Abstract
A multiplex real-time PCR assay was developed with a LightCycler instrument for detection of influenza viruses A and B and the human respiratory syncytial virus (HRSV). Detection of each viral product and of an internal control was based on determination of specific melting temperatures by the LightCycler software. The lower limit of detection in the multiplex PCR assay was found to be 50 copies for each viral target. In an evaluation of nasopharyngeal samples collected from hospitalized children (ages, 0 to 3 years) with acute respiratory tract infections during the winter of 2001 to 2002, a viral pathogen was detected by the multiplex PCR test in 139 (66.8%) of 208 cases, including 45 (21.6%) influenza A virus infections, no (0%) influenza B virus infections, 106 (51%) HRSV infections, and 12 (5.8%) coinfections. The multiplex PCR test was compared to rapid antigen detection assays for influenza viruses A and B (Directigen; Becton Dickinson, Sparks, Md.) and HRSV (RSV TestPack; Abbott Laboratories, Abbott Park, Ill.) in 172 and 204 samples, respectively. After resolution of discrepant test results by use of additional PCR assays targeting other viral genes, the sensitivity (Se) and specificity (Sp) of the multiplex PCR assay for influenza A virus were 100 and 97.7% compared to 43.6 and 98.5% for the antigenic test. Similarly, the Se and Sp of the multiplex PCR assay for HRSV were 94.5 and 98.9% compared to 81.6 and 94.7% for the antigenic test. In conclusion, our multiplex real-time PCR assay combines both rapidity and sensitivity for detecting the most important respiratory viral pathogens in children.
Acute respiratory tract infections (ARTI) are a significant cause of morbidity and mortality in all age groups but especially in young children, elderly subjects, and immunocompromised patients. Most of these infections are caused by influenza viruses A and B as well as the human respiratory syncytial virus (HRSV), which are associated with the most severe complications, i.e., bronchiolitis, pneumonitis, and occasionally death (3, 9, 19, 23, 24). Besides epidemiological considerations, the interest in rapid diagnosis of these viral pathogens resides in the availability of specific antiviral therapy (10, 16, 18, 28, 32).
Respiratory viruses have been classically identified by viral culture using a variety of permissive cell lines. However, viral culture is hampered by the need to rapidly inoculate clinical samples into multiple cell lines for optimal sensitivity. Rapid immunoenzymatic assays have been developed by many companies for detection of influenza virus A and B antigens and HRSV antigens. However, the performance of these tests is dependent on many variables, and their sensitivity has generally been lower than that of viral culture, especially for adults, because their upper respiratory tract secretions have lower viral loads than those of children (2, 5, 12, 25).
More recently, PCR assays have been developed for many respiratory viruses, allowing detection of small amounts of viral nucleic acid in clinical samples. In the so-called “multiplex” format, PCR assays have been designed to amplify more than one respiratory viral target in the same PCR test (4, 6, 8, 15, 21). However, most multiplex PCR assays reported to date require separate steps for the amplification and detection of viral genes, which greatly increases the assay's turnaround time and the risk of amplicon contamination. In order to overcome these limitations, we developed a real-time multiplex PCR assay for influenza viruses and HRSV that uses the melting-curve-analysis feature of the LightCycler instrument to rapidly distinguish viral products.
MATERIALS AND METHODS
Study population and antigenic test
Nasopharyngeal aspirates (NPA) were prospectively collected during the winter of 2001 to 2002 from children aged 0 to 3 years who were hospitalized for ARTI at a single university-based hospital in Québec City, Quebec, Canada (1). The study was approved by the institutional review board of the Centre Hospitalier Universitaire de Québec. All samples were tested by the multiplex real-time PCR assay, whereas antigen detection tests were performed on a subset of samples upon request by the treating physician. An aliquot (750 μl) of fresh NPA was first tested for the presence of HRSV and influenza A or B virus antigens by using the RSV TestPack (Abbott Laboratories, Abbott Park, Ill.) and the Directigen Flu A+B test (Becton Dickinson, Sparks, Md.), respectively. The rest of the specimen was frozen at −80°C for a maximum of 3 months before nucleic acid extraction and PCR testing.
RNA extraction and cDNA synthesis.
Viral RNA was extracted from 200 μl of NPA samples by using the QIAamp Viral RNA Mini kit (Qiagen, Mississauga, Ontario, Canada). cDNA was then synthesized by using 10 μl of the RNA preparation, 0.75 μM of random hexamer primers (Amersham Pharmacia Biotech, Baie d'Urfé, Quebec, Canada), and the Omniscript Reverse Transcriptase kit (Qiagen) in the presence of 300 copies of an internal control. The internal-control template consisted of a 558-bp transcribed region of the herpes simplex virus type 2 DNA polymerase gene flanked by influenza B virus complementary primer sequences (see below) cloned into the pDrive plasmid (Qiagen).
Real-time multiplex and conventional PCR assays.
The multiplex PCR respiratory assay was designed to amplify conserved regions of the influenza A (7) and B (14) virus matrix genes as well as the fusion gene of HRSV (17). cDNA was amplified by a real-time PCR procedure using the LC Faststart DNA Master SYBR Green 1 kit in a LightCycler instrument (both from Roche Diagnostics, Laval, Quebec, Canada). Each reaction had a total volume of 20 μl including 2 μl of cDNA and 18 μl of a reaction mixture containing 2.4 mM MgCl2, 2 μl of Faststart DNA SYBR Green 1 master mix, 3% dimethyl sulfoxide, 0.3 mM influenza A and B virus primers, and 0.8 mM HRSV primers. Cycling conditions included an initial denaturation step of 10 min at 94°C, followed by 50 cycles of 15 s at 94°C, 5 s at 58°C, and 25 s at 72°C. At the end of each cycle, the fluorescent signal was measured at a wavelength of 530 nm by using the LightCycler fluorimeter. The melting-curve-analysis program of the LightCycler was used to identify specific PCR products. Briefly, following the last amplification cycle, the reaction temperature was rapidly increased to 94°C, then decreased to 60°C for 30 s, and finally slowly increased to 94°C at a rate of 0.1°C per s, with continuous fluorescence monitoring. For resolution of discrepant results between the multiplex PCR assay and antigenic tests, conventional reverse transcription-PCR (RT-PCR) assays for detection of the glycoprotein G gene of HRSV (22) and the M2 gene of influenza A viruses (13) were performed retrospectively.
RESULTS
Assay characteristics.
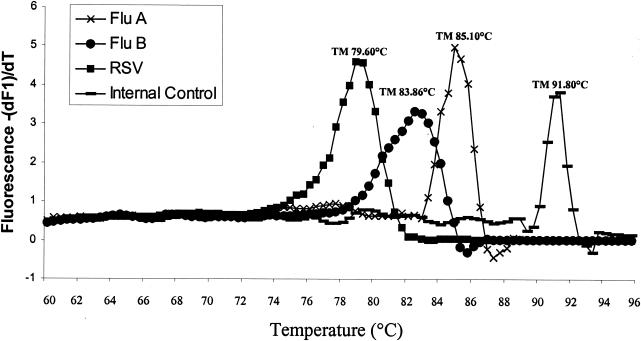
The sensitivity of each individual real-time PCR assay and of the multiplex PCR respiratory assay was determined for each target by testing serial dilutions of transcribed plasmids containing specific viral sequences. The lower limits of detection for influenza virus A, influenza virus B, and HRSV were found to be 10, 50, and 50 copies, respectively, in individual PCR assays and 50 copies for each target (10 out of 10 times for influenza virus A and 9 out of 10 times for influenza virus B and HRSV) in the multiplex PCR test. No amplification signal was detected in the real-time multiplex assay when viral DNA or RNA from adenoviruses, rhinoviruses, enteroviruses (echo 11), parainfluenza viruses 1 to 3, human metapneumovirus, and herpesviruses (herpes simplex virus types 1 and 2, cytomegalovirus, and varicella-zoster virus) were tested. Specific melting temperatures (Tm) were first determined by testing a set of clinical isolates and vaccine strains (11 influenza virus A subtype H1, 10 influenza virus A subtype H3, 10 influenza virus B, and 17 HRSV strains) from different years (Table 1). A representative set of results is shown in Fig. 1. The mean Tm were 85.27 ± 0.22 for influenza virus A, 83.47 ± 0.46 for influenza virus B, and 79.51 ± 0.30°C for HRSV, corresponding to fragments of 245, 524, and 380 bp, respectively, after gel electrophoresis (Table 1). We found no influence of influenza A virus subtype (H1 or H3) or HRSV genotype (A or B) on Tm (Table 1). The addition of 300 copies of the internal control in the assay's master mix resulted in a specific amplification peak with a Tm of 91°C (558 bp) in the absence of amplified viral products.
TABLE 1.
Evaluation of the multiplex PCR respiratory assay with temporally different strains of influenza virus and HRSV
| Virus (subtype)a | Genotypea | Yr | Type | Strain | Tm (°C) |
|---|---|---|---|---|---|
| Influenza virus A (H1) | 1999-2000 | Clinical | 85.10 | ||
| 1999-2000 | Clinical | 85.39 | |||
| 2000-2001 | Clinical | 85.12 | |||
| 2000-2001 | Clinical | 85.20 | |||
| 2000-2001 | Clinical | 85.04 | |||
| 2001-2002 | Clinical | 84.62 | |||
| 2001-2002 | Clinical | 85.07 | |||
| 2002-2003 | Clinical | 85.50 | |||
| 2002-2003 | Clinical | 85.29 | |||
| 1995 | Vaccine | A/Beijing/262/95 | 85.06 | ||
| 1933 | Vaccine | A/WSN/33 | 85.25 | ||
| Mean (H1) | 85.15 ± 0.22 | ||||
| Influenza virus A (H3) | 1998-1999 | Clinical | 85.58 | ||
| 1998-1999 | Clinical | 85.18 | |||
| 1998-1999 | Clinical | 85.35 | |||
| 1998-1999 | Clinical | 85.57 | |||
| 1998-1999 | Clinical | 85.37 | |||
| 2001-2002 | Clinical | 85.07 | |||
| 2001-2002 | Clinical | 85.65 | |||
| 2002-2003 | Clinical | 84.92 | |||
| 1999 | Vaccine | A/Panama/2007/99 | 85.18 | ||
| 1997 | Vaccine | A/Sydney/5/97 | 85.06 | ||
| Mean (H3) | 85.31 ± 0.25 | ||||
| Mean (influenza virus A) | 85.27 ± 0.22 | ||||
| Influenza virus B | 1998-1999 | Clinical | 83.87 | ||
| 1998-1999 | Clinical | 83.12 | |||
| 2000-2001 | Clinical | 83.01 | |||
| 2000-2001 | Clinical | 83.03 | |||
| 2000-2001 | Clinical | 82.88 | |||
| 2000-2001 | Clinical | 83.75 | |||
| 2000-2001 | Clinical | 83.17 | |||
| 2002-2003 | Clinical | 83.89 | |||
| 2002-2003 | Clinical | 83.84 | |||
| 1997 | Vaccine | B/Harbin/7/97 | 84.12 | ||
| Mean (influenza virus B) | 83.47 ± 0.46 | ||||
| HRSV | B | 1999-2000 | Clinical | 79.32 | |
| A | 1999-2000 | Clinical | 79.83 | ||
| A | 2000-2001 | Clinical | 79.06 | ||
| A | 2000-2001 | Clinical | 78.85 | ||
| B | 2001-2002 | Clinical | 79.34 | ||
| B | 2001-2002 | Clinical | 79.52 | ||
| B | 2001-2002 | Clinical | 79.98 | ||
| B | 2001-2002 | Clinical | 79.52 | ||
| A | 2001-2002 | Clinical | 79.84 | ||
| B | 2001-2002 | Clinical | 79.42 | ||
| B | 2001-2002 | Clinical | 79.49 | ||
| B | 2001-2002 | Clinical | 79.48 | ||
| B | 2001-2002 | Clinical | 79.55 | ||
| B | 2001-2002 | Clinical | 79.32 | ||
| A | 2001-2002 | Clinical | 79.77 | ||
| A | 2001-2002 | Clinical | 79.92 | ||
| A | 2001-2002 | Clinical | 79.40 | ||
| Mean (HRSV-A) | 79.52 ± 0.43 | ||||
| Mean (HRSV-B) | 79.49 ± 0.19 | ||||
| Mean (all HRSV) | 79.51 ± 0.30 |
FIG. 1.
Melting-curve analysis of amplified viral genes and of an internal control by the LightCycler instrument. Note that large amounts of amplified viral products may preclude detection of the internal control in some PCR runs.
Evaluation of the multiplex PCR respiratory assay in a pediatric population.
The multiplex PCR assay was evaluated by using extracted RNA from 208 NPA samples collected during a prospective pediatric study aimed at assessing the role of human metapneumovirus in hospitalized children (ages, 0 to 3 years) with ARTI (1). The numbers of samples found by multiplex PCR to be positive for influenza virus A, influenza virus B, and HRSV were 45 (21.6%), 0 (0%), and 106 (51.0%), respectively. Overall, the rate of positivity for any of the three viruses was 66.8% (139 of 208), including 5.8% (12 of 208) representing coinfections with influenza virus A and HRSV. None of the samples was considered to contain PCR-inhibitory material, as verified by amplification of the internal control in all PCR-negative samples. Ranges of Tm for influenza virus A- and HRSV-positive samples were 85.10 ± 0.39 and 79.60 ± 0.42°C, respectively. All amplified products with one of the latter Tm were confirmed as specific viral targets by visualization of the appropriate band on an agarose gel. Because none of the samples contained influenza B virus sequences (which correlates with the very occasional isolation of influenza B viruses in the Québec City area during the winter of 2001 to 2002), clinical evaluation of the multiplex PCR assay was conducted for influenza virus A and HRSV only.
Evaluation of the multiplex PCR respiratory assay for detection of influenza A virus.
Of the 208 NPA evaluated by multiplex PCR, a subset of 172 samples was also tested for the presence of influenza virus antigens by using the Directigen Flu A+B test (Table 2). Concordant results were obtained for 145 (84.3%) samples including 17 positive and 128 negative results by the two tests. A total of 27 (15.7%) samples showed discordant results: 25 were positive by the multiplex PCR assay only, and 2 were positive by the antigenic test only. In order to resolve the discrepant results, a second RT-PCR test targeting a conserved gene (M2) of influenza A viruses was performed as reported previously (13). As shown in Table 2, influenza A virus M2 sequences were detected by the conventional RT-PCR assay in 22 (81.5%) of the 27 discordant samples. By using the M2 RT-PCR assay as the reference test, the sensitivity, specificity, positive predictive value, and negative predictive value of the multiplex real-time PCR assay for influenza A virus were 100, 97.7, 92.8, and 100%, respectively, and those of the antigenic test were 43.6, 98.5, 89.5, and 85.6%, respectively. The multiplex PCR respiratory assay was positive for HRSV in 12 (44.4%) of the 27 discordant influenza A virus samples, 10 of which showed dual HRSV-influenza A virus infections (Table 3). Mean cycle threshold (CT) values, which are inversely correlated with the amounts of viral RNA, were 28.1 and 33.0 for PCR-positive-antigen-positive and PCR-positive-antigen-negative samples, respectively (Table 3). These values corresponded to approximately 5,000 and 50 copies, respectively, when a specific influenza A virus plasmid was tested in the multiplex PCR assay.
TABLE 2.
Evaluation of the multiplex PCR respiratory assay for detection of influenza A virus
| Multiplex PCRa result | Result by:
|
|||
|---|---|---|---|---|
| Antigenic test (Directigen Flu A + B) (n = 172)
|
Influenza A virus M2 PCR assayb (n = 27)
|
|||
| Positive | Negative | Positive | Negative | |
| Positive | 17 | 25 | 22 | 3 |
| Negative | 2 | 128 | 0 | 2 |
Sensitivity, 39 of 39 (100%); specificity; 130 of 133 (97.7%); positive predictive value, 39 of 42 (92.8%); negative predictive value, 130 of 130 (100%).
Performed only for samples showing discordant results with the multiplex PCR assay and the antigenic test.
TABLE 3.
Evaluation of discordant test results for influenza A virus
| Sample no. | Influenza A virus result by:
|
Other PCR-amplified virusb | ||
|---|---|---|---|---|
| Multiplex PCRa | Antigenic test | M2 PCR | ||
| 16 | − | + | − | HRSV |
| 23 | + (27) | − | + | |
| 24 | − | + | − | HRSV |
| 27 | + (28) | − | + | HRSV |
| 35 | + (28) | − | + | HRSV |
| 36 | + (32) | − | + | |
| 40 | + (34) | − | + | |
| 43 | + (36) | − | + | HRSV |
| 44 | + (35) | − | + | |
| 50 | + (34) | − | + | |
| 51 | + (33) | − | + | HRSV |
| 103 | + (35) | − | + | |
| 107 | + (35) | − | + | |
| 114 | + (37) | − | − | |
| 118 | + (38) | − | + | HRSV |
| 120 | + (36) | − | + | HRSV |
| 125 | + (38) | − | + | |
| 130 | + (35) | − | + | HRSV |
| 131 | + (36) | − | − | |
| 132 | + (24) | − | + | HRSV |
| 134 | + (36) | − | − | |
| 141 | + (28) | − | + | |
| 152 | + (34) | − | + | |
| 158 | + (32) | − | + | |
| 160 | + (33) | − | + | HRSV |
| 161 | + (28) | − | + | HRSV |
| 163 | + (32) | − | + | |
Numbers in parentheses are CT values determined by the LightCycler.
Viruses amplified by the multiplex PCR assay include influenza A and B viruses as well as HRSV.
Evaluation of the multiplex PCR respiratory assay for detection of HRSV.
A total of 204 NPA samples could be evaluated by both the multiplex real-time PCR assay and the presence of HRSV antigens by use of the RSV TestPack (Table 4). Concordant results were found in 172 (84.3%) samples representing 83 positive and 89 negative results by the two tests. Thirty-two (15.7%) samples had discordant results: 21 were positive by the multiplex PCR assay only, and 11 were positive by the antigenic test only. Conventional RT-PCR tests for HRSV glycoprotein G (A and B subtypes) were performed as previously described (22) to resolve discrepant results. Sequences specific for the HRSV glycoprotein G gene were detected by RT-PCR assays in 26 (81.2%) of the 32 discordant samples, including 11 subtype-A and 15 subtype-B strains (Table 5). By using the latter test as a “gold standard,” the sensitivity, specificity, positive predictive value, and negative predictive value of the multiplex real-time PCR assay were 94.5, 98.9, 99.0, and 94.0%, respectively, and those of the antigenic test for HRSV were 81.6, 94.7, 94.7, and 81.8%, respectively. Four (66.7%) of the six samples for which multiplex PCR gave false-negative HRSV results were positive for influenza A virus (Table 5). Mean CT values of the PCR-positive-antigen-positive and PCR-positive-antigen-negative samples were 28.6 and 32.7, respectively (Table 5). These values corresponded to approximately 1,000 and 100 copies, respectively, when a specific HRSV plasmid was tested in the multiplex PCR assay.
TABLE 4.
Evaluation of the multiplex PCR respiratory assay for detection of HRSV
| Multiplex PCRa result | Result by:
|
|||
|---|---|---|---|---|
| Antigenic test (RSV TestPack) (n = 204)
|
HRSV gG PCR assaysb (n = 32)
|
|||
| Positive | Negative | Positive | Negative | |
| Positive | 83 | 21 | 20 | 1 |
| Negative | 11 | 89 | 6 | 5 |
Sensitivity, 103 of 109 (94.5%); specificity, 94 of 95 (98.9%); positive predictive value, 103 of 104 (99.0%); negative predictive value, 94 of 100 (94.0%).
Performed only for samples showing discordant results with the multiplex PCR assay and the antigenic test.
TABLE 5.
Evaluation of discordant test results for HRSV
| Sample no. | HRSV result by:
|
Other PCR-amplified virusc | ||
|---|---|---|---|---|
| Multiplex PCRa | Antigenic test | Glycoprotein G PCRb | ||
| 16 | + (32) | − | + (B) | |
| 17 | + (27) | − | + (B) | |
| 26 | + (36) | − | + (B) | |
| 34 | − | + | − | |
| 35 | + (33) | − | + (B) | Influenza A virus |
| 43 | + (36) | − | + (A) | Influenza A virus |
| 46 | + (34) | − | + (B) | |
| 55 | + (34) | − | + (B) | |
| 58 | + (33) | − | + (B) | Influenza A virus |
| 62 | − | + | + (A) | Influenza A virus |
| 70 | + (37) | − | + (B) | |
| 71 | + (35) | − | + (B) | |
| 72 | + (34) | − | + (B) | |
| 73 | + (26) | − | + (A) | |
| 79 | + (33) | − | + (B) | |
| 80 | − | + | − | |
| 91 | + (35) | − | + (A) | |
| 95 | + (29) | − | + (B) | |
| 100 | + (30) | − | + (A) | |
| 112 | − | + | − | |
| 129 | + (34) | − | + (B) | |
| 130 | + (35) | − | + (B) | Influenza A virus |
| 134 | − | + | + (A) | Influenza A virus |
| 139 | − | + | + (A) | |
| 142 | − | + | + (A) | Influenza A virus |
| 144 | − | + | + (B) | Influenza A virus |
| 150 | + (34) | − | − | |
| 187 | + (31) | − | + (A) | |
| 191 | + (29) | − | + (A) | |
| 193 | − | + | + (A) | |
| 211 | − | + | − | |
| 212 | − | + | − | |
Numbers in parentheses are CT values as determined by the LightCycler.
Letters in parentheses are genotypes.
Viruses amplified by the multiplex PCR assay include influenza A and B viruses as well as HRSV.
DISCUSSION
In this report, we describe a new multiplex real-time PCR assay for the most commonly detected respiratory viral pathogens, namely, influenza viruses (A and B) and HRSV. The key features of this novel assay include its rapidity (turnaround time, approximately 2 h 30 min, including 30 min for sample preparation, 60 min for reverse transcription, and 60 min for real-time PCR including viral identification), and its sensitivity (50 copies per assay for each viral target). Our results clearly demonstrate that our multiplex real-time PCR assay is more sensitive than commercially available antigenic detection tests for both influenza A virus and HRSV, with the greatest difference in sensitivity noted for influenza viruses.
Multiplex RT-PCR assays targeting as many as nine different respiratory pathogens have been reported previously (6, 8, 12, 21, 26, 29). More recently, a TaqMan-based real-time PCR assay for simultaneous detection of influenza A and B viruses has also been described (31). Compared to conventional multiplex PCR assays, our real-time PCR test is much more rapid because it avoids additional nested amplification and/or hybridization steps required for identification of viral products. Furthermore, the real-time amplification procedure minimizes the chances of contamination, because there is no post-PCR processing of the samples. Although our multiplex real-time PCR assay based on melting-curve analysis of amplicons does not permit absolute quantification of the viral targets as in the TaqMan PCR procedure (31), it allows for a larger number of viral targets to be detected simultaneously, since there is no limitation related to the capability of the system to detect multiple dyes linked to detection probes with distinct emission wavelengths. Eventually, more than three viral targets could be detected simultaneously in the LightCycler assay, assuming the absence of interaction between PCR primers and a reproducible and discriminatory Tm for each viral amplicon.
Evaluation of our multiplex real-time PCR assay in a pediatric population with severe ARTI clearly illustrated its clinical potential. Indeed, our assay identified a viral pathogen (influenza virus or HRSV) in two-thirds of the hospitalized children. These numbers are similar to those previously reported by our group for the same population, which were determined by use of individual real-time assays for these viral pathogens (1). Notably, we were able to show that 6% of hospitalized children had dual viral infections, a finding that has been reported, albeit at a lower frequency, by other investigators (8, 12, 21). Rapid identification of these viral pathogens (influenza virus and HRSV) is of paramount importance for purposes of isolation in the hospital setting and early institution of specific antiviral therapy (32). We are currently designing additional multiplex assays based on the same technology to cover the whole spectrum of respiratory pathogens including parainfluenza viruses, adenoviruses, enteroviruses, and coronaviruses (27). To this list, we should also add the recently described human metapneumovirus, which has been reported by our group and others to be present in as many as 10% of hospitalized children with ARTI (1, 11, 20, 30).
A specific evaluation of the real-time multiplex PCR assay for influenza virus A revealed that it was twice as sensitive as the rapid antigen detection test in use at our institution during the winter of 2001 to 2002. None of the two PCR-negative, antigen-positive samples were confirmed as positive by a second PCR targeting another conserved gene (M2) of influenza virus A, whereas most (22 of 25 [88.0%]) multiplex PCR-positive, antigen-negative samples were. In general, the false-negative antigenic test results could be explained by small amounts of viral RNA in those NPA samples, as demonstrated by high CT values in the corresponding real-time PCR assays. Three multiplex PCR test results were not confirmed by the influenza A virus M2 PCR test and were thus considered to be false-positive results. However, since we have not formally evaluated the sensitivity of the M2 PCR test and since viral cultures were not routinely done, we cannot rule out the possibility that such discrepant results were indeed true positives. We and other investigators have shown the superiority of RT-PCR tests over conventional methods (antigenic tests and viral culture) for detecting influenza viruses in clinical samples, including those sent to the laboratory by mail (2, 4, 12, 25, 31). It is now common procedure at our institution to confirm any negative antigenic test results for influenza A and B viruses by either viral culture or RT-PCR, considering the high specificity but relatively poor sensitivity of the former assays.
For HRSV, the multiplex real-time PCR assay was also found to be more sensitive (by approximately 15%) than the rapid antigenic test when a second RT-PCR assay for the HRSV gG gene was used to resolve discrepancies. Most of the false-negative antigenic test results could be explained by small amounts of viral RNA in the NPA samples, as shown by mean CT values of 32.7 for discordant results compared to 28.6 for concordant positive results. The specificity of the multiplex assay was excellent, with only one positive result not confirmed by the second PCR test. In contrast, the multiplex PCR assay missed six cases of HRSV which tested positive in both the antigenic test and the second PCR assay for HRSV gG. Notably, four of those six samples tested positive for influenza A virus in the multiplex PCR assay. Thus, although our multiplex PCR test has the capability of detecting dual infections (as seen in 6% of our cases), it is possible that large amounts of one virus could inhibit amplification of other pathogens. It is noteworthy that the multiplex PCR assay had the ability to amplify both HRSV genotypes, as shown by the detection of 15 B and 11 A genotypes by use of specific PCR assays aimed at detecting variable regions of the HRSV gG gene (17). Although the rapid antigenic test performed relatively well (sensitivity, 82%; specificity, 95%) for detection of HRSV in young children, the real advantage of PCR probably lies in testing of the adult population, whose viral titers are lower and for whom antigenic tests with upper respiratory tract samples are not recommended (5).
Some limitations of our study are worth mentioning. First, the absence of circulation of influenza B viruses during the study period prevented adequate validation of our multiplex assay for this pathogen and in particular for its ability to detect mixed (influenza B virus and HRSV) infections. Second, the performance of our multiplex PCR assay cannot be generalized to other types of samples (e.g., throat swabs and sputum) or to other populations (e.g., outpatients and adults) at the present time. Also, the absence of serological testing in our study may have underestimated the rate of viral infections, especially for those children who presented late after the onset of symptoms. Finally, although it was relatively easy to distinguish between the different viral pathogens based on their specific Tm only (Table 1), we sometimes had to confirm viral amplicons by gel electrophoresis, which increased the assay's turnaround time.
In conclusion, we have described a rapid and sensitive multiplex real-time PCR assay for detection of influenza viruses and HRSV in children's NPA samples. This new assay is as specific as, and much more sensitive than, currently available antigen detection tests; it could complement the latter in the hospital setting when there is high clinical suspicion despite negative results. Future work is needed to expand the panel of viral pathogens detected by such rapid molecular methods in order to eventually circumvent the need for viral cultures. Also, future evaluation of the multiplex PCR assay is warranted in adult populations with severe ARTI.
REFERENCES
- 1.Boivin, G., G. De Serres, S. Côté, R. Gilca, Y. Abed, L. Rochette, M. G. Bergeron, and P. Déry. 2003. Human metapneumovirus infections in hospitalized children. Emerg. Infect. Dis. 9:634-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boivin, G., I. Hardy, and A. Kress. 2001. Evaluation of a rapid optical immunoassay for influenza viruses (FLU OIA Test) in comparison with cell culture and reverse transcription-PCR. J. Clin. Microbiol. 39:730-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowell, S. F., L. J. Anderson, H. E. Gary, Jr., D. D. Erdman, J. F. Plouffe, T. M. File, Jr., B. J. Marston, and R. F. Breiman. 1996. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J. Infect. Dis. 174:456-462. [DOI] [PubMed] [Google Scholar]
- 4.Ellis, J. S., D. M. Fleming, and M. C. Zambon. 1997. Multiplex reverse transcription-PCR for surveillance of influenza A and B viruses in England and Wales in 1995 and 1996. J. Clin. Microbiol. 35:2076-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Englund, J. A., P. A. Piedra, A. Jewell, K. Patel, B. B. Baxter, and E. Whimbey. 1996. Rapid diagnosis of respiratory syncytial virus infections in immunocompromised adults. J. Clin. Microbiol. 34:1649-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan, J., K. J. Henrickson, and L. L. Savatski. 1998. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (Hexaplex). Clin. Infect. Dis. 26:1397-1402. [DOI] [PubMed] [Google Scholar]
- 7.Fouchier, R. A., T. M. Bestebroer, S. Herfst, L. Van Der Kemp, G. F. Rimmelzwaan, and A. D. Osterhaus. 2000. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 38:4096-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grondahl, B., W. Puppe, A. Hoppe, I. Kuhne, J. A. Weigl, and H. J. Schmitt. 1999. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J. Clin. Microbiol. 37:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han, L. L., J. P. Alexander, and L. J. Anderson. 1999. Respiratory syncytial virus pneumonia among the elderly: an assessment of disease burden. J. Infect. Dis. 179:25-30. [DOI] [PubMed] [Google Scholar]
- 10.Hayden, F. G., A. D. Osterhaus, J. J. Treanor, D. M. Fleming, F. Y. Aoki, K. G. Nicholson, A. M. Bohnen, H. M. Hirst, O. Keene, K. Wightman, et al. 1997. Efficacy and safety of the neuraminidase inhibitor zanamivir in the treatment of influenza virus infections. N. Engl. J. Med. 337:874-880. [DOI] [PubMed] [Google Scholar]
- 11.Jartti, T., B. van den Hoogen, R. P. Garofalo, A. D. Osterhaus, and O. Ruuskanen. 2002. Metapneumovirus and acute wheezing in children. Lancet 360:1393-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kehl, S. C., K. J. Henrickson, W. Hua, and J. Fan. 2001. Evaluation of the Hexaplex assay for detection of respiratory viruses in children. J. Clin. Microbiol. 39:1696-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klimov, A. I., E. Rocha, F. G. Hayden, P. A. Shult, L. F. Roumillat, and N. J. Cox. 1995. Prolonged shedding of amantadine-resistant influenza A viruses by immunodeficient patients: detection by polymerase chain reaction-restriction analysis. J. Infect. Dis. 172:1352-1355. [DOI] [PubMed] [Google Scholar]
- 14.Li, J., S. Chen, and D. H. Evans. 2001. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J. Clin. Microbiol. 39:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liolios, L., A. Jenney, D. Spelman, T. Kotsimbos, M. Catton, and S. Wesselingh. 2001. Comparison of a multiplex reverse transcription-PCR-enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J. Clin. Microbiol. 39:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makela, M. J., K. Pauksens, T. Rostila, D. M. Fleming, C. Y. Man, O. N. Keene, and A. Webster. 2000. Clinical efficacy and safety of the orally inhaled neuraminidase inhibitor zanamivir in the treatment of influenza: a randomized, double-blind, placebo-controlled European study. J. Infect. 40:42-48. [DOI] [PubMed] [Google Scholar]
- 17.Mazzulli, T., T. C. Peret, A. McGeer, D. Cann, K. S. MacDonald, R. Chua, D. D. Erdman, and L. J. Anderson. 1999. Molecular characterization of a nosocomial outbreak of human respiratory syncytial virus on an adult leukemia/lymphoma ward. J. Infect. Dis. 180:1686-1689. [DOI] [PubMed] [Google Scholar]
- 18.MIST Study Group. 1998. Randomised trial of efficacy and safety of inhaled zanamivir in treatment of influenza A and B virus infections. Lancet 352:1877-1881. [PubMed] [Google Scholar]
- 19.Neuzil, K. M., B. G. Mellen, P. F. Wright, E. F. Mitchel, Jr., and M. R. Griffin. 2000. The effect of influenza on hospitalizations, outpatient visits, and courses of antibiotics in children. N. Engl. J. Med. 342:225-231. [DOI] [PubMed] [Google Scholar]
- 20.Nissen, M. D., D. J. Siebert, I. M. Mackay, T. P. Sloots, and S. J. Withers. 2002. Evidence of human metapneumovirus in Australian children. Med. J. Aust. 176:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osiowy, C. 1998. Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J. Clin. Microbiol. 36:3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peret, T. C., C. B. Hall, K. C. Schnabel, J. A. Golub, and L. J. Anderson. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J. Gen. Virol. 79:2221-2229. [DOI] [PubMed] [Google Scholar]
- 23.Simoes, E. A. 1999. Respiratory syncytial virus infection. Lancet 354:847-852. [DOI] [PubMed] [Google Scholar]
- 24.Simonsen, L., K. Fukuda, L. B. Schonberger, and N. J. Cox. 2000. The impact of influenza epidemics on hospitalizations. J. Infect. Dis. 181:831-837. [DOI] [PubMed] [Google Scholar]
- 25.Steininger, C., M. Kundi, S. W. Aberle, J. H. Aberle, and T. Popow-Kraupp. 2002. Effectiveness of reverse transcription-PCR, virus isolation, and enzyme-linked immunosorbent assay for diagnosis of influenza A virus infection in different age groups. J. Clin. Microbiol. 40:2051-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stockton, J., J. S. Ellis, M. Saville, J. P. Clewley, and M. C. Zambon. 1998. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J. Clin. Microbiol. 36:2990-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treanor, J. 2002. Respiratory infections, p. 7-26. In R. J. Whitley, D. D. Richman, and F. G. Hayden (ed.). Clinical virology, 2nd ed. ASM Press, Washington, D.C.
- 28.Treanor, J. J., F. G. Hayden, P. S. Vrooman, R. Barbarash, R. Bettis, D. Riff, S. Singh, N. Kinnersley, P. Ward, R. G. Mills, et al. 2000. Efficacy and safety of the oral neuraminidase inhibitor oseltamivir in treating acute influenza: a randomized controlled trial. JAMA 283:1016-1024. [DOI] [PubMed] [Google Scholar]
- 29.Valassina, M., A. M. Cippone, M. G. Cusi, and P. E. Valensin. 1996. Rapid detection of different RNA respiratory virus species by multiplex RT-PCR: application to clinical specimens. Clin. Diagn. Virol. 8:227-232. [DOI] [PubMed] [Google Scholar]
- 30.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Elden, L. J., M. Nijhuis, P. Schipper, R. Schuurman, and A. M. van Loon. 2001. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol. 39:196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitley, R. J., F. G. Hayden, K. S. Reisinger, N. Young, R. Dutkowski, D. Ipe, R. G. Mills, and P. Ward. 2001. Oral oseltamivir treatment of influenza in children. Pediatr. Infect. Dis. J. 20:127-133. [DOI] [PubMed] [Google Scholar]