Abstract
The splicing-factor oncoprotein SRSF1 (also known as SF2/ASF) is upregulated in breast cancers. We investigated SRSF1’s ability to transform human and mouse mammary epithelial cells in vivo and in vitro. SRSF1-overexpressing COMMA-1D cells formed tumors, following orthotopic transplantation to reconstitute the mammary gland. In 3-D culture, SRSF1-overexpressing MCF-10A cells formed larger acini than control cells, reflecting increased proliferation and delayed apoptosis during acinar morphogenesis. These effects required the first RNA-recognition motif and nuclear functions of SRSF1. SRSF1 overexpression promoted alternative splicing of BIM and BIN1 isoforms that lack pro-apoptotic functions and contribute to the phenotype. Finally, SRSF1 cooperated specifically with MYC to transform mammary epithelial cells, in part by potentiating eIF4E activation, and these cooperating oncogenes are significantly co-expressed in human breast tumors. Thus, SRSF1 can promote breast cancer, and SRSF1 itself or its downstream effectors may be valuable targets for therapeutics development.
INTRODUCTION
Most of the ~25,000 human genes express primary transcripts that undergo splicing in the nucleus to generate functional mRNAs. The majority of pre-mRNAs are alternatively spliced to yield different mRNA spliced variants, according to cell type, developmental stage, or physiological state1. The roles of spliced variants and splicing misregulation in cancer initiation and progression are incompletely understood.
The serine/arginine-rich (SR) proteins and the heterogeneous nuclear ribonucleoproteins (hnRNPs) are two important classes of factors that act, respectively, as splicing activators and repressors2. These RNA-binding proteins bind directly to pre-mRNA, eliciting concentration-dependent changes in alternative splicing (AS), and they can have antagonistic effects on AS of particular exons3,4. Thus, changes in the expression of these proteins can affect AS of many genes and are potentially involved in splicing misregulation in various diseases.
Cancer cells often display aberrant AS profiles, expressing isoforms that stimulate cell proliferation and migration, or improve resistance to apoptosis5. Some of these alterations are caused by mutations at the splice sites or splicing-regulatory elements of tumor-suppressor genes, but others reflect expression changes in splicing factors, as reported for colon6, ovarian7,8, and breast tumors9.
SRSF1 (SF2/ASF) is a prototypical SR protein that functions in constitutive and alternative splicing. SRSF1 also plays a role in nonsense-mediated mRNA decay (NMD)10, mRNA export11, and translation12–14. We previously demonstrated that SRSF1 can be oncogenic, driving transformation of murine immortal fibroblasts, and its oncogenic activity is partly mediated by controlling AS of the tumor suppressor BIN1 and the kinases Mnk2 and S6K1, inducing the expression of pro-tumorigenic isoforms15.
Although SRSF1 is frequently overexpressed in cancer, the significance and biological consequences of its overexpression in cancers of various cell types are unknown. Several lines of evidence suggest that splicing factors, including SRSF1, play a role in the development of mammary tumors in humans and mice9,16,17: i) SRSF1 is overexpressed >2-fold in 1/8 of the tumors in a breast-tumor panel15; ii) SRSF1 is upregulated in some breast cancers due to amplification of the Chr. 17q23 amplicon, which is associated with breast malignancies with poor prognosis18; iii) SRSF1 overexpression at the mRNA and protein levels was found in three breast-cancer cell lines with 17q23 amplification15.
As SRSF1 genomic amplification is preferentially observed in breast tumors, but several other genes are co-amplified on Chr. 17q23, we investigated whether SRSF1 can transform mammary epithelial cells. Our previous demonstration that SRSF1 is an oncoprotein15 relied on immunocompromised mice subcutaneously injected with fibroblasts transduced with SRSF1. To determine the potential of an oncogene to transform epithelial cells, it is necessary to use appropriate systems to measure tumorigenesis in the correct biological context, i.e., cell type, microenvironment, and differentiation program. Here we investigate the role of SRSF1 overexpression in mammary epithelial cell transformation by using a mouse orthotopic transplantation model, as well as an organotypic culture system that recapitulates the mammary-gland epithelial architecture, allowing a dissection of the underlying molecular mechanisms.
RESULTS
SRSF1 overexpression promotes mammary-gland tumorigenesis
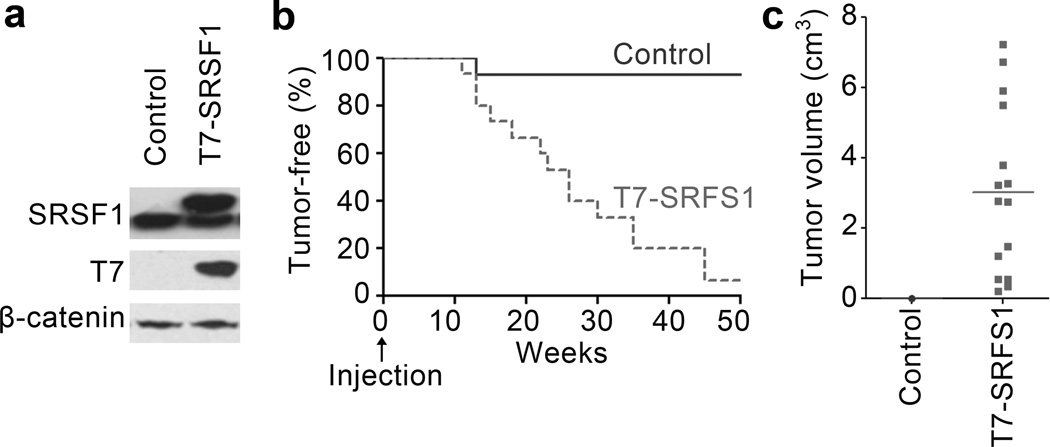
To assess SRSF1’s oncogenic effect in breast tissue in vivo, we used an orthotopic allograft mouse model based on mammary-gland reconstitution by injection of COMMA-1D cells, an immortalized, pluripotent mouse mammary epithelial cell line19,20. We generated COMMA-1D cells stably transduced with T7-SRSF1 (Fig. 1a) and injected them into the cleared mammary fat pad of 21-day old female BALB/c mice. Each transplanted mouse served as its own control, with one flank injected with control cells expressing the empty vector, and the contralateral flank injected with SRSF1-overexpressing cells, removing the effect of variations between animals. We monitored the repopulation of the mammary gland by staining and imaging the whole gland at 8 weeks post-transplantation (not shown). Starting at 12 weeks post-transplantation, SRSF1-overexpressing cells formed more tumors than the control cells (Fig. 1b). The tumors were malignant (Supplementary Fig. 1), highly proliferative, and often invaded into the skin and muscle. They exhibited many mitotic cells, very little necrosis, high angiogenesis, and well-defined borders. Only 1/15 transplants with the control vector gave rise to a very small tumor (<0.01 cm3), detected after euthanasia; in contrast, SRSF1-overexpressing cells gave tumors in 14/15 injections, and some grew quite large (3.0 cm3 average volume) (Fig. 1c). We conclude that SRSF1 promotes tumorigenesis in the mammary gland with high penetrance.
Figure 1.
SRSF1-overexpressing cells form tumors in an orthotopic allograft mouse model. (a) Western blot analysis of SRSF1 and T7 tag in COMMA-1D cells stably transduced with control (empty vector) or T7-SRSF1. (b) Tumor-free survival after injection of control and SRSF1-overexpressing COMMA-1D cells into the cleared fat pad of 21-day old female BALB/c mice (n= 15; Mantel-Cox test P<0.0005). (c) Size of tumors collected from the mice described in (b).
SRSF1 overexpression affects acinar size, proliferation and apoptosis
We also stably overexpressed T7-tagged SRSF1 cDNA in the non-transformed human mammary epithelial cell line MCF-10A, using retroviral transduction. MCF-10A cells have been extensively used to analyze the role of oncogenes in breast cancer. They undergo morphogenesis to form organized growth-arrested 3-D structures when grown in Matrigel—a basement-membrane-like extracellular matrix—recapitulating the acinar structure observed in the mammary gland21.
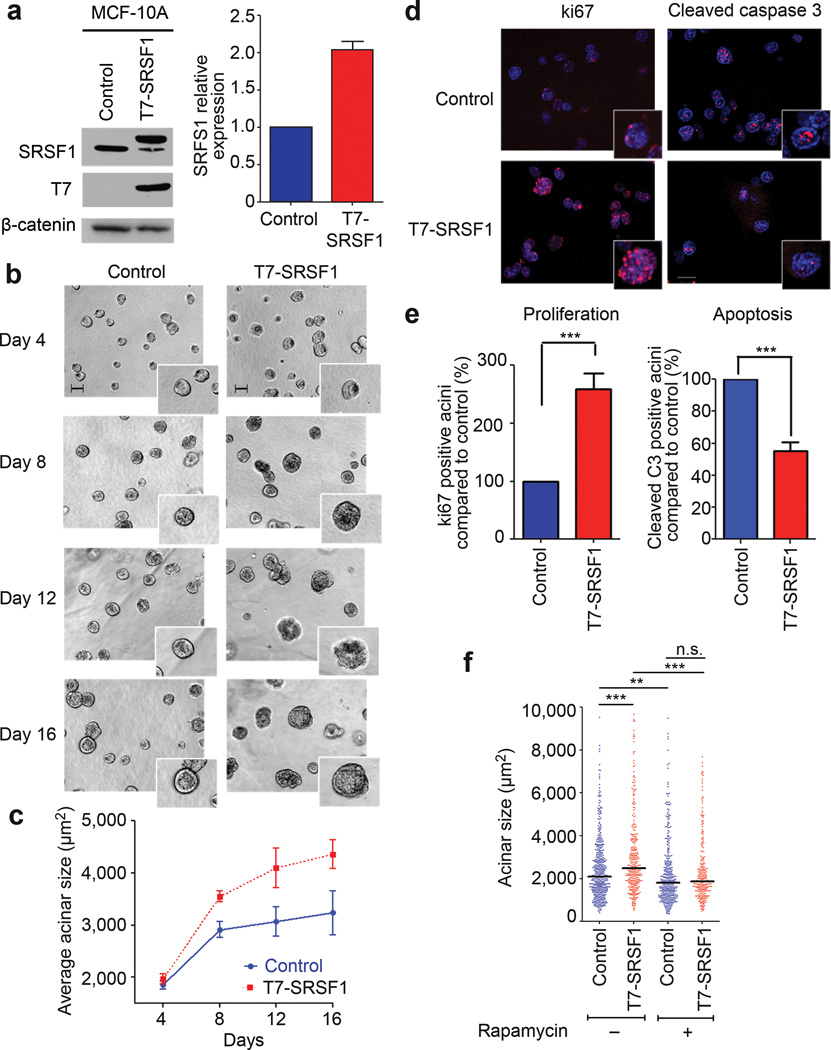
Immunoblotting of 3-D MCF-10A acini revealed that T7-SRSF1 was overexpressed 2–3 fold, compared to endogenous SRSF1 (Fig. 2a), similar to the level observed in human breast cancers15. The T7 tag has been extensively used and does not interfere with SRSF1’s known functions12–15,22. T7-SRSF1 correctly localized to the nucleus of MCF-10A cells within acini (Supplementary Fig. 2). MCF-10A cells also recapitulated the feedback inhibition of endogenous SRSF1 (Fig. 2a), which occurs at both post-transcriptional and translational levels in other cell types14,15,23.
Figure 2.
Overexpression of SRSF1 in MCF-10A cells increases acinar size in an mTOR-dependent manner. (a) Western blot analysis of SRSF1 and T7 tag in MCF-10A cells stably transduced with control (empty vector) or T7-SRSF1. SRSF1 expression was quantitated in day-8 acini by western blotting with infrared detection, and normalized to a loading control (n=5). Error bars, s.d. (b) 10× phase pictures of MCF-10A control and SRSF1-overexpressing acini from day 4 to day 16. Scale bar: 100 µm. The inserts show details of acinar morphology. (c) Average acinar size from day 4 to day 16 (n≥4, >100 acini per experiment; t-test day 8 P<2×10−9, day 16 P<5×10−19). Error bars, s.d. (d) Day-8 acini from control and SRSF1-overexpressing cells were immunostained for the proliferation marker ki67, or the apoptosis marker cleaved caspase-3 (red). Nuclei were costained with DAPI (blue). (e) Acini positive for ki67 or cleaved caspase-3 from control and SRSF1-overexpressing samples were counted on day 8. The percentage of positive SRSF1-overexpressing acini was plotted, compared to control acini (n≥3, >50 acini per experiment; Fisher test *** P<0.0001, ** P<0.001). Error bars, s.e.m. (f) The size of control and SRSF1-overexpressing acini was measured on day 16, following 12 days of treatment with rapamycin (10 nM). The dot plot shows the size distribution for each condition, with the median indicated by a horizontal line. (n=3, >100 acini per experiment; Mann-Whitney test *** P<0.0001, ** P<0.0006, n.s. not significant).
Oncogenes associated with breast cancer disrupt the highly organized architecture of MCF-10A acini24. We therefore assessed the effect of T7-SRSF1 overexpression on acinar morphology and size every four days. Cells overexpressing T7-SRSF1 formed significantly larger acini than control cells, starting on day 8 (Fig. 2b,c). However, T7-SRSF1 acini retained normal morphology after growth arrest, with a hollow lumen and no disruption of polarity markers, including α-laminin and Scribble (not shown).
The acinar-size increase could reflect an increase in either cell size or number. There was no significant difference in the size of single cells from MCF-10A 3-D (not shown) or 2-D cultures, as measured by flow cytometry (see below). MCF-10A acini undergo a proliferation phase, followed by an apoptosis phase that allows the clearing of the cells in the lumen25. Therefore, we assessed the expression of proliferation and apoptosis markers—ki67 and cleaved caspase-3, respectively—by indirect immunofluorescence in day-8 control and T7-SRSF1-overexpressing acini. T7-SRSF1 overexpression resulted in increased proliferation and decreased apoptosis (Fig. 2d,e). The difference in acinar size imparted by SRSF1 overexpression was maintained at later time points, as both control and SRSF1-overexpressing acini continued to grow. Thus, the control acini never reached the size of SRSF1-overexpressing acini, although their proliferation and apoptosis levels were comparable at day 16 (Supplementary Fig. 3), consistent with the luminal clearance observed in day-16 SRSF1-overexpressing acini.
SRSF1-induced acinar size increase involves mTOR signaling
SRSF1’s oncogenic activity in murine fibroblasts and human lung-cancer cell lines involves mTOR signaling—a major contributor to tumor growth and survival—bypassing Akt activation26. To examine the role of this pathway in mammary epithelial cells, we treated control and T7-SRSF1-overexpressing acini with the mTOR allosteric inhibitor rapamycin, starting on day 4, and followed acinar size and morphology. By day 16, rapamycin treatment resulted in a reduction in the size of T7-SRSF1-overexpressing acini to the level of control MCF-10A acini (Fig. 2f), indicating the involvement of the mTOR pathway in the acinar size-increase phenotype. SRSF1 overexpression in MCF-10A acini promoted phosphorylation of downstream components of the pathway, namely 4EBP1 and S6 (see below), as occurs in mouse fibroblasts15,26. Likewise, SRSF1 overexpression promoted eIF4E phosphorylation, bypassing upstream MAPK signaling. These changes in SRSF1-overexpressing acini are expected to result in increased translation.
SRSF1 overexpression affects alternative splicing in acini
SRSF1 affects AS of many target pre-mRNAs, some of which were previously characterized. In particular, SRSF1 regulates AS of the RON proto-oncogene (gene name: MSTR1) by promoting skipping of exon 11 to give the RONΔ11 isoform, which promotes cell motility and invasion27. SRSF1 overexpression also promotes the inclusion of exon 12a of BIN1 to generate the BIN1+12a isoform, which lacks tumor-suppressor activity because it no longer binds MYC15,28. Finally, SRSF1 enhances the inclusion of exon 13b of the kinase Mnk2 (gene name: MKNK2)15, one of two mutually exclusive 3’-terminal exons, to generate the Mnk2b isoform, which enhances eIF4E phosphorylation in a MAPK-independent manner29.
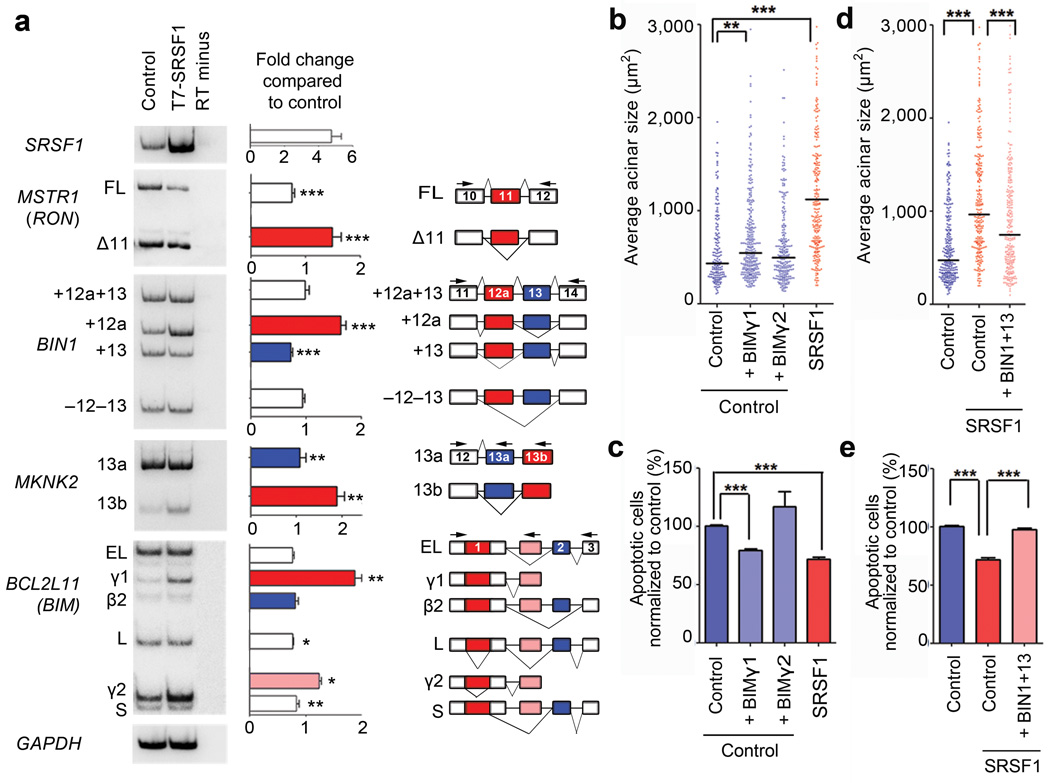
Overexpression of T7-SRSF1 in MCF-10A acini modified the AS patterns of all three of these transcripts, increasing expression of RONΔ11, BIN1+12a, and MKNK2-13b isoforms (Fig. 3a). SRSF1 overexpression in acini also induced the expression of S6K1-p31 (Supplementary Fig. 4a) a recently described isoform of the kinase S6K1 (gene name: RPS6KB1) that is involved in SRSF1’s oncogenic activity15.
Figure 3.
Alternative splicing of target genes in 3-D MCF-10A is involved in the SRSF1-induced phenotype. (a) RT-PCR analysis of SRSF1 target genes (RON, BIN1, MKNK2, and BIM) expressed in control and T7-SRSF1-overexpressing acini. Total RNA was analyzed by radioactive RT-PCR using the indicated primers (arrowheads), followed by native PAGE and autoradiography. GAPDH mRNA was used as a loading control. The exon-intron structure of each isoform is indicated (not to scale). Alternatively spliced exons are colored. The ratio of each isoform was quantified, and expressed as the fold change between control and T7-SRSF1-overexpressing acini (n≥6; t-test *** P<0.001, ** P<0.005, * P<0.01). Error bars, s.e.m. (b, d) The size of control and SRSF1-overexpressing acini overexpressing either the BIM γ1 or γ2 isoforms (b) or the BIN1+13 isoform (d) as indicated, was measured on day 8. The dot plot shows the size distribution for each condition, with the median indicated by a horizontal line. (n=3, >100 acini per experiment; Mann-Whitney test *** P<0.0001, ** P<0.001). (c, e) The level of apoptosis in control and SRSF1-overexpressing MCF-10A cells overexpressing either the BIM γ1 or γ2 isoforms (c) or the BIN1 13 isoform (e) as indicated, was measured by flow cytometry with Annexin V and PI staining. (n=3, t-test *** P<0.0001). Error bars, s.e.m.
Because SRSF1 overexpression delayed apoptosis in MCF-10A acini, we also investigated AS of BIM (gene name: BCL2L11), a pro-apoptotic Bcl-2 family member with a critical role in luminal apoptosis during acinar morphogenesis30. BIM is alternatively spliced to generate multiple isoforms with different apoptotic potential31,32, e.g., BIM EL, L, S, and β2. SRSF1 overexpression promoted the inclusion of a novel alternative 3’ exon, generating two new isoforms: BIM γ1 and γ2 (Fig. 3a and Supplementary Fig. 4). Expression of other BIM isoforms, including BIM EL, L, and S, concomitantly decreased (Fig. 3a). The γ1 and γ2 mRNAs lack exons 2 and 3 (Supplementary Fig. 5), which encode the BH3 domain of BIM; this domain binds anti-apoptotic Bcl-2 family members, and is necessary for induction of apoptosis by BIM32. Thus, the changes in BIM AS we observed are consistent with the delay in luminal apoptosis promoted by SRSF1. However, we did not detect a direct correlation between the levels of BIM γ1 and γ2 isoforms at days 4 and 16, and the delay of luminal apoptosis on SRSF1-overexpressing acini compared to control acini (Supplementary Fig. 4b)
To assess whether the new isoforms are involved in the SRSF1-induced acinar phenotype, we overexpressed them in control cells (Supplementary Fig. 4c). Expression of BIM γ1, but not γ2, promoted an increase in acinar size and a decrease in apoptosis (Fig. 3b,c), to levels intermediate between control and SRSF1-overexpressing cells. In parallel, we expressed the BIN+13 isoform in SRSF1-overexpressing cells (Supplementary Fig. 4d). Expression of BIN+13 decreased the acinar size and increased apoptosis of SRSF1-overexpressing cells (Fig. 3d,e). This suggests that both BIM γ1 upregulation and BIN+13 downregulation contribute to the SRSF1-induced phenotype.
Domain requirements for SRSF1-induced changes
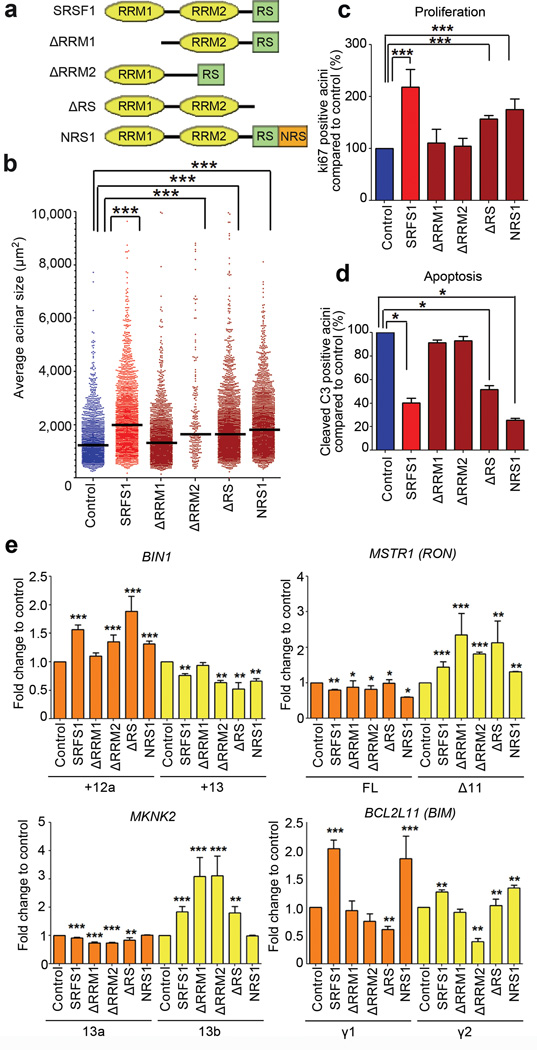
To dissect the mechanisms involved in the SRSF1-mediated acinar-size increase, we used available SRSF1 mutants33 lacking either of the two RNA-recognition motifs (ΔRRM1 or ΔRRM2 mutants) or the serine/argine-rich C-terminal domain (ΔRS mutant) (Fig. 4a and Supplementary Table 1). The RRMs recognize specific RNA sequences, and the RS domain is involved in protein-protein interactions, subcellular localization, and recruitment of spliceosome components2. We also used a C-terminal fusion of SRSF1 with the nuclear-retention signal of SRSF2, to force SRSF1 retention in the nucleus34 and assess the role of its nuclear (i.e., splicing and NMD) versus cytoplasmic (i.e., translation) functions.
Figure 4.
The SRSF1-induced increase in acinar size requires RRM1 and SRSF1 nuclear functions. (a) SRSF1 deletion mutants lack either RRM1, RRM2, or the RS domain; NRS1 consists of a C-terminal fusion to a nuclear retention signal from SRSF2. (b) Acinar sizes of control and wild-type or mutant SRSF1-overexpressing acini measured on day 8. The dot plot shows the size distribution and the median (horizontal line) for each condition. (n≥3, >150 acini per experiment; Mann-Whitney test *** P<0.001). (c, d) Proliferation (c) and apoptosis (d) in wild-type and mutant SRSF1-overexpressing acini were quantified on day 8, compared to control acini, as described in Fig. 2d (n≥3, >50 acini per condition; Fisher test *** P<0.0001; * P<0.01). Error bars, s.e.m. (e) Quantification of the level of BIN1, MKNK2, RON, and BIM AS isoforms in wild-type or mutant SRSF1-overexpressing acini on day 8, compared to control acini, as in Fig. 3a. Total RNA was analyzed by RT-PCR. Isoforms described in Fig. 3a are indicated. (n≥3; t-test *** P<0.001; ** P<0.001; * P<0.01). Error bars, s.e.m. Representative gels are shown in Supplementary Fig. 6b. See Supplementary Table 1 for summary information.
We stably transduced MCF-10A cells with each SRSF1 mutant, resulting in expression levels comparable to the wild type (Supplementary Fig. 6a). Only the ΔRRM1 mutant failed to promote the acinar-size increase (Fig. 4b). In contrast, MCF-10A cells overexpressing the ΔRRM2, ΔRS, or NRS1 mutants exhibited a similar size increase by day 8 as cells overexpressing wild-type SRSF1 (Fig. 4b and Supplementary Table 1). On day 8, the acini overexpressing ΔRRM1 or ΔRRM2 exhibited slightly decreased proliferation, compared to wild-type SRSF1-overexpressing acini; likewise, they showed increased apoptosis, similar to the levels of the control acini (Fig. 4c,d). However, either ΔRRM2 or NRS1 expression resulted in slight but significant cell-size increases compared to the control (Supplementary Fig. 6b), partly accounting for the acinar-size increase. NRS1 and ΔRS-overexpressing acini exhibited similar proliferation and apoptosis levels as wild-type SRSF1 acini (Fig. 4c,d). In addition, ΔRS- and NRS1-overexpressing cells, but not the cells overexpressing the other mutants, promoted activation of mTOR to the same or a greater extent than wild-type SRSF1—as measured by increased phosphorylation of S6 and 4EBP1 (Supplementary Fig. 6c).
The acinar-size phenotype correlated with AS changes in BIN1: the ΔRRM2, ΔRS, and NRS1 mutants promoted expression of BIN1+12a at a level comparable to wild-type SRSF1 (Fig. 4e and Supplementary Fig. 6d); in contrast, ΔRRM1-ovexepresssing cells exhibited the same BIN1 AS pattern as control cells. Interestingly, the ΔRS mutant exhibited a similar pattern as wild-type SRSF1 for AS of MKNK2-13b, whereas ΔRRM1 and ΔRRM2 behaved differently, promoting greater inclusion of exon 13b (Fig. 4e). In contrast, the NRS1 mutant did not promote the MKNK2 AS changes observed for wild-type SRSF1, suggesting that SRSF1 affects this splicing event indirectly, e.g., by enhancing translation of another splicing factor that in turn regulates MKNK2. All mutants affected RON AS in the same manner as wild-type SRSF1 (Fig. 4e). Finally, only the NRS1 mutant promoted expression of BIM γ1, and both NRS1- and ΔRS-overexpressing cells expressed BIM γ2 at the same level as wild-type SRSF1-overexpressing cells (Fig. 4e). At the protein level, ΔRRM2- and ΔRS-overexpressing cells showed expression of a new isoform of BIM to the same extent as wild-type SRSF1-overexpressing cells (Supplementary Fig. 6a). These results indicate that ΔRRM2 is not just a general loss-of-function mutant, and also suggest the existence of substrate-specific interactions between various splicing targets of SRSF1 and RRM1 and/or RRM2 of SRSF1. Among the AS changes analyzed, BIN1 correlated best with the observed acinar phenotypic changes, whereas BIM and MKNK2 showed weaker correlation (Supplementary Table 1). These data also indicate that the nuclear functions of SRSF1 are sufficient for the acinar-size increase, and that the various targets are not affected equally by each of the modular domains of SRSF1.
SRSF1 cooperates with MYC
Transformation often results from cooperation among several oncogenes35. Therefore, we investigated whether SRSF1 can cooperate in mammary epithelial cell transformation with known oncogenes associated with breast cancer and previously studied in 3-D culture. We generated MCF-10A cells overexpressing SRSF1 together with MYC20, ERBB2 (ref. 24), or HPV16 E7 (ref. 25) oncogenes (Supplementary Fig. 7a), each representing a major pathway involved in tumorigenesis. MYC and ERBB2 are frequently overexpressed in breast cancer, whereas HPV16 E7 overexpression mimics RB inactivation, another frequent event in breast cancer (for reviews see 36,37).
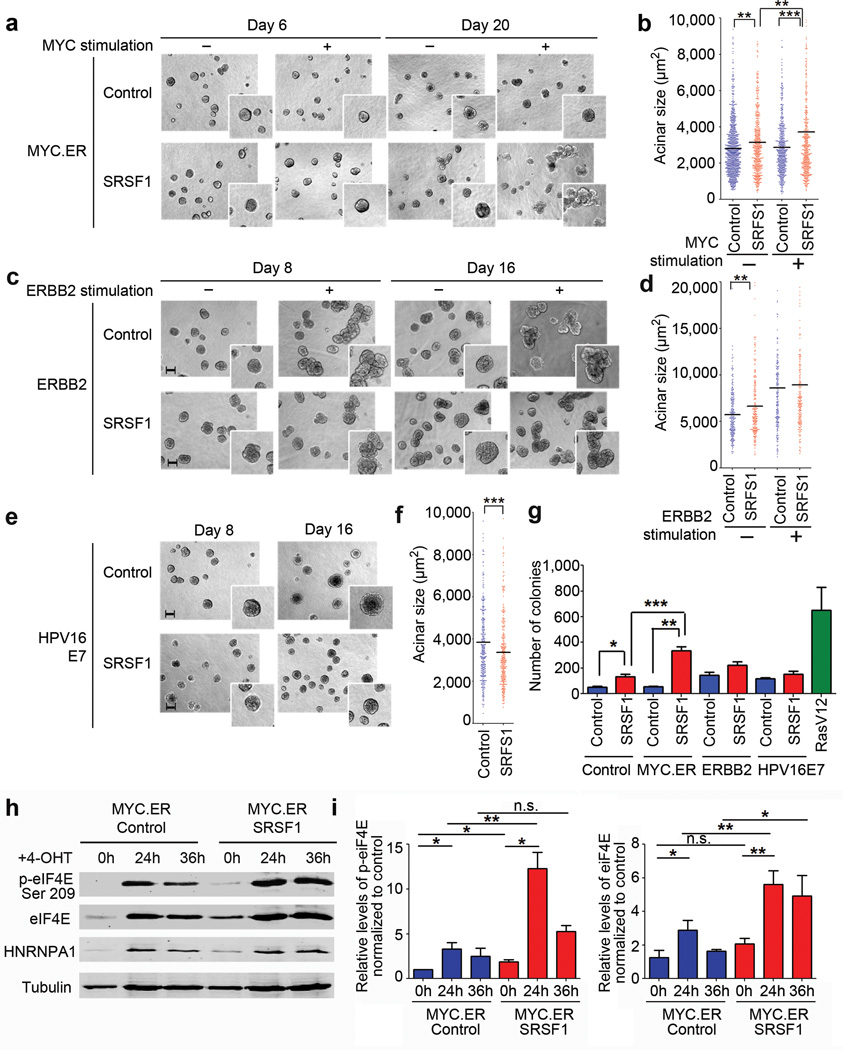
Cells overexpressing SRSF1 and MYC formed multiacinar, disorganized structures by day 20, not observed with either MYC or SRSF1 alone (Fig. 5a,b). MYC overexpression increased apoptosis, as expected20, and SRSF1 overexpression together with MYC diminished this apoptosis induction by 25% (Supplementary Fig. 7b). The NRS1 variant also cooperated with MYC to form large multiacinar structures (Supplementary Fig. 7c,d). In contrast, no significant size or morphological differences were observed for cells simultaneously overexpressing SRSF1 and ERBB2 (Fig. 5c,d). Curiously, MCF-10A cells overexpressing SRSF1 together with HPV16 E7 formed smaller acini than HPV16 E7 alone (Fig. 5e,f) and exhibited decreased proliferation and increased apoptosis compared to wild-type E7 (Supplementary Fig. 7b). We conclude that SRSF1 cooperates specifically with MYC, but not with the other oncogenes, to transform mammary epithelial cells. Consistent with the observations in 3-D culture, MCF-10A cells overexpressing both SRSF1 and MYC formed more colonies in soft agar than cells overexpressing either oncogene alone (Fig. 5g).
Figure 5.
SRSF1 cooperates with MYC, but not with ERBB2 or HPV16 E7. (a, c, e) 10× phase pictures of MCF-10A control acini or overexpressing SRSF1 together with estrogen-receptor (ER)-inducible MYC (a), inducible ERBB2 (c) or HPV16 E7 (e). The inserts show details of acinar morphology. Scale bar: 100 µm. MYC.ER acini were stimulated with 4-OHT from day 3 to day 20 to activate MYC.ER. ERBB2 acini were stimulated with AP1510 to activate ERBB2 from day 4 to day 8 or from day 8 to day 16. (b, d, f) Size of control acini or overexpressing SRSF1 together with MYC measured on day 20 (b), with ERBB2 on day 16 (d), and with HPV16 E7 on day 16 (f). The dot plot shows the size distribution and the median (horizontal line) for each condition. (n≥3, >100 acini per experiment; Mann-Whitney test *** P< 0.0001, ** P<0.001). (g) MCF-10A control cells or overexpressing SRSF1 together with MYC, HPV16 E7, or ERBB2 were plated in soft agar, and colonies were counted after 30 days. MCF-10A cells overexpressing RasV12 were used as a positive control (n=3; t-test *** P<0.001, ** P<0.005, * P<0.02). Error bars, s.d. (h) Western blot analysis of MCF-10A cells control cells or overexpressing inducible MYC.ER together with SRSF1 using phospho- and total eIF4E antibodies. hnRNPA1 was used as a control for MYC activation upon 4-OHT treatment for the indicated time points. (i) Quantification of phospho- and total eIF4E levels in cells from (h) normalized to tubulin loading control and to the level of MYC.ER control cells at 0 h (n=3; t-test **P<0.001, *P<0.05, n.s. not significant). Error bars, s.d.
We did not detect significant changes in MYC’s transcriptional activation of selected target genes upon SRSF1 overexpression (Supplementary Fig. 7e,f). To investigate how MYC and SRSF1 cooperate in transformation, we measured eIF4E activation upon MYC induction. As expected38,39, MYC induction promoted eIF4E expression (Fig. 5h,i). In addition, SRSF1 overexpression upregulated eIF4E phosphorylation (Fig. 5h,i and Supplementary Fig. 5c). However, MYC induction in SRSF1-overexpressing cells activated eIF4E phosphorylation to levels 4-fold higher that MYC alone, and 6-fold higher than SRSF1 alone (Fig. 5h,i), suggesting one mechanism of cooperation.
SRSF1 is overexpressed in human tumors with elevated MYC
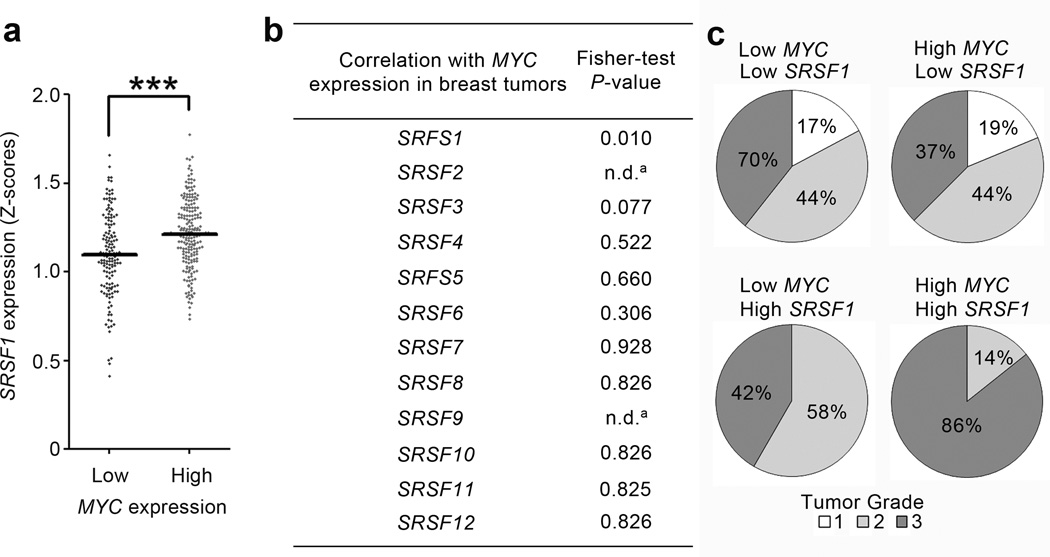
To assess the significance of MYC and SRSF1 cooperation in the context of human tumors, we used microarray data to analyze their expression in a large collection of tumors comprising 23 different cancer types. We compared the numbers of tumors expressing high levels of SRSF1 in two categories: tumors with high versus low MYC levels. High SRSF1 expression occurred significantly more often in tumors that overexpressed MYC, in 3/23 human tumor types analyzed, with breast cancer showing the strongest co-expression (Fig. 6a and data not shown). MYC expression in breast tumors was not significantly correlated with expression of the other 11 SR proteins in the same data set, indicating the specificity of the co-overexpression with SRSF1 (Fig. 6b). No correlation was observed in breast tumors for ERBB2 with SRSF1 or other SR proteins (not shown), consistent with the in vitro data from ERBB2/SRSF1 co-overexpressing acini (Fig. 5), and underscoring the significance of SRSF1/MYC co-overexpression in clinical samples. In addition, breast tumors overexpressing MYC and SRSF1 were of higher histological grade, compared to tumors with low SRSF1 and/or MYC (Fig. 6c).
Figure 6.
SRSF1 is frequently overexpressed in human breast tumors with elevated MYC. (a) Expression of SRSF1 was profiled from microarray data from a collection of 352 human breast tumors (GSE2109). The data were normalized to Z-score (see Methods) and divided into two categories: breast tumors expressing high or low MYC levels. The dot plot shows the distribution and the median (horizontal line) for each condition (Mann-Whitney test *** P<0.0001). (b) Expression of all members of the SR protein family was profiled as in (a) and divided into four categories according to SRSF and MYC RNA levels: (i) both low; (ii) both high; (iii) low MYC and high SRSF; (iv) high MYC and low SRSF. A Fisher-test was performed to compare the different categories, and P-values are shown on the right (a There were not enough samples for SRSF2 and SRSF9 with low SRSF expression to derive a P-value). (c) Expression of SRSF1 and MYC was profiled from microarray data from another collection of 196 human breast tumors with histological grading annotation (GSE7390). The data were normalized to Z-score (see Methods) and divided into four categories according to SRSF1 and MYC levels, as indicated. Histological grading of the tumors is indicated for each category, grade 3 being the most malignant. Tumors with high levels of MYC and SRSF1 have more grade-3 samples, compared to tumors with low SRSF1 and/or MYC levels (Fisher-test: P< 0.03).
DISCUSSION
The splicing factor SRSF1 is overexpressed in human tumors15. Here we investigated its involvement in breast cancer, using appropriate systems in the correct biological context, i.e., transduced mouse mammary progenitors engrafted in cleared mammary-gland fat pad of syngeneic mice, and transduced human mammary cells that differentiate into acini in 3-D culture. Overexpression of SRSF1 in transplanted murine COMMA-1D cells was sufficient to promote mammary gland tumorigenesis. It is remarkable that SRSF1 alone can robustly promote tumorigenesis in a system that does not by itself result in spontaneous tumors.
We then investigated the cellular and molecular consequences of overexpressing SRSF1 in a non-transformed human mammary epithelial cell line that forms acinar structures in organotypic culture. This 3-D model recapitulates many features of normal mammary epithelium architecture in vivo21. Moderate SRSF1 overexpression, mimicking the levels seen in many human tumors15, significantly affected proliferation and apoptosis—two pathways that are frequently deregulated during transformation. SRSF1 overexpression enhanced cell proliferation and delayed apoptosis during acinar morphogenesis, resulting in larger acini. SRSF1 overexpression was not sufficient to disrupt the acinar architecture, suggesting that additional steps are necessary for full transformation in this system. Despite the subtle in vitro phenotype, SRSF1 can clearly initiate transformation in the mammary gland, though additional events likely accelerate tumorigenesis. However, we cannot rule out that the apparent differences between in vitro and in vivo experiments reflect differences between human MCF-10A and murine COMMA-1D cells. Furthermore, the mammary gland is composed of different cell types, and SRSF1 may play a more complex role in the interactions between epithelial cells and the microenvironment, an important aspect of cancer progression that the in vitro model does not address.
SRSF1 overexpression in acini affected specific AS events in the RON proto-oncogene, the tumor suppressor BIN1, and the kinases MNK2 and S6K1, promoting the expression of pro-oncogenic isoforms. The AS changes were relatively subtle—consistent with the modest overexpression of SRSF1—suggesting that even slight variations in AS regulation can have important phenotypic consequences. It is possible that more pronounced changes in AS of as yet unidentified targets of SRSF1, and/or known targets we have not tested, contribute to the observed phenotype.
SRSF1 overexpression additionally promoted the expression of two novel BIM isoforms, dubbed γ1 and γ2, which include a novel alternative 3’ exon. BIM belongs to the Bcl-2 family, and promotes apoptosis by inhibiting anti-apoptotic Bcl-2 family members31. Several isoforms have been characterized, with varying apoptotic activity, BIM S being the most potent31,32,40. The new γ1 and γ2 isoforms are not expected to promote apoptosis, as they lack exons coding for the BH3 domain and the C-terminal hydrophobic region, similar to the β2 isoform32. Overexpression of the γ1 isoform in control cells increased acinar size and decreased apoptosis. In parallel, overexpression of BIN+13 in SRSF1-overexpressing cells decreased acinar size and increased apoptosis. In both situations, the acinar size and apoptosis increased/decreased to levels intermediate between control and SRSF1-overexpressing cells, suggesting that the increase in BIM γ1 and the decrease in BIN+13 both contribute to the SRSF1-induced phenotype. Some of the differences between the γ1 and γ2 isoforms may reflect differences in their overexpression levels. In addition, they accumulate to higher levels in the overexpression lines than in SRSF1-overexpressing cells, as the system we used does not allow titration of their expression. It is very likely that the observed acinar phenotype of SRSF1 overexpression results from changes in multiple AS events that regulate cell death and proliferation, some of which we described here, and others yet to be identified.
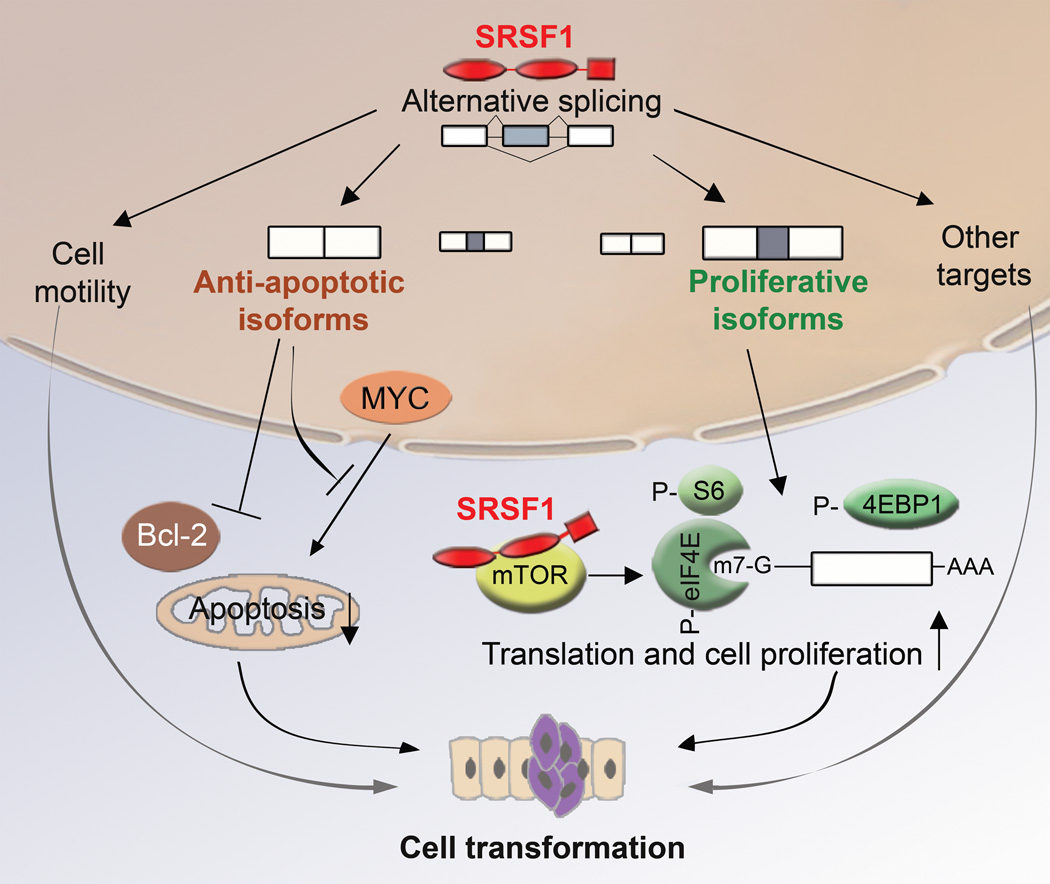
Many apoptosis factors are expressed via AS, generating isoforms with antagonistic effects41. Our study suggests that SRSF1 can regulate apoptosis by promoting the expression of BIN1 and BIM isoforms lacking pro-apoptotic activity. In addition, SRSF1 regulates AS of BCLX, MCL1 and caspases 2 and 9 (ref. 42), and its knockdown induces G2 cell-cycle arrest and apoptosis43. These data, together with our present and previous results from SRSF1-overexpressing fibroblasts15 and epithelial cells, suggest that SRSF1 plays a broad role in regulating apoptosis and cell-proliferation pathways. Thus, subtle variations in SRSF1 levels and/or activity can affect cell death and may constitute an initial step in transformation (Fig. 7).
Figure 7.
A model for SRSF1’s role in transformation. SRSF1 regulates AS of target genes involved in apoptosis, cell motility, proliferation, and other cellular functions. SRSF1 overexpression promotes expression of anti-apoptotic isoforms unable to interact with pro-apoptotic factors, or that inhibit the action of pro-apoptotic factors, such as MYC or members of the Bcl-2 family. In parallel, SRSF1 overexpression promotes expression of isoforms that stimulate translation and cell proliferation, by increasing phosphorylation of translation activators, such as S6 or eIF4E, or by inhibiting translational repressors, such as 4EBP1. SRSF1 can also interact with and activate mTOR to promote translation. By increasing proliferation and decreasing apoptosis, SRSF1 promotes cellular transformation.
We found that SRSF1’s effect on acinar size involves mTOR signaling, a pathway that controls cell proliferation and translation, and is often deregulated in cancer44. However, we cannot exclude the possibility that the effect of rapamycin on SRSF1-overexpressing acini reflects at least partly a general block of proliferation, as rapamycin also decreased the size of the control acini. In addition, rapamycin did not change the splicing patterns of BIN1, BIM, RON, or MKNK2 in SRSF1-overexpressing acini, compared to control acini (not shown), suggesting that mTOR does not affect SRSF1’s splicing functions for the tested targets. In SRSF1-overexpressing cells, proliferation is regulated at least partly by promoting AS of the MKNK2-b and S6K1-p31 isoforms. On the other hand, phosphorylation of ribosomal protein S6 and of the competitive inhibitor of cap-dependent translation, 4EBP1, are enhanced, possibly through an interaction with mTOR12; likewise, phosphorylation of the cap-binding protein eIF4E is enhanced, bypassing the activation of MAPK signaling pathways. All these changes lead to enhanced translation. Furthermore, eIF4E acts as an oncoprotein when overexpressed or hyperactivated45,46. Finally, 4EBP1 was recently implicated in cell proliferation and cell-cycle progression, whereas S6 kinase controls cell size47. Thus, SRSF1 potentially controls multiple regulators of the cell cycle and growth, via splicing-dependent, but possibly also splicing-independent functions, and it can cause deregulation of these processes in cancer (Fig. 7).
Our structure-function dissection of the modular protein domains of SRSF1 indicates that RRM1 is necessary to mediate the acinar size-increase phenotype. RRM1 deletion RRM1 prevented the delay in apoptosis and the increase in proliferation observed for intact SRSF1. Our data suggest that SRSF1’s role in regulating proliferation and apoptosis involves splicing targets recognized primarily by RRM1 (Supplementary Table 1). Surprisingly, RRM1 deletion did not affect all the tested SFRS1 target genes equally. Moreover, not all these AS events were affected by RRM1 deletion in a manner that correlated with the loss of the effect on acinar size (Supplementary Table 1). Notably, BIN1 AS showed the strongest correlation, followed by BIM and MKNK2, suggesting that only some of the observed AS changes are involved in the acinar-size increase. Furthermore, the NRS1 mutant behaved similarly to wild-type SRSF1 in almost all the assays, indicating that the nuclear functions of SRSF1 are responsible for the acinar phenotype. Finally, because the ΔRS mutant was essentially like wild-type SRSF1, but it cannot promote NMD10, we conclude that this function is not required for the acinar phenotype, whereas SRSF1’s splicing activity plays a key role in SRSF1-mediated transformation. In addition, while both ΔRRM1 and ΔRS bind mTOR12,13, only ΔRS promoted the acinar-size increase and phosphorylation of S6 and 4EBP1 as efficiently as wild-type SRSF1 (Supplementary Table 1). Therefore, physical interaction with mTOR is probably neither required nor sufficient for the acinar phenotype. However, because our system does not allow fine-tuning of the expression of wild-type and mutant proteins, we cannot exclude the possibility that some of the observed phenotypic differences reflect subtle variations in their expression levels.
The domain analysis of SRSF1 further suggests the existence of unexplored differences in RNA-binding specificity between its two RRMs. Little is known about the individual roles and splicing-target specificity of these RRMs, though their 3-D structures have been separately determined48,49. The RRMs are involved in sequence-specific RNA binding, and are required for both constitutive and alternative splicing22,33,50. RRM2, but not RRM1, is also required for SRSF1’s activities in NMD10, translational control12,13, and auto-regulation14, as well as for its interaction with mTOR12. Finally, the C-terminal RS domain is involved in nuclear localization22, nuclear-cytoplasmic shuttling51, as well as in mediating protein-protein interactions and promoting splicing by recruiting other components of the spliceosome2, but is essential only for SRSF1’s function in NMD10,52. Thus, although SRSF1 is a multifunctional protein, we demonstrate here that the nuclear functions, particularly splicing, are sufficient for transformation, and that the various targets are not equally important for tumorigenesis.
SRSF1 cooperated with MYC to disrupt acinar architecture. MYC activation may be one of the additional events required in SRSF1-overexpressing acini to promote the switch from a hyperproliferative state to a fully transformed state. SRSF1 did not cooperate with two other oncogenes—ERBB2 and HPV16 E7—indicating the specificity of the SRSF1-MYC interaction. Selective cooperation between oncogenes has already been described in breast cancer53, and is now extended to a splicing-factor oncogene. Our analysis of microarray data showed that MYC and SRSF1 are strikingly correlated in a subset of human tumors, particularly in breast cancer, suggesting that the cooperation we observed in the in vitro model is clinically relevant. Remarkably, no other SR proteins showed such a correlation.
The mechanisms of SRSF1 and MYC cooperation warrant further investigation, and will likely reveal a high level of complexity that may provide insights into SRSF1’s roles in tumorigenesis. We previously showed that SRSF1 promotes phosphorylation of eIF4E15,28, and this factor is known to enhance MYC’s translation38. MYC and eIF4E cooperate to induce tumorigenesis: eIF4E antagonizes MYC-induced apoptosis54, whereas MYC overrides eIF4E-induced senescence39. Here, we demonstrate that the molecular basis of MYC and SRSF1 cooperation includes at least in part hyper-activation of eiF4E. Furthermore, the SFRS1 promoter was identified as a putative MYC binding site in a genomewide screen for MYC targets55, and we recently demonstrated that MYC binds directly to the SFRS1 promoter and regulates its expression (Das S., Anczuków O., Akerman M., and Krainer A.R., unpublished data)—including in MCF-10A cells—suggesting a possible basis for the correlation. However, in MCF-10A cells, the total levels of SRSF1 are comparable upon MYC induction in cells overexpressing MYC alone or together with SRSF1 (Supplementary Fig. 7a). Therefore, the cooperative effects of MYC and SRSF1 probably reflect more complex mechanisms than just changes in SRSF1 levels. In addition, SRSF1 regulates AS of BIN1 (ref. 15,28), a tumor suppressor that inhibits MYC’s transforming activity56,57. Thus, SRSF1, being frequently overexpressed in breast tumors, may play an important role in regulating BIN1 to promote the expression of isoforms that fail to interact with MYC15,28, thereby abrogating its negative control. In this way, SRSF1 regulates multiple factors with roles in MYC-induced transformation.
In summary, SRSF1 promotes mammary epithelial cell transformation by specifically regulating splicing of key targets downstream of mTOR and/or functionally linked to MYC. Most likely, each of these splicing events contributes partially to SRSF1-induced tumorigenesis. Identifying additional SRSF1 splicing targets involved in transformation, and gaining a deeper understanding of how SRSF1 levels are regulated, should be important priorities towards the development of therapeutic strategies that specifically target relevant AS isoforms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mikala Egeblad, Nicholas Tonks, Victoria Aranda and Jaclyn Novatt for helpful comments on the manuscript. We thank John Erby Wilkinson for collaboration and helpful advice with histopathology, and Pamela Moody for assistance with flow cytometry. This work was supported by grants from the National Cancer Institute (grant CA13106 to A.R.K. and CA098830 to S.K.M.), postdoctoral fellowships from the Susan B. Komen Foundation for the Cure (grant KG091029 to O.A.) and from the Fondation pour la Recherche Médicale (grant SPE20070709581 to O.A.), an award grant from the Philippe Foundation to O.A. and an Era of Hope Scholar award from the Department of Defense Breast Cancer Research Program (grant BC075024 to S.K.M.).
Footnotes
AUTHOR CONTRIBUTIONS
O.A. conducted the experiments. S.D. contributed to the apoptosis assays. M.A. performed the microarray and statistical analyses. L.Z. contributed to the transplantation experiments. A.Z.R., R.K. and S.K.M. shared protocols and reagents. O.A. and A.R.K. designed the study, analyzed the data and wrote the paper. All authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 3.Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 4.Caceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 5.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 6.Ghigna C, Moroni M, Porta C, Riva S, Biamonti G. Altered expression of heterogenous nuclear ribonucleoproteins and SR factors in human colon adenocarcinomas. Cancer Res. 1998;58:5818–5824. [PubMed] [Google Scholar]
- 7.Fischer DC, et al. Expression of splicing factors in human ovarian cancer. Oncol Rep. 2004;11:1085–1090. [PubMed] [Google Scholar]
- 8.He X, Ee PL, Coon JS, Beck WT. Alternative splicing of the multidrug resistance protein 1/ATP binding cassette transporter subfamily gene in ovarian cancer creates functional splice variants and is associated with increased expression of the splicing factors PTB and SRp20. Clin Cancer Res. 2004;10:4652–4660. doi: 10.1158/1078-0432.CCR-03-0439. [DOI] [PubMed] [Google Scholar]
- 9.Stickeler E, Kittrell F, Medina D, Berget SM. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18:3574–3582. doi: 10.1038/sj.onc.1202671. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 12.Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol. 2010;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karni R, et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pind MT, Watson PH. SR protein expression and CD44 splicing pattern in human breast tumours. Breast Cancer Res Treat. 2003;79:75–82. doi: 10.1023/a:1023338718974. [DOI] [PubMed] [Google Scholar]
- 17.Watermann DO, et al. Splicing factor Tra2-beta1 is specifically induced in breast cancer and regulates alternative splicing of the CD44 gene. Cancer Res. 2006;66:4774–4780. doi: 10.1158/0008-5472.CAN-04-3294. [DOI] [PubMed] [Google Scholar]
- 18.Sinclair CS, Rowley M, Naderi A, Couch FJ. The 17q23 amplicon and breast cancer. Breast Cancer Res Treat. 2003;78:313–322. doi: 10.1023/a:1023081624133. [DOI] [PubMed] [Google Scholar]
- 19.Deugnier MA, et al. Isolation of mouse mammary epithelial progenitor cells with basal characteristics from the Comma-Dbeta cell line. Dev Biol. 2006;293:414–425. doi: 10.1016/j.ydbio.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Zhan L, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 22.Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, et al. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debnath J, et al. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 26.Karni R, Hippo Y, Lowe SW, Krainer AR. The splicing-factor oncoprotein SF2/ASF activates mTORC1. Proc Natl Acad Sci U S A. 2008;105:15323–15327. doi: 10.1073/pnas.0801376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghigna C, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Ge K, et al. Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc Natl Acad Sci U S A. 1999;96:9689–9694. doi: 10.1073/pnas.96.17.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheper GC, et al. The N and C termini of the splice variants of the human mitogen-activated protein kinase-interacting kinase Mnk2 determine activity and localization. Mol Cell Biol. 2003;23:5692–5705. doi: 10.1128/MCB.23.16.5692-5705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reginato MJ, et al. Bim regulation of lumen formation in cultured mammary epithelial acini is targeted by oncogenes. Mol Cell Biol. 2005;25:4591–4601. doi: 10.1128/MCB.25.11.4591-4601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connor L, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. Embo J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.U M, Miyashita T, Shikama Y, Tadokoro K, Yamada M. Molecular cloning and characterization of six novel isoforms of human Bim, a member of the proapoptotic Bcl-2 family. FEBS Lett. 2001;509:135–141. doi: 10.1016/s0014-5793(01)03145-3. [DOI] [PubMed] [Google Scholar]
- 33.Caceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. Embo J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cazalla D, et al. Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedraza-Farina LG. Mechanisms of oncogenic cooperation in cancer initiation and metastasis. Yale J Biol Med. 2006;79:95–103. [PMC free article] [PubMed] [Google Scholar]
- 36.Hynes NE, Stoelzle T. Key signalling nodes in mammary gland development and cancer: Myc. Breast Cancer Res. 2009;11:210. doi: 10.1186/bcr2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harb Perspect Biol. 2010;2:a003236. doi: 10.1101/cshperspect.a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin CJ, Cencic R, Mills JR, Robert F, Pelletier J. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 2008;68:5326–5334. doi: 10.1158/0008-5472.CAN-07-5876. [DOI] [PubMed] [Google Scholar]
- 39.Ruggero D, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 40.Miao J, et al. Identification and characterization of BH3 domain protein Bim and its isoforms in human hepatocellular carcinomas. Apoptosis. 2007;12:1691–1701. doi: 10.1007/s10495-007-0093-5. [DOI] [PubMed] [Google Scholar]
- 41.Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 42.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 45.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5' cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 46.Topisirovic I, Ruiz-Gutierrez M, Borden KL. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res. 2004;64:8639–8642. doi: 10.1158/0008-5472.CAN-04-2677. [DOI] [PubMed] [Google Scholar]
- 47.Dowling RJ, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ngo JC, et al. A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol Cell. 2008;29:563–576. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tintaru AM, et al. Structural and functional analysis of RNA and TAP binding to SF2/ASF. EMBO Rep. 2007;8:756–762. doi: 10.1038/sj.embor.7401031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuo P, Manley JL. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw SD, Chakrabarti S, Ghosh G, Krainer AR. Deletion of the N-terminus of SF2/ASF permits RS-domain-independent pre-mRNA splicing. PLoS One. 2007;2:e854. doi: 10.1371/journal.pone.0000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 54.Li S, et al. Translation factor eIF4E rescues cells from Myc-dependent apoptosis by inhibiting cytochrome c release. J Biol Chem. 2003;278:3015–3022. doi: 10.1074/jbc.M208821200. [DOI] [PubMed] [Google Scholar]
- 55.Mao DY, et al. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr Biol. 2003;13:882–886. doi: 10.1016/s0960-9822(03)00297-5. [DOI] [PubMed] [Google Scholar]
- 56.Elliott K, et al. Bin1 functionally interacts with Myc and inhibits cell proliferation via multiple mechanisms. Oncogene. 1999;18:3564–3573. doi: 10.1038/sj.onc.1202670. [DOI] [PubMed] [Google Scholar]
- 57.Sakamuro D, Elliott KJ, Wechsler-Reya R, Prendergast GC. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat Genet. 1996;14:69–77. doi: 10.1038/ng0996-69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.