Abstract
The post-translational modification SUMOylation is a major regulator of protein function that plays an important role in a wide range of cellular processes. SUMOylation involves the covalent attachment of a member of the SUMO (small ubiquitin-like modifier) family of proteins to lysine residues in specific target proteins via an enzymatic cascade analogous to, but distinct from, the ubiquitination pathway. There are four SUMO paralogues and an increasing number of proteins are being identified as SUMO substrates. However, in many cases little is known about how SUMOylation of these targets is regulated. Compared with the ubiquitination pathway, relatively few components of the conjugation machinery have been described and the processes that specify individual SUMO paralogue conjugation to defined substrate proteins are an active area of research. In the present review, we briefly describe the SUMOylation pathway and present an overview of the recent findings that are beginning to identify some of the mechanisms that regulate protein SUMOylation.
Keywords: post-translational modification, sentrin/small ubiquitin-like modifier-specific protease (SENP), small ubiquitin-like modifier (SUMO), small ubiquitin-like-modifier-activating enzyme (SAE), ubiquitin, ubiquitin-conjugating 9 (Ubc9), ubiquitin-like modifiers
THE SUMO (SMALL UBIQUITIN-LIKE MODIFIER) FAMILY OF PROTEINS
SUMO proteins are essential for the normal function of all eukaryotic cells. The yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe each contain one SUMO protein, Smt3 and Pmt3 respectively. Deletion of the Pmt3 gene results in severe growth impairment [1] and deletion of Smt3 causes a loss of cell viability [2,3].
In mammals there are four SUMO paralogues, designated SUMO-1 to SUMO-4. SUMO-1 (also known as UBL1 [4], PIC1 [5], sentrin [6], GMP1 [7] and Smt3c [8]) is an 11 kDa protein that was independently isolated by several groups as a binding partner of the RAD51/52 nucleoprotein filament proteins, which mediate DNA strand exchange, and PML (promyelocytic leukaemia), a component of multiprotein nuclear complexes [4–6]. The first reports that SUMO-1 functions as a covalent protein modifier described SUMOylation of the nuclear pore protein RanGAP1 (Ran-GTPase-activating protein 1) [7,9].
SUMO-2 (also known as sentrin 2 [10], Smt3b [8] and GMP-related protein [7]) and SUMO-3 (also known as sentrin 3 [10] and Smt3a [8]) were initially predicted from database searches [8] and the proteins were subsequently isolated and shown to conjugate substrate proteins [10,11]. SUMO-2 and -3 differ from each other by only three N-terminal residues and are often referred to collectively as SUMO-2/3. SUMO-2/3, however, share only approximately 50 % similarity with SUMO-1 (Figure 1). Interestingly, SUMO-2/3 are capable of forming chains on substrate proteins through internal lysine residues [12]. Although SUMO-1 chains have been reported in vitro [13], in vivo it appears that SUMO-1 may act as a chain terminator on SUMO-2/3 polymers [14].
Figure 1. SUMO family proteins.
The alignment shows the sequences of human ubiquitin, SUMO-1, SUMO-2, SUMO-3, SUMO-4, Smt3 (Smt3p) and Pmt3 (Pmt3p). Residues identical in all proteins are shown on a cyan background, and include the C-terminal di-glycine motif required for conjugation to substrate proteins (bold). Residues showing only conservative changes across all four proteins are shown in on a pink background and residues showing semi-conservative changes among each of the proteins are shown on a yellow background. Sequence alignment and determination of conservation was performed using the ClustalW program.
A SUMO-4 isoform has also been proposed from DNA sequence analysis, which predicts a 95-residue protein with 87 % amino acid similarity to SUMO-2. Conjugation of an exogenously expressed mature form of SUMO-4 has been reported in conditions of extreme cellular stress [15]. However, in contrast with the other SUMO genes the SUMO-4 gene lacks introns, raising the possibility that it may be a pseudogene [16]. In addition, despite the presence of SUMO-4 mRNA in kidney, lymph node and spleen [16,17], endogenous SUMO-4 protein has not been detected. Finally, SUMO-4 has a proline residue in a critical position, which makes it unclear, even if it is endogenously expressed, whether the precursor SUMO-4 protein could undergo maturation to conjugate to target proteins [18].
THE SUMO CYCLE
SUMO conjugation
SUMO proteins are synthesized as inactive precursors that must first undergo a C-terminal cleavage mediated by a family of SENP (sentrin/SUMO-specific protease) enzymes. This cleavage exposes a di-glycine motif that allows SUMO to be conjugated to lysine residues in target proteins (Figure 2; for an overview of the known members of the SUMO conjugation and deconjugation machinery see Table 1).
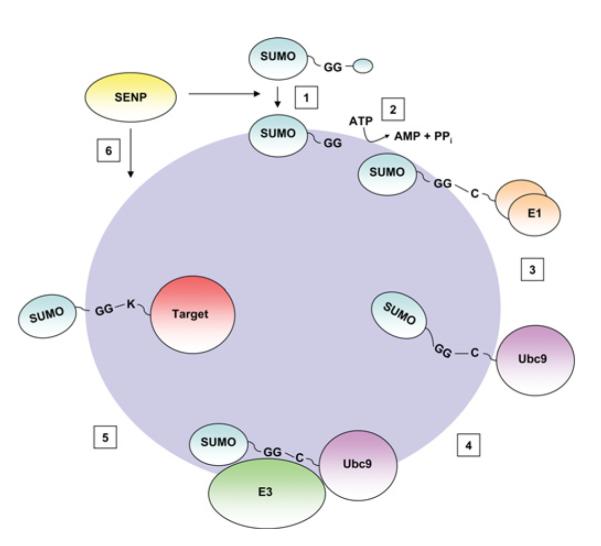
Figure 2. SUMO conjugation.
SUMO is transcribed as an inactive precursor, which is cleaved by members of the SENP family to expose a C-terminal di-glycine motif (1). This mature form of SUMO is then activated by the ATP-dependent formation of a thioester bond with the active site of the E1 enzyme, a heterodimer of SAE1 and SAE2 (2). The activated SUMO is then passed to the active site cysteine of the E2 conjugating enzyme, Ubc9 (3), which then catalyses the transfer of SUMO to the target protein, often in conjunction with an E3 enzyme (4,5). The SUMOylated substrate displays phenotypic differences to the unmodified form. DeSUMOylation is mediated by SENP family proteases (6). This releases the unmodified target protein (not shown), and mature SUMO, which is then available to undergo further rounds of conjugation to target proteins.
Table 1. SUMOylation machinery.
SUMO conjugation proceeds via the action of E1, E2 and E3 enzymes. In addition, SUMO proteins are both matured and removed from substrate proteins via the protease and isopeptidase activities respectively of the SUMO proteases. The known members of the mammalian SUMO machinery are shown alongside those of yeast (S. cerevisiae).
| Protein family | Yeast (S. cerevisiae) | Mammals |
|---|---|---|
| SUMO | Smt3 | SUMO-1 |
| SUMO-2 | ||
| SUMO-3 | ||
| SUMO E1 (activating enzyme) | Aos1 | SAE1 |
| Uba2 | SAE2 | |
| SUMO E2 (conjugating enzyme) | Ubc9 | Ubc9 |
| SUMO E3 (ligase) | Siz1 | PIAS1 |
| Siz2 | PIAS3 | |
| PIASxα | ||
| PIASxβ | ||
| PIASy | ||
| RanBP2 | ||
| Pc2 | ||
| Mms21 | Mms21 | |
| HDAC4 | ||
| HDAC7 | ||
| MUL1 | ||
| Rhes | ||
| TOPORS | ||
| TLS | ||
| TRAF7 | ||
| SUMO proteases (SUMO proteases and isopeptidases) | Ulp1 | SENP-1 |
| Ulp2 | SENP-2 | |
| SENP-3 | ||
| SENP-5 | ||
| SENP-6 | ||
| SENP-7 |
During each conjugation cycle, SUMO proteins are first activated in an ATP-dependent manner by the E1 ‘activating’ enzyme, a heterodimer of SAE1 (SUMO-activating enzyme E1) and SAE2 in mammals [19]. This step involves the formation of a thioester bond between the active-site cysteine residue of SAE2 and the C-terminal glycine residue of SUMO. SUMO is then passed to the active site cysteine of the conjugating enzyme Ubc9 (ubiquitin-conjugating 9), again via a thioester linkage [20–25]. Importantly, Ubc9 is the only known SUMO-conjugating enzyme and Ubc9 itself binds directly to the consensus SUMOylation motif on substrate proteins [26,27]. The target protein consensus motif comprises ψKxD/E (where ψ is a large hydrophobic residue). However, it should be emphasized that whereas ~75 % of known SUMO substrates are modified within a consensus motif [28], SUMOylation can also occur at lysine residues outside this motif and not all ψKxD/E motifs are SUMOylated. Thus, although useful as an initial predictor, the presence of a ψKxD/E motif on potential SUMO substrates is certainly not a definitive indicator that a protein is SUMOylated.
In the ubiquitin system, E3 ligase enzymes are generally a requirement for ubiquitination. Ubiquitin E3s can broadly be classified into two distinct types: HECT (homologous with E6-associated protein C-terminus) domain-containing E3s and RING domain-containing E3s. HECT domain E3s directly receive ubiquitin from their cognate E2s via a thioester linkage and subsequently pass ubiquitin to substrate proteins. RING domain E3s do not interact with ubiquitin directly but bind the E2 and substrate proteins to facilitate ubiquitin transfer (for a review [29]). As Ubc9 is capable of directly recognizing and SUMOylating many substrates in vitro, the requirement for E3 ligases in the SUMOylation pathway was initially a source of debate. Subsequently, numerous proteins have been reported that possess E3 ligase activity in the SUMO pathway in vivo. Each of the reported SUMO E3 ligases appear to function in a similar manner to the RING-domain E3s of the ubiquitin pathway in that they do not directly receive SUMO through a thioester linkage, but act as scaffolds bringing SUMO-loaded Ubc9 into contact with the substrate protein or holding the SUMO–Ubc9 thioester in a conformation conducive to SUMO transfer.
The first SUMO E3 ligases described were the Siz proteins in yeast, and deletion of these proteins largely abolishes SUMOylation in S. cerevisiae [30]. The mammalian homologues of the Siz proteins are the PIAS [protein inhibitor of activated STAT (signal transducer and activator of transcription)] family of proteins. The PIAS family constitutes five members, including splice variants, and each has been shown to possess SUMO E3 ligase activity ([31–35]; Table 1). The Siz and PIAS proteins contain a SP (Siz/PIAS)-RING domain that is similar to that found in many ubiquitin E3s. SP-RING ligases bind Ubc9, their substrate proteins and they also bind non-covalently to SUMO, consistent with them acting as a scaffold bringing SUMO-loaded Ubc9 together with substrate proteins. In addition to the PIAS proteins, a number of other SP-RING domain-containing proteins have been reported to function as SUMO E3 ligases, including TOPORS (topoisomerase I-binding, arginine/serine-rich) [36], MUL1 (mitochondrial E3 ubiquitin ligase 1, also known as MAPL) [37] and MMS21 [38–40]. Interestingly, TOPORS is the first-reported SUMO E3 that also functions as an E3 ligase for ubiquitin [41], suggesting it may play a central role in the cross-talk between SUMOylation and ubiquitination observed for some substrate proteins (see the SUMO and ubiquitin section).
Pc (Polycomb) proteins function as multimeric complexes involved in gene silencing and a member of this protein family, Pc2, has been reported to function as a SUMO E3 ligase [42]. Pc2 does not contain a RING domain and has no similarity to known ubiquitin ligases, but binds directly to its substrate CtBP (C-terminal-binding protein), Ubc9 and SUMO, suggesting it too may function as a scaffold to bring SUMO-loaded Ubc9 together with the substrate protein [42–44]. Similarly, HDAC4 (histone deacetylase 4) [45,46] and HDAC7 [47] (see the SUMO and acetylation section), the G-protein Rhes [48], the RNA-binding protein TLS (translocated in liposarcoma) [49] and TRAF7 (tumour-necrosis-factor-associated protein 7) [50] have also been reported as RING domain-lacking E3s that function as substrate recognition modules.
The nucleoporin RanBP2 (Ran-binding protein 2) also has SUMO E3 activity [51]. RanBP2 binds both Ubc9 and SUMO-1 and enhances the SUMOylation of Sp100, HDAC4 and PML in vitro. Interestingly, RanBP2 does not enhance SUMOylation of its binding partner RanGAP1, which is a prototypical SUMO substrate [51–53]. Structural data suggests that RanBP2 may function as a SUMO E3 through positioning SUMO-loaded Ubc9 in a conformation conducive to SUMO transfer to the substrate protein, but does not bind any of its known substrate proteins directly, suggesting it functions in a manner different to the other classes of SUMO E3 [54,55].
DeSUMOylation
Protein SUMOylation is a highly dynamic process that can be readily reversed by the action of the same SENP enzymes that are required for the maturation of pro-SUMO. In yeast, the SENP homologue Ulp1 was identified in a screen for proteins that specifically cleave Smt3 at its C-terminus [56]. Ulp1 yeast deletion strains undergo cell cycle arrest that cannot be rescued with either pro-Smt3 or mature Smt3, suggesting Ulp1 is required for both pro-Smt3 maturation and Smt3 cleavage from substrate proteins [56]. Another Smt3-specific protease, Ulp2, acts to remove Smt3 from substrate proteins but does not appear to be involved in the maturation of pro-Smt3 [57]. In addition, yeast strains lacking Ulp2 show an accumulation of Smt3 polymers, suggesting Ulp2 functions in the processing and editing of Smt3 chains [58].
In mammals there are six SENPs, designated SENP1–3 and SENP5–7, which vary in their cellular distribution, SUMO paralogue specificity and selectivity for SUMO maturation compared with deconjugation activities. The subcellular localization and activities of the SENP proteins are summarized in Table 2 (for reviews see [59,60]). Mammalian SENPs can be classified into three groups. SENP1 and SENP2 have a broad specificity for SUMO-1 and SUMO-2/3 and function both in their processing and deconjugation [61–63]. SENP3 and SENP5 favour SUMO-2/3 over SUMO-1 [64–66]. SENP6 and SENP7 also act preferentially on SUMO-2/3 [67,68]. Neither SENP6 nor SENP7 seem to be involved in maturation of pro-SUMO proteins and they show minimal activity in the deconjugation of monomeric SUMO-2/3 from substrate proteins. Rather, SENP6 and SENP7 efficiently edit and/or deconjugate poly-SUMO-2/3 chains [67,68]. Thus there are clear distinctions between the functions of different SENP enzymes. SENP1/2 are primarily responsible for cellular maturation of the SUMO proteins, and perform roles in the deconjugation of both SUMO-1 and SUMO-2/3 from substrates, SENP3/5 function in the removal of monomeric SUMO-2/3 from substrates and SENP6/7 act as editors of SUMO-2/3 chains. In addition, the paralogue specificity of the SENP proteins may be an important factor in regulating paralogue-specific SUMOylation through preferential removal of particular SUMO paralogues from substrate proteins (see the Mechanisms of SUMO paralogue specificity section).
Table 2. SENP enzymes.
SUMO is both matured and removed from substrates via the activities of the SENP family of enzymes. These enzymes differ in their tissue expression, subcellular localization, preference for SUMO-1 compared with SUMO-2/3 and in their preference for processing compared with deconjugation (removal of SUMO from substrate proteins) or chain editing (cleavage of SUMO–SUMO conjugate) activities. Alternative names are given in parentheses. NA, not applicable; ND, not determined.
| Species | Name | Tissue expression | Subcellular localization | Paralogue preference | Processing | Deconjugation | Chain editing |
|---|---|---|---|---|---|---|---|
| Yeast (S. cerevisiae) | Ulp1 | NA | Nuclear periphery | NA | Yes | Yes | No |
| Ulp2 (Smt4) | NA | Nucleoplasm | NA | No | No | Yes | |
| Mammals | SENP1 (SuPr-2) | Testes (high), pancreas, spleen, liver, ovaries, small intestine, thymus (low) | Nuclear pore and nucleoplasmic speckles | SUMO-1>SUMO-2/3 | Yes | Yes | No |
| SENP2 (AXAM2/SMT3IP1) | ND | Nuclear pore | SUMO-2/3>SUMO-1 | Yes | Yes | No | |
| SENP3 (SSP3/SMT3IP2) | ND | Nucleolus | SUMO-2/3 | ND | Yes | No | |
| SENP5 | ND | Nucleolus | SUMO-2/3 | Yes | Yes | No | |
| SENP6 (SUSP1/SSP1) | Testes (high), pancreas, ovaries, colon, peripheral blood leucocytes | Nucleoplasm | SUMO-2/3 | No | No | Yes | |
| SENP7 | ND | Nucleoplasm | SUMO-2/3 | No | No | Yes |
Given their critical roles, SENP proteins themselves are likely to be subject to tight cellular control. Although much remains unknown, some examples of SENP regulation are beginning to emerge. For example, SENP1 has been reported to be modified by SUMO-1, although the functional consequence of SENP1 SUMOylation have not yet been reported [69]. The activity of SENP3 is regulated through control of its stability. Under normal cellular conditions, SENP3 is rapidly degraded by the ubiquitin–proteasome system [70,71]. However, ROS (reactive oxygen species) cause enhanced SENP3 stability and promote its relocation from the nucleolus to the nucleoplasm. This enhanced SENP3 level leads to deSUMOylation of the protein p300, a co-activator of the transcriptional activity of the transcription factor HIF1α (hypoxia-inducible factor 1α) [71]. HIF1α functions as a master regulator of numerous cellular stress responses through transcription of stress-responsive genes. As SUMOylation of p300 inhibits its co-activator activity, the stress-induced increase in SENP3 favours deSUMOylation of p300, relieving this repression and thereby promoting the transcription of HIF1α-dependent stress-responsive genes.
KNOCKOUT STUDIES ON SUMOylation
The critical importance SUMOylation in mammals has been confirmed through knockout and knockdown studies of Ubc9. As Ubc9 is required for the conjugation of every SUMO paralogue, deletion of Ubc9 prevents all SUMO conjugation. Removal of Ubc9 in the chicken DT40 lymphocyte cell line results in abnormalities in chromosome segregation, nuclear organization and ultimately cell death by apoptosis [72]. Similarly, Ubc9-knockout mice die at an early embryonic stage due to defects in chromosomal segregation at mitosis and aberrant nuclear organization [73].
Intriguingly, Alkuraya et al. [74] identified a genomic insertion disrupting the SUMO-1 gene in a Caucasian female with cleft lip and palate. This gene insertion decreased SUMO-1 expression at both the mRNA and protein levels. In an attempt to confirm that SUMO-1 haplo-insufficiency leads to cleft lip and palate, they generated mice hetero- or homo-zygous for a β-galactosidase insertion in the SUMO-1 gene. Four of 46 heterozygote pups exhibited cleft palate and other animals died in late embryonic and early postnatal periods, indicating that SUMO-1 is also required for other developmental functions in addition to palatogenesis [74]. However, two more recent studies have reported that SUMO-1-knockout mice are viable and display an apparently normal phenotype [75,76]. These results suggest that SUMO-2/3 may act to compensate for the loss of SUMO-1. Furthermore, neither of these later studies detected a link between SUMO-1 deletion and cleft lip or palate. In support of a model in which SUMO-2/3 can compensate for a lack of SUMO-1, RanGAP1, which is preferentially modified by SUMO-1, displayed enhanced modification by SUMO-2/3 in SUMO-1-knockout embryonic extracts [76]. Thus, taken together, these results seem to indicate that, at least under extreme circumstances, SUMO-2/3 can effectively compensate for the lack of SUMO-1.
FUNCTIONAL HETEROGENEITY WITHIN THE SUMO FAMILY
Although some substrates are preferentially modified by one SUMO isoform (for example RanGAP1 and the PML-nuclear body component Sp100 are modified by SUMO-1 or SUMO-2/3 respectively [77,78]), many substrates can be modified by both SUMO-1 and SUMO-2/3 [78]. It is unclear how the SUMOylation machinery distinguishes between SUMO-1 and SUMO-2/3 for specific conjugation to protein targets. Furthermore, for most substrates that can be modified by either isoform the functional differences of SUMO-1 compared with SUMO-2/3 conjugation have yet to be defined. Nonetheless, observations that SUMO-1 and SUMO-2/3 differ in their conjugation dynamics and show discrete patterns of localization throughout the cell cycle indicate specific regulation of the SUMO paralogues [77,79]. For example, resting levels of unconjugated SUMO-1 and SUMO-2/3 differ. COS-7 cells contain a large free pool of SUMO-2/3, but there is very little free SUMO-1, suggesting SUMO-2/3 may act as a cellular reserve of SUMO [77]. Indeed, various cellular stresses invoke a massive increase in SUMO-2/3 conjugation in many cell types (for reviews see [80–85]).
The discovery of SUMO-targeted ubiquitin ligases also illustrates differences in SUMO-1 and SUMO-2/3 function and highlights the interplay between the SUMO and ubiquitin systems (for recent reviews see [86,87] and the SUMO and ubiquitin section). Ubiquitin, like SUMO, post-translationally modifies proteins at lysine residues and has been most well-characterized for its role in targeting substrate proteins for proteasomal degradation (for reviews see [88–90]). Recently, a number of ubiquitin ligases, responsible for ubiquitin conjugation to substrate proteins, have been reported that bind SUMO chains on substrate proteins, leading to the ubiquitination of the substrate protein and, ultimately, its degradation. For example, the protein PML can be modified by both SUMO-1 and SUMO-2/3. SUMO-1 modification of PML is required for of its localization to nuclear domains known as PML nuclear bodies [91]. Formation of SUMO-2/3 chains, however, leads to the degradation of PML via the recruitment of the SUMO-binding ubiquitin ligase RNF4 (ring finger protein 4), which mediates the ubiquitination of the SUMO chains, targeting the whole complex for degradation [92–94]. Thus attachment of SUMO chains to a substrate protein can act as a distinct signal from mono-SUMOylation through recruitment of proteins that specifically recognize polymeric SUMO chains.
NON-COVALENT SUMO BINDING
As mentioned briefly above, in addition to covalent SUMOylation of specific lysine residues on substrate proteins, an increasing number of proteins have been shown to bind SUMO or SUMO chains non-covalently via SIMs (SUMO-interacting motifs) (for a review see [95]). This is analogous to the ubiquitin system where the covalently coupled ubiquitin on substrate proteins can recruit ubiquitin-binding proteins that bind non-covalently. At least 20 different non-covalent ubiquitin interaction motifs have been identified [96]. Similarly, protein SUMOylation provides an interaction platform for the recruitment of SIM-containing effector proteins. The SIMs identified so far generally comprise a hydrophobic core surrounded by acidic flanking residues or phosphorylatable serine residues [97–100]. In addition to SIMs mediating the effects of SUMOylation, a growing number of proteins have been identified for which SUMOylation is dependent on the presence of the SIM in the substrate. This suggests that recruitment of SUMO-loaded Ubc9 may represent a general mechanism of substrate recognition and, possibly, paralogue specificity, through paralogue-specific binding to the substrate protein.
As discussed above, SUMOylation by SUMO-1 or SUMO-2/3 can lead to different functional consequences. SUMO paralogue-specific interacting proteins are likely to account for at least some of these differences. For example, CoREST1 (co-repressor for RE1-silencing transcription factor) is a nuclear protein that forms part of a protein complex that creates a transcriptionally repressive environment required for the silencing of neuronal-specific genes in non-neuronal cells [101–104]. CoREST contains a SIM that binds to promoter proteins which have been SUMOylated by SUMO-2, but not SUMO-1, and this interaction mediates the recruitment of the CoREST1 complex to target genes [105]. The SUMO-2-specific SIM in CoREST is similar to but distinct from other reported SIMs, containing a hydrophobic core (Ile-Asp-Ile-Glu-Val), with an N-terminal acidic stretch. Building on this discovery of a SUMO-2-specific SIM, database searches for proteins harbouring similar motifs have identified several other nuclear proteins that bind SUMO-2, but not SUMO-1 in vitro [105].
MOLECULAR CONSEQUENCES OF SUMOylation
Although the functional consequences of SUMO conjugation to a particular substrate protein are difficult to predict overall, protein modification by SUMO may lead to one of three non-mutually exclusive effects ([106]; Figure 3). First, SUMOylation may mask the binding site of a protein that interacts with the substrate protein, essentially acting to occlude the interaction in a SUMOylation-dependent manner. An example of this is SUMOylation of the ubiquitin-conjugating enzyme E2-25k, which inhibits its interaction with the ubiquitin E1 enzyme [107] leading to a decrease in its capability to conjugate ubiquitin to substrate proteins (see the SUMO and ubiquitin section). Secondly, and conversely, the covalently attached SUMO may act as an interaction ‘hub’ that recruits new interacting proteins to the substrate either by direct non-covalent interaction with the SUMO moiety, or via a novel interaction domain created at the SUMO-substrate interface. For example, SUMOylation of RanGAP1 promotes its interaction with RanBP2 and its relocation from the cytosol to the nuclear pore [7,9], poly-SUMOylation of PML can recruit the ubiquitin ligase RNF4 [92–94] and, as discussed below, SUMOylation of the transcription factor Elk1 [ETS (E twenty-six)-like kinase 1] can recruit the histone deacetylase HDAC2 [108] (see the SUMO and acetylation section). Thirdly, SUMOylation can lead to a conformational change in the SUMOylated substrate, directly regulating its function. An example of this is TDG (thymine–DNA glycosylase), which is discussed in more detail below.
Figure 3. Molecular consequences of SUMOylation.
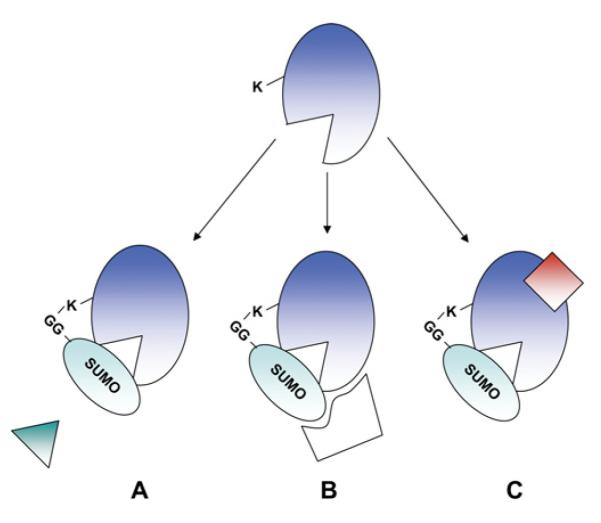
SUMOylation can have three non-mutually exclusive consequences on the substrate protein. (A) SUMO modification can inhibit interactions with substrate-interacting proteins via the occlusion of the interaction site. (B) SUMOylation can create a new binding face on the substrate protein that can recruit other binding partners in a SUMOylation-dependent manner. (C) SUMOylation can lead to a change in conformation of the substrate protein, altering its activity or revealing previously masked binding sites.
THE SUMO ENIGMA
An initially counter-intuitive observation regarding SUMOylation is that only small proportion of the available substrate protein need be SUMOylated to achieve maximal effect. This phenomenon has been referred to as the ‘SUMO enigma’ [109]. Although the underlying mechanisms for this effect on many SUMO-modified proteins have not been determined, elegant explanations based on the highly dynamic nature of the SUMOylation cycle have been proposed to account for this effect in some specific cases. For example, in neurons, the SUMO modification of GluR6 (glutamate receptor subunit 6) leads to its endocytosis at the plasma membrane [110]. Although only a small proportion of SUMOylated GluR6 can be detected at a given time point, a large proportion of GluR6 undergoes SUMO-mediated endocytosis. The low level of detectable SUMOylated GluR6 is probably due to the action of SENPs rapidly removing SUMO. Nonetheless, the functional effects of GluR6 SUMOylation persist after deSUMOylation, namely GluR6 is endocytosed from the neuronal surface. Thus once SUMOylation has mediated endocytosis, SUMO can be removed and any GluR6 SUMOylated previously will have a different cellular localization to GluR6 that has never been modified.
Similar explanations have been used to account for the potent repression of transcription by protein SUMOylation. Multiple transcription factors can undergo SUMOylation and in many cases this leads to repression of transcription (for reviews see [111,112]). For some transcription factors, such as Sp3 (stimulating protein 3) or LEF-1 (lymphoid enhancer-binding factor 1), SUMOylation causes their relocation to nuclear subdomains not associated with transcriptional activity [35,113]. Thus, as with GluR6, the SUMOylation-mediated effects persist after the removal of SUMO.
SUMO modification of promoter-occupied transcription factors has also been reported to recruit chromatin-modifying proteins, such as the HDACs, or repressor proteins, such as DAXX (death-domain-associated protein) through non-covalent interactions [108,114,115]. SUMO-mediated HDAC recruitment leads to local histone deacetylation, resulting in a more condensed chromatin structure that favours transcriptional repression. Thus recruitment of HDACs to a SUMOylated transcription factor will induce a repressive transcriptional environment that persists after removal of SUMO from the substrate. Similarly, recruitment of repressor proteins, such as DAXX, may lead to the formation of ‘repressor complexes’. Again, SUMO-mediated sequestration of transcription factors into repressive complexes would persist after removal of the SUMO.
The DNA-modifying enzyme TDG provides another example of how low-level substrate SUMOylation can result in robust effects. TDG recognizes and excises mismatched base pairs and SUMOylation is required for every enzymatic cycle [116]. When TDG removes the mismatched base, it binds strongly to the DNA as a result of its high affinity for the abasic site created by the base excision [117]. This high affinity interaction causes a break in the reaction cycle that is released by TDG SUMOylation, which decreases its affinity for DNA and releases it into the nucleoplasm [116]. Subsequent removal of SUMO by SENPs allows TDG to undergo a further round of activity. Thus, although only a very small proportion of TDG is modified at any one time due to the rapid deconjugation of SUMO in the nucleoplasm, the effects of SUMOylation are critical to every reaction cycle.
Taken together these observations suggest that although only a small proportion of a substrate may be modified at any given time point, the majority of a substrate will be SUMOylated over time and undergo a SUMO-induced functional change that remains after the protein has been deSUMOylated.
REGULATION OF SUMOYLATION
The list of SUMO substrates is increasing rapidly but how SUMOylation of the vast majority of these proteins is regulated remains unclear. However, a number of common themes are beginning to emerge. Although numerous stimuli have been reported that lead to global changes in cellular SUMOylation, it is likely that SUMOylation is controlled in a substrate-specific manner. It seems that SUMOylation of many substrates is regulated through a complex interplay between SUMOylation and other post-translational modifications of the substrate protein. Furthermore, in addition to co-regulating substrate proteins, there is also direct reciprocal interplay between SUMOylation and other post-translational modifications through modification of the proteins involved in their enzymatic pathways (Figure 4). A number of illustrative examples of this complex cross-regulation are discussed below.
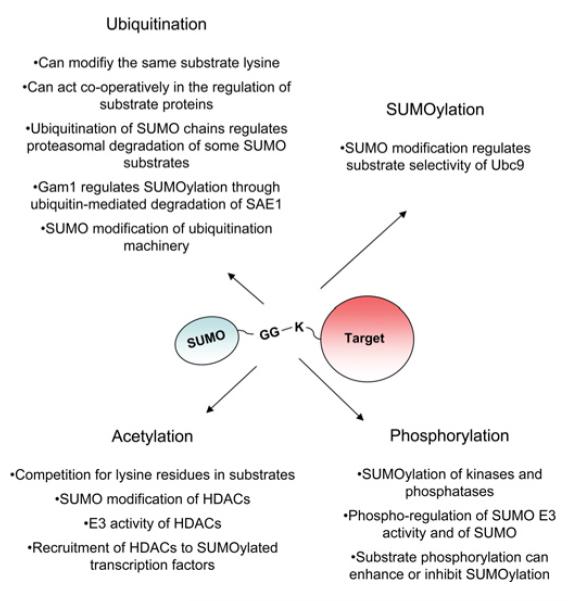
Figure 4. Complex interplay between SUMOylation and other post-translational modifications.
Summary of the cross-regulation between SUMOylation and other post-translational modifications.
Cell stress and SUMOylation
It is well-documented that global increases in protein SUMOylation occur in response to cellular stress. In COS-7 cells it has been reported that under resting conditions there is very little unconjugated SUMO-1, but a large pool of free SUMO-2/3 [77]. Following heat stress, however, large increases in SUMO-2/3 conjugation occur, whereas SUMO-1 conjugation appears unaffected. Similarly, oxidative, ethanol and osmotic stresses were each shown to cause a rapid increase in global SUMO-2/3 conjugation [77]. Stress-dependent increases in SUMO conjugation have been reported in various cellular systems including oxygen/glucose deprivation and hypothermia in neurons [118–121] (for reviews see [80–85]). This common cellular response of enhanced SUMO conjugation following stress suggests protein SUMOylation could constitute a protective response. Consistent with this it has been reported that exogenous expression of SUMO-1 or SUMO-2 make SHSY5Y human neuroblastoma cells more resistant to oxygen/glucose deprivation whereas knockdown of SUMO-1, but not SUMO-2, made SHSY5Y cells more susceptible to oxygen/glucose deprivation-mediated cell death. Similarly, overexpression of SUMO-1 has been reported to make rat cortical neurons more resistant to oxygen/glucose deprivation [122].
Although increases in SUMOylation resulting from cellular stress appear to be a widespread phenomenon, it is important to emphasize that specific substrates are differentially modified. For example, although there is a net increase in SUMOylation following heat-shock, the SUMOylation level of some substrates is unchanged and for others decreases, indicating that substrate SUMOylation under these conditions is a regulated stress-specific response, rather than a generalized non-specific increase [123]. Indeed, the requirement for enhanced SENP3 stability for the cellular response to mild oxidative stress highlights the importance of regulated deSUMOylation, even in the presence of a net increase in SUMOylation [71].
RELATIONSHIP BETWEEN SUMOylation AND OTHER POST-TRANSLATIONAL MODIFICATIONS
SUMO and ubiquitin
Co-regulation of substrate proteins
Ubiquitin performs multiple functions, but its role in targeting intracellular proteins for proteasomal degradation and membrane proteins for lysosomal degradation have been most extensively characterized (for reviews see [88–90]). Many proteins are substrates for both SUMO and ubiquitin, often at the same lysine residue, which led to the proposal that SUMO and ubiquitin act antagonistically. It is now clear, however, that the inter-relationship between the two systems is much more complex and in many cases SUMO and ubiquitin act either sequentially or in concert to regulate the substrate protein (for reviews see [87,124]).
Antagonism between SUMOylation and ubiquitination was first reported for the NFκB (nuclear factor κB) regulator IκBα (inhibitory κBα) [125]. Ubiquitination of IκBα at Lys21 and Lys22 facilitates its proteasomal degradation; Lys21 can also be SUMOylated and the SUMOylated pool of IκBα is protected from ubiquitin-mediated proteolysis. Surprisingly, however, SUMO and ubiquitin do not appear to act competitively. Ubiquitination of IκBα is dependent on its prior phosphorylation at Ser32 and Ser36 but a Ser32/Ser36 phosphomimic mutant IκBα cannot be SUMOylated. This suggests that although ubiquitin and SUMO antagonize each other they do not compete directly. Rather, the actual competition may occur between phosphorylation and SUMOylation [125].
SUMO and ubiquitin have also been reported to act sequentially in the regulation of another component of the NFκB pathway, NEMO (NF-κB essential modulator), a regulatory subunit for IKK (IκB kinase) [126]. NEMO is SUMOylated at two lysine residues in its C-terminus (Lys277 and Lys309), which results in its accumulation in the nucleus where it can be phosphorylated by the DNA-damaged-induced kinase ATM (ataxia telangiectasia mutated). Thus SUMOylation of NEMO facilitates its phosphorylation by ATM, which, in turn, is a prerequisite for NEMO ubiquitination at the same lysines originally targeted by SUMOylation. This phosphorylation-induced ubiquitination results in nuclear export of NEMO allowing it to associate with the other IKK subunits and form an active kinase [126]. Therefore, despite targeting the same lysine residues, SUMO and ubiquitin act in sequence to activate IKK via nuclear shuttling of the regulatory subunit NEMO.
Although they do not modify the same lysine residue, interplay between SUMO and ubiquitin also co-regulate the tumour suppressor p53. In this case, the RING domain E3 ligase MDM2 (murine double minute 2) mediates ubiquitination of p53, leading to recruitment of the SUMO E3 PIASy, which enhances SUMOylation of p53 at a distinct lysine residue [127].
Recently, the discovery of SUMO-targeted ubiquitin ligases, as described above, has revealed a previously unsuspected co-operation between the ubiquitination and SUMOylation pathways in regulating the proteasomal degradation of SUMO substrate proteins [87].
Cross-talk between SUMOylation and ubiquitination
In addition to co-regulating substrate proteins, ubiquitination and SUMOylation can directly cross-regulate each other by modification of components of their respective enzymatic machinery. For example, SUMOylation of E2-25k, an E2 enzyme in the ubiquitin pathway, inhibits its capability to conjugate ubiquitin. Thus SUMOylation can down-regulate the ubiquitination system [107]. Conversely, an example of ubiquitin regulating SUMOylation occurs with the viral protein Gam1. Gam1 was initially described as an anti-apoptotic protein expressed by the chicken CELO (chicken embryo lethal orphan) adenovirus type 1 [128]. Gam1 is required for viral replication [129] and subsequent work has demonstrated that it functions through the inhibition of protein SUMOylation by promoting ubiquitination of the SUMO E1 component SAE1, leading to its proteasomal degradation [130]. Complex cross-regulation between the ubiquitin and SUMO pathways also occurs via the ubiquitin E3 enzyme parkin, which is strongly implicated in the neurodegenerative Parkinson’s disease. Parkin ubiquitinates the SUMO E3 RanBP2, promoting its degradation [131] and non-covalent SUMO binding to Parkin enhances its ubiquitin-ligase activity [132] creating a negative feedback loop between the two systems.
SUMO and acetylation
Co-regulation of substrate proteins
Lysine residue acetylation can regulate multiple cellular processes (for a review see [133]). A growing number of proteins, including the transcription factors MEF2A (myocyte enhancer factor 2A) and Sp3, the transcriptional co-activator p300 and the tumour suppressor HIC1 (hypermethylated in cancer 1) can be modified by both SUMO and an acetyl group at the same lysine residue [134–137]. In each case the two modifications act antagonistically via competition for the target lysine; however, as with ubiquitin, whether this SUMO-acetyl competition is truly direct remains to be determined in most cases.
A defined motif present in many potential SUMO substrate proteins has been reported to target both SUMOylation and acetylation to a target lysine residue. This motif, the ‘SUMO-acetyl switch’ consists of a SUMOylation consensus motif flanked by a C-terminal proline residue that can also direct acetylation to the SUMOylated lysine residue [137]. In the case of MEF2A, phosphorylation of a serine residue adjacent to this proline appears to act as a switch favouring SUMOylation over acetylation, creating a ‘phosphorylation-dependent SUMO-acetyl switch’ that can regulate the exchange the acetyl group for SUMO [136].
Cross-regulation of SUMOylation and acetylation
An interesting example of complex interplay between the SUMOylation and acetylation machinery exists in the case of HDACs, which function in the removal of acetyl groups from substrate proteins and have also been reported to function as SUMO E3s for some substrates. In addition, some HDACs can also be SUMOylated, as well as binding SUMO proteins non-covalently, highlighting intricate inter-relationship between the two pathways.
Histone acetylation is generally associated with a transcriptionally active environment, and the removal of these acetyl groups by HDACs favours transcriptional repression. HDACs seem to play a broad role in SUMO-mediated repression of transcription through their recruitment to SUMOylated transcription factors. SUMOylation of the transcription factor Elk1, for example, acts to recruit HDAC2 and this has been correlated with decreased histone acetylation at Elk1-regulated promoters [108].
HDACs have also been reported to enhance the SUMOylation of some substrates. SIRT1 [sirtuin (silent mating type information regulation 2 homologue)], for example, deacetylates the co-activator p300, leading to enhanced SUMOylation at the same lysine residues [135]. Recently, a number of HDACs have also been reported to possess E3 activity in the SUMO pathway. HDAC4 enhances SUMOylation of MEF2 transcription factors in addition to the nuclear receptor LXRα (liver X receptor α), and in the case of MEF2, this effect was independent of its deacetylase activity, suggesting that this enhancement was through direct E3 activity on the SUMO pathway, as opposed to via making lysine residues available through deacetylation [45,46].
Numerous HDACs are themselves also targets of SUMOylation. The deacetylase activity of both HDAC1 and HDAC4 is enhanced by SUMO modification, correlating with enhanced transcriptional repressor activity [53,138,139]. In addition, mutation of the SUMO acceptor lysine in HDAC4 increased its ability to promote SUMOylation of MEF2. This suggests that the deacetylase and SUMO E3 functions of HDACs are both regulated by SUMOylation [45]. Thus SUMOylation can directly regulate acetylation through modification of HDACs that can, in turn, regulate protein SUMOylation through their proposed E3 activity.
Interplay between phosphorylation and SUMOylation
Co-regulation of substrate proteins
Protein phosphorylation is critical in the control of multiple cellular pathways and it is now apparent that it is a regulator of SUMOylation where it can either enhance or inhibit substrate SUMOylation, depending on the substrate protein. Phosphorylation-dependent regulation of substrate SUMOylation was first shown for the nuclear protein PML where chemically induced hyperphosphorylation of PML significantly decreased its SUMOylation [91]. Similarly, phosphorylation represses the SUMOylation of the transcription factor Elk1 [140] whereas phosphorylation of HSF1 (heat-shock factor 1) is required for its SUMOylation [141].
A specific motif present in target proteins has been reported to induce phosphorylation-mediated enhancement of substrate SUMOylation. This domain, the PDSM (phosphorylation-dependent SUMOylation motif), is defined as ψKxExxSP, where ψKxE conforms to the SUMOylation consensus motif, followed by any two residues and then a proline-directed phosphorylatable serine residue. The motif is found in a large number of proteins, including the transcription factors HSF1, GATA-1 and MEF2A. In each case the negative charge conferred by phosphorylation of the serine residue facilitates SUMO modification of the target lysine [136,142,143]. A variation of the PDSM is the NDSM (negative charge-dependent SUMOylation motif) that comprises ψKxE, followed by at least two acidic residues less than ten residues C-terminal of the target lysine residue (at least one of these must be between three and six residues C-terminal of the target lysine) [144]. Thus both the PDSM and NDSM are characterized by a negative charge C-terminal to the target lysine residue, which acts to enhance SUMOylation of that lysine residue. However, whereas in the PDSM, the presence of the negative charge is provided by phosphorylation of a nearby serine residue, the negative charge in the NDSM is a property of the primary sequence of the protein. Structural analysis indicates that the negative charge contacts a cognate basic patch on Ubc9, facilitating the recruitment of Ubc9 to the substrate [144,145].
Cross-regulation of SUMOylation and phosphorylation
Similar to ubiquitination, as well as co-regulating substrate proteins, there is a complex direct interplay between the SUMOylation and phosphorylation machineries. SUMOylation can modify the kinases FAK (focal adhesion kinase) [146], GSK3β (glycogen synthase kinase 3β) [147], HIPK2 (homeodomain-interacting protein kinase 2) [148] and ERK5 (extracellular-signal-regulated kinase 5) [149]. The phosphatase PTP1B (protein tyrosine phosphatase 1B) can also be SUMOylated [150]. Thus it seems clear that SUMOylation can regulate phosphorylation dynamics through modification of the phosphorylation machinery.
Reciprocally, phosphorylation can modify components of the SUMO pathway to directly regulate SUMO conjugation to proteins or influence the consequences of protein SUMOylation. For example, the SUMO E3s Pc2, RanBP2, TOPORS and PIAS1 have each been reported to be phosphorylated [148,151–154]. The mechanisms that enable SUMOylation to be tightly controlled with a sole E1 and E2 enzyme are unclear but these results suggest that a significant proportion of the regulation may come through post-translational modification of the more numerous E3 proteins. Phosphorylation of Pc2, for example, is mediated by the kinase HIPK2, SUMOylation of which is promoted by Pc2 [148]. In response to DNA damage, HIPK2 directly phosphorylates Pc2 and this phosphorylation event is a requirement for Pc2 E3 activity to SUMOylate HIPK2, which then mediates DNA-damage-induced transcriptional repression [148]. In addition, as discussed above, the SUMO E3 TOPORS represents the first E3 enzyme that has activity in both the ubiquitin and SUMO pathways [36,41]. Phosphorylation of TOPORS at a serine residue in its N-terminus has been reported to enhance its E3 ligase activity for ubiquitin, while leaving its activity in the SUMO pathway unchanged. Thus, for this dual-function E3, phosphorylation may regulate the balance between SUMOylation and ubiquitination activities [154].
Interestingly, the SUMO-1 protein itself can be phosphorylated. Although the functional consequence of this modification has not yet been reported, it is highly conserved amongst eukaryotes [155], suggesting it may serve an integral function in SUMO-mediated cellular processes.
Recent evidence also suggests that phosphorylation may play an important role in SUMO–SIM interactions. For example, the SIM present on the SUMO E3 PIAS1 requires phosphorylation by the serine/threonine kinase CK2 to bind SUMO-1 or SUMO-2/3 [152]. In addition, phospho-regulated SIMs were shown to be present in PML, itself a SUMO substrate, and the exosome component PMSCL1 (polymyositis/scleroderma auto-antigen 1) [152], suggesting that phosphorylation may act as a general mechanism for regulating SUMO–SIM interactions, and thus potentially mediate the downstream effects of SUMOylation or the activity of SIM-containing E3 ligases.
SUMO-REGULATION OF Ubc9
Component proteins of the SUMOylation pathway themselves are also subject to regulation by SUMOylation. The sole SUMOylation E2 enzyme Ubc9 was identified as a SUMO substrate. SUMOylation of yeast Ubc9 was initially observed in in vitro SUMOylation assays at high Ubc9 concentrations, however it was unclear if this in vitro modification was physiologically relevant [156]. Subsequent proteomic studies identified SUMO modification of Ubc9 in both yeast and mammalian cells [157–159]. In vitro SUMOylation assays followed by MS were used to identify Lys14 of Ubc9 as the SUMOylation site [160]. SUMOylation occurs at a different lysine residue (Lys153) on yeast Ubc9 indicating that SUMOylation of Ubc9 may perform distinct functions in yeast and mammals [160]. SUMOylation of Ubc9 does not appear to influence Ubc9–SUMO-1 thioester formation but rather it alters the capability of Ubc9 to modify particular SUMO substrates. For example, SUMOylation of RanGAP1 by SUMOylated Ubc9 was dramatically reduced, whereas SUMO modification of the nuclear antigen Sp100 is enhanced when Ubc9 is SUMOylated. This is consistent with the idea that SUMOylation of Ubc9 influences its target specificity towards a particular subset of substrates. Interestingly, Sp100 contains a SIM, which is required for the enhanced SUMOylation of Sp100 by SUMOylated Ubc9, suggesting that SIM-mediated recruitment of SUMOylated Ubc9 to substrates may represent a novel mechanism of substrate selection [160].
In addition to direct SUMO modification of Ubc9, other proteins in the SUMO pathway, including the deSUMOylating enzyme SENP1 and, analogous to the well-documented automodification of ubiquitin E3s, the SUMOylation E3 enzymes RanBP2, Pc2, TLS, TOPORS and the PIAS proteins are also themselves substrates of SUMOylation [32,42,49,51,69,161]. SUMOylation of RanBP2 appears to be involved in SUMO paralogue specificity (see below) but the functions of SUMOylation of SENP1 and the other E3 enzymes are currently unclear.
MECHANISMS OF SUMO PARALOGUE SPECIFICITY
Although many substrate proteins can be modified by either SUMO-1 or SUMO-2/3 it is clear that many others are SUMOylated in a paralogue-specific manner [78]. As both SUMO-1 and SUMO-2/3 use identical core conjugation machinery, a major challenge is to determine how this paralogue specificity is achieved and there has been recent progress towards understanding aspects of this for some substrates (Figure 5).
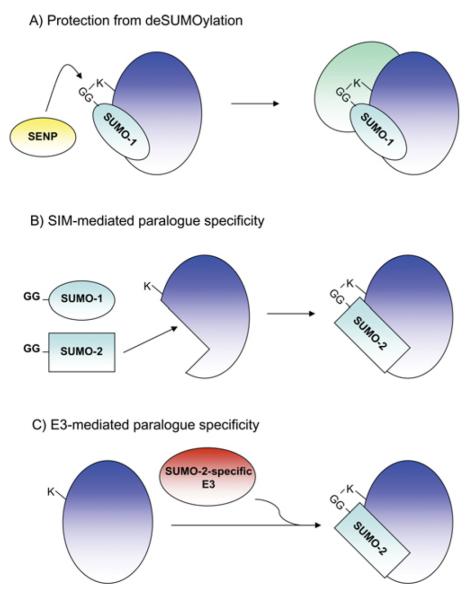
Figure 5. Mechanisms of paralogue specificity.
(A) The predominant modification by SUMO-1 of RanGAP1 has been reported to be due to the protection of RanGAP1–SUMO-1 from SENP-mediated deSUMOylation through binding of this complex to RanBP2, an interaction which only occurs weakly for RanGAP–SUMO-2, leaving it susceptible to deSUMOylation. (B) Alternatively, a growing number of SUMO substrates have been shown to contain paralogue-specific SIMs, which can mediate their paralogue-specific SUMOylation through non-covalent binding to the SUMO protein, or to SUMO-loaded Ubc9. (C) Paralogue specificity may arise through the paralogue-specific actions of E3 enzymes, such as RanBP2, which appears to preferentially conjugate SUMO-2 to substrate proteins.
Preferential deSUMOylation as a means of paralogue specificity
The nuclear pore protein RanGAP1 was the first identified SUMO substrate [7,9]. Despite the observation that RanGAP1 is equally modified by either SUMO-1 or SUMO-2 in vitro, it is predominantly modified by SUMO-1 in vivo. This paralogue preference has been attributed to specificity in the deSUMOylation of RanGAP1 [162]. SUMOylation of RanGAP1 promotes binding to the nuclear pore via an interaction with the nucleoporin RanBP2. SUMO-1 modification of RanGAP1 results in a higher-affinity interaction between SUMO-1–RanGAP1 and RanBP2 compared with SUMO-2-modified RanGAP1, which effectively protects RanGAP1 from deSUMOylation [162]. Thus SUMO-1 modification of RanGAP1 is more stable in vivo than SUMO-2-modified RanGAP1, despite the SUMO conjugation machinery showing no discernable level of specificity with respect to conjugation of SUMO-1 compared with SUMO-2/3 to RanGAP1.
SIM-mediated paralogue specificity
As discussed above, the SUMOylation of many substrates is enhanced via non-covalent interactions between the SUMO substrate and SUMO, via SIMs. For example, USP25 (ubiquitin-specific peptidase 25) binds preferentially to SUMO-2/3 in a SIM-dependent manner [163]. In addition, USP25 has been shown to be SUMOylated at a non-consensus lysine residue, which, in turn, requires an intact SIM. Similar results have also been presented in the case of the DNA helicase BLM (Bloom syndrome, RecQ helicase-like), which is SUMOylated at multiple non-consensus lysine residues; this modification is dependent on two SIM motifs in BLM. Consistent with the observed SUMO-2 covalent modification, these SIMs bind preferentially to SUMO-2 [164]. Thus preferential SIM-mediated recruitment of SUMO-loaded Ubc9 may represent a mechanism of paralogue specificity due to the ability of different SIMs to discriminate between SUMO-1 compared with SUMO-2/3.
E3-mediated paralogue specificity
E3-mediated SUMO-1 or SUMO-2/3 selection has also been suggested as a mechanism of paralogue specificity [52]. The nucleoporin RanBP2 is a capable of binding both Ubc9 and SUMO-1 at different sites. Disruption of RanBP2–Ubc9 binding inhibited the ability of RanBP2 to conjugate SUMO-2 to Sp100 and PML, but had no effect on its ability to conjugate SUMO-1 [52]. This was attributed to the ability of RanBP2 to bind and conjugate SUMO-1 independently of any interaction with Ubc9. Like many E3 enzymes of the SUMO and ubiquitin pathways, RanBP2 is also capable of autoSUMOylation [51]. Essentially, this process can be viewed as a competition between automodification and substrate modification. Thus a consequence of direct SUMO-1 binding by RanBP2 is its enhanced autoSUMOylation with SUMO-1, leading to substrate proteins being more efficiently modified by SUMO-2. Another case of E3-mediated paralogue preference has been reported for the PIASy, which appears to preferentially conjugate SUMO-2 to the transcription factor LEF-1; however, the mechanism by which this occurs is currently unclear [35].
CONCLUDING REMARKS AND FUTURE DIRECTIONS
It is clear that protein SUMOylation is a fundamentally important signalling pathway in all eukaryotic cells. There have been rapid advances in our understanding of the mechanisms and consequences of SUMOylation and recent work has begun to shed light on the complex regulation and the mechanisms of paralogue specificity. Future work will continue to delineate and refine the inter-relationships between SUMO and other post-translational modifications that act in combination to control all aspects of cell differentiation, development and function.
Acknowledgments
FUNDING We are grateful to the Medical Research Council, the Wellcome Trust and the European Research Council for financial support for our laboratory.
Abbreviations used
- ATM
ataxia telangiectasia mutated
- BLM
Bloom syndrome, RecQ helicase-like
- CoREST
co-repressor for RE1-silencing transcription factor
- DAXX
death-domain-associated protein
- Elk1
ETS (E twenty-six)-like kinase 1
- GluR6
glutamate receptor subunit 6
- HDAC
histone deacetylase
- HECT
homologous with E6-associated protein C-terminus
- HIF1α
hypoxia-inducible factor 1α
- HIPK2
homeodomain-interacting protein kinase 2
- HSF1
heat-shock factor 1
- IκB
inhibitory κB
- IKK
IκB kinase
- LEF-1
lymphoid enhancer-binding factor 1
- MEF2
myocyte enhancer factor 2
- MUL1
mitochondrial E3 ubiquitin ligase 1
- NDSM
negative charge-dependent SUMOylation motif
- NFκB
nuclear factor κB
- NEMO
NF-κB essential modulator
- Pc
polycomb
- PDSM
phosphorylation-dependent SUMOylation motif
- PIAS
protein inhibitor of activated STAT (signal transducer and activator of transcription)
- PML
promyelocytic leukaemia
- RanBP
Ran-binding protein
- RanGAP1
Ran-GTPase-activating protein 1
- RNF4
ring finger protein4
- SUMO
small ubiquitin-like modifier
- SAE
SUMO-activating enzyme
- SENP
sentrin/SUMO-specific protease
- SIM
SUMO-interacting motif
- Sp3
stimulating protein 3
- SP-RING
Siz/PIAS-RING domain
- TDG
thymine–DNA glycosylase
- TLS
translocated in liposarcoma
- TRAF7
tumour-necrosis-factor-associated protein 7
- TOPORS
topoisomerase I-binding arginine/serine-rich
- Ubc9
ubiquitin-conjugating 9
REFERENCES
- 1.Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y. Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol. Cell. Biol. 1999;19:8660–8672. doi: 10.1128/mcb.19.12.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 3.Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics. 1996;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- 5.Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 6.Okura T, Gong L, Kamitani T, Wada T, Okura I, Wei CF, Chang HM, Yeh ET. Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J. Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- 7.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapenta V, Chiurazzi P, van der Spek P, Pizzuti A, Hanaoka F, Brahe C. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21qter and defines a novel gene family. Genomics. 1997;40:362–366. doi: 10.1006/geno.1996.4556. [DOI] [PubMed] [Google Scholar]
- 9.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 10.Kamitani T, Kito K, Nguyen HP, Fukuda-Kamitani T, Yeh ET. Characterization of a second member of the sentrin family of ubiquitin-like proteins. J. Biol. Chem. 1998;273:11349–11353. doi: 10.1074/jbc.273.18.11349. [DOI] [PubMed] [Google Scholar]
- 11.Kamitani T, Nguyen HP, Kito K, Fukuda-Kamitani T, Yeh ET. Covalent modification of PML by the sentrin family of ubiquitin-like proteins. J. Biol. Chem. 1998;273:3117–3120. doi: 10.1074/jbc.273.6.3117. [DOI] [PubMed] [Google Scholar]
- 12.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 13.Yang M, Hsu CT, Ting CY, Liu LF, Hwang J. Assembly of a polymeric chain of SUMO1 on human topoisomerase I in vitro. J.Biol.Chem. 2006;281:8264–8274. doi: 10.1074/jbc.M510364200. [DOI] [PubMed] [Google Scholar]
- 14.Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei W, Yang P, Pang J, Zhang S, Wang Y, Wang MH, Dong Z, She JX, Wang CY. A stress-dependent SUMO4 sumoylation of its substrate proteins. Biochem. Biophys. Res. Commun. 2008;375:454–459. doi: 10.1016/j.bbrc.2008.08.028. [DOI] [PubMed] [Google Scholar]
- 16.Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 2004;279:27233–27238. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- 17.Guo D, Li M, Zhang Y, Yang P, Eckenrode S, Hopkins D, Zheng W, Purohit S, Podolsky RH, Muir A, et al. A functional variant of SUMO4, a new IκBα modifier, is associated with type 1 diabetes. Nat. Genet. 2004;36:837–841. doi: 10.1038/ng1391. [DOI] [PubMed] [Google Scholar]
- 18.Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 2005;337:517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 19.Gong L, Li B, Millas S, Yeh ET. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 1999;448:185–189. doi: 10.1016/s0014-5793(99)00367-1. [DOI] [PubMed] [Google Scholar]
- 20.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 21.Gong L, Kamitani T, Fujise K, Caskey LS, Yeh ET. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 1997;272:28198–28201. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- 22.Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 23.Schwarz SE, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. U.S.A. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitoh H, Sparrow DB, Shiomi T, Pu RT, Nishimoto T, Mohun TJ, Dasso M. Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 1998;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- 25.Lee GW, Melchior F, Matunis MJ, Mahajan R, Tian Q, Anderson P. Modification of Ran GTPase-activating protein by the small ubiquitin-related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J. Biol. Chem. 1998;273:6503–6507. doi: 10.1074/jbc.273.11.6503. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 27.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, He Y, Qiang B, Yuan J, Peng X, Pan XM. A novel method for high accuracy sumoylation site prediction from protein sequences. BMC Bioinformatics. 2008;9:8. doi: 10.1186/1471-2105-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickart CM. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 30.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 31.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotaja N, Karvonen U, Janne OA, Palvimo JJ. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishida T, Yasuda H. PIAS1 and PIASxα function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 2002;277:41311–41317. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- 35.Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001;15:3088–3103. doi: 10.1101/gad.944801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weger S, Hammer E, Heilbronn R. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 2005;579:5007–5012. doi: 10.1016/j.febslet.2005.07.088. [DOI] [PubMed] [Google Scholar]
- 37.Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ. Nse2, a component of the Smc5–6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajendra R, Malegaonkar D, Pungaliya P, Marshall H, Rasheed Z, Brownell J, Liu LF, Lutzker S, Saleem A, Rubin EH. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J. Biol. Chem. 2004;279:36440–36444. doi: 10.1074/jbc.C400300200. [DOI] [PubMed] [Google Scholar]
- 42.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 43.Yang SH, Sharrocks AD. The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO-interaction motif. Mol. Cell Biol. 2010 doi: 10.1128/MCB.01510-09. doi:10.1128/MCB.01510–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merrill JC, Melhuish TA, Kagey MH, Yang SH, Sharrocks AD, Wotton D. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS One. 5:e8794. doi: 10.1371/journal.pone.0008794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregoire S, Yang XJ. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell. Biol. 2005;25:2273–2287. doi: 10.1128/MCB.25.6.2273-2287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JH, Park SM, Kim OS, Lee CS, Woo JH, Park SJ, Joe EH, Jou I. Differential SUMOylation of LXRα and LXRβ mediates transrepression of STAT1 inflammatory signaling in IFN-γ -stimulated brain astrocytes. Mol. Cell. 2009;35:806–817. doi: 10.1016/j.molcel.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 47.Gao C, Ho CC, Reineke E, Lam M, Cheng X, Stanya KJ, Liu Y, Chakraborty S, Shih HM, Kao HY. Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation. Mol. Cell. Biol. 2008;28:5658–5667. doi: 10.1128/MCB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh SM, Liu Z, Okada M, Jang SW, Liu X, Chan CB, Luo H, Ye K. Ebp1 sumoylation, regulated by TLS/FUS E3 ligase, is required for its anti-proliferative activity. Oncogene. 2010;29:1017–1030. doi: 10.1038/onc.2009.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morita Y, Kanei-Ishii C, Nomura T, Ishii S. TRAF7 sequesters c-Myb to the cytoplasm by stimulating its sumoylation. Mol. Biol. Cell. 2005;16:5433–5444. doi: 10.1091/mbc.E05-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 52.Tatham MH, Kim S, Jaffray E, Song J, Chen Y, Hay RT. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat. Struct. Mol. Biol. 2005;12:67–74. doi: 10.1038/nsmb878. [DOI] [PubMed] [Google Scholar]
- 53.Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, Mathieu M, Harel-Bellan A, Kouzarides T, Melchior F, Dejean A. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 2002;21:2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pichler A, Knipscheer P, Saitoh H, Sixma TK, Melchior F. The RanBP2 SUMO E3 ligase is neither HECT- nor RING-type. Nat. Struct. Mol. Biol. 2004;11:984–991. doi: 10.1038/nsmb834. [DOI] [PubMed] [Google Scholar]
- 55.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO–RanGAP1–Ubc9–Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 57.Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 59.Yeh ET. SUMOylation and De-SUMOylation: wrestling with life’s processes. J. Biol. Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 61.Gong L, Millas S, Maul GG, Yeh ET. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- 62.Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- 63.Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nishida T, Tanaka H, Yasuda H. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 2000;267:6423–6427. doi: 10.1046/j.1432-1327.2000.01729.x. [DOI] [PubMed] [Google Scholar]
- 65.Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol. Cell. Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J. Biol. Chem. 2006;281:15869–15877. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- 67.Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, Azuma Y, Wilkinson KD, Dasso M. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J. Cell Biol. 2006;174:939–949. doi: 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen LN, Geoffroy MC, Jaffray EG, Hay RT. Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem. J. 2009;421:223–230. doi: 10.1042/BJ20090246. [DOI] [PubMed] [Google Scholar]
- 69.Bailey D, O’Hare P. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J. Biol. Chem. 2004;279:692–703. doi: 10.1074/jbc.M306195200. [DOI] [PubMed] [Google Scholar]
- 70.Kuo ML, den Besten W, Thomas MC, Sherr CJ. Arf-induced turnover of the nucleolar nucleophosmin-associated SUMO-2/3 protease Senp3. Cell Cycle. 2008;7:3378–3387. doi: 10.4161/cc.7.21.6930. [DOI] [PubMed] [Google Scholar]
- 71.Huang C, Han Y, Wang Y, Sun X, Yan S, Yeh ET, Chen Y, Cang H, Li H, Shi G, et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 2009;28:2748–2762. doi: 10.1038/emboj.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hayashi T, Seki M, Maeda D, Wang W, Kawabe Y, Seki T, Saitoh H, Fukagawa T, Yagi H, Enomoto T. Ubc9 is essential for viability of higher eukaryotic cells. Exp. Cell Res. 2002;280:212–221. doi: 10.1006/excr.2002.5634. [DOI] [PubMed] [Google Scholar]
- 73.Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 74.Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313:1751. doi: 10.1126/science.1128406. [DOI] [PubMed] [Google Scholar]
- 75.Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. Sumo-1 function is dispensable in normal mouse development. Mol. Cell. Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J. Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- 77.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 78.Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 79.Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bossis G, Melchior F. SUMO: regulating the regulator. Cell Div. 2006;1:13. doi: 10.1186/1747-1028-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Agbor TA, Taylor CT. SUMO, hypoxia and the regulation of metabolism. Biochem. Soc. Trans. 2008;36:445–448. doi: 10.1042/BST0360445. [DOI] [PubMed] [Google Scholar]
- 82.Cimarosti H, Henley JM. Investigating the mechanisms underlying neuronal death in ischemia using in vitro oxygen-glucose deprivation: potential involvement of protein SUMOylation. Neuroscientist. 2008;14:626–636. doi: 10.1177/1073858408322677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tempe D, Piechaczyk M, Bossis G. SUMO under stress. Biochem. Soc. Trans. 2008;36:874–878. doi: 10.1042/BST0360874. [DOI] [PubMed] [Google Scholar]
- 84.Yang W, Sheng H, Homi HM, Warner DS, Paschen W. Cerebral ischemia/stroke and small ubiquitin-like modifier (SUMO) conjugation – a new target for therapeutic intervention? J. Neurochem. 2008;106:989–999. doi: 10.1111/j.1471-4159.2008.05404.x. [DOI] [PubMed] [Google Scholar]
- 85.Martin S, Wilkinson KA, Nishimune A, Henley JM. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2007;8:948–959. doi: 10.1038/nrn2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends. Biochem. Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 87.Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 2009;10:564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 88.Finley D, Chau V. Ubiquitination. Annu. Rev. Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- 89.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 90.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 91.Muller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H. Arsenic degrades PML or PML–RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 93.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 94.Weisshaar SR, Keusekotten K, Krause A, Horst C, Springer HM, Gottsche K, Dohmen RJ, Praefcke GJ. Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 2008;582:3174–3178. doi: 10.1016/j.febslet.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 95.Kerscher O. SUMO junction - what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell. Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Minty A, Dumont X, Kaghad M, Caput D. Covalent modification of p73α by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 2000;275:36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 98.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 100.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 101.Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 103.Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 105.Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol. Cell. 2009;34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 107.Pichler A, Knipscheer P, Oberhofer E, van Dijk WJ, Korner R, Olsen JV, Jentsch S, Melchior F, Sixma TK. SUMO modification of the ubiquitin-conjugating enzyme E2–25K. Nat. Struct. Mol. Biol. 2005;12:264–269. doi: 10.1038/nsmb903. [DOI] [PubMed] [Google Scholar]
- 108.Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 109.Hay RT. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 110.Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–325. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 113.Ross S, Best JL, Zon LI, Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell. 2002;10:831–842. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- 114.Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT. P300 transcriptional repression is mediated by SUMO modification. Mol. Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 115.Kuo HY, Chang CC, Jeng JC, Hu HM, Lin DY, Maul GG, Kwok RP, Shih HM. SUMO modification negatively modulates the transcriptional activity of CREB-binding protein via the recruitment of Daxx. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16973–16978. doi: 10.1073/pnas.0504460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 118.Yang W, Sheng H, Warner DS, Paschen W. Transient focal cerebral ischemia induces a dramatic activation of small ubiquitin-like modifier conjugation. J. Cereb. Blood Flow Metab. 2008;28:892–896. doi: 10.1038/sj.jcbfm.9600601. [DOI] [PubMed] [Google Scholar]
- 119.Yang W, Sheng H, Warner DS, Paschen W. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J. Cereb. Blood Flow Metab. 2008;28:269–279. doi: 10.1038/sj.jcbfm.9600523. [DOI] [PubMed] [Google Scholar]
- 120.Loftus LT, Gala R, Yang T, Jessick VJ, Ashley MD, Ordonez AN, Thompson SJ, Simon RP, Meller R. Sumo-2/3-ylation following in vitro modeled ischemia is reduced in delayed ischemic tolerance. Brain Res. 2009;1272:71–80. doi: 10.1016/j.brainres.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cimarosti H, Lindberg C, Bomholt SF, Ronn LC, Henley JM. Increased protein SUMOylation following focal cerebral ischemia. Neuropharmacology. 2008;54:280–289. doi: 10.1016/j.neuropharm.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 122.Lee Y. Ja, Castri P, Bembry J, Maric D, Auh S, Hallenbeck JM. SUMOylation participates in induction of ischemic tolerance. J. Neurochem. 2009;109:257–267. doi: 10.1111/j.1471-4159.2009.05957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci. Signaling. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 124.Ulrich HD. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 2005;15:525–532. doi: 10.1016/j.tcb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 125.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 126.Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu. Rev. Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 127.Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell Biol. 2007;9:428–435. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- 128.Chiocca S, Baker A, Cotten M. Identification of a novel antiapoptotic protein, GAM-1, encoded by the CELO adenovirus. J. Virol. 1997;71:3168–3177. doi: 10.1128/jvi.71.4.3168-3177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Glotzer JB, Saltik M, Chiocca S, Michou AI, Moseley P, Cotten M. Activation of heat-shock response by an adenovirus is essential for virus replication. Nature. 2000;407:207–211. doi: 10.1038/35025102. [DOI] [PubMed] [Google Scholar]
- 130.Boggio R, Passafaro A, Chiocca S. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J. Biol. Chem. 2007;282:15376–15382. doi: 10.1074/jbc.M700889200. [DOI] [PubMed] [Google Scholar]
- 131.Um JW, Min DS, Rhim H, Kim J, Paik SR, Chung KC. Parkin ubiquitinates and promotes the degradation of RanBP2. J. Biol. Chem. 2006;281:3595–3603. doi: 10.1074/jbc.M504994200. [DOI] [PubMed] [Google Scholar]
- 132.Um JW, Chung KC. Functional modulation of parkin through physical interaction with SUMO-1. J. Neurosci. Res. 2006;84:1543–1554. doi: 10.1002/jnr.21041. [DOI] [PubMed] [Google Scholar]
- 133.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 134.Sapetschnig A, Rischitor G, Braun H, Doll A, Schergaut M, Melchior F, Suske G. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 2002;21:5206–5215. doi: 10.1093/emboj/cdf510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J. Biol. Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 136.Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 137.Stankovic-Valentin N, Deltour S, Seeler J, Pinte S, Vergoten G, Guerardel C, Dejean A, Leprince D. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol. Cell. Biol. 2007;27:2661–2675. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.David G, Neptune MA, DePinho RA. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem. 2002;277:23658–23663. doi: 10.1074/jbc.M203690200. [DOI] [PubMed] [Google Scholar]
- 139.Cheng J, Wang D, Wang Z, Yeh ET. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol. Cell. Biol. 2004;24:6021–6028. doi: 10.1128/MCB.24.13.6021-6028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang SH, Jaffray E, Senthinathan B, Hay RT, Sharrocks AD. SUMO and transcriptional repression: dynamic interactions between the MAP kinase and SUMO pathways. Cell Cycle. 2003;2:528–530. doi: 10.4161/cc.2.6.597. [DOI] [PubMed] [Google Scholar]
- 141.Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 2003;23:2953–2968. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gregoire S, Tremblay AM, Xiao L, Yang Q, Ma K, Nie J, Mao Z, Wu Z, Giguere V, Yang XJ. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J. Biol. Chem. 2006;281:4423–4433. doi: 10.1074/jbc.M509471200. [DOI] [PubMed] [Google Scholar]
- 143.Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U.S.A. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mohideen F, Capili AD, Bilimoria PM, Yamada T, Bonni A, Lima CD. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat. Struct. Mol. Biol. 2009;16:945–952. doi: 10.1038/nsmb.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kadare G, Toutant M, Formstecher E, Corvol JC, Carnaud M, Boutterin MC, Girault JA. PIAS1-mediated sumoylation of focal adhesion kinase activates its autophosphorylation. J. Biol. Chem. 2003;278:47434–47440. doi: 10.1074/jbc.M308562200. [DOI] [PubMed] [Google Scholar]
- 147.Eun Jeoung L, Sung Hee H, Jaesun C, Sung Hwa S, Kwang Hum Y, Min Kyoung K, Tae Yoon P, Sang Sun K. Regulation of glycogen synthase kinase 3β functions by modification of the small ubiquitin-like modifier. Open Biochem. J. 2008;2:67–76. doi: 10.2174/1874091X00802010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Roscic A, Moller A, Calzado MA, Renner F, Wimmer VC, Gresko E, Ludi KS, Schmitz ML. Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol. Cell. 2006;24:77–89. doi: 10.1016/j.molcel.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 149.Woo CH, Shishido T, McClain C, Lim JH, Li JD, Yang J, Yan C, Abe J. Extracellular signal-regulated kinase 5 SUMOylation antagonizes shear stress-induced antiinflammatory response and endothelial nitric oxide synthase expression in endothelial cells. Circ. Res. 2008;102:538–545. doi: 10.1161/CIRCRESAHA.107.156877. [DOI] [PubMed] [Google Scholar]
- 150.Dadke S, Cotteret S, Yip SC, Jaffer ZM, Haj F, Ivanov A, Rauscher F, III, Shuai K, Ng T, Neel BG, Chernoff J. Regulation of protein tyrosine phosphatase 1B by sumoylation. Nat. Cell Biol. 2007;9:80–85. doi: 10.1038/ncb1522. [DOI] [PubMed] [Google Scholar]
- 151.Favreau C, Worman HJ, Wozniak RW, Frappier T, Courvalin JC. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry. 1996;35:8035–8044. doi: 10.1021/bi9600660. [DOI] [PubMed] [Google Scholar]
- 152.Stehmeier P, Muller S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell. 2009;33:400–409. doi: 10.1016/j.molcel.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 153.Liu B, Yang Y, Chernishof V, Loo RR, Jang H, Tahk S, Yang R, Mink S, Shultz D, Bellone CJ, et al. Proinflammatory stimuli induce IKKα-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell. 2007;129:903–914. doi: 10.1016/j.cell.2007.03.056. [DOI] [PubMed] [Google Scholar]
- 154.Park HJ, Zheng H, Kulkarni D, Kerrigan J, Pungaliya P, Saleem A, Rubin EH. Identification of phosphorylation sites of TOPORS and a role for serine 98 in the regulation of ubiquitin but not SUMO E3 ligase activity. Biochemistry. 2008;47:13887–13896. doi: 10.1021/bi801904q. [DOI] [PubMed] [Google Scholar]
- 155.Matic I, Macek B, Hilger M, Walther TC, Mann M. Phosphorylation of SUMO-1 occurs in vivo and is conserved through evolution. J. Proteome Res. 2008;7:4050–4057. doi: 10.1021/pr800368m. [DOI] [PubMed] [Google Scholar]
- 156.Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J. Biol. Chem. 2002;277:47938–47945. doi: 10.1074/jbc.M207442200. [DOI] [PubMed] [Google Scholar]
- 157.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., III Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 158.Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 2004;279:32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhao Y, Kwon SW, Anselmo A, Kaur K, White MA. Broad spectrum identification of cellular small ubiquitin-related modifier (SUMO) substrate proteins. J. Biol. Chem. 2004;279:20999–21002. doi: 10.1074/jbc.M401541200. [DOI] [PubMed] [Google Scholar]
- 160.Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, Johnson ES, Mann M, Sixma TK, Pichler A. Ubc9 sumoylation regulates SUMO target discrimination. Mol. Cell. 2008;31:371–382. doi: 10.1016/j.molcel.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 161.Weger S, Hammer E, Engstler M. The DNA topoisomerase I binding protein topors as a novel cellular target for SUMO-1 modification: characterization of domains necessary for subcellular localization and sumolation. Exp. Cell Res. 2003;290:13–27. doi: 10.1016/s0014-4827(03)00292-1. [DOI] [PubMed] [Google Scholar]
- 162.Zhu S, Goeres J, Sixt KM, Bekes M, Zhang XD, Salvesen GS, Matunis MJ. Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1. Mol. Cell. 2009;33:570–580. doi: 10.1016/j.molcel.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 164.Zhu J, Zhu S, Guzzo CM, Ellis NA, Sung KS, Choi CY, Matunis MJ. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem. 2008;283:29405–29415. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]