Abstract
Ubiquitin (Ub) and ubiquitin-like (Ubl) modifiers such as SUMO mediate signal transduction through post-translational modification of substrate proteins in pathways that control differentiation, apoptosis, the cell cycle, and responses to stress such as the DNA damage response. In yeast, the proliferating cell nuclear antigen PCNA is modified by ubiquitin in response to DNA damage and by SUMO during S-phase. While Ub-PCNA can signal for recruitment of translesion DNA polymerases, SUMO-PCNA signals for recruitment of the anti-recombinogenic DNA helicase Srs2. It remains unclear how receptors such as Srs2 specifically recognize substrates after conjugation to Ub/Ubls. Here we show through structural, biochemical and functional studies that the Srs2 C-terminal domain harbors tandem receptor motifs that interact independently with PCNA and SUMO and that both motifs are required to specifically recognize SUMO-PCNA. The mechanism presented herein is pertinent to understanding how other receptors specifically recognize Ub/Ubl-modified substrates to facilitate signal transduction.
Keywords: PCNA, Srs2, SUMO, Signal transduction, DNA damage bypass
Post-translational modification of substrate proteins by ubiquitin (Ub) and ubiquitin-like (Ubl) modifiers such as SUMO mediates signal transduction in pathways including those that control differentiation, apoptosis, the cell cycle, and stress responses such as the DNA damage response1,2. For signal transduction to occur, downstream factors must specifically recognize the Ub/Ubl-modified substrate. While interactions between Ub/Ubl binding domains and Ub/Ubls have been addressed3, there is a paucity of data addressing how receptors specifically recognize Ub/Ubl conjugated substrates.
Proliferating cell nuclear antigen (PCNA) is a member of the β-clamp family of DNA sliding clamps and facilitates DNA replication as a processivity factor and mobile scaffold to recruit proteins that regulate DNA metabolism4. The PCNA homotrimeric ring is formed by protomers that contain two topologically related domains (domains I and II) connected by the interdomain connector loop (IDCL)5. Many proteins interact with PCNA at the interface between the IDCL and domains I and II with a motif coined the PCNA-interacting protein box (PIP box)6–8. Post-translational modification of PCNA-interacting proteins, as illustrated by p21 phosphorylation9, or of PCNA itself has been proposed as a means to dynamically modulate interactions with PCNA4.
Replication fork stalling triggers mono- or poly-ubiquitination of PCNA on an evolutionarily conserved lysine (K164 in budding yeast) leading to activation of error-prone or error-free post-replication repair (PRR) pathways, respectively10,11. MonoUb-PCNA signals for recruitment of translesion DNA polymerases such as Polη and Polζ10,11. PCNA K164 is also modified by SUMO in budding yeast during S-phase as is K127, albeit to lesser extent10. SUMO-PCNA signals for recruitment of Srs2 (refs. 12,13), a UvrD-like helicase that exerts antirecombinogenic functions by disrupting Rad51 presynaptic filaments14,15. Srs2 recruitment to SUMO-PCNA inhibits a Rad52-dependent recombinational repair pathway and deletion of Srs2 suppresses UV and chemically induced damage sensitivity of PRR deficient strains12,13,16. K127 and K164 SUMO modifications are redundant because either can function to recruit Srs2 (refs. 12,13).
Srs2 harbors a C-terminal SUMO interaction motif (SIM) that was shown to enhance interaction with SUMO-PCNA12, but it is unclear how Srs2 specifically recognizes SUMO-PCNA as it was not known to contain a canonical PCNA interaction motif. At least two models can be envisioned. In a dual interaction model distinct elements within Srs2 independently recognize PCNA and SUMO. This model is distinct, albeit not mutually exclusive, from one where the Srs2 SIM is recognized by a composite surface between SUMO and PCNA. This mode of interaction is illustrated by SUMO-modified thymine DNA glycosylase (TDG) wherein the SIM is buried in the interface between TDG and SUMO17. Srs2 recruitment to SUMO-PCNA has been touted as an ideal system for analysis because the receptor (Srs2), the SUMO-modified substrate (PCNA) and the consequence of the interaction are known12,18. Here we define determinants required for specific recognition of SUMO-PCNA by Srs2 to gain insight to how other Ubl-modified substrates may be recognized by their cognate receptors.
Srs2 interacts with SUMO and PCNA
A 139 residue C-terminal fragment of Srs2 (Srs21036–1174) was shown to interact with SUMO and PCNA by yeast two-hybrid analysis12. We found a smaller domain encompassing residues 1107–1174 that co-migrated with PCNA or SUMO using analytical gel filtration (Supplemental Fig. 1). Interactions were quantified using a fluorescence polarization (FP) assay with Srs21107–1174 N-terminally coupled to BODIPY-FL. Srs21107–1174 interacted with GST-SUMO with an apparent dissociation constant (Kd) of 877 +/− 59 nM and with PCNA with a Kd of 169 +/− 26 nM (Fig. 1; Supplemental Table 1, Fig. 2 and 3), values similar to those observed for other SIM/SUMO19,20 and PIP:PCNA interactions7,21. These data suggest that Srs21107–1174 harbors motifs that interact independently with SUMO and PCNA.
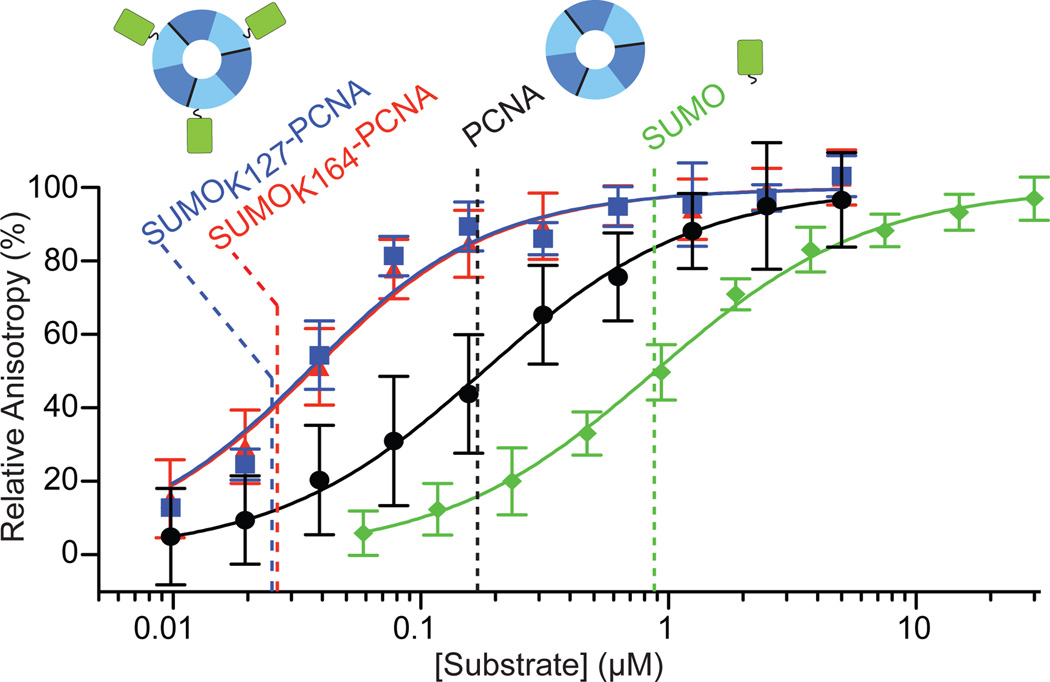
Figure 1. Srs2 C-terminal domain interacts with PCNA, SUMO, and SUMO-PCNA.
Fluorescence anisotropy curves and calculated Kd values (dashed lines) for GST-SUMO (green), PCNA (black), SUMOK127-PCNA (blue) and SUMOK164-PCNA (red) titrations against 20 nM BODIPY-FL Srs21107–1174. Cartoons of SUMO (green; right), PCNA (trimer, domain I (dark blue), domain II (light blue); middle) and SUMO-PCNA (left) above dashed lines. Assays conducted in triplicate. Error bars are ±1 standard deviation; n = 12 for SUMO, n = 12 for PCNA, n = 9 for SUMOK164-PCNA and n= 3 for SUMOK127-PCNA. See Methods.
SUMO modification of PCNA was shown to enhance interaction with Srs2 based on enrichment of SUMO-PCNA over PCNA in pull-downs12,13. To address this with a purified system we reconstituted SUMOK164-PCNA and SUMOK127-PCNA using the Siz1 E3 ligase and mutations in PCNA (K127G; K164R) to direct SUMO modification on K164 or K127, respectively22,23. K127 is conserved as glycine in most metazoan PCNA family members. FP assays show that Srs21107–1174 interacts with SUMOK164-PCNA and SUMOK127-PCNA with Kd values of 26.3 +/− 3.1 nM and 25.1 +/− 4.9 nM, respectively (Fig. 1; Supplemental Table 1, Fig. 2 and 3), values 7-fold and 33-fold tighter than for PCNA and GST-SUMO, respectively.
Structures of SUMO conjugated PCNA
A structure of reconstituted SUMOK164-PCNA was determined to a resolution of 2.6 Å (Fig. 2a; Supplemental Table 2 and Fig. 5a). PCNA adopts the archetypal trimeric ring5 and electron density and the proximity of PCNA K164 and SUMO G98 are consistent with SUMO being conjugated to PCNA K164 (Supplemental Fig. 4a and 4c). The conformation observed for SUMO and its contacts to a PCNA loop between residues 184–198 (Supplemental Fig. 5a) are similar to another structure obtained by expressing PCNA as two self-assembling polypeptides split at residues 163 and 165 with SUMO introduced as a N-terminal fusion to the C-terminal peptide24.
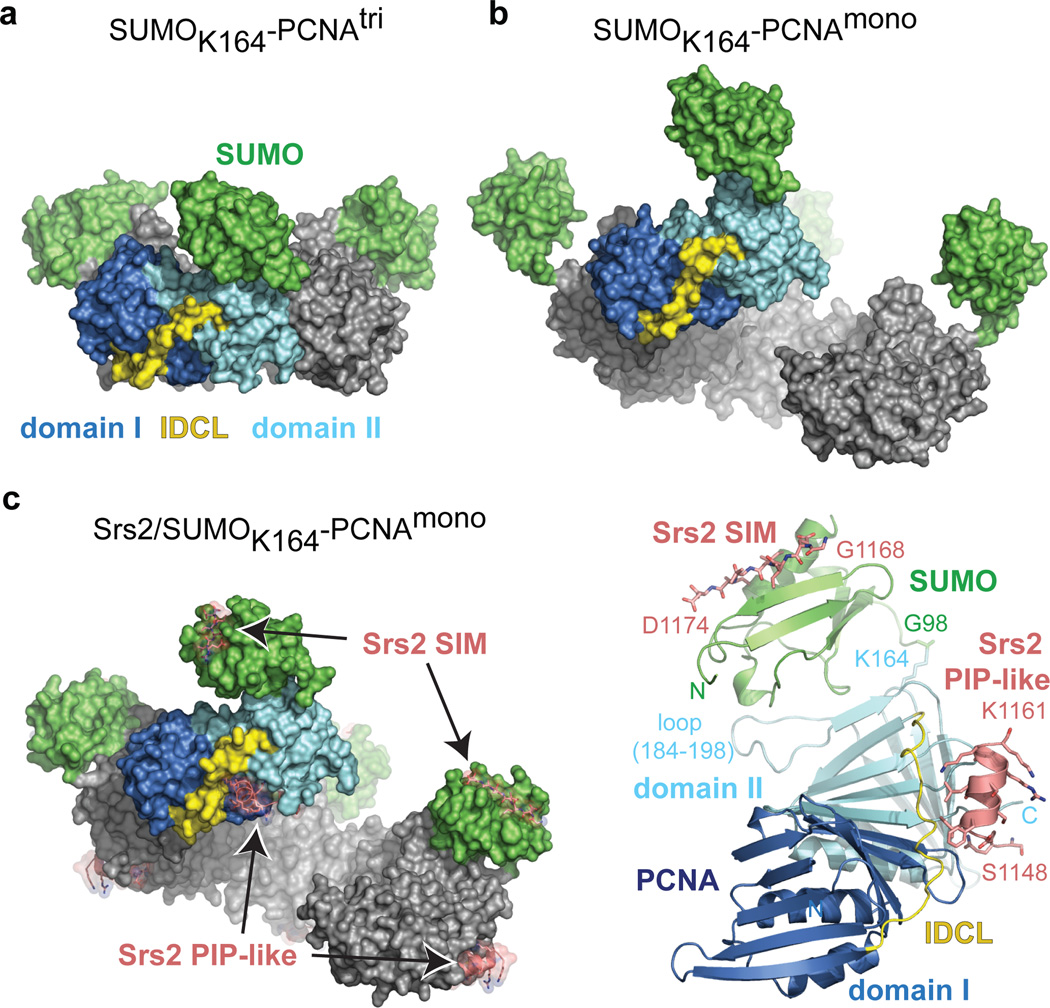
Figure 2. Structures of SUMO-PCNA alone and in complex with Srs2.
a, Surface representation for SUMOK164-PCNAtri. PCNA domain I (dark blue), domain II (light blue), IDCL (yellow) and SUMO (green) are labeled. b, SUMOK164-PCNAmono as in a. Four protomers oriented to visualize the ‘open ring’. c, Srs21107–1174/SUMOK164-PCNAmono as in a and b (left). Srs2 SIM and PIP-like motifs labeled (pink; stick and transparent surface). Cartoon representation for one protomer of Srs21107–1174/SUMOK164-PCNAmono (right) colored as in c. PCNA N- and C- termini, loop 184–198, and K164 and SUMO N-terminus and G98 are labeled. Srs2 SIM and PIP-like motifs labeled with single amino acid code and number to indicate the termini of each motif.
We found that N-ethylmaleimide (NEM) chemically modifies PCNA on cysteine side chains 22 and 81 (Supplemental Fig. 4d and 6) causing PCNA to run on gel filtration at an apparent size that is three times smaller than the PCNA trimer (Supplemental Fig. 7a). We term this preparation PCNAmono. The structure of SUMOK164-PCNAmono was determined at 2.8 Å resolution (Fig. 2b; Supplemental Table 2 and Fig. 5a). Although a monomer on gel filtration, SUMOK164-PCNAmono crystallizes as a continuous right-handed helix with four PCNA monomers comprising a single turn (Fig. 2b) through formation of a PCNA:PCNA interface similar to that observed in SUMOK164-PCNAtri and PCNAtri(ref. 5) (Supplemental Fig. 6). The right-handed helical configuration is in agreement with MD simulations of dimeric PCNA25, cryo-EM studies of the archaeal clamp-clamp loader-DNA complex26 and recent studies in which the T4 clamp loader was crystallized with an open ring27 (Supplemental Fig. 8). In this structure SUMO makes few contacts to PCNA and adopts a different orientation compared to SUMOK164-PCNAtri (Supplemental Fig. 5a). This suggests SUMO can achieve different conformations in solution although this conformation is likely stabilized by lattice contacts (Supplemental Fig. 5b and 5c).
Structure of Srs2/SUMO-PCNA
Srs21107–1174 interacts with SUMOK164-PCNAmono and SUMOK164-PCNAtri by analytical gel filtration (Supplemental Fig. 7b) and binds SUMOK164-PCNAmono and SUMOK127-PCNAmono with apparent Kd values of 79 +/− 18 nM and 14.0 +/− 3.9 nM, 3-fold worse and < 2-fold better than SUMOK164-PCNAtri or SUMOK127-PCNAtri, respectively. Our data show that Srs2 interacts with NEM treated PCNAmono 6.5-fold worse than PCNAtri. We posit that outside of its conformational constraints in the trimer PCNAmono is more flexible and this likely results in conformational heterogeneity in the PIP-box binding site. In spite of this difference Srs2 does exhibit specificity in binding SUMO-PCNA because interactions with SUMOK164-PCNAmono and SUMOK127-PCNAmono are 14-fold and 78-fold tighter than PCNAmono, respectively (Supplemental Table 1), suggesting that SUMO-PCNAmono is a reasonable surrogate for SUMO-PCNAtri in its interactions with Srs2. Furthermore, these data suggest that Srs2 recognizes SUMO-PCNA within a single SUMO-PCNA protomer and does not bridge protomers in the PCNA trimer. This hypothesis is further supported by distance constraints between the PIP-box binding site and lysine residues as intra-protomer distances to K127 or K164 are 2–3 times shorter than distances to K127 or K164 in adjacent protomers (see below).
Complexes between Srs21107–1174 and SUMOK164-PCNAtri or SUMOK164-PCNAmono were purified (Supplemental Fig. 7b). We obtained crystals and determined the structure for Srs2/SUMOK164-PCNAmono at 2.9 Å resolution (Fig. 2c and Supplemental Table 2 and Fig. 4c). In this structure, PCNA adopts a right-handed helical conformation similar to apo-SUMOK164-PCNAmono (PCNA protomers align to an RMSD of 0.98 Å) while SUMO adopts a conformation similar to SUMOK164-PCNAtri (Supplemental Fig. 5a). Electron density for Srs2 residues 1148–1161 is located in the interface between PCNA domains I, II and the IDCL, and electron density for Srs2 residues 1168–1174 is located adjacent to SUMO β2 and α1 (Fig. 2c, Supplemental Fig. 4b). Electron density for Srs2 residues 1107–1147 or 1162–1167 is not observed in our structure. To eliminate the contribution of residues N-terminal to the motifs defined in our structure to recognition of PCNA-SUMO, we prepared an Srs2 fragment encompassing residues 1137–1174. Although Srs21137–1174 interacts with PCNA and SUMO 3- and 4-fold weaker, respectively, than Srs21107–1174, interaction with PCNA-SUMO was 8-fold tighter (Supplemental Table 1 and Fig. 3).
Functional analysis of the Srs2 PIP-box
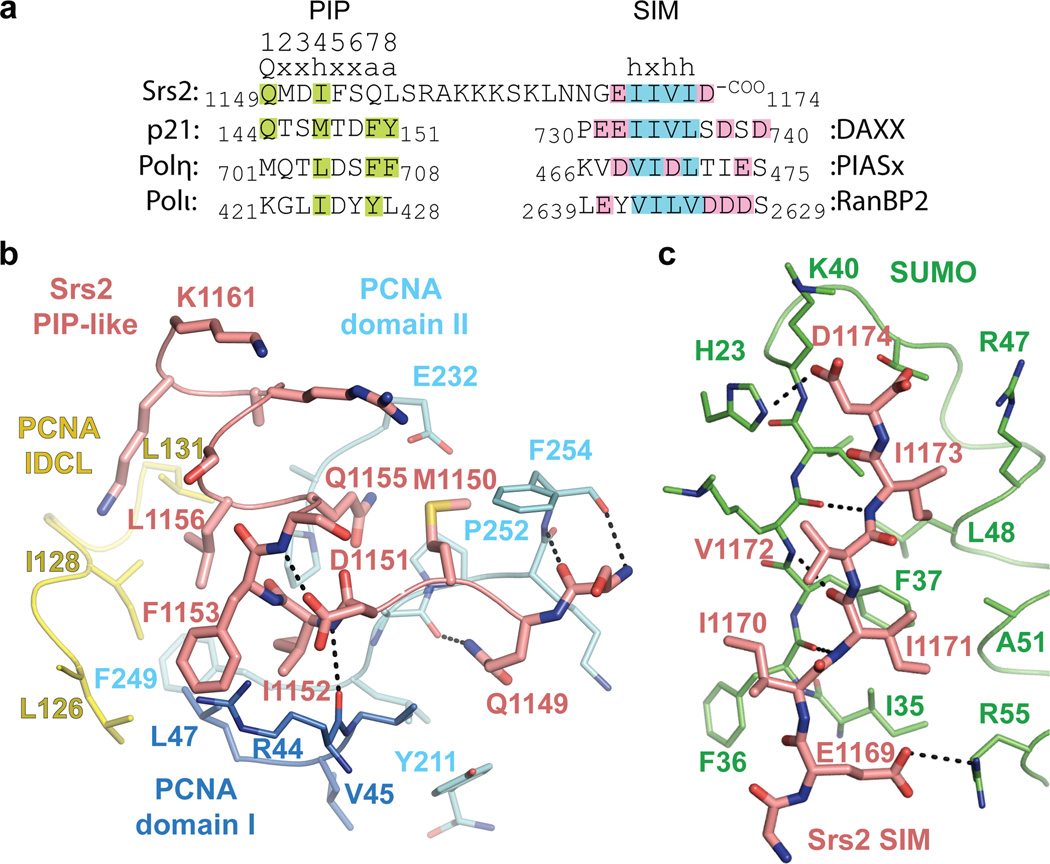
The Srs2 PCNA interaction motif differs from canonical PIP-box motifs6–8,21,28,29, perhaps consistent with its not being described previously. Most PIP-boxes contain a core QxxΨ motif followed by two conserved aromatic residues 3 and 4 residues C-terminal of the core motif (Fig. 3a and Supplemental Fig. 9). Srs2 residues 1149–1152 (QMDI) conform to a canonical PIP-box motif (QxxΨ), but Srs2 residues 1153–1161 lack the conserved aromatic residues. In addition, most PIP-boxes adopt a 310 helix after the QxxΨ motif while Srs2 residues 1153–1161 adopt two turns of α-helix that project Q1155 and L1156 into the PCNA surface (Fig 3b).
Figure 3. Srs2 PIP-like motif and SIM interactions with PCNA and SUMO.
a, Sequence alignment for Srs2 CTD (amino acids 1149–1174) above p21, Polη and Polι PIP-box motifs and DAXX, PIASx and RanBP2 SIMs. PIP-box consensus motif numbered above Srs2 (Q=glutamine, x=any amino acid, h=hydrophobic, a=aromatic). Amino acids similar to consensus are highlighted (green). Hydrophobic and acidic residues highlighted blue and pink for SIMs. b, Srs2 PIP-like motif (pink), PCNA colored as in Fig 2. PCNA, Srs2 and select amino acids labeled by numbered single letter code. Hydrogen bonds as dashed lines. c, Srs2 SIM (pink) and SUMO (green) as in b.
As the Srs2 PCNA interaction motif differs from canonical PIP-box motifs, we conducted mutational and functional studies to evaluate the importance of side chain contacts between Srs2 and PCNA. Q1149 is within hydrogen bonding distance of the backbone carbonyl oxygen of PCNA A251 (Fig. 3b), and Q1149E substitution results in a modest 3-fold defect in interaction with PCNAtri (Fig. 4a). The D1151 side chain caps the N-terminal end of the Srs2 helix with a hydrogen bond to the backbone amide of S1154 (Fig. 3b). D1151A elicits a 10-fold defect in binding PCNAtri (Fig. 4a). The I1152 side chain projects into a hydrophobic pocket on PCNA and a main chain hydrogen bond is observed between the I1152 backbone amide and carbonyl oxygen of PCNA R44 (Fig. 3b). I1152A results in a 60-fold defect in interaction with PCNAtri (Fig. 4a).
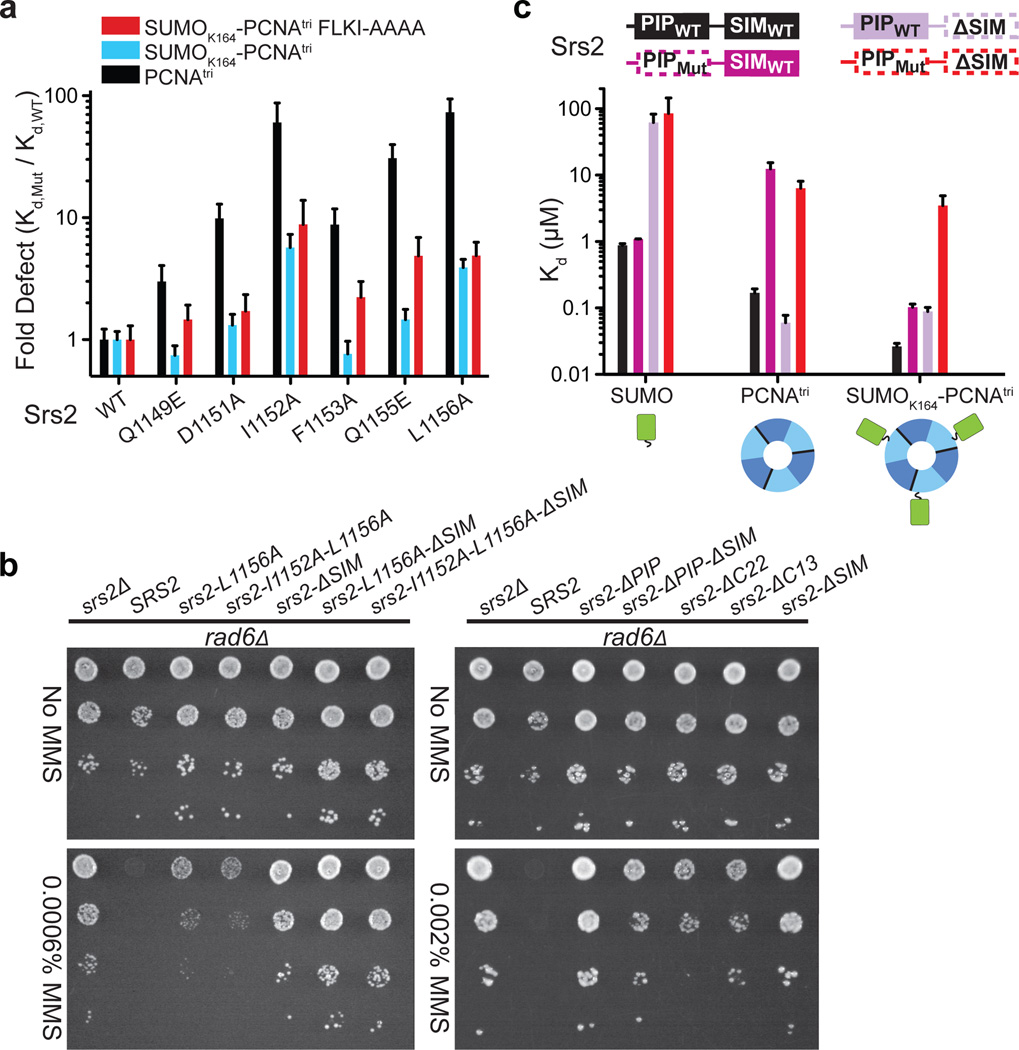
Figure 4. Srs2 PIP-like motif and SIM required for recognition of SUMO-PCNA.
a, Bar graphs showing fold-defect in Kd (y-axis) for WT and Srs21107–1174 mutants (x-axis) for PCNAtri K127G (black), SUMOK164-PCNAtri K127G (blue), or SUMOK164-PCNAtri FLKI125–128AAAA (red). b, Suppression of rad6Δ DNA damage sensitivity by srs2Δ or srs2 alleles. Shown are 10-fold serial dilutions of cells on plates containing MMS as indicated (left). c, Bar graphs showing Kd values for interaction of Srs2 containing a wild-type PIP and SIM (PIPWT-SIMWT) (black), a mutant PIP-like motif (L1156A; PIPMut) and SIMWT (dark purple; dashed line around PIPMut), a PIPWT with SIM deletion (ΔSIM) (light purple; dashed line around ΔSIM), and a PIPMut ΔSIM (red dashed line) with SUMO (left), PCNA (middle) and SUMOK164-PCNAtri (right). Cartoons colored as in Fig. 1. FP assays conducted in triplicate. Error bars are ±1 standard deviation. See Methods.
The α-helical geometry observed for Srs2 residues 1153–1161 projects side chains of F1153, Q1155 and L1156 into a mostly hydrophobic pocket on PCNA composed of residues V45 and L47 from domain I; L126, I128 and L131 from the IDCL; and P234, F249, and P252 from domain II (Fig. 3b). In addition, the Q1155 side chain Nε is within 4 Å of the PCNA E232 carboxylate. Although these Srs2 residues do not conform to side chains conserved in most other PIP-box motifs (Fig. 3a and Supplemental Fig. 9), each are important for interaction with PCNA because Srs2 containing F1153A, L1156A or Q1155E substitutions bind PCNAtri with 9-, 73- or 31-fold reduced affinity relative to wild-type Srs21107–1174, respectively (Fig. 4a).
PCNA side chains proximal to the Srs2 PIP-like motif include L126 and I128 in the IDCL (Fig. 3b), and alanine substitutions in the PCNA IDCL (FLKI125–128AAAA) weaken interaction with Srs21107–1174 by more than 60-fold (Supplemental Table 1). Furthermore, defects observed for Srs2 Q1149E, F1153A and Q1155E are exacerbated in FP assays when interactions with SUMOK164-PCNAtri FLKI125–128AAAA and SUMOK164-PCNAtri are compared (Fig. 4a). Conformations of PCNA IDCLs vary when Srs21107–1174/SUMOK164-PCNAmono and SUMOK164-PCNAmono are compared to other PCNA structures (Supplemental Fig. 10); it is possible that contacts to Srs2 elicit some of these differences.
Srs2 I1152A and L1156A substitutions result in the greatest defects in interaction with PCNA and SUMOK164-PCNAtri (Fig. 4a). To evaluate if these substitutions could impair Srs2 functions in vivo with respect to suppression of the DNA damage sensitivity of rad6Δ or rad18Δ strains, we constructed rad6Δsrs2Δ and rad18Δsrs2Δ and complemented strains with SRS2 or mutant srs2 alleles under control of the endogenous SRS2 promoter. Consistent with the importance of the PIP-like motif in vitro, strains harboring srs2-L1156A or srs2-I1152A-L1156A were more resistant to the DNA damaging agent MMS than strains containing wild-type SRS2 although both were more sensitive to MMS than strains harboring srs2 with a deletion of the PIP-like motif (Fig. 4b and Supplemental Fig. 11).
Functional analysis of the Srs2 SIM
The Srs2 SIM (residues 1168–1174) forms a β-strand that interacts with SUMO β2 and α1 (Fig. 3c). The parallel orientation of the SIM and SUMO β2 in our structure is distinct from SUMO complexes with SIMs of RanBP2 (ref. 30), thymine DNA glycosylase31 and the SUMO E1 (ref. 32) but similar to PIASx33, DAXX20 and MCAF1 (ref. 34) (Supplemental Fig. 12). Consistent with the structure and previous studies12, removal of the SIM by deletion of Srs2 residues 1168–1174 has the same effect as removing SUMO from PCNA because Srs21107–1167ΔSIM interaction with SUMO-PCNA is equivalent to PCNA alone (Fig. 4c and Supplemental Table 1).
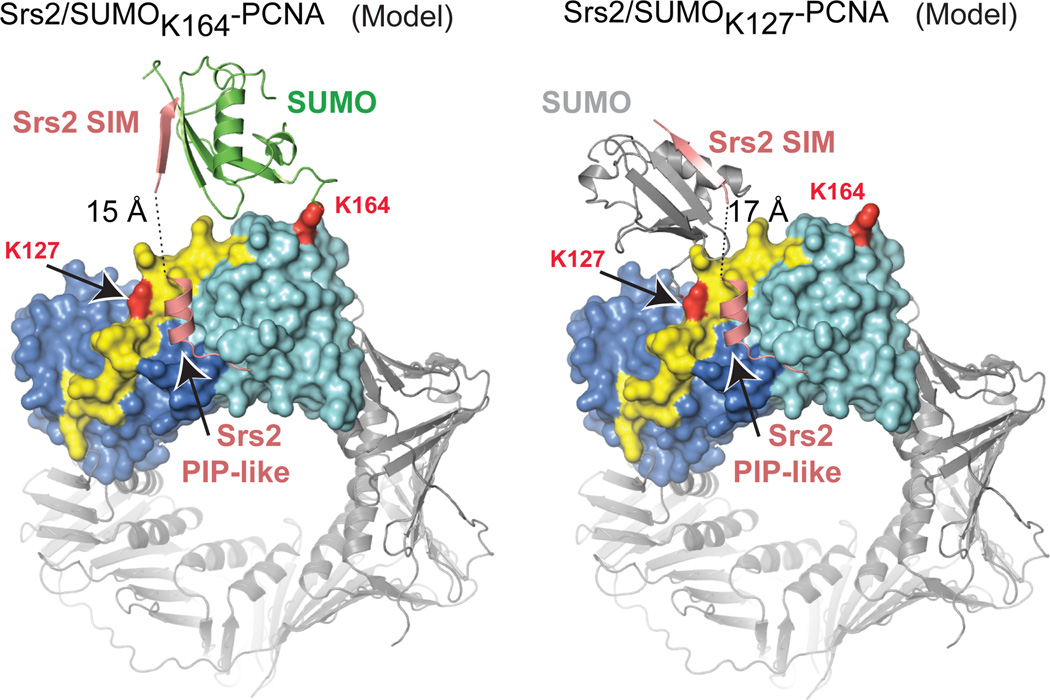
Six residues separate the Srs2 PIP-like motif (1148–1161) and SIM (1168–1174), too few to span 39 Å between these motifs in our structure even if extended (~20 Å) (Fig. 2c). We posit that the Srs2 SIM exchanged interactions with another SUMO in the lattice during crystallization as the N-termini of two symmetry related SIMs are located within 15 Å of the PIP-like motif (Supplemental Fig. 13a). It is important to note that Srs2 interacts equally well with SUMOK164-PCNAtri and SUMOK127-PCNAtri in solution (see above). For Srs2 to simultaneously engage PCNA and SUMO in these complexes SUMO would have to adopt a different conformation from that observed in SUMOK164-PCNAtri or SUMO K164-PCNAmono structures.
Models were generated for SUMOK164-PCNAtri and SUMOK127-PCNAtri to permit simultaneous engagement of the Srs2 SIM and PIP elements with PCNA and SUMO in a single protomer (Fig. 5 and Supplemental Fig. 13b). These models suggest that SUMO would adopt conformations distinct from that observed in most SUMO-PCNA structures where SUMO interacts with PCNA loop 184–198. Consistent with our models, FP data show that V186D and MEH188–190AAA mutations in PCNA loop 184–198, substitutions predicted to disrupt non-covalent contacts between SUMO and PCNA, have no impact on Srs2’s ability to interact with SUMOK164-PCNA (Supplemental Table 1).
Figure 5. Models for Srs2/SUMO-PCNA complexes.
Models for SUMO conformations to enable simultaneous interaction with the SIM and PIP-like motif when attached to PCNA K164 (left) or K127 (right). Models generated required a simple rotation of SUMO at the isopeptide linkage for SUMOK164-PCNA and a slight rotation and translation of SUMO from a symmetry related complex to mimic SUMOK127-PCNA (Supplemental Fig. 13).
The α-helical conformation of the Srs2 PIP-like motif differs from other PIP-box motifs that make more extensive contacts to the IDCL (Supplemental Fig. 9). This conformation points the Srs2 C-terminal end of the PIP-like motif away from the PCNA surface toward SUMO in our SUMO-PCNA models (Fig. 5). Furthermore, previous studies suggested that SUMO modification of PCNA antagonizes Eco1 possibly by interfering with its ability to interact with PCNA via its PIP-box35. We purified Eco1 and used it in pull-down assays to determine if Eco1 interacts with PCNA, SUMOK127-PCNA or SUMOK164-PCNA. Eco1 interacts with PCNA while PCNA FLKI125–128AAAA substitution diminished interaction (Supplemental Fig. 14). This is consistent with the IDCL mediating interactions with PIP-box proteins. We find that Eco1 binds SUMOK127-PCNA or SUMOK164-PCNA at levels similar to that observed for PCNA. Although it is clear that the SUMO pathway antagonizes PCNA-dependent Eco1 functions in vivo35, our data exclude a simple steric occlusion model because SUMO modification on PCNA K127 and K164 does not prevent Eco1 from interacting with PCNA.
Srs2 recognition of SUMO-PCNA
Our data show that Srs2 requires the SIM to recognize SUMO and the PIP-like motif to recognize PCNA and that both elements are required to specifically recognize SUMO-PCNA. Consistent with this hypothesis, interaction with SUMO is strictly dependent on the SIM but independent of mutation in the PIP-like motif while interaction with PCNA is dependent on the integrity of the PIP-like motif and independent of the SIM (Fig 4c). Finally, both PIP-like and SIM motifs are required to achieve specificity during recognition of SUMO-PCNA because mutation of the PIP-like motif or deletion of the SIM lessen interaction while mutations in both prevent interaction with SUMO-PCNA (Fig 4c).
While mutation or deletion of individual motifs results in Srs2 variants that interact with SUMO or PCNA in SUMO-PCNA in vitro, it is important to note that both mutants lack elements required to specifically recognize SUMO-PCNA. To validate this hypothesis for Srs2 function in vivo, we complemented rad6Δsrs2Δ and rad18Δsrs2Δ with Srs2 variants lacking the PIP-like motif, the SIM motif or both. As predicted, these variants behave similarly to a strain lacking SRS2 as each suppresses the DNA damage sensitivity of rad6Δ and rad18Δ strains (Fig 4b and Supplemental Fig. 11). Importantly, strains harboring a SIM deletion in conjunction with single and double point substitutions in the PIP-like motif are more resistant to DNA damage than strains containing full-length Srs2 with mutations in the PIP-like motif.
Discussion
Recognition of Ub/Ubl-modified substrates is a critical first step in Ub/Ubl-mediated signal transduction. Details of this process have been widely assumed36, but the molecular bases for these interactions have not been formally demonstrated. Here we identify one mechanism to achieve specificity during recognition of SUMO-modified PCNA. Given the modular nature of PIP-box motifs and ubiquitin binding domains in some translesion polymerase family members21,37, this mechanism may apply to their recognition of Ub-PCNA38. It is also worth noting that a recent study claims to have identified a mammalian Srs2 ortholog named PARI which contains a C-terminal canonical PIP-box and an internal SIM39, although these motifs differ in comparison to Srs2 with respect to their position in primary sequence.
It seems likely that the mechanism employed by Srs2 to recognize SUMO-PCNA, namely juxtaposition of domains or motifs in the receptor that simultaneously engage the substrate and Ub/Ubl modifier within the context of the Ub/Ubl-conjugated substrate, will be employed in other signal transduction pathways that rely on receptors to specifically recognize Ub/Ubl-modified substrates.
METHODS SUMMARY
Yeast proteins were expressed in E. coli and purified. PCNA was conjugated to SUMO using the E3 ligase Siz1. For fluorescence polarization, substrates were titrated in triplicate against 20 nM BODIPY-FL conjugated Srs2 and data fit to a single site binding model accounting for ligand depletion. Crystals were obtained by vapor diffusion. Diffraction data were collected at beam lines X29 (NSLS) and 24-IDC (APS), phases were calculated by molecular replacement. Yeast strains were constructed and complemented with plasmids containing SRS2 under the control of its endogenous promoter.
Full Methods and associated references are available in the online version of the paper at www.nature.com/nature
Methods
Cloning, Expression, and Purification of Recombinant Proteins
Expression and purification of yeast E1 (Aos1/Uba2), E2 (Ubc9), yeast SUMO (NΔ18 Smt3), Siz1112–465 and affinity tag-free yeast PCNA (Wt, K127G and FLKI125–128AAAA) have been described22,23. PCNA K164R and SUMO K19R were generated by PCR-based site-directed mutagenesis. Srs21027–1174 was obtained by PCR from S. cerevisiae genomic DNA and cloned into the pSMT3 vector encoding an N-terminal His6-Smt3 tag. Fragments, including the Srs21107–1174 construct, and mutants were generated by subcloning and by PCR-based site-directed mutagenesis. Srs2 was expressed in BL21 (DE3) RIL Codon Plus cells after induction with 0.5 mM IPTG and incubation of the cultures at 30°C for 3 hours. Cell pellets were suspended in 2.5× volume of 20% sucrose, 50 mM Tris, pH 8.0 and lysed by sonication. Srs2 was isolated from cleared lysate by immobilized metal affinity chromatography using Ni-NTA resin (Qiagen). Srs2 was eluted from the resin in buffer containing 250 mM imidazole, 350 mM NaCl, 20 mM Tris, pH 8.0, 1 mM βME, and the His6-Smt3 tag was subsequently cleaved by addition of Ulp1 to a 1:1000 mass ratio followed by overnight incubation/dialysis into 350 mM NaCl, 20 mM Tris, pH 8.0, 1 mM βME at 4°C40. Ulp1 was inactivated by adding NEM to a final concentration of 5 mM followed by incubation at RT for 30 min and 4°C for 1 hour. Srs2 was separated from other proteins in this mixture by size-exclusion chromatography (Superdex75 26/60; GE Healthcare; equilibrated with 350 mM NaCl, 20 mM Tris, pH 8.0, 1 mM βME). Fractions containing Srs2 were combined, dialyzed overnight against 50 mM NaCl, 20 mM Tris, pH 8.0, 1 mM βME and purified by anion exchange chromatography (MonoQ 10/10; GE Healthcare). Srs21107–1174 was recovered in the unbound fraction, and Srs21027–1174 was eluted using a gradient from 50 mM to 475 mM NaCl over 12 column volumes in 20 mM Tris, pH 8.0, 1 mM βME. Srs2 was concentrated using an Amicon Ultra centrifugal filter, snap frozen in liquid nitrogen and stored at −80°C. Eco1 was cloned from S. cerevisiae genomic DNA into the pET28a vector encoding an N-terminal hexahistidine tag. Eco1 was expressed in BL21 (DE3) RIL Codon Plus cells and purified through IMAC steps according to the same protocols described for expression and purification of Srs2 fragments. Upon elution from the Ni-NTA resin, Eco1 was purified by size-exclusion chromatography (Superdex75 26/60; GE Healthcare; equilibrated with 350 mM NaCl, 20 mM Tris, pH 8.0, 1 mM βME). Peak fractions were combined and Eco1 was concentrated to 8 mg/mL prior to snap freezing in liquid nitrogen for storage at −80°C.
Reductive methylation of SUMO and preparation of PCNAmono
Reductive methylation of yeast SUMO (NΔ18 K19R Smt3;*SUMO where the asterisk indicates reductively methylated) utilized paraformaldehyde and sodium borohydride as the methyl donor and reducing agent, respectively41. Monomeric PCNA or monomeric SUMO-PCNA was prepared by adding N-ethylmaleimide (NEM) to a final concentration of 5 mM to the corresponding trimeric species, incubating for 2 hours at 4°C, and then purifying by size exclusion chromatography (Superdex200 26/60).
Reconstitution of SUMO-PCNA
Yeast SUMO was conjugated to yeast PCNA using the SUMO E3 ligase Siz1 by mixing 15 µM PCNA, 65 µM SUMO (NΔ18 K19R Smt3 or reductively methylated NΔ18 K19R Smt3), 90 nM Aos1/His6-Uba2 ΔCT (E1), 300 nM Ubc9 (E2), and 2 µM Siz1112–465 (E3) and 2 mM ATP, incubated at 37°C for 2 hours, and stopped by chromatographic resolution of the reaction components.
Fluorescence Polarization Assays
A cysteine residue was added N-terminal to Srs21107–1174 which itself lacked any cysteine residues. BODIPY FL (Invitrogen) was conjugated to the thiol group via maleimide chemistry. For FP assays, substrate was titrated against a fixed concentration (20 nM) of labeled Srs21107–1174 by mixing equal volumes of Srs2 peptide and serially diluted protein. Binding reaction conditions were 50 mM NaCl, 20 mM Tris, pH 8.0, 4 mM DTT. 20 µL reaction volumes were added to the wells of a 384-well microplate, and measurements taken using a SpectraMax M5 microplate reader (Molecular Devices) employing an excitation wavelength of 485 nm, emission wavelength of 538 nm, and a cutoff of 530 nm. Experiments were performed in triplicate. Using the software package Prism 5 (GraphPad Software, Inc.) data were fit to a single site binding model accounting for ligand depletion:
where A is the measured anisotropy, [Srs2] is the fixed concentration of labeled receptor, and [Prot] is the total concentration of substrate. Ab and Af are limiting anisotropies for bound and free peptide, respectively, and Kd is the dissociation constant.
For displacement assays, BODIPY labeled Srs21107–1174 at 40 nM was pre-incubated with PCNAtri K127G, GST-SUMO, or SUMOK164-PCNAtri at 600, 2000, and 80 nM, respectively. Equal volumes of the pre-incubated binding reaction and serially diluted unlabled Srs21107–1174 were then mixed. 20 µL reaction volumes were added to the wells of a 384-well microplate, and measurements were made as for the direct FP assays. Experiments were performed in triplicate, and the data were fit using Prism 5 to a one site binding model for competition assays.
Assessment of Protein-Protein Interactions by Analytical Gel Filtration
Purified proteins were mixed at the indicated concentrations in a volume of 220 µL and centrifuged, and 200 µL was loaded onto a Superose12 HR 10/30 column (GE Healthcare) equilibrated in 50 mM NaCl, 20 mM Tris, pH 8.0, 1 mM βME. The column was eluted at a flow rate of 0.5 mL/min, and 0.5 mL fractions were collected, analyzed by SDS-PAGE and stained with Sypro Ruby Protein Gel Stain (Bio-Rad).
Assessment of Protein-Protein Interactions by Pull-downs
400 µL 750 µg/mL His6-Eco1 in binding buffer (200 mM NaCl, 20 mM Tris, pH 8.0, 40 mM imidazole) was added to 50 µL of 50% (v/v) Ni-NTA resin (Qiagen) equilibrated with the same buffer and mixed for 20 min at 4°C. Beads were subsequently washed with 3 × 400 µL of binding buffer to remove unbound His6-Eco1. 400 µL of 10 µM prey protein in binding buffer was added to Eco1 coated beads and mixed for 15 min at 4°C. Unbound prey protein was removed with 3 × 400 µL washes with binding buffer. Material was eluted from beads with successive 400 µL washes of binding buffer that contained increasing concentrations of NaCl. Eco1 and any remaining bound prey protein was eluted in a final wash with 100 µL 350 mM NaCl, 20 mM Tris, pH 8.0, 400 mM imidazole.
Crystallization and Structure Determination
Crystals were grown using the hanging drop vapor diffusion method by mixing equal volumes of sample and reservoir solution. SUMOK164-PCNAtri K127G: Purified SUMOK164-PCNAtri was crystallized at 18°C in 4% PEG 8000, 500 mM LiSO4 (pH 4.5). Crystals were transferred to cryoprotectant (reservoir supplemented with 15% ethylene glycol) prior to flash freezing in LN2. Diffraction data were collected at beamline 24-ID-C at the Advanced Photon Source at a wavelength of 0.979 Å. Crystals belong to space group F432 with one protomer of SUMO-PCNA in the asymmetric unit. SUMOK164-PCNAmono K127G: Srs21027–1174 was added to 8 mg/mL SUMOK164-PCNA in 3-fold molar excess and dialyzed against 100 mM NaCl, 20 mM Tris pH 8.0, 1 mM βME at 4°C for 16 hrs. The Srs21027–1174:SUMOK164-PCNA complex was purified on a Superdex 200 column and concentrated to ~8 mg/mL. Crystals were obtained at 4°C in 21% MPD, 100 mM BaCl2, 100 mM Bis-Tris, pH 6.5 and transferred to cryoprotectant (reservoir supplemented with 12% glycerol) prior to flash freezing in LN2. Diffraction data were collected at beamline 24-ID-C at the Advanced Photon Source at a wavelength of 0.979 Å. Crystals belong to space group I41 with one protomer of SUMOK164-PCNAmono in the asymmetric unit. It was subsequently determined that these crystals lacked the Srs21027–1174 protein component, and that contaminating NEM, presumably present in earlier Srs1027–1174 preparations, had modified PCNA C22 and C81. SUMOK164-PCNA K127G:Srs21107–1174: Srs21107–1174 was added in 3-fold molar excess to SUMOK164-PCNAtri at a protomer concentration of 60 µM. NEM was added to a final concentration of 5 mM, and the sample incubated for 3 hours at 4°C prior to co-purification of the complex by gel filtration (Superdex200 26/60; GE Healthcare; equilibrated with 100 mM NaCl, 20 mM Tris, pH 8.0, 1 mM βME). The complex was concentrated to ~10 mg/mL and crystallized at 6°C in 1.9 M ammonium sulfate, 4% PEG400, 100 mM HEPES, pH 7.5 by the sitting drop vapor diffusion method. Crystals were washed and cryoprotected in 2 M ammonium sulfate, 4% PEG400, 100 mM HEPES, pH 7.5, 20% glycerol. Diffraction data were collected at beamline X29 of the National Synchrotron Light Source at a wavelength of 1.075 Å. Crystals belong to space group C2 with two Srs2/SUMOK164-PCNA complexes in the asymmetric unit.
Crystallographic data were indexed, integrated, and scaled using HKL200042 and data reduced using programs in CCP443. Data obtained for SUMOK164-PCNAmono were scaled maintaining separation of anomalous pairs to maintain the anomalous signal from the associated barium ions. The programs MOLREP44 and PHASER45 were used to find molecular replacement solutions using the coordinates of a yeast PCNA protomer from PDB 1PLQ and yeast SUMO from PDB 1EUV (Supplemental Table 2 and Methods). Models were manually inspected and rebuilt prior to refinement using the program O46 and refined using CCP4’s REFMAC47. Models have reasonable geometry at the respective resolution as assessed by MolProbity48. SUMOK164-PCNAtri has 95.5% and 3.9% of modeled residues in favored and allowed Ramachandran regions, respectively with 0.6% outliers. SUMOK164-PCNAmono has 95.7% of modeled residues in favored regions, 3.4% in allowed regions and 0.9% as outliers, and Srs21107–1174/SUMOK164-PCNAmono has 91.5% in favored regions, 7.6% in allowed regions and 0.9% as outliers. Structures of SUMOK164-PCNAtri, SUMOK164-PCNAmono and Srs21107–1174/SUMOK164-PCNAmono have Clash Scores in the 93rd, 95th and 87th percentiles, respectively, and MolProbity Scores in the 70th, 90th and 50th percentiles respectively. All graphical representations of structure were generated using PYMOL49.
Yeast complementation and growth assays
Yeast strains used in this study include srs2Δrad6Δ (MATa, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0, srs2Δ::kanMX, rad6Δ::kanMX) and srs2Δrad18Δ (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, srs2Δ::kanMX, rad18Δ::kanMX). Double deletion strains were constructed by mating srs2Δ (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) to rad6Δ or rad18Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) knockout strains from the Saccharomyces Genome Deletion Project (Open Biosystems). Diploids were selected on media lacking lysine and methionine then sporulated in 2% potassium acetate. The desired double deletion strains were selected by screening spores by PCR to establish the genotype and by replica plating spore colonies on selective media. Double deletion strains were transformed with the pRS416 plasmid (URA3) or the the pRS416 plasmid (URA3) containing SRS2 or srs2 alleles flanked by 500 bp of its endogenous 5’ and 3’ UTRs. These constructs were generated by cloning the SRS2 5' UTR along with the SRS2 coding sequence corresponding to the first 1115 residues into the XhoI and SalI sites of the pRS416 vector and the SRS2 3' UTR into the XmaI and BamHI sites. Use of the SalI site introduced a DNA mutation that is silent with respect to the encoded Srs2 amino acid. The remainder of the SRS2 coding sequence was placed between the SalI and XmaI sites to produce the desired alleles including SRS2, srs2-ΔPIPΔSIM (Srs21–1148), srs2-ΔC22 (Srs21–1152), srs2-ΔC13 (Srs21–1161), and srs2-ΔSIM (Srs21–1167). srs2-ΔPIP (Srs21–1148+PG+1168–1174) was generated by placing the SRS2 coding region corresponding to the C-terminal 7 residues along with the 3' UTR into XmaI and BamHI sites. The SRS2 coding sequence corresponding to residues 1115–1148 was then placed between the SalI and XmaI sites. This strategy introduced two non-native amino acids (Pro-Gly) between Srs2 residues 1148 and 1168. Yeast transformants were selected on media lacking uracil. For spotting assays to assess growth defects and drug sensitivity, cultures were grown overnight and diluted to an A600 of 1.0. 10-fold serial dilutions were made and 3 µl aliquots were spotted onto SD -Ura plates or SD -Ura + MMS plates containing the indicated concentration of DNA damage agent. Plates were incubated at 30°C for ~3 days with photographs taken every 6–8 hours.
Supplementary Material
Acknowledgements
We thank John Kuriyan and Brian Kelch for coordinates of the T4 clamp prior to publication. NE-CAT beamlines (Advanced Photon Source) supported by RR-15301 (NIH NCRR). APS supported by U.S. Dept. of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Beamline X29 (National Synchrotron Light Source) supported by U.S. Dept. of Energy, Office of Basic Energy Sciences and P41RR012408 (NIH NCRR). A.A.A., F.M. and C.D.L. supported by NIH R01 GM065872 to C.D.L. and F32 GM086066 to A.A.A.
Footnotes
Supplemental Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Experiments performed and analyzed by A.A.A., F.M. and C.D.L. Manuscript prepared by A.A.A and C.D.L.
Author Information Atomic coordinates and structure factors deposited in RCSB under accession codes 3V60, 3V61 and 3V62.
The authors declare no competing financial interests.
References
- 1.Kirkin V, Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr Opin Cell Biol. 2007;19:199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 6.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 7.Bruning JB, Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure. 2004;12:2209–2219. doi: 10.1016/j.str.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 8.Vijayakumar S, et al. The C-terminal domain of yeast PCNA is required for physical and functional interactions with Cdc9 DNA ligase. Nucleic Acids Res. 2007;35:1624–1637. doi: 10.1093/nar/gkm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott MT, Morrice N, Ball KL. Reversible phosphorylation at the C-terminal regulatory domain of p21(Waf1/Cip1) modulates proliferating cell nuclear antigen binding. J Biol Chem. 2000;275:11529–11537. doi: 10.1074/jbc.275.15.11529. [DOI] [PubMed] [Google Scholar]
- 10.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 11.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 12.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 13.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Krejci L, et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 15.Veaute X, et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence CW, Christensen RB. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J Bacteriol. 1979;139:866–876. doi: 10.1128/jb.139.3.866-876.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baba D, et al. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 18.Ulrich HD. PCNASUMO and Srs2: a model SUMO substrate-effector pair. Biochem Soc Trans. 2007;35:1385–1388. doi: 10.1042/BST0351385. [DOI] [PubMed] [Google Scholar]
- 19.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang CC, et al. Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol Cell. 2011;42:62–74. doi: 10.1016/j.molcel.2011.02.022. [DOI] [PubMed] [Google Scholar]
- 21.Hishiki A, et al. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J Biol Chem. 2009;284:10552–10560. doi: 10.1074/jbc.M809745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yunus AA, Lima CD. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol Cell. 2009;35:669–682. doi: 10.1016/j.molcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yunus AA, Lima CD. Purification of SUMO conjugating enzymes and kinetic analysis of substrate conjugation. Methods Mol Biol. 2009;497:167–186. doi: 10.1007/978-1-59745-566-4_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freudenthal BD, Brogie JE, Gakhar L, Kondratick CM, Washington MT. Crystal Structure of SUMO-Modified Proliferating Cell Nuclear Antigen. J Mol Biol. 2011;406:9–17. doi: 10.1016/j.jmb.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kazmirski SL, Zhao Y, Bowman GD, O'Donnell M, Kuriyan J. Out-of-plane motions in open sliding clamps: molecular dynamics simulations of eukaryotic and archaeal proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2005;102:13801–13806. doi: 10.1073/pnas.0506430102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyata T, et al. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc Natl Acad Sci U S A. 2005;102:13795–13800. doi: 10.1073/pnas.0506447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelch BA, Makino DL, O'Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334:1675–1680. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowman GD, O'Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 29.Sakurai S, et al. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baba D, et al. Crystal structure of SUMO-3-modified thymine-DNA glycosylase. J Mol Biol. 2006;359:137–147. doi: 10.1016/j.jmb.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 32.Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 34.Sekiyama N, et al. Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J Biol Chem. 2008;283:35966–35975. doi: 10.1074/jbc.M802528200. [DOI] [PubMed] [Google Scholar]
- 35.Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 37.Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Ai Y, Wang J, Haracska L, Zhuang Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat Chem Biol. 2010;6:270–272. doi: 10.1038/nchembio.316. [DOI] [PubMed] [Google Scholar]
- 39.Moldovan GL, Dejsuphong D, Petalcorin MIR, Hofmann K, Takeda S, Boulton SJ, D’Andrea AD. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 40.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 41.Rayment I. Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 1997;276:171–179. [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. In: Methods Enzymol. Carter CW Jr, Sweet RM, editors. Vol. 276. Academic Press; 1997. pp. 307–326. [Google Scholar]
- 43.Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Vagin A, Teplyakov A. MOLREP: an automated program from molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 45.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 47.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 48.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delano W. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.