Abstract
mirEX is a comprehensive platform for comparative analysis of primary microRNA expression data. RT–qPCR-based gene expression profiles are stored in a universal and expandable database scheme and wrapped by an intuitive user-friendly interface. A new way of accessing gene expression data in mirEX includes a simple mouse operated querying system and dynamic graphs for data mining analyses. In contrast to other publicly available databases, the mirEX interface allows a simultaneous comparison of expression levels between various microRNA genes in diverse organs and developmental stages. Currently, mirEX integrates information about the expression profile of 190 Arabidopsis thaliana pri-miRNAs in seven different developmental stages: seeds, seedlings and various organs of mature plants. Additionally, by providing RNA structural models, publicly available deep sequencing results, experimental procedure details and careful selection of auxiliary data in the form of web links, mirEX can function as a one-stop solution for Arabidopsis microRNA information. A web-based mirEX interface can be accessed at http://bioinfo.amu.edu.pl/mirex.
INTRODUCTION
MicroRNAs are the key post-transcriptional regulators of gene expression in Eukaryota. They control gene expression by targeting the cleavage of cognate mRNAs or by inhibiting their translation (1–3). Therefore, when studying the biology of any organism, it is of utmost importance to quantify precisely the expression level of each particular microRNA gene during organ development. Northern hybridization, microarray analysis, deep sequencing approaches and real-time quantitative PCR (RT–qPCR) are standard techniques used to accomplish this task as described previously (4–7). The RT–qPCR is considered a gold standard method in precise quantification of gene transcript levels (8).
MicroRNA genes that encode transcripts which are processed to the same or similar mature microRNA species are grouped in families. In Arabidopsis thaliana, the number of such family members varies between 1 (e.g. miR163, miR173) and 14 (miR169) [miRBase release 17 (9)]. MicroRNA genes from the same family, although represented by identical or almost identical mature microRNAs, differ considerably in gene organization and sequence. In many cases it is only possible to observe the expression of all family members as a group, rather than the individual members due to the large sequence conservation when using northern hybridization or deep sequencing approaches. However, individual members of a given microRNA family may be expressed in different developmental stages or in response to various biotic/abiotic stimuli (10–12).
Our resource contains information regarding the expression of 190 A. thaliana microRNA genes at the level of primary microRNA (pri-miRNA) in different developmental stages obtained using a RT–qPCR high-throughput platform. Many databases that store and allow access to microRNA gene expression profiles from variety of organisms already exist [e.g. (13–18)], however, they have various limitations and restrictions. One of the limitations includes the lack of tools and/or data for comparisons between developmental stages and various genes at the same time. With this in mind, we developed the mirEX platform as a comprehensive starting point for a comparative investigation of microRNA genes expression. Our database offers to the scientific community easily accessible data and is of interest to researchers working on the specific microRNA function, the expression profile of entire microRNA family members during a particular organ/developmental stage, or on microRNA biogenesis. Additionally, by creating the mirEX interface, we hope to propose a new database interface standard for comparative microRNA gene expression data mining.
WEB TOOLS AND DATA
Expression datasets
In the current release, the mirEX database includes expression data of microRNA genes from A. thaliana. To collect the data, we prepared a real-time PCR platform for 190 primary microRNA sequences. For each microRNA, a pair of specific primers amplifying a single product was designed; 10 microRNAs primer sequences were taken from Pant et al. (19). The qPCR reactions for one biological replicate included 12 different controls and standards, were carried out in parallel as described previously (20) using RNA template isolated from six developmental stages of A. thaliana Columbia-0 wild-type plants. RNA from seeds and siliques was isolated according to Ref. (21). The detailed procedure of running the platform can be found in the mirEX database documentation.
The plant material used in expression profiling experiments includes: seeds, 10-day-old seedlings, 14-day-old seedlings, 25-day-old plants, 35-day-old plants, 42-day-old plants (rosette leaves and stems) and 53-day-old plants (rosette leaves, stems, inflorescence and siliques). For better visualization of the individual developmental stages, we included pictures of actual plants used in our experiments. Our high-throughput platform contains an original and novel set of data on microRNA gene expression in dormant seeds. The PP2A (At1g13320) and actin (At3g18780) cDNAs were used as an expression reference (22). The mean measurements of three replicas were used to calculate the fold-change value and presented in a form of log10. In a case when the value of correlation coefficient of the three replicas was <0.995, such data was labeled as ‘low quality’ and is not shown by default on the graphs, but indicated in gray in the data tables. Since the level of expression of most of the primary microRNAs is lower than the reference genes, the data shown on the graphs in mirEX is rescaled in order to avoid presenting it as a negative value. Rescaling shifts the zero value of the graph's y-axis to the basal expression level of the whole experiment. This allows showing the expression profile in a positive data range, but does not change the actual values.
Web interface
The mirEX database interface is designed to be used by a bench scientist on an everyday basis. Following a simplicity rule, the interface of mirEX has been built on only two types of result windows and a simple two-step querying system.
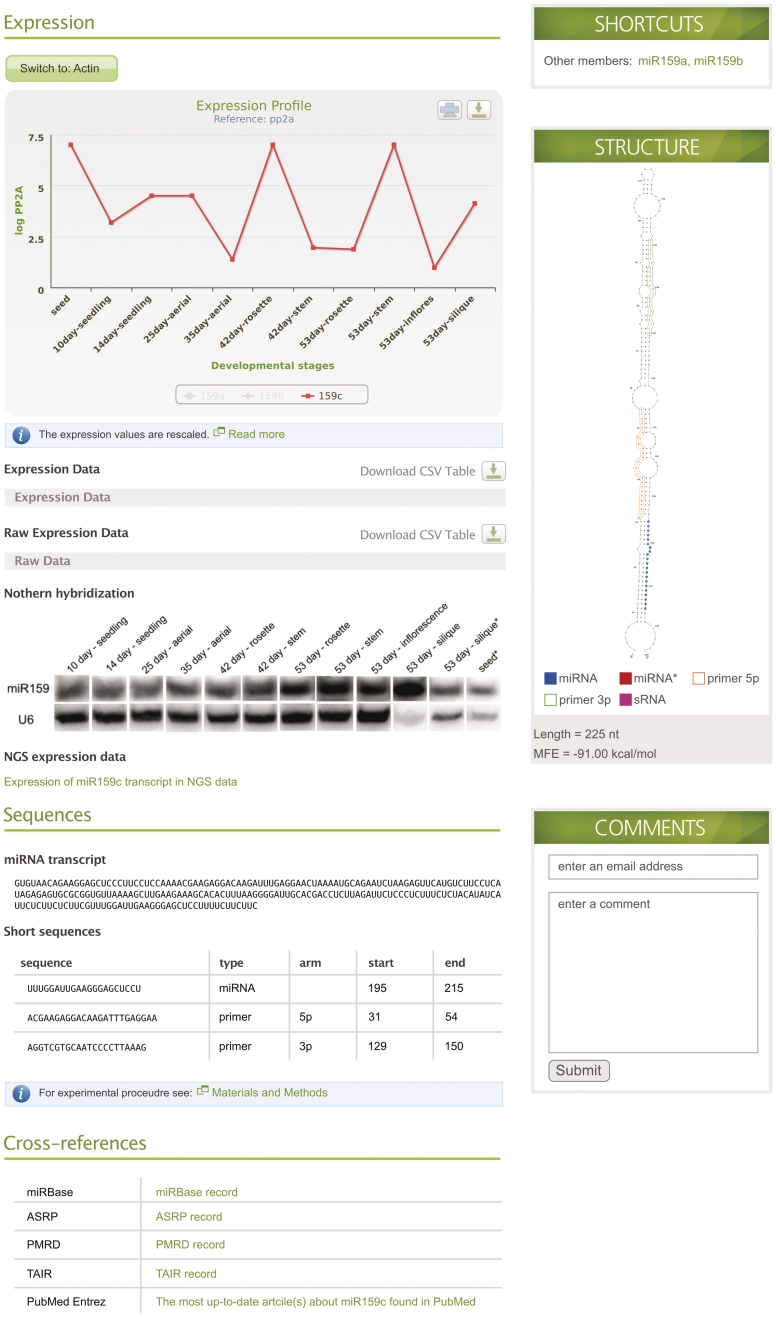
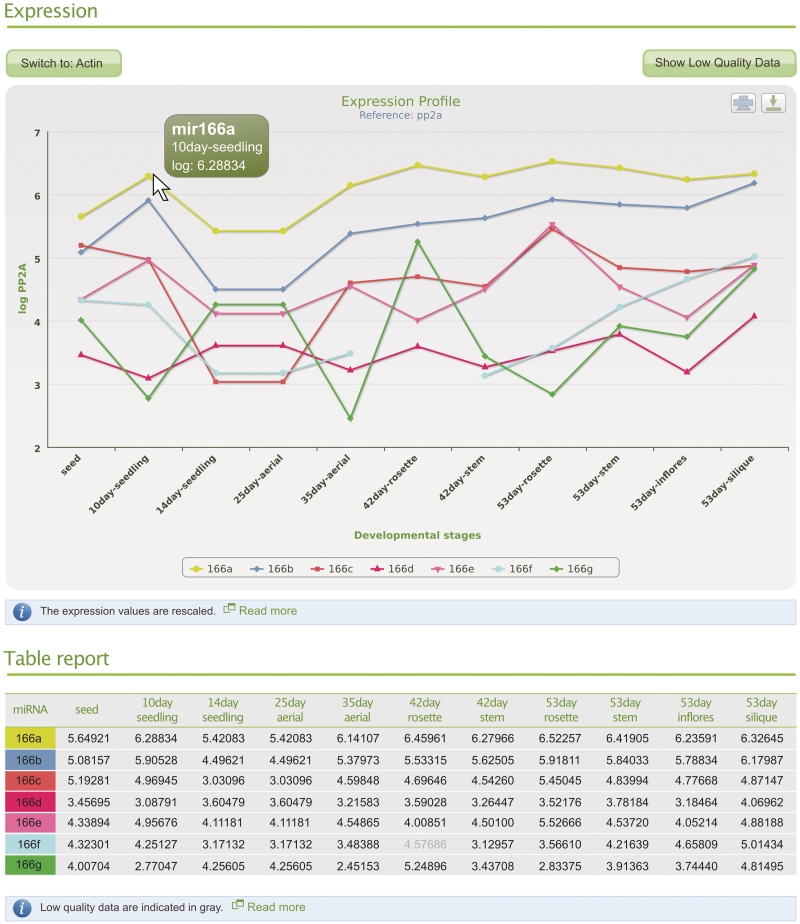
There are two ways to access data in mirEX: by searching for a particular microRNA or microRNA family, or by browsing the database content. A single input window allows searching for an individual or a family of microRNA by providing the numerical part of its identification (ID). When entered ID corresponds to a record of a single microRNA, the resulting page shows all of the details for this particular sequence (Figure 1). In the case of whole family ID, the search results are presented in the form of a line graph, showing expression data for all stages available in the database (Figure 2). The table located below the graph contains numerical values for data presented in the graph, including low quality measurements. The IDs shown in the table allow quick access to detailed information about particular microRNA.
Figure 1.
Example of mirEX record window for ath-miR159c. This window is divided into distinct sections representing: expression data, sequences and external references. The right panel contains shortcuts to microRNA family members' records, structure of the RNA transcript molecule with labeled mature sequences and RT–qPCR primers and webform for user comments.
Figure 2.
Example of mirEX report window for ath-miR166 gene family. The report window contains graphical presentation of the expression levels, as well as actual numerical values presented in tabular format. Holding a mouse pointer over any data point allows access to details of expression measurements. The datasets for a particular microRNA can be dynamically turned on and off by clicking ID in the legend of the graph.
Browsing the data in mirEX starts at the two-step selection process. The first step includes selection of developmental stages, which is followed by the selection of microRNAs. In mirEX, there are three ways to select microRNAs: (i) by typing their IDs in the input window, (ii) by uploading a file with a coma-separated list of microRNA IDs (that can be created in the first step) and (iii) by selecting individual or groups of sequences from the tree-like expandable menu. During the process of entering information using a keyboard, the mirEX interface will provide the list of available microRNAs. It is possible to mix all of the input methods—the resulting page will refer to all entered or selected IDs.
Presentation of search/browse results may differ depending on the number of selected developmental stages or microRNA genes (see ‘Data mining’ section). By default, the graph of expression profiles for various stages and microRNAs shows only high quality data. However, it is a user-defined option in mirEX whether the low quality measurements are presented on the graph. The expression values can be displayed using two reference genes: actin and PP2A.
The results for a single microRNA are presented in a record window in a form that is divided into distinct sections (Figure 1). The upper part of the record is dedicated to expression information presented in the form of a graph and two tables. When applicable, the graph contains information about the expression of other members of the microRNA family, that can be dynamically turned on and off. Additionally, specific shortcuts allow quick access to the records of any member of such family. The two tables, containing either reference-calculated or raw data, can be hidden to reduce the amount of information presented at any given moment on the single record window. Each graph or table in the mirEX database can be saved for later use by selecting an appropriate button.
Additionally, below the quantitative expression data, each record contains a web link to a text-based map of publicly available high-throughput Next-Generation Sequencing (NGS) short reads aligned to primary microRNA transcript. The information included here contains sequence, location, length and number of the mapping reads. On a separate window, it is also possible to access the original record of the used NGS data.
The record window contains a graphical presentation of the structure of microRNA transcript with designations of mature molecule(s) and primers used to assess its expression level by RT–qPCR. When available, the record data will be modified to include results from northern hybridization with mature microRNAs. However, it has to be noted that in most cases, such hybridization represents the level of the whole microRNA gene family expression, and will be not specific for an individual microRNA gene. Moreover, the record window contains a table with sequences relevant to particular microRNA and a link to experimental procedures.
The last part of the record window includes references to external databases. Our aim was to avoid replication of the data available in other databases. For information such as gene structure or available papers, we direct the user to specialized databases, for example TAIR (23) or PubMed (24). The number of external references will grow in time, when new resources for microRNA biology are available, and in response to user requests.
To maintain a close relation with users of our database, we provide in the record window a simple tool for entering comments. The comment may be of any nature: concerning the biological aspect of the presented data or any general issue.
Although the interface is very intuitive and simple, at the main page we provide a video tutorial on all of the capabilities and various ways available to explore the mirEX tools and data. Moreover, every window in the mirEX interface contains context-specific help guides.
A complete ‘road map’ of the mirEX interface is included as Supplementary Figure S1.
Data mining
The usefulness of the mirEX database lies in the ability to compare expression between different developmental stages and various microRNA genes. By using the ‘browse’ button, the user can, in two simple steps, select available developmental stages and a single or sets of microRNAs. Depending on the number of selected stages, the expression results are presented in the form of a bar graph (for one-stage analysis) or a line-based graph (for many stages). The bar graph presentation allows analysis of all pri-miRNAs at once with a zoom-in on an individual gene.
The line graph displays by default, data for only 24 microRNAs selected from the list. However, this restriction can be turned off. The line graph offers an additional option to dynamically remove and/or add any of the selected microRNA genes.
Another, quick way to access comparative expression profiles are shortcuts. In general, the shortcuts represent a new and easy way to explore data stored in the mirEX database. They are located in the same area on every page, and their content follows user selections. The use of shortcuts allows quick access to specific sets of data, e.g. the most expressed genes, or records representing microRNAs belonging to the same family. Periodically, we will modify the shortcuts in response to comments and readapt them according to the most frequently issued queries.
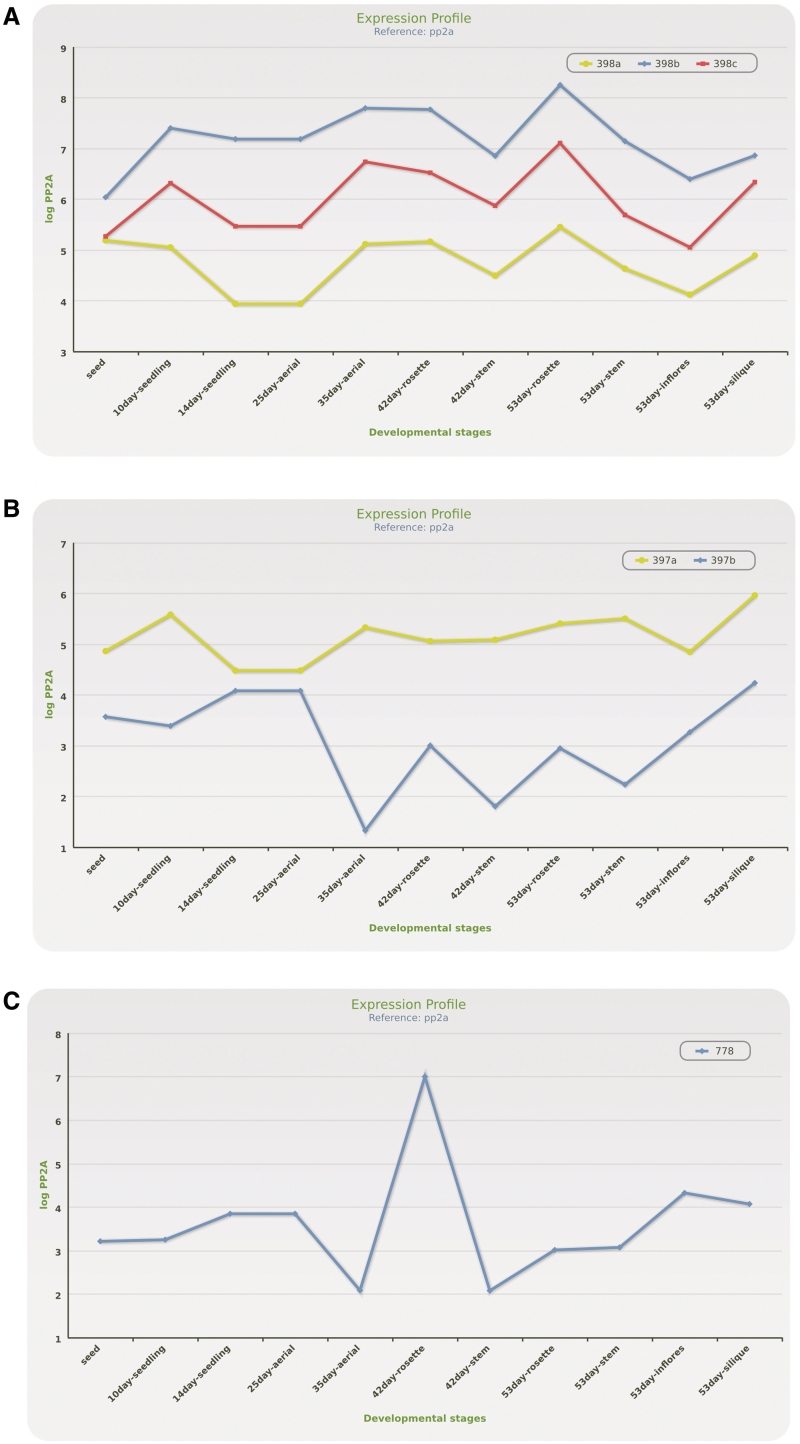
The strategy to use primary microRNA expression data allows fine exploration of gene activities of members of microRNA families. In some families, the expression pattern of individual genes in all developmental stages and organs is similar. However, the expression level of the individual pri-miRNAs may differ considerably: the fold change in the primary transcript levels within one microRNA gene family may range between 10 and 105 (e.g. miR160, miR162, miR164, miR165, miR167, miR168, miR394, miR396, miR398, miR447 families, Figure 3A). Moreover, a few individual pri-miRNAs show profound differences in their expression depending on the developmental stage and/or organ studied (e.g. miR156, miR157, miR158, miR159, miR166, miR169, miR397, miR399, miR404, Figure 3B). These differences may reflect either the existence of promoter regulatory elements that are responsive to developmental stimuli, or the various rate of pri-miRNA maturation during plant growth and organ formation. MicroRNA families containing a single representative also may exhibit dramatic changes in their expression levels (reaching even 106-fold change) during plant growth and organ development (e.g. miR173, miR774, miR776, miR778, miR780, miR783, Figure 3C). Conversely, there are also pri-miRNAs that show relatively small fluctuations of expression levels during plant growth and organ formation. In conclusion, our data shows that each microRNA gene has its own characteristic expression profile reflecting its spatial and temporal regulation that can be followed and comparatively analyzed using tools implemented in the mirEX platform.
Figure 3.
Examples of various microRNA gene expression profiles. The presented graphs were created with options available in mirEX interface and represent (A) miR398 and (B) miR397 gene family (see text for details). (C) Example of miR778 showing dramatic changes in expression level during growth and organ development.
CONCLUSIONS
By creating the mirEX platform, we provide the scientific community with a high quality pri-miRNA expression data in seven developmental stages represented by 11 distinct organs of A. thaliana. This is a new and user-friendly platform designed to explore expression data in various developmental stages for a large number of related genes. The querying system has been limited to two simple steps, which allow access to any type of data stored in mirEX. Additionally, the mirEX interface does not require the use of a keyboard—the database user interface is fully mouse operated. This makes the process of selection and comparing data very easy and effective. Moreover, graphs presenting expression data are designed to accommodate user selection dynamically, which makes exploration of the mirEX content even more efficient.
The data currently incorporated in mirEX represents the starting point in our database development. The modular character of the database design makes it possible for further mirEX expansion to incorporate new species and datasets. Following, we plan to include Arabidopsis microRNA genes discovered in the future, various mutants and microRNA expression profiles from other plant species. The work on barley microRNA expression profiling is already underway. Incorporation of data from other species will broaden the available tool set allowing comparative analyses within and between species.
By designing the mirEX interface, we would like to propose a new trend in biological databases for simplicity and user-friendliness. Additionally, we put special attention to the web browser compatibility issue of the interface by testing all of the most popular tools. Careful selection of informatics techniques resulted in the platform that can be accessed even via iOS on mobile devices without loosing any of the functionality and interface features.
Carefully selected links to external databases, prevent the user interface from overloading with data, yet creates the opportunity to easy access-related information from the most significant databases in the field. In every day laboratory work, this approach proved to be very efficient, and made the mirEX platform a one-stop information center for Arabidopsis microRNA data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure S1.
FUNDING
The Polish Ministry of Science and Higher Education (grant 3011/B/P01/2009/37); the Faculty of Biology Adam Mickiewicz University in Poznan, Poland; the Foundation for Polish Science (FNP) within the International PhD Program co-financed from European Union Regional Development Fund (MPD 2010/3 to D.B. and J.D.). Funding for open access charge: The Faculty of Biology Adam Mickiewicz University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Professor Eva Czarnecka for the helpful suggestions in writing this manuscript and Lance Verner for proofreading it.
REFERENCES
- 1.Reinhart B, Weinstein E, Rhoades M, Bartel B, Bartel D. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 3.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 4.Chambers C, Shuai B. Profiling microRNA expression in Arabidopsis pollen using microRNA array and real-time PCR. BMC Plant Biol. 2009;9:87. doi: 10.1186/1471-2229-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee H, Yoo SJ, Lee JH, Kim W, Yoo SK, Fitzgerald H, Carrington JC, Ahn JH. Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 2010;38:3081–3093. doi: 10.1093/nar/gkp1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 2004;14:1641–1653. doi: 10.1101/gr.2275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graeber K, Linkies A, Wood ATA, Leubner-Metzger G. A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. Plant Cell. 2011;23:2045–2063. doi: 10.1105/tpc.111.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2010;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y. Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 2007;354:585–590. doi: 10.1016/j.bbrc.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 11.Moldovan D, Spriggs A, Yang J, Pogson BJ, Dennis ES, Wilson IW. Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. J. Exp. Bot. 2010;61:165–177. doi: 10.1093/jxb/erp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao M, Ding H, Zhu J-K, Zhang F, Li W-X. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011;190:906–915. doi: 10.1111/j.1469-8137.2011.03647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Backman TWH, Sullivan CM, Cumbie JS, Miller ZA, Chapman EJ, Fahlgren N, Givan SA, Carrington JC, Kasschau KD. Update of ASRP: the Arabidopsis small RNA project database. Nucleic Acids Res. 2008;36:D982–D985. doi: 10.1093/nar/gkm997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng Y, Gou L, Chen D, Mao C, Jin Y, Wu P, Chen M. PmiRKB: a plant microRNA knowledge base. Nucleic Acids Res. 2010;39:D181–D187. doi: 10.1093/nar/gkq721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho S, Jun Y, Lee S, Choi H-S, Jung S, Jang Y, Park C, Kim S, Lee S, Kim W. miRGator v2.0: an integrated system for functional investigation of microRNAs. Nucleic Acids Res. 2010;39:D158–D162. doi: 10.1093/nar/gkq1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaya KD, Karakülah G, Yakicier CM, Acar AC, Konu O. mESAdb: microRNA expression and sequence analysis database. Nucleic Acids Res. 2011;39:D170–D180. doi: 10.1093/nar/gkq1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie W, Flamant S, Rasko JEJ. mimiRNA: a microRNA expression profiler and classification resource designed to identify functional correlations between microRNAs and their targets. Bioinformatics. 2010;26:223–227. doi: 10.1093/bioinformatics/btp649. [DOI] [PubMed] [Google Scholar]
- 19.Pant B, Musialak-Lange M, Nuc P, May P, Walther D, Scheible W. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time PCR profiling and small RNA sequencing. Plant Physiol. 2009;150:1541–1555. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szarzynska B, Sobkowiak L, Pant B, Balazadeh S, Scheible W, Mueller-Roeber B, Jarmolowski A, Szweykowska-Kulinska Z. Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 2009;37:3083–3093. doi: 10.1093/nar/gkp189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oñate-Sánchez L, Vicente-Carbajosa J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes. 2008;1:93. doi: 10.1186/1756-0500-1-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamesch P, Dreher K, Swarbreck D, Sasidharan R, Reiser L, Huala E. Using the Arabidopsis information resource (TAIR) to find information about Arabidopsis genes. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi0111s30. Chapter 1, Unit1.11. [DOI] [PubMed] [Google Scholar]
- 24.Lu Z. PubMed and beyond: a survey of web tools for searching biomedical literature. Database. 2011;2011 doi: 10.1093/database/baq036. doi: 10.1093/database/baq036. [DOI] [PMC free article] [PubMed] [Google Scholar]