Abstract
Summary: MicroRNAs (miRNAs) are ∼20- to 22-nt long endogenous RNA sequences that play a critical role in the regulation of gene expression in eukaryotic genomes. Confident identification of miRNA targets is vital to understand their functions. Currently available computational algorithms for miRNA target prediction have diverse degrees of sensitivity and specificity and as a consequence each predicted target generally requires experimental confirmation. miRNAs and other small RNAs that direct endonucleolytic cleavage of target mRNAs produce diagnostic uncapped, polyadenylated mRNA fragments. Degradome sequencing [also known as PARE (parallel analysis of RNA ends) and GMUCT (genome-wide mapping of uncapped transcripts)] samples the 5′-ends of uncapped mRNAs and can be used to discover in vivo miRNA targets independent of computational predictions. Here, we describe a generalizable computational pipeline, CleaveLand, for the detection of cleaved miRNA targets from degradome data. CleaveLand takes as input degradome sequences, small RNAs and an mRNA database and outputs small RNA targets. CleaveLand can thus be applied to degradome data from any species provided a set of mRNA transcripts and a set of query miRNAs or other small RNAs are available.
Availability: The code and documentation for CleaveLand is freely available under a GNU license at http://www.bio.psu.edu/people/faculty/Axtell/AxtellLab/Software.html
Contact: mja18@psu.edu
1 INTRODUCTION
Small silencing RNAs guide Argonaute (AGO)-containing complexes to regulate target RNA sequences based upon Watson–Crick base pairing (Bartel, 2004). Small RNAs are expressed by most eukaryotes and have key roles in developmental timing, antiviral defense, genome rearrangement and chromatin modification. Classes of small silencing RNAs include microRNAs (miRNAs), short interfering RNAs (siRNAs), trans-acting siRNAs (tasiRNAs) and Piwi-interacting RNAs (piRNAs). miRNAs are 20- to 22-nt long and are derived from the stem-loop structures of folded precursor RNA sequences and are particularly critical for gene regulation in plants and animals.
In contrast to animals, plant miRNAs tend to have perfect or near perfect complementarity to their target mRNAs and the mode of regulation typically involves AGO-catalyzed target cleavage. Current in silico miRNA-target prediction methods in plants generally search for mRNAs with perfect or near perfect complementarity to a mature miRNA (reviewed by Mallory and Bouche, 2008), and have been of enormous value in guiding experimentation. However, these predictions require experimental confirmation to eliminate false positives, and may in some cases also miss some bona fide targets (false negatives).
Experimental studies have shown that small RNA-guided, AGO-mediated cleavage of mRNA targets occurs exactly between the 10th and 11th nucleotide of complementarity relative to the small RNA 5′-end. The resulting upstream fragment of the cleaved target rapidly degrades, while the downstream fragment is stable in vivo (Llave et al., 2002). Deep sequencing of the 5′-ends of uncapped, polyadenylated mRNAs thus captures these downstream cleavage fragments (Addo-Quaye et al., 2008; German et al., 2008; Gregory et al., 2008). This technique has been referred to as PARE (parallel analysis of RNA ends) (German et al., 2008) and GMUCT (genome-wide mapping of uncapped transcripts) (Gregory et al., 2008), but for simplicity, we shall refer to it as degradome sequencing herein. Degradome data can be scrutinized to find evidence of cleaved small RNA targets without resorting to computational predictions. Here we describe CleaveLand, a general pipeline for detecting fragments diagnostic of small RNA-mediated cleavage from degradome sequencing experiments. CleaveLand is not limited in applicability to plant miRNAs: coupled with degradome sequencing, CleaveLand will find cleaved small RNA targets from any organism.
2 METHODS
2.1 Formulation
If a decapped mRNA is the consequence of small RNA-mediated cleavage, then its 5′-end must contain the first 10 nt of the small RNA complementary region. This is because AGO-mediated cleavage occurs between the 10th and 11th nucleotide of complementarity. Hence, mapping the 5′-ends of uncapped mRNAs to the relevant transcriptome and extending 13 nt upstream captures the entire region of potential complementarity. Aligning these extended sequences to a set of small RNA queries allows discovery of cleaved targets. Subsequent quality filters and signal-to-noise analyses then assess the confidence with which targets have been identified.
2.2 Input data
The pipeline requires three FASTA-formatted input datasets: degradome sequences (sequences are trimmed to the 5′-most 20 nts), a set of query small RNAs and a target database (typically mRNAs). An optional fourth input is a FASTA file containing any known structural RNAs (e.g. rRNAs) for which matching degradome tags should be ignored. FASTA headers for the degradome and small RNA files require special formatting as described in the software documentation.
2.3 Processing
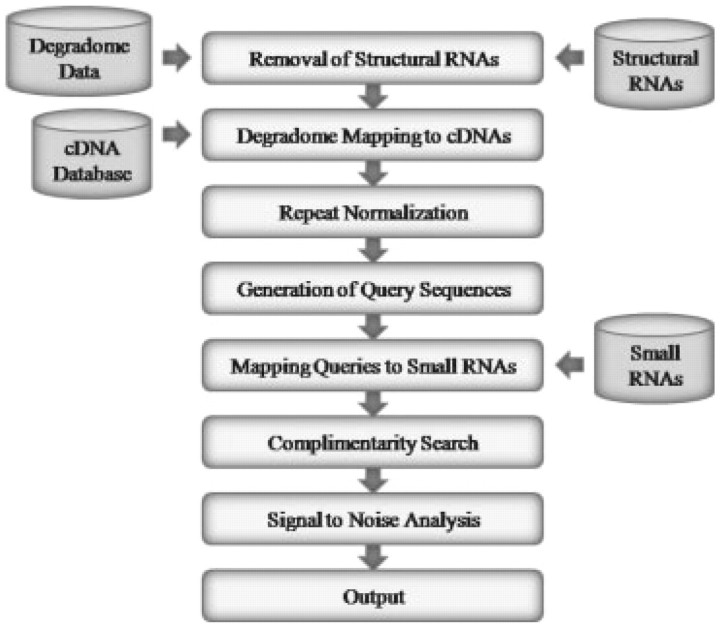
Figure 1 shows the various stages of the pipeline. Degradome sequences are matched to the structural RNAs using the Oligomap short reads aligner (Berninger et al., 2008). Raw sequence counts for degradome sequences are scaled to ‘reads per million’ (RPM) to enable comparisons across different sized datasets. All degradome sequences with exact sense matches to the structural RNAs are removed and the filtered dataset is mapped to the transcriptome, again using Oligomap. The RPM abundances of any degradome sequences with multiple transcriptome hits are repeat normalized; the abundance is divided by the total number of hits to give ‘normalized reads per million’ (NRPM). For each exact match to the sense strand of an mRNA transcript, a 26-nt long ‘query’ mRNA subsequence is generated by extracting 13-nt long sequences upstream and downstream of the location of the 5′-end of the matching degradome sequence. All query sequences are aligned to each small RNA sequence using the Needle program in the EMBOSS package (Rice et al., 2000). Alignments are then scored according to a previously described scheme developed for plant miRNA/target pairings (Allen et al., 2005). All alignments with scores not exceeding the user-set threshold and having the 5′-end of the degradome sequence coincident with the 10th nucleotide of complementarity to the small RNA are retained. Most of the degradome is not the result of small RNA-mediated cleavage. Thus, to differentiate spurious results from real targets, the pipeline re-runs using randomly shuffled small RNA sequences to estimate signal-to-noise ratios; shuffled sequences have dinucleotide and trinucleotide compositions consistent with those of the input transcriptome. All hits are categorized based on the abundance of the diagnostic cleavage tag relative to the overall profile of degradome tags matching the target. Optionally, users may limit the search to the highest confidence (‘Category I’) targets where the cleavage tag is the most abundant degradome sequence matching the target.
Fig. 1.
A schematic description of the CleaveLand pipeline.
2.4 Output
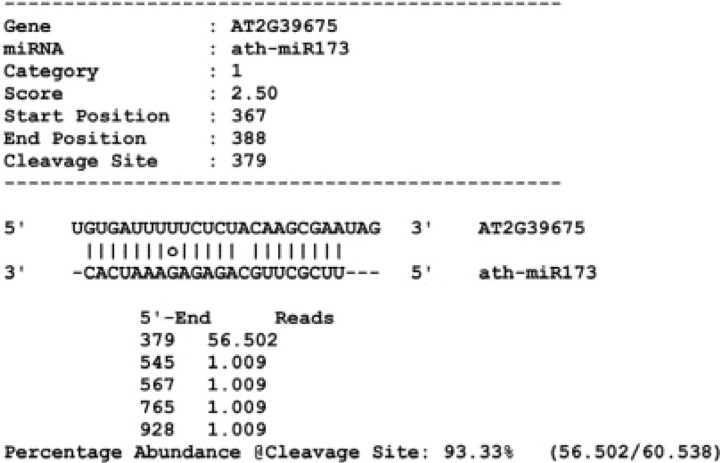
The pipeline generates a list of all confidently detected mRNA targets along with the corresponding alignments for the small RNA–mRNA pairs (Figure 2). In addition, complete information on the degradome profile of each target mRNA and signal-to-noise information is provided. Optionally, complete degradome mapping data, independent of small RNA alignments, are also produced.
Fig. 2.
Sample output from CleaveLand.
3 IMPLEMENTATION
The pipeline programs were written in C and run on a linux machine with a 3.0 GHz processor and 4 GB RAM. The pipeline requires the installation of the EMBOSS package and the Oligomap program. Considering a single query small RNA with 10 shuffles, a typical runtime for processing ∼1 million degradome sequences is about 1 h. A web interface to CleaveLand will soon be made available via Galaxy (galaxy.psu.edu) (Giardine et al., 2005).
Funding: US NSF (MCB 0718051 to M.J.A.).
Conflict of Interest: none declared.
REFERENCES
- Addo-Quaye C, et al. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, et al. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berninger P, et al. Computational analysis of small RNA cloning data. Methods. 2008;44:13–21. doi: 10.1016/j.ymeth.2007.10.002. [DOI] [PubMed] [Google Scholar]
- German MA, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- Giardine B, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Llave C, et al. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bouche N. MicroRNA-directed regulation: to cleave or not to cleave. Trends Plant Sci. 2008;13:359–367. doi: 10.1016/j.tplants.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Rice P, et al. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]