significant upregulation of CDH1 (epithelial cadherin), 2, 4, 11, 15, 22, and 24 and downregulation of CDH5 and 13. This indicates that, unlike the three Australian skinks studied to date, adherens junctions actually proliferate in C. ocellatus during pregnancy and apparently do not inhibit uterine growth.
Our transcriptomic analyses reveal some significant differences between our study and previous studies of cell adhesion molecules in the uteri of viviparous skinks. These differing results could be due to the higher resolution of gene expression possible with transcript sequencing as opposed to immunological assays or western blots used in previous studies (Biazik et al. 2007, 2008, 2010; Wu et al. 2011). However, rather than any methodological problem, a much more plausible explanation for the difference in uterine cell adhesion gene expression is that the study organisms represent ancient independent derivations of viviparity. Although also a skink, C. ocellatus and Australian lygosomine skinks share a common ancestor approximately 95 Ma (Brandley et al. 2008, 2011), about the same age as the common ancestor of primates, ungulates, carnivores, and bats (Murphy et al. 2007). It is therefore reasonable to hypothesize that these two skink lineages evolved different molecular processes and suggests that there is a diversity of genetic processes that act to maintain pregnancy within viviparous squamate reptiles.
Angiogenesis and Vascular Physiology
As the embryo matures, its growing oxygen demand is accommodated by increasing placental vascularization (Guillette and Jones 1985; Masson and Guillette 1987; Murphy, Belov, et al. 2010; Murphy, Parker, et al. 2010; Parker, Manconi, et al. 2010; Parker, Murphy, et al. 2010). Like mammals, increasing expression of vascular endothelial growth factor A (VEGFA) in pregnant squamate reptiles is correlated with vascular growth in the uterus (Murphy, Belov, et al. 2010). VEGFA is also significantly upregulated in the uterus of pregnant C. ocellatus. VEGFA is upregulated in addition to a suite of other genes with angiogenic properties. Most notable are the endothelial PAS domain protein 1 (EPAS1) and hypoxia-inducible factor 1α (HIF1A) genes that activate VEGF expression in response to hypoxic conditions (Forsythe et al. 1996; Peng et al. 2000; Takeda et al. 2004); interleukin 20 receptor β (IL20RB), an inducer of endothelial cell proliferation (Gaspar et al. 2002; Hsieh et al. 2006); angiogenin (ANG) and angiopoietin 4 (ANGPT4), and general cell proliferation genes with angiogenic properties such as ENPP2/Autotaxin. As in remodeling the structure of the uterus, proteolytic enzymes such as cathepsins are also associated with angiogenesis (Joyce et al. 2004).
In addition to genes that promote the development of blood vessels, several genes that control blood physiology are highly expressed in the C. ocellatus pregnant uterus. Prostacyclin (PTGIS), a vasodilator, is downregulated. Renin, a part of the renin–angiotensin system (RAS) that regulates blood pressure, is highly upregulated suggesting intense vasoconstriction of the uterine vasculature and consequently high blood pressure (perhaps facilitating the diffusion of oxygen and nutrients to the embryo across the placenta). Renin is expressed in the uteroplacental tissues of mammals where it participates in a local RAS (Cooper et al. 1999; Nielsen et al. 2000; Vinson et al. 1997) that may also stimulate cell proliferation and angiogenesis (Hagemann et al. 1994). However, renin does not directly increase blood pressure but rather converts angiotensinogen (AGT) to angiotensin I, which is then converted to the much more potent vasoconstrictor angiotensinogen II via action of angiotesin-converting enzyme (ACE). Angiotensinogen (AGT) is not significantly regulated in the pregnant C. ocellatus uterus, and we found no expression of ACE. Moreover, prolylcarboxypeptidase/angiotensinase C (PRCP) and angiotensin II receptor–associated protein (AGTRAP), two inhibitors of angiotensin II, are upregulated and therefore suggest that blood pressure is locally regulated by the expression of vasoconstrictors and dilators.
Regulation of the Maternal Innate Immune System
The embryo is an allograft of both maternal and paternal tissue and is therefore at risk of attack by the maternal immune system. As such, a variety of mechanisms that “hide” the embryo have evolved in mammals by downregulating elements of the maternal immune system (Moffett and Loke 2006). This immune response is expected to be much higher in mammals with invasive placentation because the maternal epithelia are breached. With a single possible exception (Trachylepis [Mabuya] ivensii; Blackburn and Flemming 2011), all viviparous squamate reptiles studied to date, including C. ocellatus, possess an epitheliochorial placenta with no breaching of the maternal epithelia. Nonetheless, as pregnancy progresses in squamate reptiles, the remnants of the ancestral shell membrane commonly disintegrate, thereby creating direct contact between the embryonic and the maternal epithelia (Blackburn 1993). Although the epithelia remain intact, studies of the mammalian epitheliochorial placentae demonstrate adaptations to hide from the maternal immune system (Moffett and Loke 2006) by regulating the maternal innate immune system and embryonic major histocompatability (MHC) loci.
The innate immune system comprises the general mechanisms by which the body defends against infection in a nonspecific manner. Its components consist of leukocytes (e.g., macrophages, natural killer cells), general antimicrobial proteins (e.g., bactericidal/permeability-increasing protein 2), and molecules that promote inflammation (e.g., cytokines, chemokines). It differs from the adaptive immune system that recognizes and remembers specific pathogens and thereby provides long-term immunity. Inflammation is an innate immune response critical to fighting infection, but excessive uterine inflammation may also lead to spontaneous abortion (Challis et al. 2009). Therefore, regulation of the proinflammatory and anti-inflammatory molecules in the pregnant uterus is essential to the maintenance of pregnancy while maintaining the maternal immune response to pathogens (Walker et al. 2010). Inflammation is regulated by a suite of genes, many of them cytokines—molecules that initiate cascades of intracellular signaling that are particularly active in the innate immune system. In squamate reptiles, the interleukin 1 cytokine system has received particular attention. IL1A and IL1B, two proinflammatory cytokines that also initiate immune response and regulate cellular growth and differentiation, are expressed in the uterine epithelia of C. chalcides, a close relative of C. ocellatus, and this expression continues throughout pregnancy (Paulesu et al. 1995, 2005; Paulesu 1997; Romagnoli et al. 2003; Paulesu, Romagnoli, et al. 2005). Thus, it was assumed that IL1A and IL1B expression was involved in the maintenance of pregnancy in both C. chalcides and mammals.
Surprisingly, our transcriptomic analysis reveals no expression of either IL1A or IL1B in both pregnant and nonpregnant C. ocellatus uteri and only low (but significant) upregulation of an interleukin 1 receptor (IL1R1). If IL1R1 expression indicates IL1 activity, we cannot locate the source of transcripts, and the lack of IL1A or IL1B expression in the uterus C. ocellatus starkly contrasts with that detected in C. chalcides. A potential explanation for these conflicting results could be that we were unable to identify IL1A and IL1B transcripts using our sequence identification strategy. That is, IL1A and IL1B may evolve quickly enough that the Chalcides transcripts of these genes cannot be reliably aligned to those in our reference genomes. An alternate explanation is that uterine IL1 expression was lost in the evolutionary history of the lineage leading to extant C. ocellatus. However, C. chalcides and C. ocellatus share a common ancestor only ∼10.5 Ma (Carranza et al. 2008); that uterine expression of IL1 is also detected in Zootoca (Lacerta) vivipara (Paulesu et al. 2005), a lacertid lizard that shares a common ancestor with scincid lizards (including Chalcides) ∼175 Ma (Brandley et al. 2008, 2011), and mammals, suggests that uterine IL1 expression is evolutionarily conserved. Although we do not detect IL1 expression, we nonetheless find extensive regulation of other genes that regulate the inflammatory response. The anti-inflammatory cytokine IL11 (Zourbas et al. 2001) is upregulated, whereas the proinflammatory IL8 and IL15 (Zourbas et al. 2001; Challis et al. 2009) are significantly downregulated. Finally, the proinflammatory cytokine high-mobility group box 1 (HMGB1) is significantly downregulated.
Significant upregulation and downregulation of other immune system genes also suggest active regulation of the inflammatory response in the uterus. There is significant downregulation of complement component 3 (C3). C3 plays a key role in activating the immune complement system, a component of the innate immune system that has roles in promoting local inflammation and stimulating a cascade of immune response including elements of the adaptive immune system (Sahu and Lambris 2001; Janssen et al. 2005; Walker et al. 2010). Excessive complement activation may lead to inhibited fetal growth or pregnancy termination in mammals (Caucheteux et al. 2003; Girardi et al. 2006).
However, not all inflammatory immune response genes are regulated to prevent inflammation in the pregnant C. ocellatus uterus. For example, phospholipase A2 IB is highly upregulated (PLA2G1B). PLA2G1B is typically expressed in the pancreas to digest dietary phospholipids in the duodenum, but in mammals, it is also expressed in nonpancreatic tissues where it likely plays a role in promoting inflammation by stimulating arachidonic acid release (Nevalainen et al. 2000). Phospholipase A2 expression is also critical for proper implantation in mammals (Dey et al. 2004). On the other hand, function of PLA2G1B may be mediated by the expression of its receptor (Moses et al. 1998) that is not differentially expressed between the pregnant and nonpregnant uteri (P = 0.91; data not shown). Regardless, even if PLAS2G1B expression promotes inflammation, its actions are likely regulated by the highly upregulated annexin A4 (ANXA4), a phospholipase A2 inhibitor.
Major histocompatibility loci encode molecules that present antigens to the host's immune system and are a key component in the body's recognition of “self” from “nonself.” Downregulating MHC expression in the embryonic tissues is therefore one mechanism by which the embryo can hide from the maternal immune system (Moffett and Loke 2006). However, both MHC Class I and II loci are significantly downregulated in the pregnant C. ocellatus uterus (the embryonic tissues were not sampled). Uterine downregulation of MHC Class I expression also occurs in cattle, pigs, and sheep, species with noninvasive epitheliochorial placentae, but in these cases, trophoblast cells merge with those of the uterine epithelia to form binucleate syncytia of embryonic and maternal origin (Davies et al. 2000; Choi et al. 2003; Joyce et al. 2008); downregulation of MHC Class I expression may be a mechanism by which the maternal immune system ignores these allograft cells in mammals. However, there is no evidence of syncytia formation in C. ocellatus (but they may form in the very late stages in pregnancy in C. chalcides [Blackburn et al. 1998] and some Mabuya group skinks [Ramírez-Pinilla et al. 2006; Vieira et al. 2007; Blackburn and Flemming 2011]), and thus, the function of uterine MHC downregulation in C. ocellatus remains unclear.
Transport across the Placenta
A majority of the water, organic and inorganic ions, and nutrients used by the embryo in oviparous and lecithotropic viviparous species is provided as yolk within the ovulated egg. By contrast, there is substantial transport of these molecules from the mother to the embryo via the placenta in lizard species with complex placentation (“placentotrophy”) (Thompson et al. 2000; Ramírez-Pinilla 2006; Thompson and Speake 2006). This resource allocation strategy must therefore involve substantial changes in the expression of molecular transporter genes, and our transcriptomic analysis of C. ocellatus reveals upregulation of many such loci. Placentotrophy has not been directly measured in C. ocellatus, and the species ovulates larger eggs (10 mm; Corso et al. 2000) with a larger yolk mass than highly placentrophic species such as C. chalcides (3 mm; Giacomini 1891), but light and scanning electron microscopic analysis of placental morphology imply that some placentotrophy occurs in C. ocellatus (Corso et al. 2000). Moreover, some placentotrophy has even been documented in primarily lecithotrophic species (Thompson et al. 2000; Thompson and Speake 2006).
Mothers of viviparous species provide much more water to their embryos than oviparous species (Thompson et al. 2000). Water transport is facilitated by aquaporins, cell membrane proteins that selectively regulate water transport. Aquaporins (AQP) are upregulated in the pregnant rat uterus (Lindsay and Murphy 2007) and indeed, AQP1 and AQP3 are also expressed in the uterus of the highly placentotrophic South American scincid lizard, Mabuya sp. (Wooding et al. 2010). AQP3, AQP5, and AQP11 are upregulated in the pregnant C. ocellatus uterus.
The last third of pregnancy in reptiles is particularly crucial for the provision of nutrients and ions such as calcium (Stewart and Thompson 2000; Herbert et al. 2006) and sodium (Thompson et al. 2001), and most of these ions are transported via transcellular routes (Loffing et al. 2001). The upregulation of 136 genes in 35 families of solute carrier proteins (SLCs; a majority not shown in table 2 but see supplementary information 1, Supplementary Material online) in the pregnant C. ocellatus uterus demonstrates a significant shift to increased inorganic and organic molecular transport during pregnancy (see “transport” in fig. 1). These upregulated SLC genes include transporters for inorganic ions (includes Ca2+, Cl−, CO32−, H+, HCO3−, K+, Na+, and PO43−), metals (includes Cu2+, Fe2/3+, Mg2+, and Zn2+), and organic molecules, including glucose, amino acids, peptides, fatty acids, and carboxylic acids. The three highest upregulated SLC genes are SLC15A1, an oligopeptide transporter, SLC22A17, an organic cation transporter, and SLC2A3, a glucose transporter, which suggest a great transport of organic molecules by the embryo in late pregnancy—a finding supported by physiological studies of other placentotrophic skinks (Thompson et al. 2000; Thompson and Speake 2006).
FIG. 1.—
Aggregated GO biological processes for significantly regulated genes in the pregnant and nonpregnant uterine transcriptomes. For group descriptions, see text.
Lipids represent the primary source of nutrition to the developing embryo, and physiological studies have inferred a net uptake of lipids across the placenta in numerous viviparous species with relatively complex placentae (Thompson et al. 2000; Speake et al. 2004; Thompson and Speake 2006). Lipid transport is likely facilitated by upregulated fatty acid packaging and transport gene families in the uterus of pregnant C. ocellatus. The primary functions of fatty acid binding proteins are to facilitate intracellular transport of fatty acids (Chmurzyńska 2006) and are active in the mammalian placenta (Haggarty 2002). Similarly, FABP genes (FABP1—5, 7) are upregulated in pregnant C. ocellatus, with FABP1 and FABP7 showing a log2-fold expression change of 15.1 and 12.8, respectively. Apolipoproteins are transporters of fatty acids, cholesterol, and phospholipids, and five apolipoproteins (APOA1, APOA2, APOA4, APOE, and APOM) are significantly upregulated in the pregnant uterus of C. ocellatus.
Uterine Metabolism and Possible Histotrophy Mediated by Lysosomes in the Omphaloplacenta
Uterine Metabolism
As pregnancy progresses, the energy demands to fuel embryonic growth, placental remodeling, and active transport increase sharply, which is reflected in the pregnant Chalcides uterus where genes involved with ATP production are highly upregulated. Most of these genes are associated with carbohydrate metabolism, including genes involved in glycolysis (ALDOA, ALDOB, ALDOC, GAPDH, LDHA, PKM2, PGAM1), the citric acid cycle (MDH1, MDH2), electron transport chain (ATP synthases, cytochromes b and c), gluconeogenesis (PCK2), ketogenesis (HMGCS2), and ATP regeneration (CK). One highly upregulated metabolic gene of particular note is glutamate–ammonia ligase (GLUL, also called glutamine synthetase). Glutamine is critical for the synthesis of nucleic acids and sugars as well as cell growth and differentiation (Self et al. 2004), and upregulation of GLUL mirrors the pattern in numerous mammal species where glutamine concentrations in the fetal plasma are far higher than those in the maternal circulation (e.g., Wu et al. 1995; Cetin 2001; Kwon et al. 2003; Manso Filho et al. 2009). Thus, the upregulation of GLUL is probably critical to maintaining both reptilian and mammalian pregnancies. The peroxidases catalase (CAT) and glutathione peroxidases 1, 3–5 (GPX1, GPX3–5) are upregulated in the pregnant C. ocellatus uterus. These enzymes function to scavenge reactive oxygen species that are the by-product of oxidative metabolism (Myatt and Cui 2004) and are thus markers of increased uterine metabolic activity during pregnancy.
No study has inferred the precise mechanism by which carbon dioxide produced by the embryo is eliminated by the mother, yet the upregulation of carbonic anhydrase II (CA2) offers an intriguing potential mechanism (Sterling et al. 2001). Carbonic anhydrase II catalyzes the reversible reaction of converting CO2 and H2O to HCO3− and H+. In oviparous amniotes, expression of CA2 in the chorioallantois promotes acidification of the eggshell to release calcium carbonate for absorption by the embryo (Stewart and Ecay 2010). Thus, the significant upregulation of this gene in the viviparous C. ocellatus uterus suggests that it may play a different role during pregnancy. We hypothesize that CA2 has been co-opted into the role of eliminating CO2 produced by the embryo in viviparous squamates.
Histotrophy
Viviparous squamate reptiles with complex placentae experience a net uptake of macromolecules during pregnancy (especially lipids; Thompson and Speake 2006). Thus, these species must have the catabolic machinery to digest lipid and protein macromolecules into smaller units that can be actively transported to the embryo via the placenta. Microscopic examination and alkaline phosphatase assays suggest that catabolism and transport of macromolecules occur in the omphaloplacenta (also called the “yolk sac” placenta) of placentrophic lizards (Corso et al. 2000; Adams et al. 2005; Thompson and Speake 2006; Biazik et al. 2009). The omphaloplacenta lies at the abembryonic pole of the embryo/yolk mass and typically consists of well-developed columnar epithelial tissue usually associated with secretory epithelia (Corso et al. 2000; Adams et al. 2005; Thompson and Speake 2006). An extensive array of electron dense structures, interpreted to be lysosomes, occur in the omphaloplacenta of two viviparous Pseudemoia skinks (Biazik et al. 2009) and in C. ocellatus (Corso et al. 2000, fig. 3B in that paper). Although many of the nutrients transported across the placenta to the embryo may derive from the mother's digestive system, the massive upregulation of lysosomal proteases (e.g., cathepsins, LGMN, TPP1) supports the morphological interpretation that histotrophic activity occurs in the omphaloplacenta. We also detected upregulation of lysosomal hydrolases, including hexosaminidases, fucosidases, galactosidases, and sialidase (table 2). Lysosomal hydrolases break down the glycosidic bonds of glycolipids and glycoproteins and frequently process macromolecules digested by proteolytic enzymes (Winchester 2005). The putative presence of lysosomes and the upregulation of lysosomal genes collectively suggest that the omphaloplacenta plays a key role in placentotrophy in C. ocellatus but testing this hypothesis will require studies of tissue and cell-specific expression of these genes.
Steroid Hormones
Concentrations of circulating estrogen and progesterone during gestation are extremely varied across both oviparous and viviparous reptile species (Girling and Jones 2003; Murphy and Thompson 2011). In C. ocellatus, we found significant downregulation of progesterone (PGR) receptor and estrogen receptor 1 (ESR1) (log2-fold change = −3.1 and −4.0, respectively). This finding is puzzling because in C. chalcides, blood serum progesterone concentrations increase over the course of pregnancy with the highest concentration before birth (Guarino et al. 1998). We did not measure serum levels of progesterone in C. ocellatus and thus cannot determine whether the downregulated activity of PGR corresponds to a lower concentration of the hormone during late pregnancy or whether PGR downregulation is a mechanism of functional progesterone withdrawal.
Although serum estrogen has not been measured during pregnancy in any Chalcides species, concentrations of circulating estradiol rise during vitellogenesis, peak at the periovulation stage, and then decrease to previtellogenesis concentrations in late pregnancy in the distantly related skink Niveoscincus metallicus (Jones and Swain 1996). Thus, low ESR1 expression in the pregnant C. ocellatus uterus is not wholly unexpected. The absence or downregulation of PGR and ESR1 in the uterine epithelia is not unprecedented. In mammals, implantation is preceded by a loss of PGR and ESR1 expression in the uterine epithelia (Spencer and Bazer 2002; Spencer et al. 2007; Bazer et al. 2008), and expression in the luminal epithelium and superficial glandular epithelium of PGR and ESR1 is also absent through most of sheep and pig pregnancy, but PR is expressed in endometrial stroma (Geisert et al. 1994; Spencer and Bazer 2002). Mammalian uterine epithelia proliferate in response to progesterone and estrogen, but the actions of these hormones are mediated by paracrine signaling from endometrial stromal cells that do exhibit PGR and ESR1 expression (see Bazer et al. 2008 for review; Cooke et al. 1997). However, this mammalian model of stromal cell–mediated effects of progesterone and estrogen may not be applicable to C. ocellatus because our pregnant uterus sample contained stromal tissue, yet PGR and ESR1 expression were downregulated. Sustained PGR and ESR expression also occur in the myometrium of mice and sheep (Spencer and Bazer 2002), but this too appears to differ with C. ocellatus given that our tissue sample contained both myometrium and endometrium. In summary, PGR and ESR expression are extremely low in the uterine epithelia of mammals and in the entire uterus of C. ocellatus, but the mechanism that explains this phenomenon, whether low progesterone and estrogen levels or some other compensatory mechanism, remains unknown.
Conclusion
This study is the first successful use of RNA-Seq methods to characterize changes in pregnancy-associated gene expression in any squamate reptile and a significant step toward uncovering the genetic mechanisms behind the evolution of viviparity in reptiles. Our results serve as a baseline for future comparative gene expression analyses. Chalcides represents only one of the 100+ origins of viviparity in squamate reptiles, so we are not able assess the generality of our gene expression results. Nonetheless, there are clear parallels between the genetic processes associated with pregnancy in mammals and Chalcides in terms of gene expression related to tissue remodeling, angiogenesis, immune system regulation, and nutrient provisioning to the embryo. Although estrogen and progesterone receptors are downregulated in mammalian uterine epithelia, downregulation of these genes in the entire C. ocellatus uterus is evidence that the gene expression profiles of pregnant mammalian and reptilian uteri are not identical. Future RNA-Seq analyses of other viviparous and oviparous squamate reptiles will reveal the extent to which our results represent the generality of genetic processes associated with pregnancy in mammals and reptiles and potentially explain why the transition to viviparity is so easy in squamates. Moreover, future studies should localize the expression of highly regulated genes in both maternal and embryonic tissues.
Supplementary Material
Supplementary information 1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank O. Griffith, V. Lynch, B. Murphy, A. Seago, Y. Surget-Groba, Z. Wang, the MBT laboratory, and three anonymous reviewers for advice and/or comments on the manuscript; A. Dornburg and G. Watkins-Colwell for care of laboratory animals; J. Mai for computational assistance; and Intersect Australia Ltd. for supercomputing resources. This work was supported by the Yale Institute for Biospheric Studies Gaylord Donnelley Postdoctoral Fellowship to M.C.B., the American Association of University Women American Fellowship to R.L.Y., National Science Foundation postdoctoral fellowship DBI-0905701 to D.L.W., an Australian Research Council grant awarded to M.B.T. and C. Murphy, and the John Templeton Foundation grant # 12793 to G.P.W. The opinions expressed in this article are not those of the John Templeton Foundation. Animal care and procedures were approved by the Yale Institutional Animal Care and Use Committee (#2008-11242).
References
- Adams SM, Biazik JM, Thompson MB, Murphy CR. Cytoepitheliochorial placenta of the viviparous lizard Pseudemoia entrecasteauxii: a new placental morphotype. J Morphol. 2005;264:264–276. doi: 10.1002/jmor.10314. [DOI] [PubMed] [Google Scholar]
- Afonso S, Romagnano L, Babiarz B. The expression and function of cystatin C and cathepsin B and cathepsin L during mouse embryo implantation and placentation. Development. 1997;124:3415–3425. doi: 10.1242/dev.124.17.3415. [DOI] [PubMed] [Google Scholar]
- Bazer FW, Burghardt RC, Johnson GA, Spencer TE, Wu G. Interferons and progesterone for establishment and maintenance of pregnancy: interactions among novel cell signaling pathways. Reprod Biol. 2008;8:179–211. doi: 10.1016/s1642-431x(12)60012-6. [DOI] [PubMed] [Google Scholar]
- Biazik JM, Thompson MB, Murphy CR. The tight junctional protein occludin is found in the uterine epithelium of squamate reptiles. J Comp Physiol B Biochem Syst Environ Physiol. 2007;177:935–943. doi: 10.1007/s00360-007-0192-1. [DOI] [PubMed] [Google Scholar]
- Biazik JM, Thompson MB, Murphy CR. Claudin-5 is restricted to the tight junction region of uterine epithelial cells in the uterus of pregnant/gravid squamate reptiles. Anat Rec. 2008;291:547–556. doi: 10.1002/ar.20677. [DOI] [PubMed] [Google Scholar]
- Biazik JM, Thompson MB, Murphy CR. Lysosomal and alkaline phosphatase activity indicate macromolecule transport across the uterine epithelium in two viviparous skinks with complex placenta. J Exp Zool B Mol Dev Evol. 2009;312:817–826. doi: 10.1002/jez.b.21297. [DOI] [PubMed] [Google Scholar]
- Biazik JM, Thompson MB, Murphy CR. Desmosomes in the uterine epithelium of non-invasive skink placentae. Anat Rec. 2010;293:502–512. doi: 10.1002/ar.21093. [DOI] [PubMed] [Google Scholar]
- Blackburn DG. Chorioallantoic placentation in squamate reptiles: structure, function, development, and evolution. J Exp Zool A Ecol Genet Physiol. 1993;266:414–430. [Google Scholar]
- Blackburn DG. Squamate reptiles as model organisms for the evolution of viviparity. Herpetol Monogr. 2006;20:131–146. [Google Scholar]
- Blackburn DG, Flemming AF. Invasive implantation and the intimate placental associations in a placentotrophic African lizard, Trachylepis ivensi (Scincidae) J Morphol. 2011;273:137–159. doi: 10.1002/jmor.11011. [DOI] [PubMed] [Google Scholar]
- Blackburn DG, Kleis-San Francisco S, Callard IP. Histology of abortive egg sites in the uterus of a viviparous placentrophic lizard, the skink Chalcides chalcides. J Morphol. 1998;235:97–108. doi: 10.1002/(SICI)1097-4687(199802)235:2<97::AID-JMOR1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Blackburn DG, Vitt LJ, Beuchat CA. Eutherian-like reproductive specializations in a viviparous reptile. Proc Natl Acad Sci U S A. 1984;81:4860–4863. doi: 10.1073/pnas.81.15.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandley MC, Huelsenbeck JP, Wiens JJ. Rates and patterns in the evolution of snake-like body form in squamate reptiles: evidence for repeated re-evolution of lost digits and long-term persistence of intermediate body forms. Evolution. 2008;62:2042–2064. doi: 10.1111/j.1558-5646.2008.00430.x. [DOI] [PubMed] [Google Scholar]
- Brandley MC, et al. Accommodating heterogenous rates of evolution in molecular divergence dating methods: an example using intercontinental dispersal of Plestiodon (Eumeces) lizards. Syst Biol. 2011;60:3–15. doi: 10.1093/sysbio/syq045. [DOI] [PubMed] [Google Scholar]
- Carranza S, Arnold EN, Geniez PH, Roca J, Mateo JA. Radiation, multiple dispersal and parallelism in the skinks, Chalcides and Sphenops (Squamata: Scincidae), with comments on Scincus and Scincopus and the age of the Sahara Desert. Mol Phylogenet Evol. 2008;46:1071–1094. doi: 10.1016/j.ympev.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Caucheteux SM, Kanellopoulos-Langevin C, Ojcius DM. At the innate frontiers between mother and fetus: linking abortion with complement activation. Immunity. 2003;18:169–172. doi: 10.1016/s1074-7613(03)00028-1. [DOI] [PubMed] [Google Scholar]
- Cetin I. Amino acid interconversions in the fetal-placental unit: the animal model and human studies in vivo. Pediatr Res. 2001;49:148–154. doi: 10.1203/00006450-200102000-00004. [DOI] [PubMed] [Google Scholar]
- Challis JR, et al. Inflammation and pregnancy. Reprod Sci. 2009;16:206–215. doi: 10.1177/1933719108329095. [DOI] [PubMed] [Google Scholar]
- Chmurzyńska A. The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J Appl Genet. 2006;47:39–48. doi: 10.1007/BF03194597. [DOI] [PubMed] [Google Scholar]
- Choi Y, Johnson GA, Spencer TE, Bazer FW. Pregnancy and interferon tau regulate major histocompatibility complex class I and β2-microglobulin expression in the ovine uterus. Biol Reprod. 2003;68:1703–1710. doi: 10.1095/biolreprod.102.012708. [DOI] [PubMed] [Google Scholar]
- Cooke PS, et al. Stromal estrogen receptors mediate mitogenic effects of estradiol on uterine epithelium. Proc Natl Acad Sci U S A. 1997;94:6535–6540. doi: 10.1073/pnas.94.12.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AC, Robinson G, Vinson GP, Cheung WT, Pipkin FB. The localization and expression of the renin-angiotensin system in the human placenta throughout pregnancy. Placenta. 1999;20:467–474. doi: 10.1053/plac.1999.0404. [DOI] [PubMed] [Google Scholar]
- Corso G, Delitala GM, Carcupino M. Uterine morphology during the annual cycle in Chalcides ocellatus tiligugu (Gmelin) (Squamata: Scincidae) J Morphol. 2000;243:153–165. doi: 10.1002/(SICI)1097-4687(200002)243:2<153::AID-JMOR4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Corso G, Pala M, Pinna AM, Carcupino M. Aspetti morfofunzionali dell'ovidutto di Chalcides ocellatus tiligugu (Gmelin) (Squamata, Scincidae) Arch Ital Anat Embriol. 1988;93(4):237–251. [Google Scholar]
- Davies CJ, Fisher PJ, Schlafer DH. Temporal regional regulation of major histocompatibility complex class I expression in the bovine uterine/placental interface. Placenta. 2000;21:194–202. doi: 10.1053/plac.1999.0475. [DOI] [PubMed] [Google Scholar]
- Defaure JP, Hubert J. Table de development de lezard vivipare: Lacerta vivipara Jacquin. Arch Anat Micros Morphol Exp. 1961;50:309–328. [Google Scholar]
- Dey SK, et al. Molecular cues to implantation. Endocr Rev. 2004;25:341–373. doi: 10.1210/er.2003-0020. [DOI] [PubMed] [Google Scholar]
- Divya CP, Mahajan VS, Datta GS, Chauhan SS. Differential activity of cathepsin L in human placenta at two different stages of gestation. Placenta. 2002;23:59–64. doi: 10.1053/plac.2001.0748. [DOI] [PubMed] [Google Scholar]
- Enders AC, Carter AM. What can comparative studies of placental structure tell us? Placenta. 2004;25:S3–S9. doi: 10.1016/j.placenta.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Forsythe JA, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar J, et al. Opposing functions of the Ets factors NERF and ELF-1 during chicken blood vessel development. Arterioscler Thromb Vasc Biol. 2002;22:1106–1112. doi: 10.1161/01.atv.0000023427.92642.cd. [DOI] [PubMed] [Google Scholar]
- Geisert RD, Pratt TN, Bazer FW, Mayes JS, Watson GH. Immunocytochemical localization and changes in endometrial progestin receptor protein during the porcine oestrous cycle and early pregnancy. Reprod Fertil Dev. 1994;6:749–760. doi: 10.1071/rd9940749. [DOI] [PubMed] [Google Scholar]
- Giacomini E. Materiali per la storia dello sviluppo del Seps chalcides (Cuv.) Bonap. Monitore Zool. 1891;2:179–192. , 198–211. [Google Scholar]
- Girardi G, Bulla R, Salmon JE, Tedesco F. The complement system in the pathophysiology of pregnancy. Mol Immunol. 2006;43:68–77. doi: 10.1016/j.molimm.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Girling JE, Jones SM. In vitro progesterone production by maternal and embryonic tissues during gestation in the southern snow skink (Niveoscincus microlepidotus) Gen Comp Endocrinol. 2003;133:100–108. doi: 10.1016/s0016-6480(03)00147-3. [DOI] [PubMed] [Google Scholar]
- Gracheva EO, et al. Molecular basis of infrared detection by snakes. Nature. 2010;464:1006–1011. doi: 10.1038/nature08943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino FM, et al. Endocrine activity of the corpus luteum and placenta during pregnancy in Chalcides chalcides (Reptilia, Squamata) Gen Comp Endocrinol. 1998;111:261–270. doi: 10.1006/gcen.1998.7098. [DOI] [PubMed] [Google Scholar]
- Guillette LJ, Jones RE. Ovarian, oviductal and placental morphology of the reproductively bimodal lizard, Sceloporus aeneus. J Morphol. 1985;184:85–98. doi: 10.1002/jmor.1051840109. [DOI] [PubMed] [Google Scholar]
- Hagemann A, Nielson AH, Poulsen K. The uteroplacental renin-angiotensin system: a review. Exp Clin Endocrinol. 1994;102:252–261. doi: 10.1055/s-0029-1211289. [DOI] [PubMed] [Google Scholar]
- Haggarty P. Placental regulation of fatty acid delivery and its effect on fetal growth—a review. Placenta. 2002;16:S28–S38. doi: 10.1053/plac.2002.0791. [DOI] [PubMed] [Google Scholar]
- Herbert JF, Lindsay LA, Murphy CR, Thompson MB. Calcium transport across the uterine epithelium of pregnant lizards. Herpetol Monogr. 2006;20:205–211. [Google Scholar]
- Hsieh MY, Chen WY, Jiang MJ, Cheng BC, Huang TY, Chang MS. Interleukin-20 promotes angiogenesis in a direct and indirect manner. Genes Immun. 2006;7:234–242. doi: 10.1038/sj.gene.6364291. [DOI] [PubMed] [Google Scholar]
- Janssen BJC, et al. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- Jerez A, Ramírez-Pinilla MP. The allantoplacenta of Mabuya mabouya (Sauria: Scincidae) J Morphol. 2001;249:132–146. doi: 10.1002/jmor.1045. [DOI] [PubMed] [Google Scholar]
- Jones SM, Swain R. Annual reproductive cycle and annual cycles of reproductive hormones in plasma of female Niveoscincus metallicus. J Herpetol. 1996;30:140–146. [Google Scholar]
- Joyce J, et al. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Joyce MM, et al. Uterine MHC class I molecules and β2-microglobulin are regulated by progesterone and conceptus interferons during pig pregnancy. J Immunol. 2008;181:2494–2505. doi: 10.4049/jimmunol.181.4.2494. [DOI] [PubMed] [Google Scholar]
- Kirschke H, Barrett AJ, Rawlings ND. Lysosomal cysteine proteases. 2nd ed. Oxford: Oxford University Press; 1997. [Google Scholar]
- Kwon H, Spencer TE, Bazer FW, Wu G. Developmental changes in amino acids in ovine fetal fluid. Biol Reprod. 2003;68:1813–1820. doi: 10.1095/biolreprod.102.012971. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lindsay LA, Murphy CR. Aquaporins are upregulated in glandular epithelium at the time of implantation in the rat. J Mol Histol. 2007;38:87–95. doi: 10.1007/s10735-007-9083-8. [DOI] [PubMed] [Google Scholar]
- Loffing J, et al. Distribution of transcellular calcium and sodium transport pathways. J Comp Physiol B. 2001;177:935–943. doi: 10.1152/ajprenal.0085.2001. [DOI] [PubMed] [Google Scholar]
- Manso Filho HC, Costa HE, Wu G, McKeever KH, Watford M. Equine placenta expresses glutamine synthetase. Vet Res Commun. 2009;33:175–182. doi: 10.1007/s11259-008-9167-2. [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-Seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson GR, Guillette LJ. Changes in oviducal vascularity during the reproductive cycle of three oviparous lizards (Eumeces obsoletus, Sceloporus undulatus and Crotaphytus collaris) J Reprod Fertil. 1987;80:361–371. doi: 10.1530/jrf.0.0800361. [DOI] [PubMed] [Google Scholar]
- Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- Moses EK, Freed KA, Brennecke SP, Rice GE. Distribution of the phospholipase A2 receptor messenger RNA in human gestational tissues. Placenta. 1998;19:35–40. doi: 10.1016/s0143-4004(98)90096-0. [DOI] [PubMed] [Google Scholar]
- Murphy BF, Belov K, Thompson MB. Evolution of viviparity and uterine angiogenesis: vascular endothelial growth factor (VEGF) in oviparous and viviparous skinks. J Exp Zool B Mol Dev Evol. 2010;314:148–156. doi: 10.1002/jez.b.21317. [DOI] [PubMed] [Google Scholar]
- Murphy BF, Brandley MC, Murphy CR, Thompson MB. Morphology and development of the placentae in Eulamprus quoyii group skinks (Squamata: Scincidae) J Anat. 2012 doi: 10.1111/j.1469-7580.2012.01492.x. Advance Access published Mar 16, 2012, doi:10.1111/j.1469-7580.2012.01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy BF, Parker SL, Murphy CR, Thompson MB. Angiogenesis of the uterus and the chorioallantois in the eastern water skink Eulamprus quoyii. J Exp Biol. 2010;213:3340–3347. doi: 10.1242/jeb.046862. [DOI] [PubMed] [Google Scholar]
- Murphy BF, Thompson MB. A review of the evolution of viviparity in squamate reptiles: the past, present and future role of molecular biology and genomics. J Comp Physiol B Biochem Syst Environ Physiol. 2011;181:575–594. doi: 10.1007/s00360-011-0584-0. [DOI] [PubMed] [Google Scholar]
- Murphy BF, Thompson MB, Belov K. Evolution of viviparity and the maternal immune system: major histocompatibility complex (MHC) class I genes in skinks. Orbit. 2009;1:1–16. [Google Scholar]
- Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myatt L, Cui X. Oxidative stress in the placenta. Histochem Cell Biol. 2004;122:369–382. doi: 10.1007/s00418-004-0677-x. [DOI] [PubMed] [Google Scholar]
- Nevalainen TJ, Haapamäki MM, Grönroos JM. Roles of secretory phospholipases A2 in inflammatory diseases. Biochim Biophys Acta. 2000;1488:83–90. doi: 10.1016/s1388-1981(00)00112-8. [DOI] [PubMed] [Google Scholar]
- Nielsen AH, Schauser KH, Poulsen K. The uteroplacental renin-angiotensis system. Placenta. 2000;21:468–477. doi: 10.1053/plac.2000.0535. [DOI] [PubMed] [Google Scholar]
- Parker SL, Manconi F, Murphy CR, Thompson MB. Uterine and placental angiogenesis in the Australian skinks, Ctenotus taeniolatus and Saiphos equalis. Anat Rec. 2010;293:829–838. doi: 10.1002/ar.21052. [DOI] [PubMed] [Google Scholar]
- Parker SL, Murphy CR, Thompson MB. Uterine angiogenesis in squamate reptiles: implications for the evolution of viviparity. Herpetol Conserv Biol. 2010;5:330–334. [Google Scholar]
- Paulesu L. Cytokines in mammalian reproduction and speculation about their possible involvement in nonmammalian viviparity. Microsc Res Tech. 1997;38:188–194. doi: 10.1002/(SICI)1097-0029(19970701/15)38:1/2<188::AID-JEMT19>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Paulesu L, Romagnoli R, Bigliardi E. Materno-fetal immunotolerance: is interleukin-1 a fundamental mediator in placental viviparity? Dev Comp Immunol. 2005;29:409–415. doi: 10.1016/j.dci.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Paulesu L, et al. Cytokines in the viviparous reproduction of squamate reptiles: interleukin-1a (IL-1a) and IL-1b in placental structures of a skink. Placenta. 1995;16:193–205. doi: 10.1016/0143-4004(95)90008-x. [DOI] [PubMed] [Google Scholar]
- Paulesu L, et al. Evidence of Hβ58, a gene involved in mammalian placental development, in the three-toed skink, Chalcides chalcides (Squamata: Scincidae), a viviparous placentotrophic reptile. Placenta. 2001;22:735–741. doi: 10.1053/plac.2001.0714. [DOI] [PubMed] [Google Scholar]
- Paulesu L, et al. Cytokines in the oviparity/viviparity transition: evidence of the interleukin-1 system in a species with reproductive bimodality, the lizard Lacerta vivipara. Evol Dev. 2005;7:282–288. doi: 10.1111/j.1525-142X.2005.05034.x. [DOI] [PubMed] [Google Scholar]
- Peng J, Zhang L, Drysdale L, Fong GH. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc Natl Acad Sci U S A. 2000;97:8386–8391. doi: 10.1073/pnas.140087397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Pinilla MP. Placental transfer of nutrients during gestation in an Andean population of the highly matrotrophic lizard genus Mabuya (Squamata: Scincidae) Herpetol Monogr. 2006;20:194–204. [Google Scholar]
- Ramírez-Pinilla MP, De Pérez G, Carreño-Escobar JF. Allantoplacental ultrastructure of an Andean population of Mabuya (Squamata, Scincidae) J Morphol. 2006;267:1227–1247. doi: 10.1002/jmor.10471. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-Seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnoli R, Cateni C, Guarino FM, Bigliardi E, Paulesu LR. Potential role of interleukin-1 at the peri-ovulation stage in a species of placental viviparous reptile, the three-toed skink, Chalcides chalcides (Squamata: Scincidae) Reprod Biol Endocrinol. 2003;1:60–65. doi: 10.1186/1477-7827-1-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu A, Lambris JD. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol Rev. 2001;180:35–48. doi: 10.1034/j.1600-065x.2001.1800103.x. [DOI] [PubMed] [Google Scholar]
- Salamonsen LA, Nie G. Proteases at the endometrial-trophoblast interface: their role in implantation. Rev Endocr Metab Disord. 2002;3:133–143. doi: 10.1023/a:1015407012559. [DOI] [PubMed] [Google Scholar]
- Schwartz TS, et al. A garter snake transcriptome: pyrosequencing, de novo assembly, and sex-specific differences. BMC Genomics. 2010;11:694. doi: 10.1186/1471-2164-11-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self JT, et al. Glutamine synthesis in the developing porcine placenta. Biol Reprod. 2004;70:1444–1451. doi: 10.1095/biolreprod.103.025486. [DOI] [PubMed] [Google Scholar]
- Song G, Bazer FW, Spencer TE. Differential expression of cathepsins and cystatin C in ovine uteroplacental tissues. Placenta. 2007;28:1091–1098. doi: 10.1016/j.placenta.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Song G, Spencer TE, Bazer FW. Cathepsins in the ovine uterus: regulation by pregnancy, progesterone, and interferon tau. Endocrinology. 2006;146:4825–4833. doi: 10.1210/en.2005-0768. [DOI] [PubMed] [Google Scholar]
- Song G, et al. Cathepsin B, cathepsin L, and cystatin C in the procine uterus and placenta: potential roles in endometrial/placental remodeling and in fluid-phase transport of proteins secreted by uterine epithelia across placental areolae. Biol Reprod. 2010;82:854–864. doi: 10.1095/biolreprod.109.080929. [DOI] [PubMed] [Google Scholar]
- Speake BK, Herbert JF, Thompson MB. Evidence for placental transfer of lipids during gestation in the viviparous lizard, Pseudemoia entrecasteauxii. Comp Biochem Physiol A Mol Integr Physiol. 2004;139:213–220. doi: 10.1016/j.cbpb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Bazer FW. Biology of progesterone action during pregnancy recognition and maintenance of pregnancy. Front Biosci. 2002;7:1879–1898. doi: 10.2741/spencer. [DOI] [PubMed] [Google Scholar]
- Spencer TE, et al. Pregnancy recognition and conceptus implantation in domestic ruminants: roles of progesterone, interferons and endogenous retroviruses. Reprod Fertil Dev. 2007;19:65–78. doi: 10.1071/rd06102. [DOI] [PubMed] [Google Scholar]
- Sterling D, Reithmeier RAF, Casey JR. A transport metabolon: functional interaction of carbonic anhydrase II and chloride/bicarbonate exchangers. J Biol Chem. 2001;276:47886–47894. doi: 10.1074/jbc.M105959200. [DOI] [PubMed] [Google Scholar]
- Stewart JR. Yolk sac placentation in reptiles: structural innovation in a fundamental vertebrate nutritional system. J Exp Zool. 1993;266:431–449. [Google Scholar]
- Stewart JR, Ecay TW. Patterns of maternal provision and embryonic mobilization of calcium in oviparous and viviparous squamate reptiles. Herpetol Conserv Biol. 2010;5:341–359. [Google Scholar]
- Stewart JR, Thompson MB. Evolution of placentation among squamate reptiles: recent research and future directions. Comp Biochem Physiol A Mol Integr Physiol. 2000;127:411–431. doi: 10.1016/s1095-6433(00)00273-7. [DOI] [PubMed] [Google Scholar]
- Surget-Groba Y, Montoya-Burgos JI. Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res. 2010;20:1432–1440. doi: 10.1101/gr.103846.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, et al. Endothelial PAS domain protein 1 gene promotes angiogenesis through the transactivation of both vascular endothelial growth factor and its receptor, Flt-1. Circ Res. 2004;95:146–153. doi: 10.1161/01.RES.0000134920.10128.b4. [DOI] [PubMed] [Google Scholar]
- Thompson MB, Speake BK. A review of the evolution of viviparity in lizards: structure, function and physiology of the placenta. J Comp Physiol B Biochem Syst Environ Physiol. 2006;176:179–189. doi: 10.1007/s00360-005-0048-5. [DOI] [PubMed] [Google Scholar]
- Thompson MB, Speake BK, Russell KJ, McCartney RJ. Utilisation of lipids, protein, ions, and energy during embryonic development of Australian oviparous skinks in the genus Lampropholis. Comp Biochem Physiol A Mol Integr Physiol. 2001;129:313–326. doi: 10.1016/s1095-6433(00)00349-4. [DOI] [PubMed] [Google Scholar]
- Thompson MB, Stewart JR, Speake BK. Comparison of nutrient transport across the placenta of lizards differing in placental complexity. Comp Biochem Physiol A Mol Integr Physiol. 2000;127:469–479. doi: 10.1016/s1095-6433(00)00277-4. [DOI] [PubMed] [Google Scholar]
- Varanou A, et al. The importance of cysteine cathepsin proteases for placental development. J Mol Med. 2006;84:305–317. doi: 10.1007/s00109-005-0032-2. [DOI] [PubMed] [Google Scholar]
- Vieira S, De Pérez G, Ramírez-Pinilla MP. Invasive cells in the placentome of Andean populations of Mabuya: an endotheliochorial contribution to the placenta? Anat Rec. 2007;290:1508–1518. doi: 10.1002/ar.20609. [DOI] [PubMed] [Google Scholar]
- Vinson GP, Saridogan E, Puddefoot JR, Djahanbakhch O. Tissue renin-angiotensin systems and reproduction. Hum Reprod. 1997;12:651–652. doi: 10.1093/humrep/12.4.651. [DOI] [PubMed] [Google Scholar]
- Vitt LJ, Blackburn DG. Ecology and life history of the viviparous lizard Mabuya bistriata (Scincidae) in the Brazilian Amazon. Copeia. 1991;1991:916–927. [Google Scholar]
- Walker CG, et al. Modulation of the maternal immune system by the pre-implantation embryo. BMC Genomics. 2010;11:474. doi: 10.1186/1471-2164-11-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall CE, et al. Whole transcriptome analysis of the fasting and fed Burmese python heart: insights into extreme physiological cardiac adaptation. Physiol Genomics. 2011;43:69–79. doi: 10.1152/physiolgenomics.00162.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-Seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Weekes HC. A review on placentation among reptiles with particular regard to function and evolution of placenta. Proc Linn Soc Lond. 1935;2:625–645. [Google Scholar]
- Winchester B. Lysosomal metabolism of glycoproteins. Glycobiology. 2005;15:1–15. doi: 10.1093/glycob/cwi041. [DOI] [PubMed] [Google Scholar]
- Wooding FBP, Ramirez-Pinilla MP, Forhead AS. Functional studies of the placenta of the lizard Mabuya sp. (Scincidae) using immunocytochemistry. Placenta. 2010;31:675–685. doi: 10.1016/j.placenta.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Wu G, Bazer FW, Tuo W. Developmental changes of free amino acid concentration in fetal fluids in the pig. J Nutr. 1995;125:2859–2868. doi: 10.1093/jn/125.11.2859. [DOI] [PubMed] [Google Scholar]
- Wu Q, Thompson MB, Murphy CR. Changing distribution of cadherins during gestation in the uterine epithelium of lizards. J Exp Zool B Mol Dev Evol. 2011;316:440–450. doi: 10.1002/jez.b.21419. [DOI] [PubMed] [Google Scholar]
- Zourbas S, Dubanchet S, Martal J, Chaouat G. Localisation of pro-inflammatory (IL-12, IL-15) and anti-inflammatory (IL-11, IL-13) cytokines at the foetal-maternal interface during murine pregnancy. Clin Exp Immunol. 2001;126:519–528. doi: 10.1046/j.1365-2249.2001.01607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]