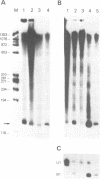
Abstract
B1 (Alu-equivalent) is a murine short interspersed element whose amplification probably involved an RNA intermediate. B1-homologous RNA comprise a population of heterogenous transcripts of questionable function. A cloned B1 is expressed in the injected frog oocyte by RNA polymerase III transcription, ribonucleoprotein formation, post-transcriptional 3'-processing, and nucleocytoplasmic transport. The present study characterizes small cytoplasmic B1 transcripts of mouse cells. Analyses of ten cDNA clones revealed a subset of a high degree of sequence identity (98%) from which a novel consensus was developed. Structural analyses of these RNAs demonstrated a conserved Alu domain originally identified as part of the 7SL RNA within the translational control domain of the signal recognition particle, while this structure was not conserved in the majority of B1s in the sequence database. Furthermore, it was demonstrated that 3'-processing occurred in only a subset of B1 transcripts in-vitro using homologous nuclear extracts, and in the injected oocyte. The data demonstrate that a limited set of B1 sequences are expressed as processed RNA polymerase III-transcripts of a high degree of structural conservation. Although this subset is transcriptionally active, the selective expression may be due to regulation at the levels of processing and cytoplasmic accumulation. Their lack of Poly-(A) or 3'-oligo-(U) tracts argue that these RNAs are unlikely to represent transposition intermediates. Rather, their cytosolic compartmentalization and conservation of a biologically recognized structure, suggests potential involvement in other aspects of cellular metabolism.
Full text
PDF


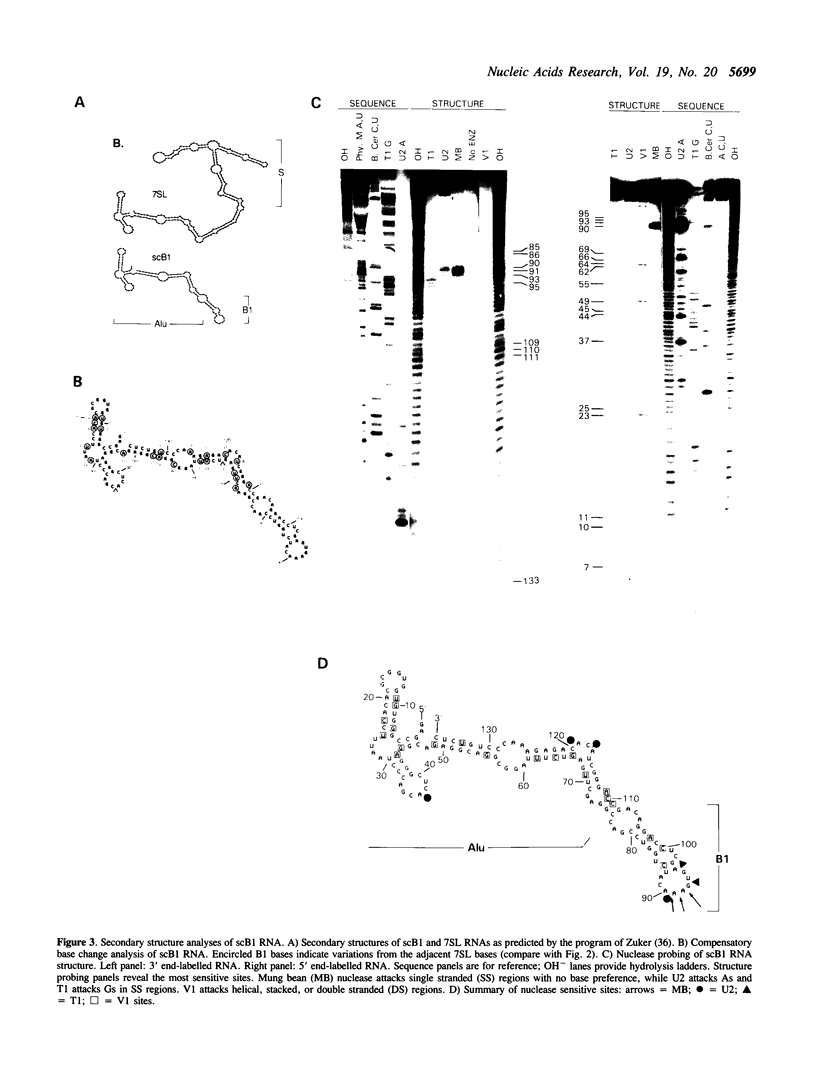



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adeniyi-Jones S., Zasloff M. Transcription, processing and nuclear transport of a B1 Alu RNA species complementary to an intron of the murine alpha-fetoprotein gene. Nature. 1985 Sep 5;317(6032):81–84. doi: 10.1038/317081a0. [DOI] [PubMed] [Google Scholar]
- Balmain A., Krumlauf R., Vass J. K., Birnie G. D. Cloning and characterisation of the abundant cytoplasmic 7S RNA from mouse cells. Nucleic Acids Res. 1982 Jul 24;10(14):4259–4277. doi: 10.1093/nar/10.14.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer M. A., Kilroy G. E., Richard P. E., Shaikh T. H., Desselle T. D., Hoppens C. L., Deininger P. L. Structure and variability of recently inserted Alu family members. Nucleic Acids Res. 1990 Dec 11;18(23):6793–6798. doi: 10.1093/nar/18.23.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett K. L., Hill R. E., Pietras D. F., Woodworth-Gutai M., Kane-Haas C., Houston J. M., Heath J. K., Hastie N. D. Most highly repeated dispersed DNA families in the mouse genome. Mol Cell Biol. 1984 Aug;4(8):1561–1571. doi: 10.1128/mcb.4.8.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake M. C., Jambou R. C., Swick A. G., Kahn J. W., Azizkhan J. C. Transcriptional initiation is controlled by upstream GC-box interactions in a TATAA-less promoter. Mol Cell Biol. 1990 Dec;10(12):6632–6641. doi: 10.1128/mcb.10.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Baron W. F., Stout D. B., Davidson E. H. Sources and evolution of human Alu repeated sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4770–4774. doi: 10.1073/pnas.85.13.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M. F., Singh K., Botchan M., Cozzarelli N. R. Induction of specific transcription by RNA polymerase III in transformed cells. Mol Cell Biol. 1986 Sep;6(9):3068–3076. doi: 10.1128/mcb.6.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amore M. A., Gallagher P. M., Korfhagen T. R., Ganschow R. E. Complete sequence and organization of the murine beta-glucuronidase gene. Biochemistry. 1988 Sep 6;27(18):7131–7140. doi: 10.1021/bi00418a070. [DOI] [PubMed] [Google Scholar]
- Daniels G. R., Deininger P. L. Characterization of a third major SINE family of repetitive sequences in the galago genome. Nucleic Acids Res. 1991 Apr 11;19(7):1649–1656. doi: 10.1093/nar/19.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington G. J., Papaconstantinou J., Sammons D. W., Brown P. C., Wong E. Y., Esterman A. L., Kang J. Generation and chracterization of variants of mouse hepatoma cells with defects in hepato-specific gene expression. I. Albumin synthesis variants. Somatic Cell Genet. 1982 Jul;8(4):451–464. doi: 10.1007/BF01538707. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev G. P., Kramerov D. A., Ryskov A. P., Skryabin K. G., Lukanidin E. M. Dispersed repetitive sequences in eukaryotic genomes and their possible biological significance. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1109–1121. doi: 10.1101/sqb.1983.047.01.125. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Collins K. W., Hernandez E. M., Garrard W. T. Vacuum blotting: a simple method for transferring DNA from sequencing gels to nylon membranes. Gene. 1988 Dec 30;74(2):347–356. doi: 10.1016/0378-1119(88)90168-0. [DOI] [PubMed] [Google Scholar]
- Gundelfinger E. D., Di Carlo M., Zopf D., Melli M. Structure and evolution of the 7SL RNA component of the signal recognition particle. EMBO J. 1984 Oct;3(10):2325–2332. doi: 10.1002/j.1460-2075.1984.tb02134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E. D., Krause E., Melli M., Dobberstein B. The organization of the 7SL RNA in the signal recognition particle. Nucleic Acids Res. 1983 Nov 11;11(21):7363–7374. doi: 10.1093/nar/11.21.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B., Klanner A., Ramm K., Sänger H. L. The 7S RNA from tomato leaf tissue resembles a signal recognition particle RNA and exhibits a remarkable sequence complementarity to viroids. EMBO J. 1988 Dec 20;7(13):4063–4074. doi: 10.1002/j.1460-2075.1988.tb03300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard B. H., Sakamoto K. Alu interspersed repeats: selfish DNA or a functional gene family? New Biol. 1990 Sep;2(9):759–770. [PubMed] [Google Scholar]
- Jagadeeswaran P., Forget B. G., Weissman S. M. Short interspersed repetitive DNA elements in eucaryotes: transposable DNA elements generated by reverse transcription of RNA pol III transcripts? Cell. 1981 Oct;26(2 Pt 2):141–142. doi: 10.1016/0092-8674(81)90296-8. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R., Schmid C. W. Repetitive sequences in eukaryotic DNA and their expression. Annu Rev Biochem. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- Kalb V. F., Glasser S., King D., Lingrel J. B. A cluster of repetitive elements within a 700 base pair region in the mouse genome. Nucleic Acids Res. 1983 Apr 11;11(7):2177–2184. doi: 10.1093/nar/11.7.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp G. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- Kramerov D. A., Grigoryan A. A., Ryskov A. P., Georgiev G. P. Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: evidence from DNA cloning experiments. Nucleic Acids Res. 1979 Feb;6(2):697–713. doi: 10.1093/nar/6.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov D. A., Lekakh I. V., Samarina O. P., Ryskov A. P. The sequences homologous to major interspersed repeats B1 and B2 of mouse genome are present in mRNA and small cytoplasmic poly(A) + RNA. Nucleic Acids Res. 1982 Dec 11;10(23):7477–7491. doi: 10.1093/nar/10.23.7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev A. S., Kramerov D. A., Skryabin K. G., Ryskov A. P., Bayev A. A., Georgiev G. P. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res. 1980 Mar 25;8(6):1201–1215. doi: 10.1093/nar/8.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuda D., Sinnett D., Richer C., Deragon J. M., Striker G. Evolution of mouse B1 repeats: 7SL RNA folding pattern conserved. J Mol Evol. 1991 May;32(5):405–414. doi: 10.1007/BF02101280. [DOI] [PubMed] [Google Scholar]
- Larsen N., Zwieb C. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 1991 Jan 25;19(2):209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia R., Zasloff M., Plotz P., Adeniyi-Jones S. Pathway of B1-Alu expression in microinjected oocytes: Xenopus laevis proteins associated with nuclear precursor and processed cytoplasmic RNAs. Mol Cell Biol. 1988 Oct;8(10):4433–4440. doi: 10.1128/mcb.8.10.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G., Hellmann U., Hintz M. F., Schmid C. W. Recently transposed Alu repeats result from multiple source genes. Nucleic Acids Res. 1990 Oct 25;18(20):6019–6023. doi: 10.1093/nar/18.20.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G., Hellmann U., Schmid C. W. A transpositionally and transcriptionally competent Alu subfamily. Mol Cell Biol. 1990 Oct;10(10):5424–5432. doi: 10.1128/mcb.10.10.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell G. A., Labuda D., Fontaine G., Saudubray J. M., Bonnefont J. P., Lyonnet S., Brody L. C., Steel G., Obie C., Valle D. Splice-mediated insertion of an Alu sequence inactivates ornithine delta-aminotransferase: a role for Alu elements in human mutation. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):815–819. doi: 10.1073/pnas.88.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Stable C., Shen C. K. Competitive and cooperative functioning of the anterior and posterior promoter elements of an Alu family repeat. Mol Cell Biol. 1986 Jun;6(6):2041–2052. doi: 10.1128/mcb.6.6.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poritz M. A., Bernstein H. D., Strub K., Zopf D., Wilhelm H., Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990 Nov 23;250(4984):1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- Quentin Y. Successive waves of fixation of B1 variants in rodent lineage history. J Mol Evol. 1989 Apr;28(4):299–305. doi: 10.1007/BF02103425. [DOI] [PubMed] [Google Scholar]
- Quentin Y. The Alu family developed through successive waves of fixation closely connected with primate lineage history. J Mol Evol. 1988;27(3):194–202. doi: 10.1007/BF02100074. [DOI] [PubMed] [Google Scholar]
- Ribes V., Römisch K., Giner A., Dobberstein B., Tollervey D. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell. 1990 Nov 2;63(3):591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- Siegel V., Walter P. Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell. 1988 Jan 15;52(1):39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- Singer M. F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982 Mar;28(3):433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Strub K., Moss J., Walter P. Binding sites of the 9- and 14-kilodalton heterodimeric protein subunit of the signal recognition particle (SRP) are contained exclusively in the Alu domain of SRP RNA and contain a sequence motif that is conserved in evolution. Mol Cell Biol. 1991 Aug;11(8):3949–3959. doi: 10.1128/mcb.11.8.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Walter P. Assembly of the Alu domain of the signal recognition particle (SRP): dimerization of the two protein components is required for efficient binding to SRP RNA. Mol Cell Biol. 1990 Feb;10(2):777–784. doi: 10.1128/mcb.10.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Murphy S., Melli M. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell. 1982 May;29(1):195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Van Arsdell S. W., Denison R. A., Bernstein L. B., Weiner A. M., Manser T., Gesteland R. F. Direct repeats flank three small nuclear RNA pseudogenes in the human genome. Cell. 1981 Oct;26(1 Pt 1):11–17. doi: 10.1016/0092-8674(81)90028-3. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982 Oct 21;299(5885):691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- Walter P., Gilmore R., Blobel G. Protein translocation across the endoplasmic reticulum. Cell. 1984 Aug;38(1):5–8. doi: 10.1016/0092-8674(84)90520-8. [DOI] [PubMed] [Google Scholar]
- Weiner A. M. An abundant cytoplasmic 7S RNA is complementary to the dominant interspersed middle repetitive DNA sequence family in the human genome. Cell. 1980 Nov;22(1 Pt 1):209–218. doi: 10.1016/0092-8674(80)90169-5. [DOI] [PubMed] [Google Scholar]
- White R. J., Stott D., Rigby P. W. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell. 1989 Dec 22;59(6):1081–1092. doi: 10.1016/0092-8674(89)90764-2. [DOI] [PubMed] [Google Scholar]
- White R. J., Stott D., Rigby P. W. Regulation of RNA polymerase III transcription in response to Simian virus 40 transformation. EMBO J. 1990 Nov;9(11):3713–3721. doi: 10.1002/j.1460-2075.1990.tb07584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard C., Nguyen H. T., Schmid C. W. Existence of at least three distinct Alu subfamilies. J Mol Evol. 1987;26(3):180–186. doi: 10.1007/BF02099850. [DOI] [PubMed] [Google Scholar]
- Young P. R., Scott R. W., Hamer D. H., Tilghman S. M. Construction and expression in vivo of an internally deleted mouse alpha-fetoprotein gene: presence of a transcribed Alu-like repeat within the first intervening sequence. Nucleic Acids Res. 1982 May 25;10(10):3099–3116. doi: 10.1093/nar/10.10.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- Zwieb C. The secondary structure of the 7SL RNA in the signal recognition particle: functional implications. Nucleic Acids Res. 1985 Sep 11;13(17):6105–6124. doi: 10.1093/nar/13.17.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]