Summary
DNA methylation is a major epigenetic mechanism for gene silencing. While methyltransferases mediate cytosine methylation, it is less clear how unmethylated regions in mammalian genomes are protected from de novo methylation and whether an active demethylating activity is involved. Here we show that either knockout or catalytic inactivation of the DNA repair enzyme Thymine DNA Glycosylase (TDG) leads to embryonic lethality in mice. TDG is necessary for recruiting p300 to retinoic acid (RA)-regulated promoters, protection of CpG islands from hypermethylation, and active demethylation of tissue-specific, developmentally- and hormonally-regulated promoters and enhancers. TDG interacts with the deaminase AID and the damage-response protein GADD45a. These findings highlight a dual role for TDG in promoting proper epigenetic states during development and suggest a two-step mechanism for DNA demethylation in mammals, whereby 5-methylcytosine and 5-hydroxymethylcytosine are first deaminated by AID to thymine and 5-hydroxymethyluracil, respectively, followed by TDG-mediated thymine and 5-hydroxymethyluracil excision repair.
Keywords: embryonic lethality, base excision repair, CpG dinucleotides, DNA demethylation, promoter methylation profiles
Introduction
Cytosine methylation - the formation of 5-methylcytosine (5mC) at CpG sites - is an important epigenetic modification used by mammals to mediate transcriptional regulation, including transcriptional repression, X-chromosome inactivation, imprinting, and suppression of parasitic sequences (Bird, 1992; Kass et al., 1997; Siegfried and Cedar, 1997). The establishment and maintenance of the correct DNA methylation patterns at CpG sites is essential in mammals during development, gametogenesis and differentiation of somatic tissues. Indeed alterations in DNA methylation patterns, with the associated chromatin changes, have profound consequences, as demonstrated by embryonic lethality in the absence of DNA methylation (Li et al., 1992; Okano et al., 1999), developmental defects and accelerated aging in cloned mammals (Rideout et al., 2001), and characteristic epigenetic changes in cancer, such as global genome hypomethylation and tumor suppressor gene hypermethylation (Feinberg and Tycko, 2004; Jones and Laird, 1999).
While DNA methylation is mediated by de novo DNA methyltransferases (DNMT3a and DNMT3b) that act on unmethylated DNA and maintenance DNA methyltransferases (DNMT1) that act on newly replicated, transiently hemimethylated DNA, the demethylating activities or processes that remove methylation marks in mammals are largely unknown. Indeed, it has been controversial as to whether demethylation is an active process in mammals (Ooi and Bestor, 2008) and which mechanisms are involved (Wu and Zhang, 2010).
Demethylation can occur passively due to replication in the absence of re-methylation, with consequent dilution of this modification. However, there is evidence supporting the occurrence of active demethylation in mammals, including demethylation of the paternal genome shortly after fertilization (Mayer et al., 2000; Oswald et al., 2000), demethylation to erase and reset imprinting in primordial germ cells (Reik et al., 2001; Surani et al., 2007), demethylation during somatic differentiation of the developing embryo to establish tissue-specific gene expression patterns (Kress et al., 2006; Niehrs, 2009) and during gene activation in adult kidney (Kim et al., 2009) and brain (Ma et al., 2009). In addition, it is generally thought that active transcription contributes to the maintenance of the unmethylated state of promoter-associated CpG-rich sequences known as CpG islands, but the molecular details of protection from hypermethylation and the potential involvement of an active demethylation process are unknown (Illingworth and Bird, 2009).
Accumulating evidence in non-mammalian model organisms point to the involvement of DNA repair mechanisms in active demethylation (Gehring et al., 2009; Niehrs, 2009). In Arabidopsis, the base excision repair (BER) proteins Demeter and ROS1 affect demethylation by directly removing 5mC through their glycosylase activities (Gehring et al., 2006; Morales-Ruiz et al., 2006). In Xenopus, demethylation has been reported to be initiated by the genome stability protein Gadd45a (growth arrest and DNA damage-inducible protein 45 alpha) in a process dependent on the nucleotide excision repair protein XPG (Barreto et al., 2007); however the role of mammalian GADD45 in demethylation (Barreto et al., 2007; Schmitz et al., 2009) has been challenged (Jin et al., 2008). In zebrafish embryos, rapid demethylation of exogenous and genomic DNA occurs in two coupled steps: enzymatic 5mC deamination to thymine by Activation Induced deaminase (AID) or Apolipoprotein B RNA-editing catalytic component 2b and 2a (Apobec2b, 2a), followed by removal of the mismatched thymine by the zebrafish thymine glycosylase MBD4, with Gadd45 promoting the reaction (Rai et al., 2008). Recently, 5-hydroxymethylcytosine (5hmC), an oxidative product of 5mC generated by the Tet hydroxylases (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009), has been proposed as a demethylation intermediate (Globisch et al., 2010; Wu and Zhang, 2010). During gene activation in the adult mouse brain, demethylation by TET1-mediated hydroxylation of 5meC to 5hmC was promoted by AID/Apobec deaminases, in a process that generates 5-hydroxymethyluracil (5hmU) and ultimately requires BER, although the specific glycosylases involved were not identified (Guo et al., 2011).
Numerous in vitro studies have documented a potential role of the BER enzyme TDG (thymine DNA glycosylase) in transcriptional regulation and demethylation. Indeed, TDG interacts with several transcription factors, including retinoic acid receptor (RAR), retinoid X receptor (RXR) (Um et al., 1998), estrogen receptor α (ERα) (Chen et al., 2003), thyroid transcription factor 1 (TTF1) (Missero et al., 2001) and histone acetyl-transferases p300 and CBP (Tini et al., 2002). It has been proposed that TDG may be responsible for demethylation, either through a direct 5mC glycosylase activity (Zhu et al., 2000), or indirectly, by acting on G:T mismatches originated by a controlled deaminase activity of DNMT3a and DNMT3b (Metivier et al., 2008). Very recently, TDG was shown to be involved in maintaining active and bivalent chromatin marks in mouse embryo fibroblasts and ES cells undergoing neuronal differentiation, respectively, but the mechanism for such epigenetic effects and the requirement of its catalytic activity were not clarified (Cortazar et al., 2011). To investigate the functional role of TDG in epigenetic regulation, DNA demethylation and mammalian development, we generated mice with targeted inactivation of the TDG gene. Tdg-null embryos die in midgestation and exhibit a complex developmental phenotype that appears to derive from the failure to establish and maintain proper DNA methylation patterns at promoters and enhancers. A knock-in mutation that inactivates the glycosylase function of TDG is also embryonically lethal, and TDG is found in a complex with AID and GADD45a. These findings suggest a two-step catalytic mechanism for DNA demethylation that is essential for mammalian development.
Results
Generation of Tdg-null mice
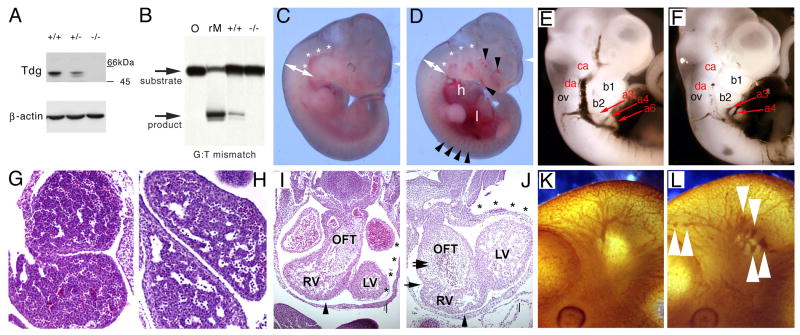
The Tdg gene was deleted in mice by homologous recombination, using a targeting construct that removed exons 3-7, corresponding to most of the catalytic domain (Figure S1). The resulting Tdg- allele does not produce detectable protein by western blotting (Figure 1A), suggesting that it is null. TDG is dispensable for efficient uracil removal from G:U mismatches (Figure S2). However, removal of the mispaired thymine from G:T mismatch-containing oligonucleotides is virtually abrogated in Tdg-/- homozygous mouse embryo fibroblast (MEF, vide infra) nuclear extracts (Figure 1B), thus confirming that Tdg- is a null allele and suggesting that TDG is the predominant G:T mismatch repair activity in MEFs.
Figure 1. Developmental defects in Tdg-null embryos.
(A) The expression of Tdg was monitored by western blot. No expression was detected in MEFs homozygous for the Tdg- allele, whereas heterozygous MEFs showed reduced expression compared to wild type cells.
(B) Tdg is the foremost G:T mismatch repair activity at CpG dinucleotides in MEFs. Repair of a double stranded oligonucleotide containing a G:T mismatch by nuclear extracts of MEFs with different genotypes, in comparison to a substrate neither exposed to lysate nor enzyme (O). Reaction with recombinant MBD4/MED1 (rM) was used as a size marker for cleavage at the mismatched thymine.
(C and D) Gross phenotype of wild type and Tdg-/- littermate embryos at embryonic day E11.5: double-arrows show constriction in the cervical region of Tdg-null embryos (D) compared to wild type embryos (C); white asterisks mark the carotid artery that is stenotic in Tdg-null embryos; the enlarged heart with pericardial effusion (h) and hemorrhagic liver (l) are apparent; arrowheads point to hemorrhagic lesions in the cranium, and enlarged and irregular segmental arteries.
(E and F) Cardiac perfusion with India ink in wild type and Tdg-mutant embryos at E11; in Tdg-null embryo (F), circulatory insufficiency is demonstrated by reduced perfusion of the dorsal aorta (da) and carotid artery (ca), whereas the third (a3) and fourth (a4) branchial arch arteries are enlarged in comparison to wild type embryos (E); the first (b1) and second (b2) branchial arches, as well as the otic vesicle (ov), are indicated.
(G and H) Transverse sections of the liver at E11; compared to wild type (G), the mutant liver has enlarged hepatic sinusoids, the likely proximal causes of abdominal hemorrhage.
(I and J) Transverse sections of the heart at E11.5 show a patterning defects in mutant embryos; the conal part of the OFT is severely hyperplastic in mutant embryos (twin arrows in J) when compared to the heterozygous specimen (I), generating an atypical indentation between the right ventricle (RV) and OFT (arrow head in J); the characteristic “dog-leg bend” of the OFT, which is responsible for correctly positioning the OFT over the midline of left ventricle (LV) and RV is not observed in mutant hearts; instead, the OFT is situated right above the RV; as a result, the left part of the body wall is pushed out by the LV (asterisks).
(K and L) Immunostaining of the vascular labyrinth with a PECAM/CD31 antibody in wild type (K) and Tdg-null (L) embryos at E11 reveal a generalized disorganization of the vascular network in the latter; arrowheads point to irregular branches of the internal carotid with varicosities, bulges and ectasias.
See also Figures S1, S2 and S3.
Lethality and complex developmental phenotype of Tdg-null embryos
While heterozygous Tdg+/- mice are viable, fertile and show no obvious phenotype, homozygosity for the null Tdg allele leads to embryonic lethality. In fact, when heterozygous Tdg+/- mice were interbred, no live birth Tdg-/- homozygotes were derived. Specifically, the numbers of wild type, heterozygous and homozygous mutant pups obtained were 11, 29 and 0, respectively, which is significantly different from the expected Mendelian ratios (p < 0.0008 by χ2). To examine in detail the embryonic lethality, timed matings were set up between Tdg+/-heterozygotes, and pregnant mice were sacrificed at different gestation times, ranging from embryonic day (E) 10.5 to E14.5. This analysis revealed an arrest of development associated with Tdg nullizygosity at E11.5; at later gestation times, homozygous mutant embryos are beginning to be resorbed (E12.5-13.5), or are completely resorbed and never detected (E14.5)(Table S1). At E11.5, developmentally arrested Tdg-null embryos manifest a complex phenotype characterized macroscopically by abdominal (liver) hemorrhage, pericardial edema/hemorrhage, hypoplastic branchial arches, delayed limb development, prominent telencephalic vesicles and diffuse hemorrhagic lesions (Figure 1C,D,G,H).
Microscopically, Tdg-null embryos exhibit specific patterning defects of the developing heart, with the most significant abnormalities seen in the outflow tract (OFT) (Figure 1I,J and Figure S3A-D); vasculogenesis defects of dorsal aortae, carotid arteries and branchial arteries (Figure 1E,F); and generalized defect of angiogenesis, particularly evident as altered branching of the internal carotid (Figure 1F) and the coronaries (Figure S3E,F).
TDG has two proposed roles, in mutational avoidance (DNA repair) and transcriptional regulation (Cortazar et al., 2007). The embryo phenotype was fully penetrant and reproducible, which is inconsistent with an anti-mutagenic, DNA repair defect that would be expected to yield a variable, heterogeneous phenotype caused by stochastic secondary mutations at different target genes. We therefore focused on the role of TDG in transcription as a possible mechanistic explanation of the phenotype.
Remarkably, many features of the phenotype of Tdg-null embryos (altered vasculogenesis and angiogenesis, hemorrhagic lesions, heart abnormalities with thinning of the myocardium and pericardial effusion) have been previously described for either Cbp-/- or p300-/- embryos (Tanaka et al., 2000; Yao et al., 1998). Similarly, some other specific phenotypic features in Tdg-null embryos (OFT septation defects, hypoplastic myocardium, abnormal great arteries derived from branchial arches, delayed limbs) resemble those of embryos deficient in various RAR and RXR genes (Mark et al., 2006), or hypomorphic for retinaldehyde dehydrogenase, the enzyme involved in RA biosynthesis (Vermot et al., 2003). We infer that the lethality phenotype is likely related to the inactivation of a developmentally relevant, transcription-related function of TDG. In order to investigate this issue directly, we established MEF lines from Tdg-null embryos.
Attenuated RA-dependent transcription and altered p300 recruitment in Tdg-null MEFs
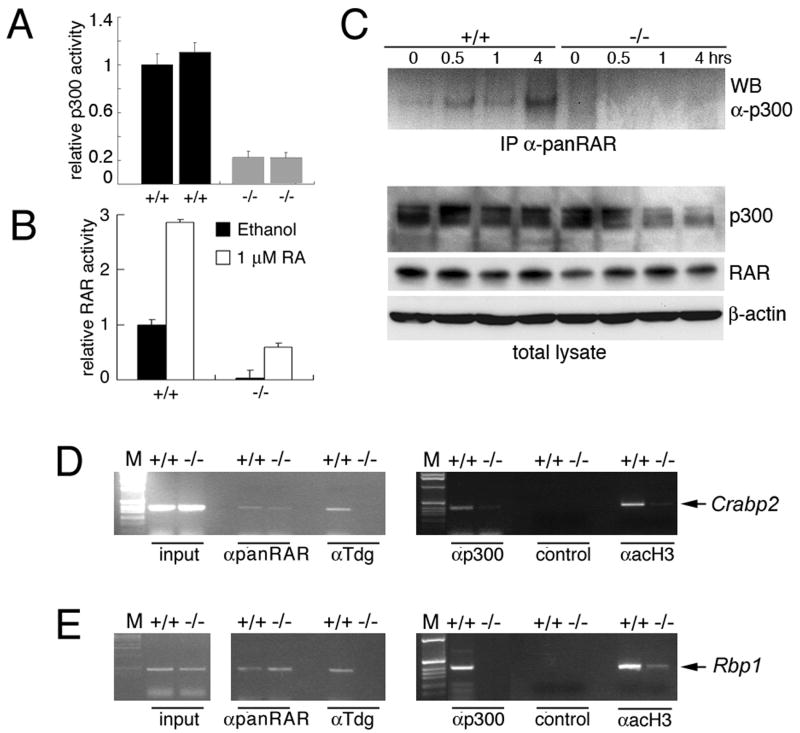
We hypothesized that, given the phenotypic features described above, it is possible that RAR/RXR and p300 activity might be reduced in the absence of TDG. Indeed, assay of p300 activity with reporter constructs indicated that transcriptional co-activation by this acetyltransferase is significantly reduced in two independent Tdg-/- MEF lines in comparison to wild type MEF lines (Figure 2A). RA-dependent RAR/RXR transcriptional activity is also attenuated in Tdg-null MEFs (Figure 2B).
Figure 2. Involvement of TDG in transcription and composition of RAR-p300 complexes.
(A) Reduced p300-induced transcriptional activation in Tdg-/- MEFs. Luciferase activity of a Gal4 operator luciferase reporter co-transfected with a Gal4 DNA binding domain-p300 fusion construct, and normalized to transfection efficiency using β-galactosidase expression. Since the transfected plasmids are unmethylated, this assay reflects only the co-activator role of TDG.
(B) Reduced retinoic-dependent RAR/RXR transcriptional activity in Tdg-/- MEFs. CAT activity of a RARE-containing reporter was normalized to transfection efficiency using β-galactosidase expression.
(C) Co-IP with an antibody capable of recognizing all the RARs shows lack of association between RAR and p300 in Tdg-/- MEFs. Wild type and mutant MEFs were stimulated with 1 μM RA for the indicated time. Approximately equal levels of p300 and RAR are present in wild type and mutant cells. Detection of β-actin is shown as a loading control.
(D and E) Chromatin immunoprecipitation shows that Tdg binds directly to the promoter of two differentially expressed, RAR-RXR target genes, Crabp2 (D) and Rbp1 (E), and is required for p300 recruitment and histone H3 acetylation. Approximately equal amounts of input chromatin were used for immunoprecipitation. As negative control, immunoprecipitation with non-specific immunoglobulins was performed.
Data are presented as mean ± standard error of the mean (SEM).
See also Figure S4.
To clarify the role of TDG in transcription, we compared the transcriptome of wild type and Tdg-null MEFs. In keeping with a role of TDG in transcriptional activation, of the 108 differentially expressed genes from 120 probe sets, approximately 3/4 of the genes were down-regulated in its absence (Table S2). The differentially expressed genes were analyzed with gene ontology and pathway analysis applications. Remarkably, the pathway/network with the highest score was centered around RA (Figure S4) and comprised retinol biosynthesis and RA-dependent target genes downregulated in Tdg-null MEFs, including those encoding cellular retinoic acid binding protein 2 (Crabp2, 15.3-fold), retinol binding protein 1 (Rbp1, 9.3-fold), Igfbp6 (16.3-fold), embryonal fyn-associated substrate (Efs, 6-fold) and Rai14 (1.9-fold). These observations indicate that TDG is a positive regulator of transcription, particularly p300- and RAR/RXR-dependent transcription.
To define the role of TDG in RA-dependent transcription, we examined the composition of RAR-containing complexes by co-immunoprecipitation (co-IP) and found that p300 is in a complex with RAR/RXR in wild type MEFs but not in Tdg-null MEFs, despite the presence of approximately equal levels of p300 and RAR in total lysates (Figure 2C). In addition, RARs occupy retinoic acid response elements (RARE) on the Crabp2 and Rbp1 promoters in both wild type and Tdg-null cells, but in the absence of TDG there is little recruitment of p300 and a reduction in the presence of its product, acetylated histone H3 (Figure 2D,E). These findings are consistent with the differential expression of Crabp2 and Rbp1 and indicate that TDG has an obligatory, direct role in their proper transcriptional regulation.
Altered DNA methylation patterns in Tdg-null cells and tissues
Given the possible role of TDG in demethylation (Metivier et al., 2008; Zhu et al., 2000), we examined the DNA methylation patterns of promoters of select genes that were differentially expressed between wild type and Tdg-null MEFs using sodium bisulfite/DNA sequencing. In Tdg-null MEFs, the downregulated genes contain a CpG island within 2 kb of sequence upstream of the transcriptional start site and are hypermethylated, including Efs, Crabp2, Hoxa5 and H19, (Figure 3A-D). Since these CpG islands and the maternal allele of the imprinted H19 gene are unmethylated in zygotes, ES cells and the soma (Mohn et al., 2008; Reese and Bartolomei, 2006)(data not shown), this observation suggests that, in the absence of TDG, sequences that are normally kept unmethylated succumb to hypermethylation, likely as a consequence of unscheduled de novo methylation.
Figure 3. Hypermethylation of CpG islands in the absence of TDG.
(A-E) DNA methylation analysis by sodium bisulfite modification and sequencing of cloned PCR products in wild type and Tdg-null cells. Open and closed circles represent unmethylated and methylated CpGs, respectively. Crabp2 (A), Efs (C) and Hoxa5 (D) promoters are unmethylated at various degrees in wild type cells, whereas their CpG islands are hypermethylated in Tdg-null MEFs. For the H19 promoter (B), the first three clones in wild type MEFs are likely derivatives of the inactive paternal allele, and the remaining ones are probably of maternal origin (H19 is maternally expressed), whereas in Tdg-null MEFs both alleles are hypermethylated. Analysis of the 3′ half of the Igf2 DMR2 (E): the first five clones in wild type PGCs are likely derived from the paternal (methylated) allele, and the remaining ones are probably of maternal origin, whereas in Tdg-null PGCs, all the alleles are hypermethylated. See also Figure S4.
To rule out the possibility that hypermethylation was caused by the in vitro culture stress of MEFs (Pantoja et al., 2005), we analyzed the methylation pattern of the imprinted gene Igf2 in wild type and Tdg-mutant primordial germ cells (PGCs) isolated from E11 embryos (prior to the onset of lethality). While wild type E11 PGCs show the typical methylation profile of maternal unmethylated and paternal methylated alleles at the Igf2 differentially methylated region 2 (DMR2), all the alleles sequenced in Tdg-mutant PGCs, presumably including those of maternal origin, are methylated (Figure 3E). Although it is currently unclear whether TDG has any specific role in the establishment or maintenance of imprinting, these data confirm a TDG-dependent protection from hypermethylation in early development.
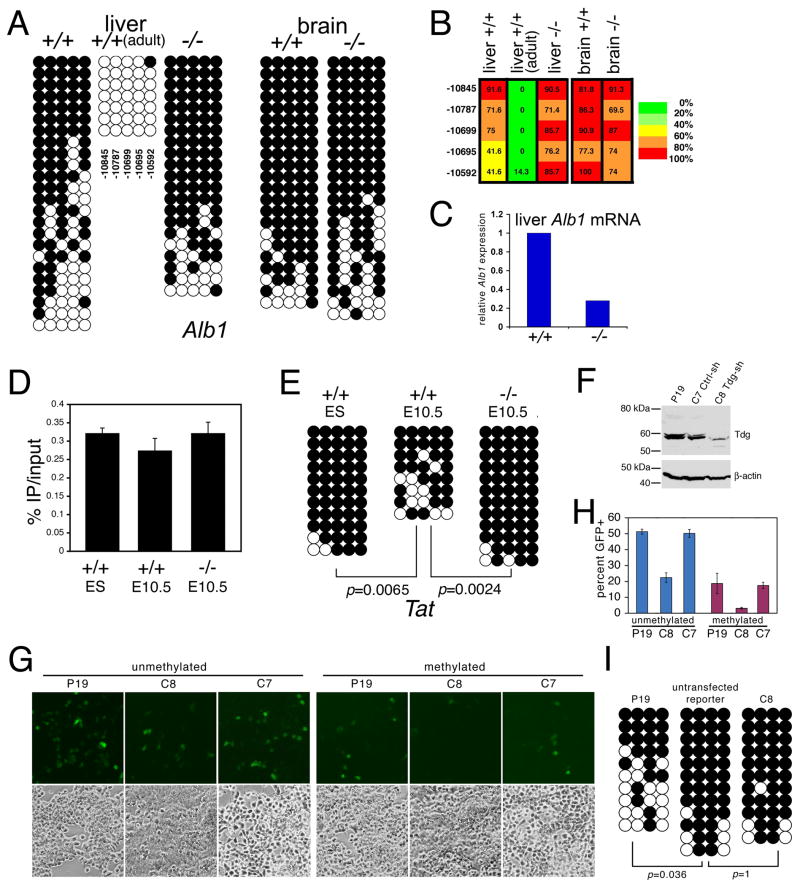
The observed protective function begs the question of whether TDG might also have a role in DNA demethylation. During development, highly conserved non-coding elements and enhancers undergo demethylation in a process linked to tissue-specific gene expression and differentiation (Kress et al., 2006; Niehrs, 2009). One such example is the albumin gene (Alb1) enhancer, whose five CpG dinucleotides are progressively demethylated during liver development and are associated with Alb1 mRNA transcription (Xu et al., 2007). Analysis of these sites revealed that they remain methylated in Tdg-null liver at midgestation, in a configuration similar to that of a non-albumin producing organ, e.g. brain (Figure 4A-B). This correlated with inefficient Alb1 mRNA transcription in the Tdg-null liver (Figure 4C).
Figure 4. TDG is involved in DNA demethylation.
(A) DNA methylation analysis of five CpG dinucleotides of the Alb1 enhancer in liver and brain of wild type and Tdg-null embryos at E11; the numbers refer to the position of CpGs relative to the transcription start site (TSS).
(B) Corresponding quantification and color-coded display of percent DNA methylation at each CpG dinucleotide of the Alb1 enhancer.
(C) Real-time RT-PCR quantification of Alb1 mRNA expression, normalized to Hprt mRNA expression, in wild type and Tdg-null livers at E11.
(D) Methylated DNA immunoprecipitation-quantitative PCR (MeDIP-qPCR) analysis of methylation levels at the Tat gene GRU, expressed as percent immunoprecipitated DNA relative to input DNA, in ES cells, wild type and Tdg-null embryos at E10.5 (headless embryo body dissected to enrich for liver).
(E) Methylation analysis by sodium bisulfite modification and sequencing of five CpG dinucleotides of the Tat gene GRU (at -2520, -2485, -2473, -2390, and -2386 bp relative to TSS) in ES cells, wild type, and Tdg-null embryos at E10.5 (dissected to enrich for liver); two-sided Fisher's Exact Test at the 5% significance level.
(F) Western blot analysis showing effective downregulation of TDG in P19 C8 cells expressing a short hairpin RNA (shRNA) directed against murine Tdg mRNA in comparison to parental P19 cells and control shRNA P19 C7 cells.
(G) Detection by fluorescence of GFP+ cells (top) in cultures of parental P19, TDG-shRNA-containing P19 C8 cells and control shRNA C7 cells, transfected with unmethylated and SssI-methylated human Oct4∷EGFP reporter. Cells were plated at approximately equal density, as evidenced by phase contrast microscopy (bottom).
(H) Quantitation of expression of unmethylated and SssI-methylated human Oct4∷EGFP reporter in P19, TDG-shRNA C8 and control shRNA C7 cells.
(I) DNA methylation analysis by sodium bisulfite modification and sequencing of the proximal region (region 8)(Deb-Rinker et al., 2005) of the human Oct4 promoter from the untransfected methylated Oct4∷EGFP reporter and the same reporter transfected and recovered from P19 and C8 cells; two-sided Fisher's Exact Test at the 5% significance level.
Data are presented as mean ± standard error of the mean (SEM).
The glucocorticoid-responsive unit (GRU) of the Tyrosine aminotransferase (Tat) gene enhancer undergoes demethylation at midgestation in the developing rat liver in a process stimulated by the prenatal peak of glucocorticoids (Thomassin et al., 2001). Demethylation of this enhancer is associated with single-strand nicks 3′ to the 5mC, leading to the suggestion that a demethylating activity initiates base or nucleotide excision repair at these sites (Kress et al., 2006). We found that demethylation of five CpG sites at the GRU of the murine Tat enhancer begins at midgestation and is dependent on TDG (Figure 4D-E).
Taken together, these observations indicate that TDG is required for the establishment of proper DNA methylation patterns that are conducive to transcription of developmentally- and hormonally-regulated genes and of tissue-specific genes, both by guarding from CpG island hypermethylation and promoting selective demethylation events.
Involvement of TDG in active DNA demethylation
While protection against CpG island hypermethylation might be, in principle, a reflection of the co-activator function of TDG, the involvement of this enzyme in DNA demethylation suggests an active catalytic role. In order to determine directly whether TDG is involved in active DNA demethylation, we studied the transcriptional reactivation of a heterologous in vitro-methylated Oct4 pluripotency gene in embryonic carcinoma P19 cells or in the same cells expressing either an shRNA (C8) targeting TDG or a control shRNA (C7)(Figure 4F). We used an Oct4 promoter∷EGFP reporter assay in which reactivation of EGFP expression is due to demethylation of the heterologous Oct4 promoter (Barreto et al., 2007). We found that the unmethylated Oct4∷EGFP reporter was expressed within 12 hours of transfection in both parental P19 cells and its derivative line C8 bearing the TDG knock-down (the 2.3-fold expression differential between P19 and C8 likely reflects the co-activator function of TDG). In contrast, the methylated reporter is efficiently expressed only in parental P19 cells but not C8 cells (6-fold expression differential). The C7 cells expressing a control shRNA and P19 cells infected with a scrambled shRNA behaved similarly to the parental P19 line (Figure 4G,H and data not shown). A bisulfite sequencing analysis of the transgene recovered after 12 hours from transfected cell lines revealed that demethylation of the proximal region of the Oct4 promoter is compromised in the Tdg knockdown cells, thus establishing a direct effect of TDG on demethylation (Figure 4I). The short time frame of the demethylation and the fact that the reporter plasmid used lacks an origin of replication rules out any potential effect of passive demethylation and confirms that TDG is involved in an active demethylation process.
The DNA glycosylase activity of TDG is required for development and DNA demethylation
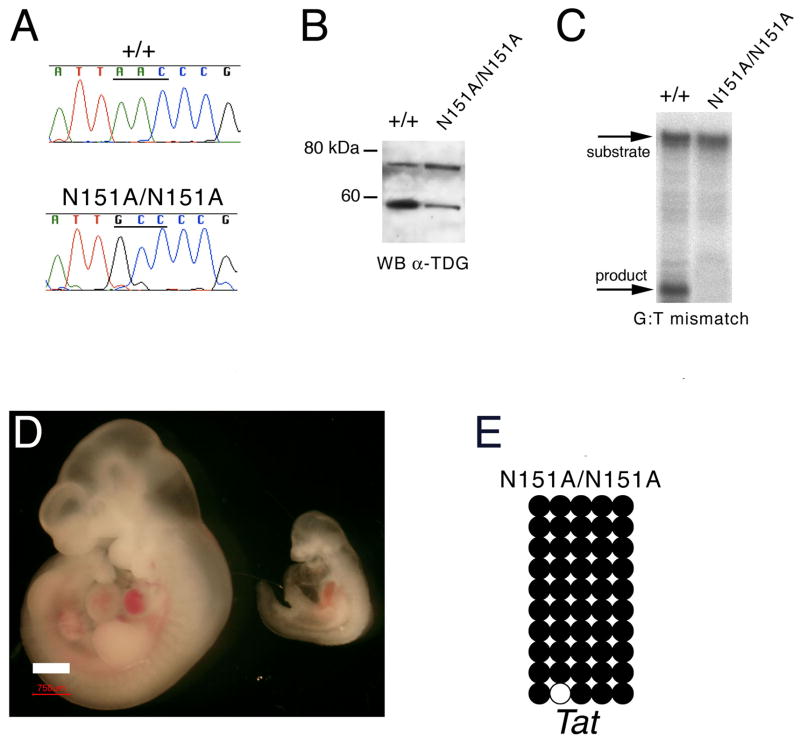
If TDG has a catalytic role in DNA demethylation, the prediction is that an inactivating point mutation at the glycosylase active site would reproduce the embryonic lethality. On the other hand, lack of lethality of such mutation would suggest that TDG affects methylation patterns and development as a reflection of its co-activator function. We tested these two possibilities by generating a knock-in mouse strain (Figure S1) expressing a point mutation (N151A) in the TDG glycosylase domain (Figure 5A-B) that eliminates the obligatory asparagine residue at the TDG active site (Hardeland et al., 2000) and abrogates glycosylase activity (Figure 5C). Live birth TdgN151A/N151A homozygotes were never derived from the breeding of heterozygotes, indicating that indeed TDG glycosylase activity is required for embryonic development. Remarkably, analysis of timed matings revealed that lethality occurs even earlier than in knock-out embryos, i.e. at approximately E10.5. TdgN151A/N151A embryos are much smaller than wild type littermates and show general developmental delay, turning defect, and pericardial effusion (Figure 5D). Importantly, DNA demethylation of the Tat enhancer was abrogated in TdgN151A/N151A embryos (Figure 5E). We conclude that the catalytic activity of TDG is essential for development and DNA demethylation.
Figure 5. The DNA glycosylase activity of TDG is required for development and DNA demethylation.
(A) Sequence analysis of a cDNA fragment encompassing the relevant Tdg exon 4 region from wild type and TdgN151A/N151A E10.5 embryo total RNA confirms expression of the knock-in allele.
(B) Western blot analysis with an anti-TDG antibody reveals expression of the wild type and TDGN151A protein in E10.5 embryo lysates of corresponding genotypes.
(C) Repair of a double stranded oligonucleotide containing a G:T mismatch by nuclear extracts of whole E10.5 embryo extracts of the indicated genotypes.
(D) Gross phenotype of wild type (left) and TdgN151A/N151A (right) littermate embryos at embryonic day E10.5. Size bar corresponds to 750 μm.
(E) Methylation analysis by sodium bisulfite modification and sequencing of five CpG dinucleotides of the Tat gene GRU in a TdgN151A/N151A E10.5 embryo (headless embryo body preparations dissected to enrich for liver); comparison of methylation levels with the wild type embryo in Figure 4E was made using the two-sided Fisher's Exact Test at the 5% significance level and revealed a p-value equal to 0.0004.
See also Figure S1.
TDG is in a complex with AID and GADD45a
A direct 5mC glycosylase activity of TDG has been reported (Zhu et al., 2000), but we were unable to detect such activity using a preparation of recombinant TDG that was extremely active on its cognate G:T mismatched substrates (Figure S5A). In zebrafish embryos, demethylation is initiated by enzymatic deamination of 5mC to T by AID, Apobec2a or Apobec2b, followed by MBD4 glycosylase removal of the mismatched T in a reaction promoted by GADD45 (Rai et al., 2008). We therefore set out to determine whether TDG may mediate DNA demethylation in a similar two-step mechanism by interacting with AID/Apobec members and GADD45a in mammalian cells.
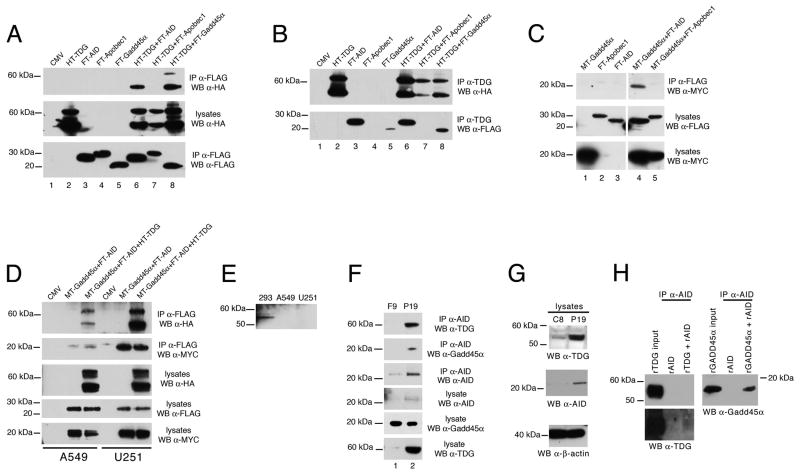
Co-IP experiments conducted in HEK-293 cells transfected with cDNAs encoding tagged versions of TDG, AID, Apobec1 and GADD45a revealed that TDG forms a complex with AID (Figure 6A-B, lane 6) and GADD45a (Figure 6A-B, lane 8). In addition, transfected AID and GADD45a co-IP with endogenous TDG (Figure 6B, lanes 3 and 5). The interaction of TDG with AID is specific, as TDG does not co-IP with the AID-related family member Apobec1 (Figure 6A-B, lane 7). Furthermore, AID interacts with GADD45a (Figure 6C, lane 4) in a TDG-independent manner, as shown by co-IP in two cell lines having undetectable levels of TDG (Figure 6D-E).
Figure 6. TDG is in a complex with AID and GADD45a.
(A-D) Immunoprecipitates of lysates of HEK-293 (A-C) or A549 and U251 (D) cells transfected with hemagglutinin (HA)-, FLAG- or MYC-tagged expression constructs were resolved by PAGE and detected by western blotting with the indicated antibodies. Western blotting of lysates shows that identical tagged constructs were expressed at approximately equal levels. In these and other Western blots, the presence of additional TDG bands is the result of sumoylation or other post-translational modifications (Cortazar et al., 2007). Note that in (C) FLAG-tagged AID and FLAG-tagged GADD45a immunoprecipitate not only with exogenous, HA-tagged TDG (lanes 6 and 8) but also with endogenous TDG (lanes 3 and 5).
(E) Western blotting with an anti-TDG antibody reveals TDG expression in HEK-293 cells but not in A549 or U251 cells.
(F) Co-immunoprecipitation experiments with the indicated antibodies show that AID forms a complex with TDG and GADD45a at endogenous levels of expression in P19 embryonic carcinoma cells. A western with anti-AID antibody confirms that AID was actually immunoprecipitated.
(G) AID levels are reduced by shRNA in the P19 derivative, TDG knockdown cell line C8, as evidenced by western blotting of lysates with the indicated anti-TDG and anti-AID antibodies; western blotting with an anti-β-actin antibody acts as a loading control.
(H) The indicated recombinant proteins were pre-mixed and the mixtures were immunoprecipitated with an anti-AID antibody; immunoprecipitates along with the input recombinant TDG or GADD45a were detected by western blotting with an anti-TDG or anti-GADD45a antibody, as indicated. GADD45a is readily detected, whereas only a small amount of TDG (visible in the longer exposure, bottom panel, left) precipitates with AID, suggesting a low-affinity interaction.
See also Figures S5 and S6.
We further tested whether these interactions are taking place at endogenous levels of expression, in the developmentally relevant context of embryonic carcinoma P19 cells and teratocarcinoma F9 cells, exhibiting high and low levels, respectively, of TDG and AID. Co-IP experiments conducted on endogenous lysates demonstrated that in P19 cells TDG interacts with AID (Figure 6F, top panel lane 2) and that AID and GADD45a also interact (Figure 6F, second panel, lane 2). These interactions were not detected in F9 cells, likely due to the very low levels of TDG and AID expression. Interestingly, shRNA-mediated down-regulation of TDG in the P19-derived C8 cells leads to reduction of AID expression (Figure 6G), suggesting that TDG may regulate the levels and/or the stability of AID. Recombinant AID and recombinant GADD45a purified proteins directly interact, as do recombinant AID and recombinant TDG, albeit with lower affinity (Figure 6H). One possible difference to explain the stronger interaction detected in cells is that the latter may be mediated by post-translational modifications not present in the recombinant proteins. We conclude that the TDG-AID-GADD45a interaction occurs in vivo and has functional consequences for AID levels.
Interestingly, the TDG N151A protein retains interaction with AID and GADD45a (Figure S6), suggesting that the more severe phenotype of the TdgN151A/N151A compared to the Tdg knockout embryos might be due to a dominant-negative action of the catalytically dead mutant protein in sequestering AID and GADD45 or other interactors in non-functional, non-productive complexes.
TDG has glycosylase activity on 5hmU
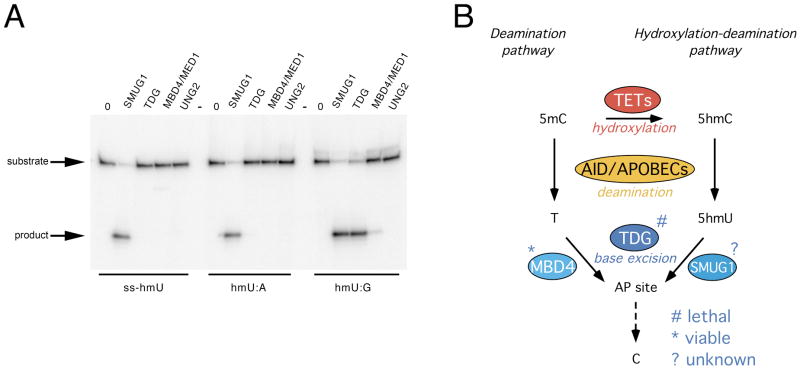
Since 5hmC, the hydroxylation product of 5mC, has been proposed as an intermediate in demethylation, we assayed but failed to detect 5hmC glycosylase activity for TDG or any other related glycosylases (Figure S5B). However, during active DNA demethylation in mammalian cells, AID/Apobec deaminases convert 5hmC to 5hmU for subsequent processing by BER (Guo et al., 2011). We therefore assayed the 5hmU glycosylase activity of TDG and related glycosylases. As previously described (Haushalter et al., 1999), SMUG1 has activity on 5hmU in single-strand DNA or when paired with adenine in double-strand DNA to resemble an expected product of thymine oxidation (Fig. 7A). Remarkably, similar to a sequence-unrelated thermophilic Tdg (Baker et al., 2002), mammalian TDG exhibits robust activity, comparable to SMUG1, on a 5hmU:G mismatch in double-strand DNA, the expected product of deamination following hydroxylation of 5mC (Fig. 7A). This result expands the suggested role of TDG downstream of deamination to include initiation of BER following hydroxylation of 5mC (Fig. 7B).
Figure 7. TDG glycosylase activity on 5hmU and model of the role of TDG in DNA demethylation pathways.
(A) Recombinant TDG and related glycosylases were incubated with 5hmU-containing single-strand oligonucleotide or double-strand oligonucleotides bearing 5hmU:A pairing or 5hmU:G mismatch, all 32P-labeled on the 5hmU strand. The resulting AP site was cleaved with alkali at high temperature. (B) Schematic of the involvement of TDG in both the deamination and hydroxylation-deamination pathways of DNA demethylation. Lethality/viability of the glycosylase knock-out mice is indicated.
See also Figures S5 and S7.
Lack of promoter mutations in Tdg-mutant cells
Deamination of 5mC or 5hmC in the absence of TDG-mediated repair of the resulting G:T or G:5hmU mismatch is expected to increase G:C to A:T transition mutations. For this reason, we conducted a sequence analysis of non-bisulfite modified DNA of promoters undergoing TDG-dependent protection from hypermethylation or DNA demethylation. No mutation was found in H19, Efs, HoxA5 and Crabp2 promoters in Tdg-mutant MEFs, or at the Oct4 promoter of the Oct4∷EGFP transgene recovered from the C8 Tdg-knockdown cells (data not shown). Although a possible compensation by MBD4, SMUG1 or mismatch repair cannot be ruled out, this result suggests the possibility that the deamination and glycosylase steps are coordinated, such that deamination does not occur in the absence of TDG.
Discussion
Our results demonstrate that TDG is required for normal mammalian development and the establishment of proper promoter/enhancer DNA methylation patterns that are conducive to transcription during embryogenesis. Failure to establish and maintain correct DNA methylation patterns is likely the cause of lethality and the observed complex developmental phenotype, as it is known that even small changes in DNA methylation cause abnormalities or lethality (Gaudet et al., 2003).
In maintaining the proper epigenetic states, TDG apparently has a dual role: protection from aberrant hypermethylation (examples include the CpG islands of Efs, HoxA5 and Crabp2 in MEFs and the maternal alleles of H19 and Igf2 DMR2 in PGCs) and promotion of demethylation (exemplified by the Alb1 and Tat enhancers in hepatoblasts and the Oct4∷EGFP reporter in P19 cells).
Our combined biochemical and developmental data suggest that demethylation is an active process requiring TDG catalytic activity immediately downstream of the deaminase-catalyzed conversion of 5mC into thymine and/or of 5hmC into 5hmU. Thus, both the 5mC deamination and hydroxylation-deamination pathways may converge on TDG (Figure 7B) and perhaps this convergence may explain the absolute requirement of TDG during embryogenesis, although the relative prevalence of each pathway in different developmental contexts is currently unknown. The extent of possible partial compensation by activities of MBD4 and SMUG1 is also unknown, although the observed lethality of the Tdg knockout suggests that it is not sufficient to maintain embryogenesis.
Similarity of the lethal phenotype of the TDG knock-in and knock-out embryos, and in turn their resemblance to the p300/CBP and RAR/RXR mutant embryos, strongly suggests the possibility that protection from aberrant de novo methylation may also involve an active process, requiring TDG glycosylase activity to constantly antagonize methylation. However, a non-catalytic role of TDG cannot be ruled out completely and it is possible that protection from hypermethylation may occur at least in part via TDG inhibition of de novo DNA methyltransferases (Li et al., 2007).
Our observation that TDG is required for the interaction of RAR/RXR with p300 both on and off the DNA suggests a model in which transcription factor binding (which is TDG-independent, Fig 2D,E) may be responsible for tethering TDG to the promoter/enhancer (Figure S7). Thus, it is possible that the TDG-provoked demethylation of differentiation-associated promoters/enhancers depends on TDG tethering by tissue-specific transcription factors. Of note, GADD45 is known to bind to nuclear hormone receptors (Yi et al., 2000) and preferentially to hyperacetylated nucleosomes (Carrier et al., 1999), suggesting that GADD45 may also be involved in targeting promoters for demethylation. In this model, proper targeting of AID may in turn depend on its interaction with GADD45 and TDG. Future binding studies, conducted on a genomic scale in different tissues and at various developmental stages, will further define the promoters/enhancers that are direct targets of the TDG-AID-GADD45a complex.
The identification of a ternary complex containing TDG, AID and GADD45a is consistent with the recent recognition of AID as a factor required for DNA demethylation during reprogramming of somatic cells (Bhutani et al., 2010) and erasure of DNA methylation at imprinted and other loci in PGCs (Popp et al., 2010), as well as with the role of GADD45a (Barreto et al., 2007; Schmitz et al., 2009) and the related GADD45b (Ma et al., 2009) in demethylation of specific promoters. It is also consistent with the role of BER in genome-wide active DNA demethylation in PGCs (Hajkova et al., 2010). However, AID-deficient mice are viable and fertile (Muramatsu et al., 2000; Revy et al., 2000), which suggests that TDG may also function downstream of Apobec deaminases and possibly engage different GADD45 proteins. In fact, various TDG-deaminase-GADD45 complexes, and possibly different TET proteins, may be utilized for demethylation associated with distinct developmental processes and reprogramming events.
Our observation of the lack of deamination-induced transition mutations in Tdg-mutant MEFs and Tdg-knockdown cells at promoters that undergo TDG-dependent protection from hypermethylation and demethylation suggests that TDG has a role not only in repairing deamination products, but also in initiating the DNA demethylation process, thus controlling the potentially mutagenic deaminase activity of AID. It is possible that initiation is regulated by targeting of the AID-TDG-GADD45a complex to relevant promoter/enhancers or by the optimal reciprocal amounts of these proteins in the complex, as TDG affects AID levels and/or stability (Figure 6G). Given the frequent CpG transition mutations and hypermethylation of tumor suppressor gene promoters in human cancer, inactivation of TDG and its demethylating complex or altered relative amounts of TDG, AID and GADD45a may play a role in tumor formation.
Experimental Procedures
Derivation of Tdg-null and Tdg knock-in mice
Tdg-null and Tdg knock-in mice were generated by homologous recombination of positive-negative selection targeting constructs in R1 ES cells; ES clones carrying the targeted Tdg locus were injected into C57/BL6 blastocysts to generate chimeric mice. The chimeric mice were backcrossed into the C57/BL6 line for at least eight generations.
Isolation of MEFs
Mouse embryo fibroblasts (MEFs) were prepared as previously described (Cortellino et al., 2003) from embryos harvested at E10.5, and grown in DMEM supplemented with 15% fetal bovine serum.
Isolation of primordial germ cells
PGCs were isolated from E11 germinal ridges dissected by pipetting up-down and trypsin digestion. PGCs stained with anti-SSEA1 antibody were sorted by FACS. The morphology of the collected PGCs was evaluated after phosphatase alkaline staining to confirm cellular specificity.
Analysis of DNA methylation by bisulfite modification-sequencing
Genomic DNA was subjected to the sodium bisulfite modification reaction, as previously described (Howard et al., 2009). Products from the bisulfite reactions were amplified by PCR using primers designed with the MethPrimer software at http://www.urogene.org/methprimer/. Purified PCR products were subcloned into pGEM T-Easy vector (Invitrogen), and individual inserts from 10-15 clones were sequenced. Comparisons of methylation levels were made using the two-sided Fisher's Exact Test at the 5% significance level.
Analysis of active DNA demethylation
P19, P19 C8, P19 C7 and P19 scrambled shRNA cells were transfected with unmethylated or SssI in vitro methylated Oct4∷EGFP reporter plasmid. Cells were visualized for GFP expression 12 hours after transfection. Transfected plasmid was recovered with QIAGEN Midikit and then processed for bisulfite treatment.
Microarray data were submitted to ArrayExpres with accession number E-MEXP-2610.
Supplementary Material
Acknowledgments
We thank Drs. F. De Angelis, H.-Y. Fan, R. Katz, O. Segatto, K. Zaret and R. Zhang for critical reading of the manuscript; Drs. P.K. Cooper, F. Roegiers, and K. Soprano for comments and advice; G. Albergo, K. Brewer, C. Garnier, B. Lurie and M. Oliver for mouse genotyping; Z.P. Zhang for help with CAT assays; Dr. J. Kulkosky, A. Kowalski, T. Stulkivska and W. Schroeder for Crabp2 and Oct4 methylation analysis; Dr. M. Xu for the targeting plasmid (with permission from Dr. G. Martin); Dr. A.J. Furnace for recombinant GADD45a; Dr. K. Sugasawa for anti-TDG antibody; Drs. L. Bagella and P.L. Puri for p300 reporters; Drs. S. Peri and Y. Zhou for Ingenuity pathway analysis; Dr. F. Alt for AID cDNA; Dr. N. Davidson for Apobec-1 cDNA; Dr. W. Cui for human Oct4∷EGFP reporter; Drs. C. Niehrs and A. Schafer for advice on transfected plasmid recovery; Dr. J. Thorvaldsen for advice on bisulfite modification of PGC DNA; and R. Sonlin for secretarial assistance. We thank the following core facilities at the Fox Chase Cancer Center: Genotyping, Cell Culture, Transgenic and Knock-out, Laboratory Animal, and Fannie E. Rippel Biotechnology Facility. This study was supported by NIH grants CA78412 and CA06927, an appropriation from the Commonwealth of Pennsylvania to the Fox Chase Cancer Center, and the Italian Association for Cancer Research (AIRC). S.C. was supported in part by an American-Italian Cancer Foundation Post-Doctoral Research Fellowship. LKA was supported by NIH training grant T32GM008216.
A.B. would like to dedicate this article to the memory of his father Pancrazio who taught him the importance of hard work, perseverance and dedication.
Footnotes
Supplemental Information: Supplemental information includes Extended Experimental Procedures, seven figures and two tables and can be found with this article online at.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker D, Liu P, Burdzy A, Sowers LC. Characterization of the substrate specificity of a human 5-hydroxymethyluracil glycosylase activity. Chem Res Toxicol. 2002;15:33–39. doi: 10.1021/tx010113b. [DOI] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Carrier F, Georgel PT, Pourquier P, Blake M, Kontny HU, Antinore MJ, Gariboldi M, Myers TG, Weinstein JN, Pommier Y, et al. Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol Cell Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Lucey MJ, Phoenix F, Lopez-Garcia J, Hart SM, Losson R, Buluwela L, Coombes RC, Chambon P, Schar P, et al. T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor alpha. J Biol Chem. 2003;278:38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, Macdougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Turner D, Masciullo V, Schepis F, Albino D, Daniel R, Skalka AM, Meropol NJ, Alberti C, Larue L, et al. The base excision repair enzyme MED1 mediates DNA damage response to antitumor drugs and is associated with mismatch repair system integrity. Proc Natl Acad Sci USA. 2003;100:15071–15076. doi: 10.1073/pnas.2334585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb-Rinker P, Ly D, Jezierski A, Sikorska M, Walker PR. Sequential DNA methylation of the Nanog and Oct-4 upstream regions in human NT2 cells during neuronal differentiation. J Biol Chem. 2005;280:6257–6260. doi: 10.1074/jbc.C400479200. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, Carell T. Tissue distribution of 5-Hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming Gl, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland U, Bentele M, Jiricny J, Schar P. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J Biol Chem. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- Haushalter KA, Todd Stukenberg MW, Kirschner MW, Verdine GL. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr Biol. 1999;9:174–185. doi: 10.1016/s0960-9822(99)80087-6. [DOI] [PubMed] [Google Scholar]
- Howard JH, Frolov A, Tzeng CD, Stewart A, Midzak A, Majmundar A, Godwin AK, Heslin MJ, Bellacosa A, Arnoletti JP. Epigenetic downregulation of the DNA repair gene MED1/MBD4 in colorectal and ovarian cancer. Cancer Biol Ther. 2009;8(1) doi: 10.4161/cbt.8.1.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. CpG islands--‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- Kress C, Thomassin H, Grange T. Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci U S A. 2006;103:11112–11117. doi: 10.1073/pnas.0601793103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic Acids Res. 2007;35:390–400. doi: 10.1093/nar/gkl1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M, Ghyselinck NB, Chambon P. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol. 2006;46:451–480. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- Martienssen RA, Richards EJ. DNA methylation in eukaryotes. Curr Opin Genet Dev. 1995;5:234–242. doi: 10.1016/0959-437x(95)80014-x. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Missero C, Pirro MT, Simeone S, Pischetola M, Di Lauro R. The DNA glycosylase T:G mismatch-specific thymine DNA glycosylase represses thyroid transcription factor-1-activated transcription. J Biol Chem. 2001;276:33569–33575. doi: 10.1074/jbc.M104963200. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci U S A. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Niehrs C. Active DNA demethylation and DNA repair. Differentiation. 2009;77:1–11. doi: 10.1016/j.diff.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Pantoja C, de Los Rios L, Matheu A, Antequera F, Serrano M. Inactivation of imprinted genes induced by cellular stress and tumorigenesis. Cancer Res. 2005;65:26–33. [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese KJ, Bartolomei MS. Establishment and maintenance of H19 imprinting in the germline and preimplantation embryo. Cytogenet Genome Res. 2006;113:153–158. doi: 10.1159/000090827. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Rideout WM, 3rd, Eggan K, Jaenisch R. Nuclear cloning and epigenetic reprogramming of the genome. Science. 2001;293:1093–1098. doi: 10.1126/science.1063206. [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Siegfried Z, Cedar H. DNA methylation: a molecular lock. Curr Biol. 1997;7:R305–R307. doi: 10.1016/s0960-9822(06)00144-8. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Naruse I, Hongo T, Xu M, Nakahata T, Maekawa T, Ishii S. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech Dev. 2000;95:133–145. doi: 10.1016/s0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- Thomassin H, Flavin M, Espinas ML, Grange T. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J. 2001;20:1974–1983. doi: 10.1093/emboj/20.8.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- Um S, Harbers M, Benecke A, Pierrat B, Losson R, Chambon P. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J Biol Chem. 1998;273:20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- Vermot J, Niederreither K, Garnier JM, Chambon P, Dolle P. Decreased embryonic retinoic acid synthesis results in a DiGeorge syndrome phenotype in newborn mice. Proc Natl Acad Sci USA. 2003;100:1763–1768. doi: 10.1073/pnas.0437920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, Zaret KS, Weissman IL, Smale ST. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:12377–12382. doi: 10.1073/pnas.0704579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- Yi YW, Kim D, Jung N, Hong SS, Lee HS, Bae I. Gadd45 family proteins are coactivators of nuclear hormone receptors. Biochem Biophys Res Comm. 2000;272:193–198. doi: 10.1006/bbrc.2000.2760. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, Siegmann M, Thiry S, Jost JP. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc Natl Acad Sci USA. 2000;97:5135–5139. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.