Abstract
Pfam is a widely used database of protein families, currently containing more than 13 000 manually curated protein families as of release 26.0. Pfam is available via servers in the UK (http://pfam.sanger.ac.uk/), the USA (http://pfam.janelia.org/) and Sweden (http://pfam.sbc.su.se/). Here, we report on changes that have occurred since our 2010 NAR paper (release 24.0). Over the last 2 years, we have generated 1840 new families and increased coverage of the UniProt Knowledgebase (UniProtKB) to nearly 80%. Notably, we have taken the step of opening up the annotation of our families to the Wikipedia community, by linking Pfam families to relevant Wikipedia pages and encouraging the Pfam and Wikipedia communities to improve and expand those pages. We continue to improve the Pfam website and add new visualizations, such as the ‘sunburst’ representation of taxonomic distribution of families. In this work we additionally address two topics that will be of particular interest to the Pfam community. First, we explain the definition and use of family-specific, manually curated gathering thresholds. Second, we discuss some of the features of domains of unknown function (also known as DUFs), which constitute a rapidly growing class of families within Pfam.
INTRODUCTION
Pfam is a database of protein families, where families are sets of protein regions that share a significant degree of sequence similarity, thereby suggesting homology. Similarity is detected using the HMMER3 (http://hmmer.janelia.org/) suite of programs.
Pfam contains two types of families: high quality, manually curated Pfam-A families and automatically generated Pfam-B families. The latter are derived from clusters produced by the ADDA algorithm (1), followed by the subtraction of overlapping Pfam-A regions at each release. Pfam-A families are built following what is, in essence, a four-step process:
building of a high-quality multiple sequence alignment (the so-called seed alignment);
constructing a profile hidden Markov model (HMM) from the seed alignment (using HMMER3);
searching the profile HMM against the UniProtKB sequence database (2) and
choosing family-specific sequence and domain gathering thresholds (GAs); all sequence regions that score above the GAs are included in the full alignment for the family (GAs are described in detail in a later section of this paper).
In addition to providing matches to UniProtKB, Pfam also provides matches for the NCBI non-redundant database, as well as a collection of metagenomic samples. We generate a variety of data downstream, including, among others, a family sequence-conservation logo based on the HMM, a description of domain architectures, where all co-occurrences with other domains are reported, and a species tree summarizing the taxonomic range in the family.
The quality of the seed alignment is the crucial factor in determining the quality of the Pfam resource, influencing not only all data generated within the database but also the outcome of external searches that use our profile HMMs, e.g. to assign domains to proteins which are part of newly sequenced genomes. For this reason, a considerable curatorial effort goes into seed alignment generation.
Members of the same Pfam family are expected to share a common evolutionary history and thus at least some functional aspect. Ideally, our families should represent functional units, which, when combined in different ways, can generate proteins with unique functions. The ultimate goal of Pfam is to create a collection of functionally annotated families that is as representative as possible of protein sequence-space, such that our families can be used effectively for both genome-annotation and small-scale protein studies. It must be stressed, however, that homology is no guarantee of functional similarity and transfer of functional annotation based solely on family membership should always be undertaken with caution. On the other hand, additional data that are available from Pfam, such as conservation of family signature residues or conservation of common domain architectures, can increase confidence in a given functional hypothesis. For more background on how to query and use our web interface please refer to Coggill et al. (3).
In this paper, we report on the most recent Pfam release (26.0) as well as on important changes that have been introduced over the last 2 years, since our 2010 NAR database issue paper (where we presented release 24.0) (4). Arguably, the change carrying the most significant philosophical implications has been the decision to follow the lead of the Rfam database (5) and out-source functional annotation of Pfam families to Wikipedia. We will discuss the background to this decision and give details of the progress towards Wikipedia coverage of Pfam families. Another important development has been the adoption of the iterative sequence-search program jackhmmer (6) as our principal tool for generating new families. In addition, we have extended our mechanism for family curation, which now allows trained and trusted external collaborators to create and add their own families to Pfam. Finally, we will take this opportunity to address and present fresh analysis on two topics that we consider of particular importance: family-specific GAs and Domains of Unknown Function (DUFs).
WHAT'S NEW
Community annotation
Using Wikipedia as a repository for protein family annotation
Historically, Pfam has provided only a basic level of textual annotation for each family. This has included a few sentences, with references, designed to give users an overview of the function(s) of the family or domain. However, rather than have the Pfam curators describe our families, we would strongly prefer to have annotations written by those who know the proteins and families best, namely the biologists and informaticians who work with them on a day-to-day basis. Harnessing the knowledge of these experts remains a significant challenge.
One recent approach has been to use Wikipedia as a source of scientific information (7,8). Wikipedia is the world's largest online encyclopedia, with over 3.7 million English language articles, and is widely acknowledged to be the most popular general reference work on the internet. A cornerstone of Wikipedia is that anyone can edit the content.
The Rfam database moved all of its family annotation into articles in Wikipedia in 2009, thereby allowing anyone to freely edit and improve their content. This experiment has proved successful, engaging the wider scientific community to provide expert annotations and improving the overall quality of the Rfam annotation (7). In light of this positive experience, we decided to adopt the same approach and use Wikipedia as the primary source of Pfam annotation (Figure 1A).
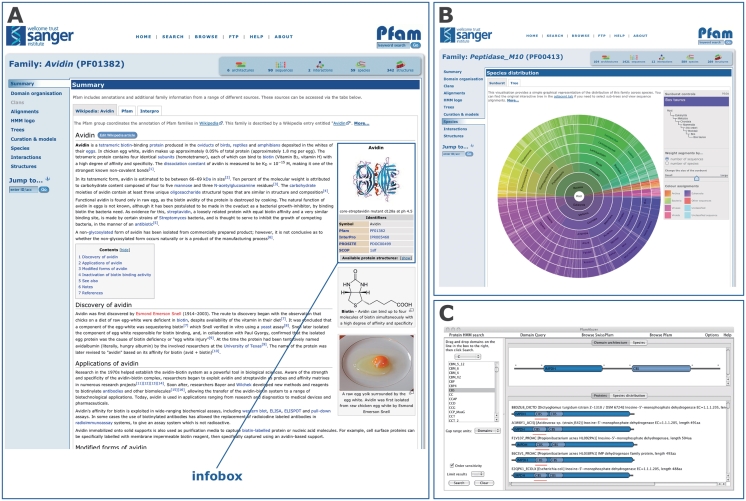
Figure 1.
New Pfam features since release 24.0. (A) The Pfam-A family page for Avidin (PF01382), showing the embedded contents of the associated Wikipedia article. The ‘infobox’ is highlighted. (B) The ‘sunburst’ representation of the tree showing the species distribution of the Pfam-A family Peptidase_M10 (PF00413). (C) The PfamAlyzer applet, showing the results of searching for all architectures that include the domains IMPDH and CBS. The PfamAlyzer applet allows querying of Pfam for proteins with particular domains, domain combinations or architectures.
The Rfam database included around 600 families when the switch to Wikipedia was made. This made it feasible to assign existing articles where possible and to generate new ‘stub’ articles in Wikipedia for any family that still lacked any relevant article. These stubs have since been gradually expanded and improved by the Rfam and Wikipedia communities. At the time when Pfam began using Wikipedia for annotations, release 25.0, there were 12 273 Pfam-A families. We initially identified existing articles that described protein domains or families and which provided useful information about the Pfam family. These articles are now assigned as the primary annotation for the appropriate families. Given the number of families that remain without an article, however, it is simply not feasible to manually generate articles for all of them. For these families we continue to show the original annotation comments, which were written by the curator of the family, while encouraging our users to tell us about appropriate Wikipedia articles or to create them on our behalf. Furthermore, it is likely that there will be some families that are not sufficiently notable for inclusion in Wikipedia and we anticipate that many of these will remain without Wikipedia annotations for the long term, perhaps indefinitely.
As of release 26.0, there are 4909 Pfam families that link to 1016 Wikipedia articles. We invite readers and users of the Pfam website to edit and improve these articles in Wikipedia. Mapping of Pfam-A families to Wikipedia articles is available in JSON format, from: http://pfamsrv.sanger.ac.uk/cgi-bin/mapping.cgi?db=pfam.
Some Wikipedia articles cover multiple Pfam families, such as the Zinc finger article or the Interleukin article. Pfam contains a large series of 3526 families noted as DUFs. Virtually all of these DUF families link to a single Wikipedia article, which describes DUFs in general, and will do so until such time as their function is determined and they have an article of their own.
Although the process of manually creating a new Wikipedia article can be time-consuming and difficult, we are keen to increase the number of Wikipedia-annotated families in Pfam as much as possible. We have therefore developed a pipeline to generate stub articles automatically in a Wikipedia ‘sandbox’, often taking existing family annotations from the InterPro database (9) as the basis for the article. These stubs can then be reviewed and edited by our curators, before being moved out of the sandbox into Wikipedia proper and used to annotate families. We have implemented several automated procedures for augmenting the basic annotation text and expanding the content as far as possible before its final publication in Wikipedia.
A particularly useful feature of Wikipedia is the highlighting of terms within an article that are themselves described by another Wikipedia article. This network of linked terms allows readers to quickly understand the background to the article they are reading and, as such, they are crucial to the success of any article. To assist with the cross-linking of our new, automatically generated Wikipedia articles, we took the initial set of ∼700 Pfam Wikipedia articles and computationally collected a broad set of common terms. These terms were then automatically marked as links in the stub articles. Another essential feature of a Wikipedia page is the reference list. We used the TemplateFiller Perl module (http://search.cpan.org/dist/WWW-Wikipedia-TemplateFiller/) to retrieve and include the full details of the references cited in the InterPro annotation that we used as our starting point. Finally, we have automatically populated the ‘infobox’ (http://en.wikipedia.org/wiki/Help:Infobox) in our stub articles. This infobox (Figure 1A), located on the right-hand side of Wikipedia protein family pages, shows images of the relevant three-dimensional structures, where available, and additional database links. When an image of the protein structure was available from Wikimedia commons (http://commons.wikimedia.org/wiki/Main_Page), this was added to the top of the infobox, along with a caption. Further information was extracted from the Pfam database and added to the infobox, such as the Pfam clan accession and links to other database sites such as PROSITE (10), SCOP (11) and CAZy (12).
Altogether, the automatic article creation process generated 7823 articles in our Wikipedia sandbox. We continue to review, edit and move these stub articles into the main part of Wikipedia. In order to prioritize the best articles arising from the generation process, we calculated for each one a heuristic score, based on the size of the annotation, availability or otherwise of an image, the number of references and the number of links out to other databases. The score gave an overall measure of the level of information in the page and thus an indication of its potential for addition to Wikipedia. Already >200 of the highest scoring articles have been moved across to generate new Wikipedia pages.
One of the major concerns about Wikipedia generally is the risk of vandalism and deliberate errors being introduced into publicly edited articles. This is of particular concern to both the Pfam and Rfam projects, since we ‘scrape’ and re-display Wikipedia contents within our respective websites. In order to reduce the likelihood of blatant vandalism or egregious errors propagating through to our websites, we include an additional approval process before displaying newly edited Wikipedia articles. Our curators review and, if necessary, revert changes to articles on a daily basis and only after an article has been reviewed it is flagged for update and presentation within the Pfam website. In our experience, almost every case of vandalism is reverted by the Wikipedia community before we come to review the changes. Overall we have found that ∼1% of all edits are reverted by the Wikipedia community, suggesting an upper bound on the possible number of vandalism edits.
It is important to stress that the Wikipedia content displayed in Pfam family pages is an exact copy of the article that can be found on the main Wikipedia website, subject to a delay of a day or so for the approval process described above.
Family function annotation via the Pfam helpdesk
Val Wood of the Schizosaccharomyces pombe database, PomBase, routinely reports new findings from the literature to the Pfam helpdesk (pfam-help@sanger.ac.uk). Over the last 12 months, 74 such communications have been received, of which at least four provided evidence for the function of a DUF. A further 16 concerned hits of newly characterized S. pombe sequences to Pfam-B families, thus leading to the building of at least that number of new families.
One good example of a family that has been characterized in this way is DUF1709, in which a fission yeast anillin protein was characterized. Anillin proteins are actin-binding proteins involved in septin-organization, which are localized to the cleavage-furrow during cell division (13). The DUF has been re-named Anillin (PF08174).
Similarly, Pfam-B family PB008473 from Pfam release 24.0 was found to contain a fission yeast protein, Mtr4 (UniProtKB: Q9P795), which had been determined experimentally to be an essential RNA helicase that performs a critical role as an activator of the nuclear exosome in RNA processing and degradation. From this finding (14), family rRNA_proc-arch (PF13234) was built and described.
Other contributions from the community over the last year have included 51 direct annotation submissions (received via a web form available from the Pfam family pages) with suggestions for improvements and updates to the Pfam annotations; of these, 14 offered information about the function of DUFs, 4 of these coming from the InterPro team. A good example of how the functionality of InterPro benefits Pfam was a case where a team member flagged up one of our DUFs, DUF3462 (PF11945), as being the WASH subunit of the WASH complex (Wiskott–Aldrich Syndrome Protein and SCAR Homolog) that acts as an Arp2/3 activator necessary for Golgi-directed trafficking (15). The DUF was re-named as the WAHD domain of WASH complex.
Examples of cases where the determination of the three-dimensional structure of sequences from bacterial DUFs has led to the discovery of function are detailed in a dedicated issue of Acta Crystallographica, Section F (16).
Extending the community of Pfam curators
Although Wikipedia offers a mechanism for external scientists to contribute annotations using an established mechanism, it is restricted purely to functional annotation. As outlined above, the helpdesk provides a way of making more substantial contributions to the database, but the fraction of new families derived from helpdesk submissions is relatively small. Furthermore, contributions from the helpdesk are often in a different format and/or use a different sequence database to that used by Pfam. More often than not, a Pfam curator has to spend time understanding and modifying the submission to conform to the Pfam data model, which makes the helpdesk a far from ideal interface for bulk submissions.
Pfam is run by an international consortium of three groups, but until very recently our fundamental family data could be modified only at our Cambridge, UK, site. This has meant that even full consortium members have been unable to add their own families to Pfam. In order to remove this restriction, and with the goal of making it easier for members of the wider community to add families, we have developed a system that allows Pfam families to be added by registered users anywhere in the world. The distributed system involves the local installation of our family building pipeline (a set of Perl scripts and modules) and various quality control procedures. It allows the addition of new families and clans, as well as the modification of existing entries. Data are sent backwards and forwards between the user and a central, master server using HTTPS, and we are able to authenticate all traffic that results in changes to the database. Files are maintained using the widely used Subversion revision control system (http://subversion.apache.org/), thereby preventing the inadvertent conflicts that could occur when multiple users wish to make changes in a distributed environment.
Owing to the organizational changes detailed above, we have been able to embrace two external groups who work with data relevant to Pfam, giving them direct access to the Pfam submission pipeline. The Protein and Genome Evolution Research Group, run by L. Aravind at the National Center for Biotechnology Information (NCBI, USA), are experts in protein evolution, routinely publishing articles on large evolutionary related superfamilies. The second external contributing group is the one of Adam Godzik at the Burnham Institute (USA), part of the Joint Center for Structural Genomics (JCSG).
Members of both of these groups are now able to submit families and clans directly to Pfam, allowing them to improve Pfam data and to extend its reach to new communities and a broader audience. We see the introduction of this distributed curation model, in combination with the use of Wikipedia as a source of our annotations, as two important steps in making our database a community-based resource. Our goal is to provide an infrastructure that empowers scientists to contribute using whichever mechanism they feel most comfortable, while still allowing us to maintain oversight and control of the quality of our fundamental data.
Generating new families using jackhmmer iterative searches
The HMMER3 package (http://hmmer.janelia.org/) includes the jackhmmer (6) program for running iterative, profile HMM-based searches against a sequence database (PSI-BLAST-like) starting with a single sequence. This, in parallel with curation of Pfam-B alignments, has become our main protocol for generating families. Sequences of interest are used as queries for a 3-iteration jackhmmer search. Seed alignments for new Pfam families are produced from the resulting jackhmmer multiple sequence alignment. In particular, we have applied this protocol for family mining in a set of complete proteomes drawn from a wide taxonomic range (>50 proteomes overall). For a given proteome, every sequence lacking a match to a Pfam entry was used to initiate a jackhmmer search.
Website changes
Sunburst representation of the taxonomic tree
For each Pfam-A family we provide an interactive taxonomic tree, showing the species distribution of sequences in the family. However, due to the size of many families, this tree can be very large, making it difficult to gain a clear impression of the species distribution of the family. In order to address this problem, we have introduced a ‘sunburst’ representation of the species trees, as shown in Figure 1B. Sunbursts are a commonly used method of visualizing tree-like data sets, whereby the root of a tree is plotted as a circle, surrounded by concentric rings representing child nodes. In Pfam, each node of the taxonomic tree is drawn as an arc, whose distance from the centre corresponds to the taxonomic level of the node and whose length (or, equivalently, the angle subtended by the arc) is scaled to represent either the number of sequences or the number of species belonging to that node in the tree. The switch between scaling according to numbers of sequences or species may be changed interactively using a control in the page. Arcs are coloured according to kingdom. As the mouse pointer is moved across the sunburst, a tool-tip shows a summary of the current node, giving the species name for that node, along with the number of species and number of sequences beneath it. A summary panel also shows a simple graphical representation of the lineage of the relevant node. The overall size of the plot may be adjusted using a simple slider.
The sunburst tree is generated by mapping the UniProtKB assigned NCBI taxonomy identifiers onto the standard NCBI taxonomy. Unfortunately, there is not a perfect equivalence between taxonomy trees used by UniProtKB and NCBI, due simply to the fluid nature of the data and the different update cycles of the two resources. This mis-match inevitably generates cases where the mapping between the taxonomy identifiers in UniProtKB and NCBI breaks down. Species that cannot be assigned an exact node in the NCBI tree are shown as ‘Unclassified’ in the sunburst. Furthermore, because the NCBI taxonomy contains numerous levels that are not present across all species, we have attempted to normalize taxonomic levels to the eight major ones (domain, kingdom, phyla, class, order, family, genus, species). For example, the lineage of Bos taurus contains the sub-family level Bovinae, which we skip over and connect the genus directly to the family level, Bovidae. Some lineages also omit one or more of the major levels. Again, in the case of B. taurus, the level ‘order’ is omitted and the missing level is flagged with ‘No order’. We perform a node merger in the case of sub-species so that, for example, all sub-species of Escherichia coli are merged up to the species level and presented as E. coli sequences. These normalization steps allow us to draw every species with the same eight levels, making the outer ring of the sunburst complete and allowing the plot to represent more intuitively the distribution of sequences at each level.
Reinstatement of the PfamAlyzer tool for complex architecture queries
PfamAlyzer (17) is a Java applet that provides a user-friendly graphical interface to Pfam (Figure 1C). It was available in a previous version of the Pfam website (18) but was removed during development of the new website. It has now been reinstated and can be accessed through the search page. PfamAlyzer enables complex domain architecture queries to be specified using a simple drag-and-drop interface. The user can select a set of domains from drop-down lists of Pfam-A families or Pfam clans and drag and arrange them to build a query architecture. PfamAlyzer use has been described in detail elsewhere (17,18).
PFAM STATISTICS
In our last NAR database paper (4), we reported on statistics from Pfam release 24.0. Here, we compare those numbers to our latest release, 26.0.
General
Pfam 26.0 comprises 13 672 Pfam-A families, an increase of ∼15% with respect to Pfam 24.0. The total number of clans is now 499, up 18% since Pfam 24.0. Of the added families, 40% belong to clans. This brings the total number of families in clans to 31%, compared with 26% in release 24.0. Added families that are not in a clan are on an average much smaller than those in release 24.0 (average size of non-clan family in release 24.0 was 832 members, compared to 337 for those added after release 24.0). Finally, 34% of all new families are DUFs (8% of these belong to clans) bringing the total number of DUFs in release 26.0 to 3526.
UniProtKB coverage
Pfam uses UniProtKB as its reference sequence database. Between Pfam releases 24.0 and 26.0, UniProtKB has increased in size by 69% (9.4 million sequences in UniProtKB in August 2009 versus 15.9 million sequences in June 2011). Pfam seems to have coped well with the increase in number of sequences (Table 1), with UniProtKB sequence coverage up >4% since release 24.0. Amino acid coverage has followed a very similar trend. In addition, the coverage of the redundancy-reduced sequence dataset UniRef50 (19) (redundancy reduced version of a dataset including UniProt and a number of other additional sequences from UniParc), decreased only slightly between releases 24.0 and 26.0. It is important to note, however, that coverage of UniRef50 is ∼20% lower with respect to coverage of a non-redundancy reduced UniProtKB database. This data indicates that Pfam has good coverage of the large, densely populated regions of protein space. The numerous less well-populated regions represent a significant challenge to all protein family databases, if the whole of protein space is ever to be completely represented by such databases.
Table 1.
UniProtKB and UniRef50 coverage comparison between Pfam release 24.0 and 26.0
| Pfam release 24.0 | Pfam release 26.0 | |
|---|---|---|
| UniProtKB sequence coverage (%) | 75.1 | 79.4 |
| UniProtKB amino acid coverage (%) | 53.2 | 57.1 |
| UniRef50 sequence coverage (%) | 58.2 | 57.7 |
| UniRef50 amino acid coverage (%) | 36.9 | 36.6 |
Release 24.0 coverage is calculated on UniProtKB 15.6 (August 2009) version and corresponding UniRef50; release 26.0 coverage is calculated on UniProtKB 2011_06 version and corresponding UniRef50.
Coverage of complete proteomes
An alternative way to measure Pfam growth is to assess the sequence and amino acid coverage of ‘complete’ genomes. Completed genomes provide relatively stable protein data sets, making it easier to assess changes in growth from release to release. Proteome sets are derived from the list of proteome FASTA files provided by Ensembl Genomes (20). Table 2 lists the Pfam 26.0 coverage of proteomes from a diverse set of organisms. The list is the same as that reported in 2010 (4), except that Bacillus subtilis has also been included. Generally, there has been an increase of 2–4 percentage points in both amino acid and sequence coverage since release 24.0. However, this trend is not observed in the large eukaryotic genomes Homo sapiens, Gallus gallus, Mus musculus and Danio rerio, where sequence and/or amino acid coverage has remained similar or has even become lower compared to that reported previously. This observation is explained by the fact that these four proteomes have substantially increased in size (number of proteins) over the past 2 years (increasing 25–71%), due to better integration of Ensembl data into UniProt. This has allowed us to improve cross-referencing between Pfam 26.0 and Ensembl Genomes. We will use such coverage analysis to drive the selection of proteomes for family mining using jackhmmer, as described previously.
Table 2.
Residue and sequence coverage of a number of complete proteomes in Pfam 26.0
| Species | Sequence coverage (%) | Amino acid coverage (%) |
|---|---|---|
| Archaea | ||
| Methanococcus vannielii (strain SB/ATCC 35089) | 86.7 | 65.9 |
| Methanosphaera stadtmanae (strain DSM 3091) | 80.8 | 56.3 |
| Thermofilum pendens (strain Hrk 5) | 73.1 | 54.7 |
| Bacteria | ||
| Bacillus subtilis | 85.7 | 67.8 |
| Escherichia coli (strain MG1655) | 93.4 | 71.8 |
| Helicobacter pylori (strain HPAG1) | 80.4 | 60.3 |
| Pseudomonas aeruginosa (strain UCBPP-PA14) | 87.6 | 66.2 |
| Salmonella typhi (strain CT18) | 85.7 | 68.3 |
| Staphylococcus aureus (strain MW2) | 85.5 | 68.6 |
| Streptococcus pyogenes (strain MGAS9429) | 81.9 | 67.4 |
| Thermus thermophilus (strain HB8) | 84.0 | 64.5 |
| Yersinia pestis (strain Pestoides F) | 89.0 | 67.7 |
| Eukaryota | ||
| Anopheles gambiae | 77.9 | 42.8 |
| Arabidopsis thaliana | 75.3 | 43.9 |
| Caenorhabditis elegans | 67.2 | 40.2 |
| Danio rerio | 85.1 | 45.8 |
| Dictyostelium discoideum (strain AX4) | 60.4 | 28.7 |
| Drosophila melanogaster (strain Berkeley) | 74.3 | 38.0 |
| Gallus gallus | 79.7 | 45.6 |
| Homo sapiens | 69.7 | 44.4 |
| Leishmania braziliensis | 55.2 | 21.7 |
| Mus musculus | 73.9 | 44.1 |
| Paramecium tetraurelia | 54.6 | 24.8 |
| Saccharomyces cerevisiae (strain ATCC 204508) | 81.6 | 44.2 |
| Schizosaccharomyces pombe (strain ATCC 38366) | 88.3 | 48.5 |
| Toxoplasma gondii (strain RH) | 57.3 | 19.1 |
| Tetraodon nigroviridis | 70.2 | 42.0 |
Website usage
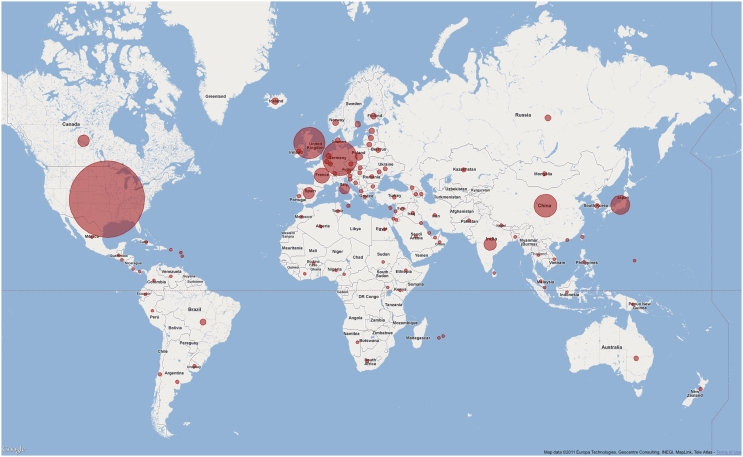
The Pfam website continues to be widely used, both in terms of the geographic spread of users (see Figure 2) and in terms of the breadth of information retrieved from it. The various sequence search tools provided in the website are also heavily used. Taking as an example the period from 1 to 30 June 2011, a broadly representative month in terms of overall Pfam usage, we performed a total of 97 853 sequence searches across the three mirror sites. Of these, 93 871 were single-sequence searches for Pfam-A matches, while 3472 were single-sequence searches for Pfam-B matches. We also ran a total of 510 offline multiple-sequence searches, which were submitted by >100 different users; offline search results are emailed back to the user once the search completes.
Figure 2.
Pfam users in the world. A world map showing the usage of Pfam website at the Wellcome Trust Sanger Institute, UK. Usage statistics were obtained from our Google Urchin tracking database and plotted using the Google map API. Circle size is proportional to number of visits from each country for those with >5000 visits. Countries contributing <5000 visits are all shown with the same sized marker. Data refer to the period between 1 and 30 June 2011.
A MORE DETAILED VIEW OF GATHERING THRESHOLDS AND DUF
In this section, we discuss two topics. First, we address issues concerning statistical significance levels for inclusion of sequences into Pfam families; we regularly receive questions on this subject, which may indicate that the meaning of our family-specific gathering thresholds/cutoffs is not widely understood. Second, we continue (16) our analysis of DUFs, pointing to cases that may be of more interest for experimental functional characterization.
Pfam gathering thresholds
What are sequence and domain gathering thresholds?
The gathering thresholds, or GAs, are manually curated, family-specific, bit score thresholds that are chosen by Pfam curators at the time a family is built. Every family is given two GAs, a ‘sequence’ threshold, and a ‘domain’ threshold. In HMMER, the sequence bit score is the sum of all scoring matches between the sequence and the profile HMM. The domain bit score is the score assigned to each reported match between the sequence and the profile HMM. For a protein region to be considered as part of a family, both its sequence and domain bit scores must be equal to, or greater than, the corresponding GA. In families that contain sequences with multiple matches to the profile HMM, domain thresholds can be set to a value lower than sequence thresholds, in order to increase sensitivity. This is based on the assumption that finding multiple copies of a domain on the same sequence increases the chance of those instances being genuine matches, even when their E-values are not very significant when taken in isolation. This is particularly true for Pfam families assigned the type ‘repeat’, where instances of the repeating unit within a sequence diverge substantially from the consensus. In practice, only 2.3% of all families have different sequence and domain GAs.
Criteria for gathering threshold assignment
Family GAs are chosen with the goal of maximizing coverage while excluding any false positive matches. Although the number of false positives for a given threshold is generally unknown, one way to monitor the false positive-rate indirectly is to check for overlaps between one Pfam family and another. If the same region of a sequence matches two Pfam families, it should be considered a false positive in one of them. This holds true unless the two families are found to be in the same clan, i.e. the observed overlap is believed to reflect an evolutionary relationship between them.
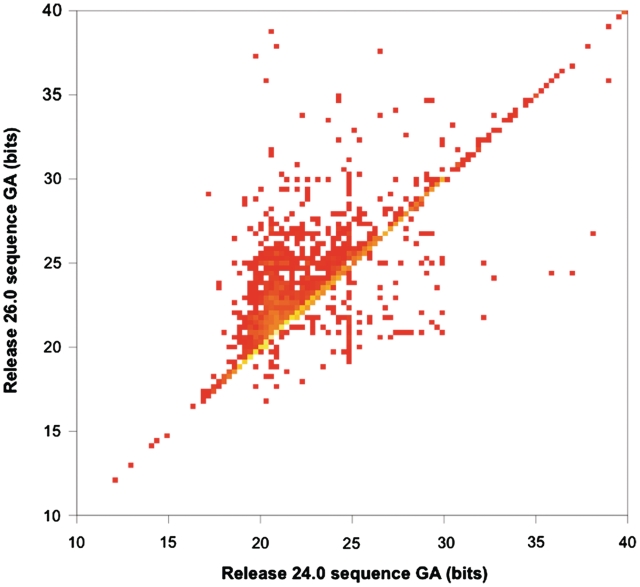
When building a new family, therefore, the GA choice is often influenced by overlaps with other families. In general, overlap-resolution between old and new families leads to GAs being raised over time, since one way to resolve the overlap is to raise the GA in one or the other of the families. This means, for example, that when the UniProtKB dataset underlying Pfam is updated, i.e. at every new release, numerous GAs need to be modified. This is because new sequences will have introduced many new overlaps and, as stated above, Pfam does not allow overlaps between families that are not in the same clan. In a few cases, these sequences will indeed be judged ‘transitional’ between two families and the families will be added to a same clan. In Figure 3, we compare the values of sequence GAs for families in Pfam release 24.0 with GAs of the same families in release 26.0. Overall, 13% of GAs have changed, of which 91% have been raised.
Figure 3.
Heat map showing sequence gathering threshold (GA) changes between Pfam releases 24.0 and 26.0. Yellow squares represent high density; red squares represent low density. Squares on the diagonal correspond to GAs that are unchanged; squares in the region above the diagonal are GAs that have increased; and squares below the diagonal are GAs that have decreased. For the sake of clarity, we chose to show a zoomed-in version of the complete plot, which also includes a number of points outside of the range seen here. The plot was created using R (21).
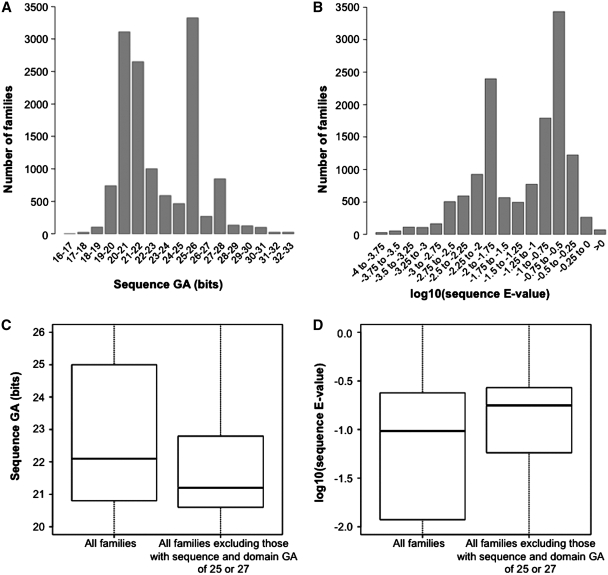
Distribution of gathering thresholds of Pfam families and their relationship to E-values
The distributions of Pfam family GAs and corresponding E-values are shown in Figures 4A and B, respectively. The two GA peaks observed for intervals 25.0–26.0 and 27.0–28.0 (Figure 4A) are due to the fact that numerous Pfam GAs (∼27%) are set to fixed integer values of either 25.0 or 27.0. This is also the cause of the bimodal E-value distribution seen in Figure 4B. Historically, a large number of Pfam families were assigned a reference GA of 25.0. More recently, we have used a higher (guidance) reference threshold of 27.0. These values correspond roughly to ‘safe’ E-value thresholds of ∼10−2 and their increase (from GA 25.0 to GA 27.0) reflects the increase in the size of the UniProtKB database (any particular bit score value will become less significant as the database size increases). In the absence of overlaps with other families, these thresholds are often left unchanged. In retrospect, however, these choices look too conservative, since most families that do not have thresholds of 25.0 or 27.0 have a distribution of GAs that is strongly shifted toward lower bit score values (median = 21.2, 25th percentile = 20.6, 75th percentile = 22.8) (Figure 4C, right side). This seems to indicate that most reference thresholds could be lowered, thereby increasing coverage.
Figure 4.
Distribution of sequence gathering (GA) thresholds and of corresponding E-values. (A) Distribution of sequence GAs for all Pfam-A families. Note that intervals are such that, for example, ‘25–26’ translates into 25 ≤ sequence GA(bits) < 26. (B) Same as the histogram in panel (A), with log10(E-values) in place of GAs. E-values are calculated from GAs according to the following formula: E = N × exp[−λ·(x − τ)], where x is the bit score GA, λ and τ are parameters derived from the HMM model (λ is the slope parameter, τ is the location parameter) and N is the database size (in this case the size of UniProtKB) (22). (C) Box-plot of all Pfam families’ GAs (left side; median = 22.1, 25th percentile = 20.8, 75th percentile = 25.0), and for all families excluding those where both sequence and domain thresholds equal 25.0 or 27.0 (right side; median = 21.2, 25th percentile = 20.6, 75th percentile = 22.8). (D) Same as (C) with log10(E-values) in place of GAs. E-values calculated as in panel (B). Left side: median = 0.096, 25th percentile = 0.012, 75th percentile = 0.24. Right side: median = 0.18, 25th percentile = 0.057, 75th percentile = 0.27. Note that values reported here for median and percentiles are for E-values and not log10(E-values).
Figure 4D reports the distribution of E-values that correspond to family GAs for all families (left side) or excluding those families with GA 25.0 or 27.0 (right side). In the latter case the distribution has a median of 0.18, a 25th percentile of 0.057 and a 75th percentile of 0.27. A handful of families have E-values that, at first sight, appear to be either too high or too low. In particular, there are 72 families with E-value >1, and 82 with E-value <10−6. A survey of these families indicates the following. High E-values arise for two possible reasons. Firstly, the E-value may have been set high because the model is very short and a more ‘realistic’ E-value would result in no matching sequences being reported. Alternatively, the high E-value may have been chosen because that was the relevant value for the size of the sequence database when the model was first built and this family has not been revised subsequently because no overlaps have been introduced. Low E-values, which often correspond to very long profile HMMs, are likely to have been set low in order to avoid inclusion of sequences from other families (overlaps). These overlaps frequently originate from the biased distribution of amino acids in these particular profile HMMs, such that the profile HMMs are too generic and capture equally biased but unrelated sequences (one example is coiled-coil families). We will revisit some of the families that may need re-thresholding in the near future, as part of a larger scale analysis of manually set GAs, and their discriminatory power versus using a uniform fixed threshold.
DUFs and Uncharacterized Protein Families
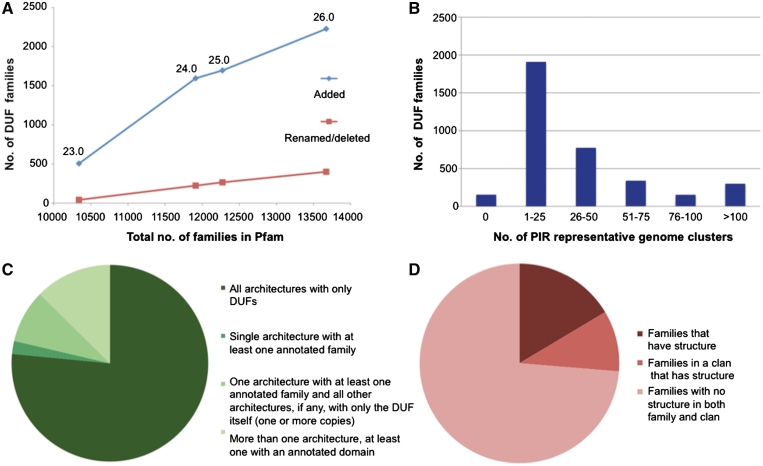
DUFs and uncharacterized protein families (UPFs) (hereafter simply referred to as ‘DUFs’) are families that lack any functional annotation in Pfam. They currently constitute more than a quarter of all Pfam families and their number has been steadily increasing over the last few years (Figure 5A, blue line, and Bateman et al. (16)). As previously reported (16), DUFs are, on an average, less widely distributed on the evolutionary tree than functionally annotated families. For this reason, despite representing 26.5% of all families, they account for only 6.7% of Pfam 26.0 sequence coverage of UniProtKB.
Figure 5.
DUF families’ statistics. (A) Comparison between number of DUFs added (blue) and number of DUFs renamed or otherwise removed (red) since Pfam 22.0 (data shown for releases 23.0–26.0, as indicated by labels on the graph). (B) Number of PIR representative clusters of genomes (23) in DUF families. We used Representative Proteomes version 2.0, comprising a total of 671 clusters for a 35% membership cut-off. (C) Co-occurrence between DUFs and other families. The term ‘architecture’ refers to a combination of families occurring within the same protein sequence. Note that we only considered architectures with at least five member sequences. (D) DUF families and protein structure. ‘Families that have structure’ means that a PDB structure is available for a member of the family; ‘families in a clan that has structure’ means that a PDB structure is available for a member of the same clan.
Normally, when function for at least one protein in a DUF has been experimentally determined, the family is renamed. Although the number of functionally characterized DUFs that have been renamed is on the rise (Figure 5A, red line), this rise is easily outpaced by the number of DUFs that are being newly generated (Figure 5A, blue line). To compound the problem, we have struggled to keep up with published functional studies for such a large number of DUFs. We are therefore in the process of reviewing the scientific literature, with the aim of improving our annotation of DUFs.
Pfam also contains numerous domains, e.g. YbbR (PF07949) or YfcL (PF08891), that have been named after representative proteins from bacteria such as E. coli and B. subtilis, but whose function remains uncharacterized. We would like to make Pfam users (especially those using Pfam for large scale studies) aware of the fact that these families with an assigned name, i.e. a name other than DUF/UPF, still remain in the category of domain of unknown function. We estimate the number of uncharacterized Pfam families that are not named DUF to be around 700, although this figure is likely to be an under-estimate.
DUFs include numerous families that are potentially of great interest for experimental characterization of function. Among these are ∼300 DUFs that are found in >100 representative genome clusters (using the set of clusters with 35% cut-off from PIR (23)) (Figure 5B). The wide taxonomic distribution of these DUFs suggests that they are likely to be associated with important functions in the cell. Furthermore, the Pfam DUF list includes >400 families with at least one human protein.
Two interesting examples of former DUF families that have been characterized in recent years are DUF26 and DUF1017. DUF26 (PF01657), now annotated as salt stress response/antifungal family, has been found as a duplicated domain in the Oryza sativa root meander curling (OsRMC) protein, where it plays a role in salt stress response (24). It is also found in ginkbilobin-2 from Ginkgo biloba, which possesses anti-fungal activity (25). The crystal structure of ginkbilobin-2 has been determined (26) and, as a result of this, we have been able to extend the boundaries of PF01657 to encompass the entire domain. In the second example, E. coli GfcC belongs to DUF1017 (PF06251). This protein is known to play a role in group 4 capsule (G4C) polysaccharide biosynthesis (27), so the family has been re-annotated as capsule biosynthesis GfcC. The crystal structure of GfcC has recently been published (28) and, based on this structure, we have again extended the boundaries of this family.
Part of our motivation for creating DUF families is that we hope to provide information that can guide or accelerate functional characterization of these domains. Data that may be retrieved from the Pfam website for such families include alignments that pinpoint sequence conservation, species distribution and domain co-occurrence. These can help elucidate the evolutionary origin of the family and, in some cases, reduce the number of functional hypotheses. Information about co-occurrence with annotated domains, for example, is of value because it points to functional processes in which DUF families may be involved. In our latest release (26.0), we find that 23% of all DUFs co-occur with at least one annotated domain and that 76 of them are found in a single architecture in combination with at least one annotated domain (note: we only consider architectures with at least five members) (Figure 5C). In the case of DUFs for which the structure of one or more members is available, structural information can be effectively combined with sequence conservation, for example, to highlight putative binding sites for small ligands, proteins or DNA. Some 26% of DUFs have at least one structurally determined protein within the family or within the clan to which the family belongs [Figure 5D; see also Jaroszewski et al. (29)]. Taken alone, this information is unlikely to be enough to confidently assign function to a family, but it can be sufficient to identify interesting targets for experimental characterization.
CONCLUDING REMARKS
Pfam is a database of protein sequence families. Each Pfam family is represented by a statistical model, known as a profile hidden Markov model, which is ‘trained’ using a curated alignment of representative sequences. These models can be searched against protein sequences in order to find occurrences of Pfam families, thereby aiding the identification of evolutionarily related (or homologous) sequences. As homologous proteins are more likely to share structural and functional features, Pfam families can aid in the annotation of uncharacterized sequences and guide experimental work. Despite the continued growth of the sequence databases, Pfam has maintained and even increased its coverage of UniProtKB. Over the coming years we will continue to add new families to Pfam. These, as ever, will come from a variety of sources, in particular, the Protein Data Bank (PDB) and the analysis of complete proteomes for sequences not matched by Pfam. As new data become available, we will also re-visit existing families, to improve their annotation, sequence diversity and domain boundaries as necessary. Use of structural information, in particular, will help us improve domain definitions and increase coverage of UniProtKB at the amino acid level. At the same time, we plan to revise clan organization in order to further increase representation in dense areas of sequence-space. Finally, we hope that the systems that we have put in place to allow external contributions, be it via Wikipedia or directly into the Pfam database, will engage scientists and motivate them to contribute their knowledge and experimental results to Pfam, a community resource for all.
FUNDING
Wellcome Trust (grant numbers WT077044/Z/05/Z); BBSRC Bioinformatics and Biological Resources Fund (grant numbers BB/F010435/1); Howard Hughes Medical Institute (to G.C., J.C., S.R.E and R.D.F.); Stockholm University, Royal Institute of Technology and the Swedish Natural Sciences Research Council (to K.F. and E.L.L.S.) and Systems, Web and Database administration teams at Wellcome Trust Sanger Institute (WTSI) (infrastructure support). Funding for open access charge: Wellcome Trust (grant numbers WT077044/Z/05/Z); BBSRC Bioinformatics and Biological Resources Fund (grant numbers BB/F010435/1).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful for the infrastructure support provided by the Systems, Web and Database administration teams at Wellcome Trust Sanger Institute (WTSI). We also would like to thank L. Aravind of the National Center for Biotechnology Information (NCBI, USA), Adam Godzik of the Burnham Institute (USA) and part of the Joint Center for Structural Genomics (JCSG) and Val Wood of the S. pombe database, PomBase, for their contributions to Pfam. M.P. would like to thank Lars Barquist of WTSI for useful discussions. Finally, we would to like to thank all of the users of Pfam who have submitted new families and/or annotation updates for existing entries.
REFERENCES
- 1.Heger A, Holm L. Exhaustive enumeration of protein domain families. J. Mol. Biol. 2003;328:749–767. doi: 10.1016/s0022-2836(03)00269-9. [DOI] [PubMed] [Google Scholar]
- 2.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coggill P, Finn RD, Bateman A. Identifying protein domains with the Pfam database. Curr. Protoc. Bioinformatics. 2008;Chapter 2 doi: 10.1002/0471250953.bi0205s23. Unit 2 5. [DOI] [PubMed] [Google Scholar]
- 4.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daub J, Gardner PP, Tate J, Ramskold D, Manske M, Scott WG, Weinberg Z, Griffiths-Jones S, Bateman A. The RNA WikiProject: community annotation of RNA families. RNA. 2008;14:2462–2464. doi: 10.1261/rna.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huss JW, 3rd, Orozco C, Goodale J, Wu C, Batalov S, Vickers TJ, Valafar F, Su AI. A gene wiki for community annotation of gene function. PLoS Biol. 2008;6:e175. doi: 10.1371/journal.pbio.0060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 12.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao WM, Fang G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J. Biol. Chem. 2005;280:33516–33524. doi: 10.1074/jbc.M504657200. [DOI] [PubMed] [Google Scholar]
- 14.Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bateman A, Coggill P, Finn RD. DUFs: families in search of function. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:1148–1152. doi: 10.1107/S1744309110001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollich V, Sonnhammer EL. PfamAlyzer: domain-centric homology search. Bioinformatics. 2007;23:3382–3383. doi: 10.1093/bioinformatics/btm521. [DOI] [PubMed] [Google Scholar]
- 18.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 20.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kahari A, et al. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Development CoreTeam. Vienna, Austria: R Foundation for Statistical Computing; 2011. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 22.Eddy SR. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput. Biol. 2008;4:e1000069. doi: 10.1371/journal.pcbi.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, Natale DA, Finn RD, Huang H, Zhang J, Wu CH, Mazumder R. Representative proteomes: a stable, scalable and unbiased proteome set for sequence analysis and functional annotation. PLoS One. 2011;6:e18910. doi: 10.1371/journal.pone.0018910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 2009;149:916–928. doi: 10.1104/pp.108.131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawano Y, Miyakawa T, Yamazaki H, Tanokura M, Hatano K. Purification, characterization, and molecular gene cloning of an antifungal protein from Ginkgo biloba seeds. Biol. Chem. 2007;388:273–280. doi: 10.1515/BC.2007.030. [DOI] [PubMed] [Google Scholar]
- 26.Miyakawa T, Miyazono K, Sawano Y, Hatano K, Tanokura M. Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases. Proteins. 2009;77:247–251. doi: 10.1002/prot.22494. [DOI] [PubMed] [Google Scholar]
- 27.Peleg A, Shifrin Y, Ilan O, Nadler-Yona C, Nov S, Koby S, Baruch K, Altuvia S, Elgrably-Weiss M, Abe CM, et al. Identification of an Escherichia coli operon required for formation of the O-antigen capsule. J. Bacteriol. 2005;187:5259–5266. doi: 10.1128/JB.187.15.5259-5266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sathiyamoorthy K, Mills E, Franzmann TM, Rosenshine I, Saper MA. The crystal structure of Escherichia coli group 4 capsule protein GfcC reveals a domain organization resembling that of Wza. Biochemistry. 2011;50:5465–5476. doi: 10.1021/bi101869h. [DOI] [PubMed] [Google Scholar]
- 29.Jaroszewski L, Li Z, Krishna SS, Bakolitsa C, Wooley J, Deacon AM, Wilson IA, Godzik A. Exploration of uncharted regions of the protein universe. PLoS Biol. 2009;7:e1000205. doi: 10.1371/journal.pbio.1000205. [DOI] [PMC free article] [PubMed] [Google Scholar]