Abstract
Hypotrichosis with juvenile macular dystrophy (HJMD) and ectodermal dysplasia, ectrodactyly and macular dystrophy (EEM) are both caused by mutations in the CDH3 gene. In this report, we describe a family with EEM syndrome caused by a novel CDH3 gene mutation and review the mutation spectrum and limb abnormalities in both EEM and HJMD. A protein structure model showing the localization of different mutations causing both syndromes is presented. The CDH3 gene was sequenced and investigation of the mutations performed using a protein structure model. The conservation score was calculated by ConSurf. We identified a novel CDH3 gene mutation, p.G277V, which resides in a conserved residue located on a β-strand in the second cadherin domain. Review of the data on previously published mutations showed intra-familial and inter-familial variations in the severity of the limb abnormalities. Syndactyly was the most consistent clinical finding present in all the patients regardless of mutation type. The results of our study point to a phenotypic continuum between HJMD and EEM. It is important for genetic counseling to keep in mind the possible clinical/phenotypic overlap between these 2 syndromes and to be aware of the possible risk of limb abnormalities in future pregnancies in families with HJMD syndrome. CDH3 gene mutation screening is recommended in patients with both these syndromes as part of the work-up in order to offer appropriate genetic counseling.
Key Words: Ectrodactyly, Hypotrichosis, Macular dystrophy, Syndactyly
Introduction
Hypotrichosis with juvenile macular dystrophy (HJMD; OMIM 601553) is a rare autosomal recessive disorder characterized by sparse scalp hair and progressive macular degeneration resulting in severe visual impairment. The disease is caused by mutations in the CDH3 gene (MIM 114021), encoding the classic cadherin molecule P-cadherin [Sprecher et al., 2001]. Cadherins are integral membrane glycoproteins responsible for calcium-dependent cell-cell adhesion at adherens junctions. β-Catenin physically links the actin cytoskeleton to the cytoplasmic tail of P-cadherin [Yagi et al., 2000]; it is associated with the P-cadherin promoter and activates its expression in a cell-type specific manner [Faraldo et al., 2007]. P-cadherin is highly expressed in retinal pigment epithelium and in the hair matrix at various stages of development of these tissues [Shimoyama et al., 1989; Müller-Röver et al., 1999]. Mutations in the CDH3 gene also cause autosomal recessive ectodermal dysplasia, ectrodactyly and macular dystrophy (EEM; OMIM 225280), a syndrome characterized by hypotrichosis, macular dystrophy and limb defects such as ectrodactyly, syndactyly and camptodactyly. Mutations have been identified in only 2 families with EEM syndrome [Kjaer et al., 2005]. No genotype-phenotype correlations have been established for either EEM or HJMD syndromes. As opposed to some ectodermal dysplasia syndromes, such as hypohidrotic (or anhidrotic) ectodermal dysplasia (HED or EDA), which is characterized by sparse hair, missing or scanty eye brows and lashes, severe oligodontia, absent or reduced sweating, as well as defects in a number of other ectodermal organs, patients with EEM and HJMD syndromes do not have sweating disorders but may have dental problems [Balarin Silva et al., 1999].
In this study, we describe a family with EEM syndrome caused by a novel CDH3 gene mutation and review the mutation spectrum and limb abnormalities in both EEM and HJMD syndromes. A protein structure model showing localization of different point mutations causing both EEM and HJMD syndromes is presented.
Materials and Methods
CDH3 Gene Sequencing
All 16 exons and exon-intron border regions in the CDH3 gene were sequenced on both strands as described previously [Kjaer et al., 2005].
Prediction of the Structure of Human Cadherin 3
A template structure for human cadherin 3 was sought using HHpred [Söding, 2005; Söding et al., 2005] against the protein database (PDB). The result with the highest score was that of mouse EP-cadherin (PDB 1Q55) [He et al., 2003]. The sequence identity between human cadherin 3 and mouse EP-cadherin is 54%. The structure model was then built using NEST [Petrey and Honig, 2000] and validated with ProsA [Sippl, 1993; Wiederstein and Sippl, 2007], ProQ [Wallner and Elofsson, 2003] and Ramachandran plot analysis using the RAMPAGE webserver [Lovell et al., 2003]. The conservation score calculated by ConSurf [Landau et al., 2005], based on 21 unique sequences of various cadherins all containing 5 cadherin domains, was projected onto the 2 structure models in order to determine whether the conserved amino acids were in the core of the protein structure and the variable amino acids on its periphery. The ConSurf results also support the mutation analysis (for details, see online supplementary figure 1, www.karger.com/doi/10.1159/000327156). The figures were generated using PyMol (PyMol Molecular Graphics System; DeLano Scientific, Palo Alto, Calif., USA).
Results
Clinical Evaluation
The clinical details on this family have been described previously [Senecky et al., 2001]. The affected individuals from this family were recently re-evaluated in the genetics clinic. Briefly, the healthy nonconsanguineous parents are of Yemenite Jewish origin and there is no positive family history of eye, limb or hair problems.
Patient 1
The first affected child, a girl, was born at term after a normal pregnancy with a birth weight of 2,750 g. Her current cognitive development is normal and no dysmorphic features were noted on physical examination at the age of 20 years. Her weight was 46.5 kg (−2 SD), height 158 cm (−1 SD) and occipitofrontal head circumference 51.5 cm (−2 SD). Her hair was sparse and slow growing with normal eyebrows and eyelashes. The teeth were normal. Examination revealed a mildly hypoplastic nail on the second finger of the right hand (fig. 1A). At the age of 9 years, bilateral macular degeneration was diagnosed by retinoscopy. Fundoscopy at the age of 18 years showed bilateral symmetrical atrophy and scarring of the macula and hemorrhages around the macula. In addition, macular hyperpigmentation was present as detected by photographic fluorescein angiogram and color fundus photography. On visual field examination at the age of 20 years, right central and cecocentral scotoma and left central and temporal scotoma were detected. On skeletal X-ray, hypoplasia of the distal phalanx of the right second finger was present (fig. 1B). Microscopic examination of the hair showed pili torti (fig. 2A–F).
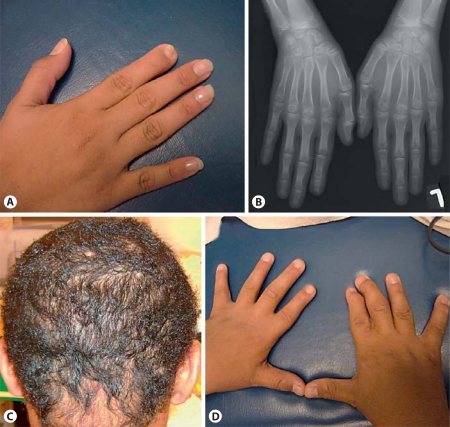
Fig. 1.
Clinical and radiographic features. A Right hand of patient 1 showing the hypoplastic distal phalanx of the second finger. B Anteroposterior radiograph of the right hand of patient 1 at the age of 12½ years showing the hypoplastic distal phalanx of the second finger. C Sparse and curly hair of patient 2. D Right hand of patient 2 showing a hypoplastic nail on the second finger with camptodactyly and the presence of scarring between the second and third fingers.
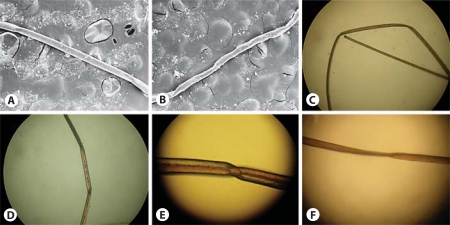
Fig. 2.
Scanning electron microscopy of the hair. Scanning electron microscopy of the hair of patient 1 with original magnification ×50 (A, B) and light microscopy with original magnification of ×40 (C, D), ×70 (E) and ×50 (F). A The periodic twists of the hair shaft (pili torti). B Hair shaft with sharp angle folding. C Flattening of the hair shaft and a straight band. D Flattening and angulations along a hair shaft. E Polarized light microscopy of pili torti and hair shaft twisting of 180°. F Flattening and angulations along a hair shaft.
Patient 2
The second affected child, a boy, was born at term after a normal pregnancy with a birth weight of 3,100 g (−1 SD). His cognitive development is completely normal. On examination at the age of 16 years, he was nondysmorphic. Limb examination showed a hypoplastic nail on the second finger of the right hand with camptodactyly and scarring between the right second and third fingers resulting from surgery to correct cutaneous syndactyly (fig. 1D). His hair was sparse, curly and slow growing (fig. 1C). Teeth were normal. He was referred for ERG and examination of the eye fundus and visual fields but refused to undergo these tests. However, at the age of 7 years, he was found to have visual disturbance, and, at 10 years 9 months, VEP and ERG revealed unilateral neural optic dysfunction of the right eye and unilateral retinal impairment in the left eye. Microscopic examination of the hair showed the same findings as in patient 1.
Molecular Studies
Both the affected brother and sister are homozygous for a missense mutation (c.830G→T; p.G277V) in the CDH3 gene. The father, mother and an unaffected older brother are all heterozygous for the mutation. This mutation has not been reported before in either EEM or HJMD syndrome. The amino acid is highly conserved between species and between different human cadherins (ConSurf results, online suppl. figure 1), and the mutation was not found in 178 control chromosomes of Yemenite Jewish origin.
Bioinformatics
The CDH3 gene resides on 16q22.1 [NM_001793] and has 16 exons (all coding), spanning 54,807 genomic basepairs (68,678,151–68,732,957). The mRNA consists of 4,276 bps, and it is translated to a large protein of 829 residues [NP_001784]. CDH3 is a calcium-dependent cell adhesion protein found in the cell membrane. It has 2 known isoforms, P22223-1 and P22223-2, the latter differing from the canonical sequence between residues 761 and 829 [data from Uniprot: P22223 (CADH3_HUMAN)]. CDH3 consists of 5 domains (cadherin 1–5). The localization of the p.G277V mutation on the University of California Santa Cruz genome browser (UCSC) is shown in figure 3.
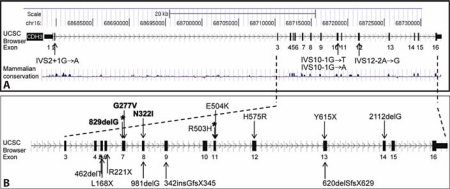
Fig. 3.
CDH3 gene mutations. ACDH3 gene structure according to the UCSC genome browser. Splice-site variations are marked. B Enlargement of the region including exons 3–16, where most of the variations are located. Variations in bold mark EEM syndrome-causing mutations. Variations in plain text mark mutations causing HJMD syndrome. Asterisks mark the variations found in both syndromes.
The mutations so far reported in EEM and HJMD syndromes and their genomic localization are depicted in figure 3A and enlarged in figure 3B. A mammalian conservation scheme is depicted, as determined by the UCSC genome browser (fig. 3A), showing high conservation, especially in the exon regions. Fourteen out of 18 mutations lie between exons 5 and 14 (fig. 3B).
The structure model of human cadherin 3 (fig. 4) was constructed based on the EP-cadherin X-ray structure (PDB 1Q55). G277 is located on the fifth β-strand of the second cadherin domain (fig. 4A). It is involved in the hydrogen bond network between the second and fifth β-strands (fig. 4B) and is buried in the core of the domain (fig. 4E). This position is highly conserved (ConSurf results, online suppl. figure 1).
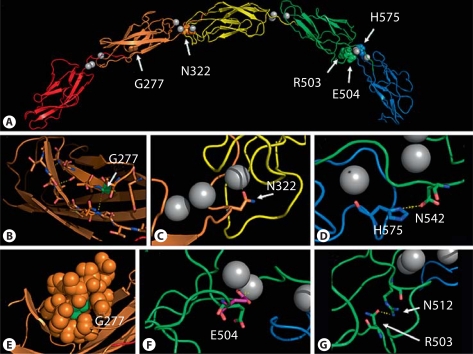
Fig. 4.
A structure model of human cadherin 3. A The 5 cadherin domains are colored red, orange, yellow, green, and blue. Calcium ions, as found in EP-cadherin crystal, are shown as gray spheres. The missense mutations are shown as colored spheres and marked in the figure. B G277 is shown as sticks and colored according to the elements (oxygen in red, nitrogen in blue, carbon in green). It is located on a β-strand in the second cadherin domain. The residues of the second and fifth β-strands are shown as sticks. Three hydrogen bonds between the main chains of the 2 β-strands are shown as dashed yellow lines (T232–V278, S233–I279 and V238–G277). C N322 is shown as sticks and colored according to the elements (as described above). It is located close to a calcium ion (2.4 angstroms). D H575 is shown as sticks and colored according to the elements (as described above). It is located in the fifth domain, in close proximity to N542 in the fourth domain (3.5 angstroms). E van der Waals spheres representation of G277 and residues 5 angstroms away. G277 is shown in green. It is located on the second domain (orange). F E504 is shown as sticks and colored according to the elements (green carbons). It is located in the fourth domain. In the X-ray structure of EP-cadherin, this residue (shown in purple) is in close proximity to a calcium ion. G R503 is shown as sticks and colored according to the elements. It is located in the fourth domain, in close proximity to N512 (3 angstroms).
Discussion
We report on a family with EEM syndrome and a novel missense CDH3 gene mutation. Table 1 shows limb abnormalities in patients with EEM or HJMD syndrome and identified CDH3 mutations, and table 2 shows the clinical features in reported patients with EEM and HJMD and the CDH3 mutations.
Table 1.
Limb abnormalities in patients with EEM or HJMD syndrome and identified CDH3 mutations
| Clinical diagnosis | Mutation | Hands | Feet | Reference |
|---|---|---|---|---|
| EEM | 829delG/829delG | Absent proximal, middle and distal phalanges, interstitial polydactyly; syndactyly of fingers | Syndactyly of toes | Albrectsen and Svendsen, 1956; Kjaer et al, 2005 |
| EEM | 965A→T/965A→T | Different degrees of syndactyly of 2–3, 2–4 or 1–4 fingers | Normal | Balarin et al, 1999; Kjaer et al, 2005 |
| HJMD | IVS2+1G→A/E504K | Normal | Bilateral bipartite patella and left foot syndactyly | Indelman et al, 2007 |
| EEM | 830G→T/830G→T | Hypoplastic nails, hypoplasia of distal phalanges, camptodactyly, fingers cutaneous syndactyly | Normal | Present study |
Table 2.
Clinical features in reported patients with EEM and HJMD and CDH3 mutations
| Mutation | Clinical diagnosis | Scalp hypotrichosis | Macular pigment degeneration | Limb abnormalities | Additional clinical findings | Reference |
|---|---|---|---|---|---|---|
| 981delG/981delG | HJMD | + | + | − | Sprecher et al., 2001 | |
| R503H/R503H | HJMD | + | + | − | Indelman et al., 2002 | |
| L168X/2112delG | HJMD | + | + | − | Atopic dermatitis | Indelman et al., 2003 |
| 462delT/462delT | HJMD | + | + | − | Indelman et al., 2003 | |
| 829delG/829delG | HJMD | + | + | − | Keratosis pilaris | Indelman et al., 2003 |
| R503H/R503H | HJMD | + | + | − | Indelman et al., 2003 | |
| R503H/R503H | HJMD | + | + | − | Centrofacial lentigines | Indelman et al., 2003 |
| N322I/N322I | EEM | + | + | + | Kjaer et al., 2005 | |
| Y615X/Y615X | HJMD | + | Symmetrical marked central pigmentary scar | − | Indelman et al., 2005 | |
| IVS2+1G→A/E504K | HJMD | + | + | + | Indelman et al., 2007 | |
| R221X/H575R | HJMD | + | + | Discolored primary teeth, nail dystrophy (resolved with time) | Indelman et al., 2007 | |
| IVS10-1G→T/IVS10-1G→T | HJMD | + | + | − | Jelani et al., 2009 | |
| C.IVS10-1G→A/IVS10-1G→A | HJMD | + | + | − | Kamran-ul-Hassan et al., 2010 | |
| IVS12-2A→G/IVS12-2A→G | HJMD | + | NR, severe visual impairment | − | Shimomura et al., 2010 | |
| IVS10-1G→T/IIVS10-1G→T | HJMD | + | NR, severe visual impairment | − | Shimomura et al., 2010 | |
| 342insGfsX345/342insGfsX345* | AR hypotrichosis | + | − | Sparse eyebrows/eyelashes | Basit et al., 2011 | |
| 620delSfsX629/620delSfsX629* | AR hypotrichosis | + | − | Sparse eyebrows/eyelashes | Basit et al., 2011 | |
| G277V/G277V | EEM | + | + | + | Present study |
NR = Not reported; AR = autosomal recessive.
Suspected digenic inheritance.
We investigated this and other previously reported missense mutations using the protein structure model. The G277V mutation lies within a conserved site in the fifth β-strand of the second cadherin domain, a location that may be critical for the protein's structure. Glycine and valine are both non-polar, aliphatic amino acids. However, as the side-chain of valine requires a larger space than that of glycine, the substitution of G to V may hamper the original structure of the protein, which may break the hydrogen bond between V238 and G277.
All 5 previously published missense mutations causing EEM and HJMD are located in loops that connect the cadherin domains. These loops are known to be calcium-binding sites, which are an essential property of the function and stability of the cadherins [van Roy and Berx, 2008]. More specifically, N322 and R503 are both located close to a calcium ion (fig. 4C, G, respectively). They are highly conserved among various cadherins. Substitutions of these residues may prevent the protein from binding the Ca2+ ions due to the change in charge [Indelman et al., 2002; Kjaer et al., 2005]. H575 is located in the fifth domain, in close proximity to D542 in the fourth domain (fig. 4D). H575 is moderately conserved, varying between serine, histidine, asparagine and glutamate. D542 is highly conserved among various cadherins. These 2 amino acids may form a hydrogen bond in the wild-type cadherin. Substitution of H575 to arginine might break this bond [Indelman et al., 2007]. E504 is located in the fourth domain and is well conserved. In the X-ray structure of EP-cadherin, which was used to model the cadherin 3 structure, this residue (shown in purple in fig. 4F) is in close proximity to a calcium ion. Ions and other ligands were not taken into account during the modeling process, and this caused E504 to rotate away from the calcium ion in the cadherin 3 structure model [Indelman et al., 2007]. To summarize, various CDH3 substitutions reported and reviewed above are located within highly conserved positions and may cause abnormal functioning of the protein. The impaired function may be due to calcium binding (e.g. N322) or to structural instability (e.g. G277).
In addition to the missense mutations, 4 splice-site mutations are known; these are shown in figure 3A [Indelman et al., 2007; Jelani et al., 2009; Kamran-ul-Hassan et al., 2010; Shimomura et al., 2010]. A large number of the deletion and substitution mutations are known to cause premature termination of the protein (829delG, 462delT, L168X, R221X, 981delG, Y615X, 2112delG) [Indelman et al., 2003, 2005, 2007; Kjaer et al., 2005]. In one previously described family with a frameshift mutation, ectrodactyly was severe; however, another patient from the same family displayed mild limb abnormalities – syndactyly only with no ectrodactyly [Kjaer et al., 2005].
Recently, two homozygous frameshift mutations, p.342insGfsX345 and p.620delSfsX629, have been reported in two consanguineous families with AR hypotrichosis [Basit et al., 2011]. Linkage analysis was compatible with digenic inheritance, but no mutation was found in candidate genes studied in an additional locus on 12q21.2–q22. Interestingly, in contrast to all previously reported patients with CDH3 mutations (table 2), none of the patients described in this article had visual problems; fundus examination, electroretinography and electrophysiological tests of affected members of both families had normal macular pigment epithelium and retinas.
Although the number of reported patients is small, it seems that there is no obvious genotype-phenotype correlation for the severity of limb abnormalities, and other genetic factors may also play a role in the development of ectrodactyly in EEM syndrome. Syndactyly was the most consistent clinical finding present in all the patients regardless of mutation type (table 1). In most of the patients described in the literature, hands were affected more than feet. Ectrodactyly of the feet has been described in only one patient [Ohdo et al., 1983]. It should be emphasized that mild limb abnormalities such as syndactyly in patients with EEM syndrome might be overlooked; this will lead to a diagnosis of HJMD syndrome. In addition, the same mutation, c.829delG, has been described as causative in both syndromes. No limb abnormalities were observed in individuals with HJMD who were homozygous for c.829delG [Indelman et al., 2007; E. Sprecher, pers. commun.], while in patients with this same mutation reported as having EEM syndrome severe phalangeal abnormalities have been described (table 1).
Pili torti have been previously reported in HJMD [Indelman et al., 2002, 2003], and, although not found in all patients with EEM and/or HJMD syndrome, they might be a useful diagnostic marker for these syndromes. Other reported hair abnormalities include pseudomoniletrix [Sprecher et al., 2001; Indelman et al., 2007] and telogen hairs [Indelman et al., 2007].
Conclusions
The EEM and HJMD syndromes most probably represent a continuous phenotypic spectrum related to mutations in the CDH3 gene rather than different genetic syndromes. Bearing this in mind, the possibility of a clinical overlap between the EEM and HJMD syndromes is of the utmost importance for genetic counseling since there may be a risk of limb abnormalities in future pregnancies in families with presumed HJMD syndrome. Fetal limb ultrasound examination should be recommended for parents of patients with a clinical diagnosis of either syndrome. Further studies including more patients are required in order to determine more precisely possible genotype-phenotype correlations regarding the different clinical features of these 2 syndromes.
Supplementary Material
ConSurf conservation analysis of CDH3. ConSurf conservation analysis of CDH3 was performed using 21 mammalian, avian and amphibian homologues. Conservation analysis was performed on residues 258 to 577, where all substitution mutations reside. Substitution mutations are shown by an asterisk below the residue. The conservation scale is shown in the lower left region of the figure; turquoise residues are highly variable and magenta residues are highly conserved.
Acknowledgement
We thank Dr. Gabrielle Halpern for her help with editing the manuscript. Wilhelm Johannsen Centre was established by the National Danish Research Foundation.
References
- Albrectsen B, Svendsen IB. Hypotrichosis, syndactyly, and retinal degeneration in two siblings. Acta Derm Venereol. 1956;36:96–101. [PubMed] [Google Scholar]
- Balarin Silva V, Simões AM, Marques-de-Faria AP. EEM syndrome: report of a family and results of a ten-year follow-up. Ophthalmic Genet. 1999;20:95–99. doi: 10.1076/opge.20.2.95.2290. [DOI] [PubMed] [Google Scholar]
- Basit S, Wali A, Aziz A, Muhammad N, Jelani M, Ahmad W. Digenic inheritance of an autosomal recessive hypotrichosis in two consanguineous pedigrees. Clin Genet. 2011;79:273–281. doi: 10.1111/j.1399-0004.2010.01455.x. [DOI] [PubMed] [Google Scholar]
- Faraldo MM, Teulière J, Deugnier MA, Birchmeier W, Huelsken J, et al. beta-Catenin regulates P-cadherin expression in mammary basal epithelial cells. FEBS Lett. 2007;581:831–836. doi: 10.1016/j.febslet.2007.01.053. [DOI] [PubMed] [Google Scholar]
- He W, Cowin P, Stokes DL. Untangling desmosomal knots with electron tomography. Science. 2003;302:109–113. doi: 10.1126/science.1086957. [DOI] [PubMed] [Google Scholar]
- Indelman M, Bergman R, Lurie R, Richard G, Miller B, et al. A missense mutation in CDH3, encoding P-cadherin, causes hypotrichosis with juvenile macular dystrophy. J Invest Dermatol. 2002;119:1210–1213. doi: 10.1046/j.1523-1747.2002.19528.x. [DOI] [PubMed] [Google Scholar]
- Indelman M, Hamel CP, Bergman R, Nischal KK, Thompson D, et al. Phenotypic diversity and mutation spectrum in hypotrichosis with juvenile macular dystrophy. J Invest Dermatol. 2003;121:1217–1220. doi: 10.1046/j.1523-1747.2003.12550_1.x. [DOI] [PubMed] [Google Scholar]
- Indelman M, Leibu R, Jammal A, Bergman R, Sprecher E. Molecular basis of hypotrichosis with juvenile macular dystrophy in two siblings. Br J Dermatol. 2005;153:635–638. doi: 10.1111/j.1365-2133.2005.06734.x. [DOI] [PubMed] [Google Scholar]
- Indelman M, Eason J, Hummel M, Loza O, Suri M, et al. Novel CDH3 mutations in hypotrichosis with juvenile macular dystrophy. Clin Exp Dermatol. 2007;32:191–196. doi: 10.1111/j.1365-2230.2006.02335.x. [DOI] [PubMed] [Google Scholar]
- Jelani M, Salman Chishti M, Ahmad W. A novel splice-site mutation in the CDH3 gene in hypotrichosis with juvenile macular dystrophy. Clin Exp Dermatol. 2009;34:68–73. doi: 10.1111/j.1365-2230.2008.02933.x. [DOI] [PubMed] [Google Scholar]
- Kamran-ul-Hassan Naqvi S, Azeem Z, Ali G, Ahmad W. A novel splice-acceptor site mutation in CDH3 gene in a consanguineous family exhibiting hypotrichosis with juvenile macular dystrophy. Arch Dermatol Res. 2010;302:701–703. doi: 10.1007/s00403-010-1035-6. [DOI] [PubMed] [Google Scholar]
- Kjaer KW, Hansen L, Schwabe GC, Marques- de-Faria AP, Eiberg H, et al. Distinct CDH3 mutations cause ectodermal dysplasia, ectrodactyly, macular dystrophy (EEM syndrome) J Med Genet. 2005;42:292–298. doi: 10.1136/jmg.2004.027821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, et al. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Müller-Röver S, Tokura Y, Welker P, Furukawa F, Wakita H, et al. E- and P-cadherin expression during murine hair follicle morphogenesis and cycling. Exp Dermatol. 1999;8:237–246. doi: 10.1111/j.1600-0625.1999.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Ohdo S, Hirayama K, Terawaki T. Association of ectodermal dysplasia, ectrodactyly, and macular dystrophy: the EEM syndrome. J Med Genet. 1983;20:52–57. doi: 10.1136/jmg.20.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrey D, Honig B. Free energy determinants of tertiary structure and the evaluation of protein models. Protein Sci. 2000;9:2181–2191. doi: 10.1110/ps.9.11.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senecky Y, Halpern GJ, Inbar D, Attias J, Shohat M. Ectodermal dysplasia, ectrodactyly and macular dystrophy (EEM syndrome) in siblings. Am J Med Genet. 2001;101:195–197. doi: 10.1002/ajmg.1361. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Wajid M, Kurban M, Christiano AM. Splice site mutations in the P-cadherin gene underlie hypotrichosis with juvenile macular dystrophy. Dermatology. 2010;220:208–212. doi: 10.1159/000275673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama Y, Yoshida T, Terada M, Shimosato Y, Abe O, Hirohashi S. Molecular cloning of a human Ca2+-dependent cell-cell adhesion molecule homologous to mouse placental cadherin: its low expression in human placental tissues. J Cell Biol. 1989;109:1787–1794. doi: 10.1083/jcb.109.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher E, Bergman R, Richard G, Lurie R, Shalev S, et al. Hypotrichosis with juvenile macular dystrophy is caused by a mutation in CDH3, encoding P-cadherin. Nat Genet. 2001;29:134–136. doi: 10.1038/ng716. [DOI] [PubMed] [Google Scholar]
- van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65:3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Takeichi M. Cadherin superfamily genes: functions, genomic organization and neurological diversity. Genes Dev. 2000;14:1169–1180. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ConSurf conservation analysis of CDH3. ConSurf conservation analysis of CDH3 was performed using 21 mammalian, avian and amphibian homologues. Conservation analysis was performed on residues 258 to 577, where all substitution mutations reside. Substitution mutations are shown by an asterisk below the residue. The conservation scale is shown in the lower left region of the figure; turquoise residues are highly variable and magenta residues are highly conserved.