Abstract
Nascent RNA structures may regulate RNA chain elongation either directly through interaction with RNA polymerase or indirectly by disrupting nascent RNA contacts with polymerase or DNA. To distinguish these mechanisms we tested whether the effects of the his leader pause RNA hairpin could be mimicked by pairing of antisense DNA or RNA oligonucleotides to the nascent transcript. The his pause hairpin inhibits nucleotide addition when it forms 11 nucleotides from the transcript 3′ end. It also can terminate transcription when base changes extend its stem to ≤8 nucleotides from the 3′ end. All oligonucleotides that disrupted the pause hairpin reduced the dwell time of RNA polymerase at the pause site dramatically, even when they mimicked the 11-nucleotide 3′-proximal RNA spacing or created a suitably positioned RNA loop. Oligonucleotides that paired ≤8 nucleotides from the pause RNA 3′ end could trigger transcript release, but only when added to an already paused complex. These results argue that direct interaction of a nascent RNA hairpin with RNA polymerase delays escape from a pause, but that indirect effects of a hairpin may trigger transcript release from a paused complex. Resistance of the paused complex to pyrophosphorolysis and its reversal by antisense oligonucleotides further suggest that interaction of the pause hairpin with RNA polymerase disengages the RNA 3′ end from the active site.
Keywords: RNA polymerase, RNA hairpins, pausing, termination
Nascent RNA hairpins are components of certain classes of intrinsic pause and termination signals that regulate RNA chain elongation (traditionally called ρ-independent terminators and hairpin-dependent pause sites; for review, see Richardson and Greenblatt 1996; Uptain et al. 1997). Either pausing or termination can be stimulated by these RNA structures, depending on their distance from the pause or terminated RNA 3′ end and the surrounding RNA and DNA sequences (Chan et al. 1997). A central question about the effects of nascent RNA hairpins is whether they influence transcription through direct interactions with RNA polymerase (RNAP) or if their principal effects on the transcription complex (TC) are indirect and reflect removal of the nascent RNA chain from other interactions.
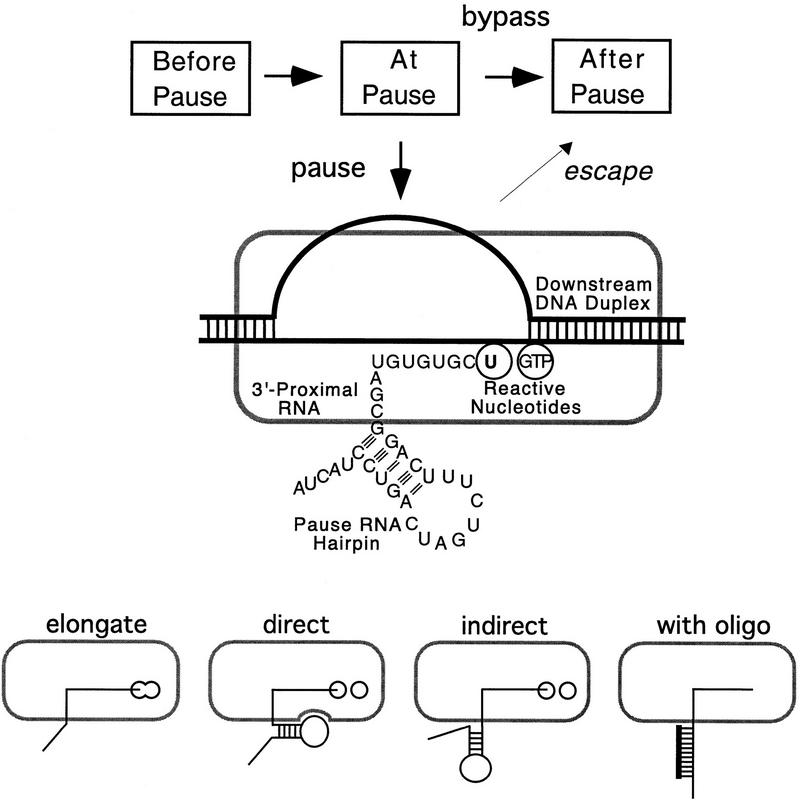
At the well-characterized his leader pause site, a nascent RNA hairpin is one of four components of a multipartite pause signal that directs pausing in a two-step mechanism (Fig. 1). The pause signal consists of the 5-bp stem, 8-nucleotide loop pause hairpin, the 11-nucleotide 3′-proximal region between the pause hairpin and the pause RNA 3′ end, the two bases in the active site, and the DNA duplex downstream from the active site (Chan and Landick 1993). In the first step of the mechanism, isomerization to a paused conformation competes with bypass of the pause site. In the second step, the paused RNAP escapes slowly back to a rapidly elongating conformation by nucleotide addition. The isomerization versus bypass competition is controlled principally by the 3′-proximal and downstream DNA sequences, which probably slow bypass by causing transient backtracking of the TC (Wang et al. 1995; Landick 1997; see Discussion). All four components inhibit pause escape from a single kinetic intermediate, probably by interfering with proper alignment of the 3′ OH group and the incoming GTP (Chan et al. 1997; illustrated in Fig. 1 by separation of the 3′ OH and NTP-binding subsites; see Discussion).
Figure 1.
Possible effects of antisense oligonucleotides on hairpin-dependent pausing. (Top) The mechanism of hairpin-dependent transcriptional pausing. The four components of the his pause signal are labeled on the paused TC, which is formed in competition with bypass of the site and then slowly escapes back into the elongation pathway. How the pause hairpin inhibits nucleotide addition is unknown, but it presumably disrupts reactive alignment of the RNA 3′OH and incoming NTP (depicted here by separation of the 3′ OH and NTP-binding subsites, i and i + 1; see Fig. 7). (Bottom) In the direct model of hairpin-dependent pausing, a specific interaction between the RNA hairpin and its binding site on RNAP disrupts nucleotide addition in the active site of RNAP (Chan et al. 1997; Wang and Landick 1997). In the indirect model, the hairpin merely defines a particular length of 3′-proximal, single-stranded RNA transcript and thus could both disrupt RNA–RNAP interactions required for elongation or TC stability and prevent backtracking of RNAP along the DNA template (Komissarova and Kashlev 1997b; Nudler et al. 1997). Annealing of antisense oligonucleotides to the nascent RNA would be able to recapitulate indirect effects of hairpins, but not direct effects.
On the basis of RNA cross-linking experiments, the pause hairpin appears to contact the 904–950 region of the β subunit in the paused TC and to disrupt nascent RNA contact to the amino-terminal region of β′ (Wang and Landick 1997). The effects of the pause hairpin can be eliminated at ⩾0.5 m Cl− ion, which, together with other salt effects, suggests that the pause hairpin slows nucleotide addition through a direct, Cl−-sensitive interaction with an easily disordered region of RNAP (Chan et al. 1997). However, it remains possible that the pause hairpin affects pausing indirectly by interfering with the β′ interaction or by defining a particular length of 3′-proximal RNA.
ρ-Independent terminators also are multipartite signals, in which a G/C-rich nascent RNA hairpin is located 7–9 nucleotides from the point of release (d’Aubenton Carafa et al. 1990). The intervening 3′-proximal RNA is shorter than at the his pause site, is typically U rich, and invariably contains three Us immediately after the hairpin. At least in vitro, termination efficiency is increased when nucleotide addition is slowed (Reynolds and Chamberlin 1992; McDowell et al. 1994; Wilson and von Hippel 1995) and by the presence of certain downstream DNA sequences if the 3′-proximal RNA is not mostly Us (Telesnitsky and Chamberlin 1989; Reynolds et al. 1992). It is likely that the U-rich 3′-proximal RNA and the downstream sequences, when important, cause transient backtracking of RNAP that allows proper timing of terminator hairpin formation and thus of a termination-prone complex (Nudler et al. 1995; Landick 1997).
Pausing and termination may pass through a common paused intermediate (Chan et al. 1997; Landick 1997). Destabilization of the paused complex by invasion of the terminator hairpin closer to the RNA 3′ end and weak rU · dA base-pairing may favor transcript release, whereas a stable paused intermediate may allow escape back to the elongation pathway. In support of this hypothesis, the his pause signal can be converted into a weak terminator by base changes that extend the hairpin stem to ≤8 nucleotides from the RNA 3′ end (the his perfect hairpin; Chan et al. 1997). Furthermore, some observations also favor a role for direct interactions of terminator hairpins with RNAP: (1) The sizes of stems (7 ± 3 bp) and loops (4 ± 1 nucleotides) in terminator hairpins are weakly conserved (d’Aubenton Carafa et al. 1990); (2) increasing stem length decreases termination in some cases (Wilson and von Hippel 1995); (3) certain sequences in terminator hairpins are weakly conserved (d’Aubenton Carafa et al. 1990); and (4) base substitutions that preserve base-pairing and predicted hairpin stability can reduce termination (Cheng et al. 1992). Alternatively (Fig. 1), a terminator hairpin could act indirectly by blocking backtracking of the TC (Komissarova and Kashlev 1997a; Nudler et al. 1997), by disrupting the RNA–DNA hybrid (Yager and von Hippel 1991), or by blocking interaction of the nascent transcript with RNAP (Chamberlin 1995).
In principle, direct versus indirect mechanisms of nascent hairpin action can be distinguished by the effects of antisense RNA or DNA oligonucleotides that hybridize to the nascent transcript (Fig. 1). The indirect effect model predicts that an antisense oligonucleotide that leaves an 11-nucleotide 3′-proximal region should mimic the pause hairpin and stimulate pausing relative to other antisense oligonucleotides that are known to decrease pausing by destroying the hairpin (Fisher and Yanofsky 1983). Likewise, antisense oligonucleotides that invade to within 9 nucleotides of the 3′ end should stimulate termination, whereas others that disrupt the terminator hairpin should decrease it. On the other hand, if the RNA hairpin affects elongation directly through interaction with RNAP, no oligonucleotide should be able to substitute for a pause or terminator hairpin as its annealing to the hairpin would destroy the secondary structure. Yanofsky and co-workers showed more than a decade ago that antisense oligonucleotides could inhibit either ρ-independent termination or hairpin-dependent pausing (Winkler et al. 1982; Fisher and Yanofsky 1983, 1984), but they could not easily conduct systematic experiments owing to the difficulty of obtaining oligonucleotides at the time. The availability of oligonucleotides and recent reports that antisense oligonucleotides can inhibit transcriptional arrest (Reeder and Hawley 1996; Komissarova and Kashlev 1997a) encouraged us to conduct the experiments reported here. Our results suggest that RNA hairpins play distinct roles in pausing and termination: a direct RNAP–RNA hairpin interaction is required to slow escape from the his pause site, but is dispensable for transcript release from preformed paused complexes.
Results
Antisense oligonucleotide pairing to nascent RNA cannot stimulate pausing but can release transcripts from already paused complexes
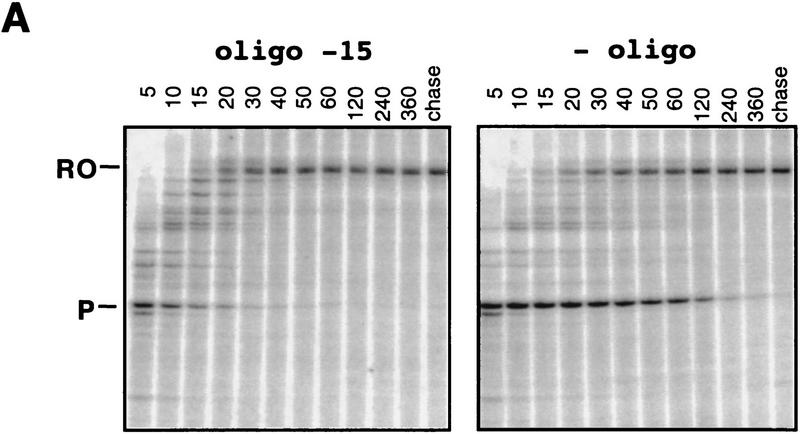
To determine whether pairing of an optimal antisense oligonucleotide to the nascent RNA could substitute for the function of a nascent RNA hairpin in pausing or transcript release, we tested the effects of 22-mer antisense oligonucleotides whose 5′ ends approached the pause RNA 3′ end in one-nucleotide increments on the fraction of elongating complexes that enters the paused state (pause efficiency), the rate of escape from the paused state (pause half-life), and tendency of the paused complex to dissociate (termination). We designated the antisense oligonucleotides by the length of nascent RNA remaining 3′ proximal to the antisense DNA–nascent RNA heteroduplex. For example, the 5′-most nucleotide of oligonucleotide −11 hybridizes to the 3′-most G in the pause hairpin (G-12, see Fig. 1) and leaves an 11-nucleotide 3′-proximal nascent RNA. We measured oligonucleotide effects on efficiency and half-life at 10 μm GTP (the next nucleotide added at the pause site) after addition of oligonucleotides in 500-fold molar excess and then NTPs to halted elongation complexes that were formed at position A29 of the template (see Materials and Methods). None of the oligonucleotides significantly altered pause efficiency, which we estimated to be 70 ± 10% on this template (data not shown; see Wang et al. 1995; Landick et al. 1996, for description of pause efficiency). Oligonucleotides that paired far upstream from the RNA hairpin had no effect on pause half-life, whereas oligonucleotides that paired to bases within the pause hairpin decreased it by factors of up to 15 (e.g., ∼4 sec for −15 oligonucleotide vs. ∼60 sec for −49 or no oligonucleotide; Fig. 2A,B). The large effect on half-life of oligonucleotides annealing to pause hairpin bases was relatively constant for oligonucleotides −11 to −27, but gradually diminished as oligonucleotide 5′ ends were shifted further upstream. Importantly, even an antisense oligonucleotide that should create the same 11-nucleotide 3′-proximal RNA as the pause hairpin failed to stimulate pausing (oligonucleotide −11; Fig. 2B).
Figure 2.
Effect of DNA oligonucleotides on the his pause half-life. (A) Preformed [α32P]CTP-labeled A29 complexes were chased with 10 μm GTP, 150 μm ATP, CTP, UTP in the presence (left) or absence (right) of the −15 oligonucleotide. Samples were taken at the times (in sec) indicated above each lane. The chase lanes contain samples that were incubated for an additional 5 min with 250 μm each NTP after completion of the time course. (P) Pause RNA transcript; (RO) run-off RNA transcript. Nonlinear regression yielded pseudo-first order half-lives of 5 and 60 sec for the −15-oligonucleotide and no oligonucleotide experiments, respectively (see Materials and Methods). (B) Pause half-lives were plotted (as a fraction of a ‘no oligo’ control from the same experiment) by the 5-most nucleotide of the nascent RNA that remains outside the RNA–oligonucleotide duplex (equivalent to the length of nascent RNA between the RNA–oligonucleotide duplex and the nascent RNA 3′ end). Each value is an average of at least two independent measurements.
We confirmed that the DNA oligonucleotides hybridized to the RNA transcript where expected by assaying cleavage of the nascent transcript in the presence of oligonucleotides and RNase H, which cuts RNA only in an RNA–DNA hybrid (data not shown; RNA–template DNA base pairs within the TC are protected from RNase H, presumably by steric exclusion). However, it was formally possible that the effects of oligonucleotides on pause half-life resulted from additional hybridization to the nontemplate DNA strand, rather than to the RNA transcript. Such effects seemed unlikely to explain all our results because some oligonucleotides that reduced the pause half-life annealed entirely upstream from the region corresponding to the ∼17-bp transcription bubble (i.e., oligonucleotides whose 5′ ends are more that 15 nucleotides from RNA 3′ end, the typical upstream end of the transcription bubble, Fig. 2B; see Gamper and Hearst 1982; Lee and Landick 1992; Zaychikov et al. 1995, for transcription bubble measurements). To confirm this interpretation, we adopted the approach of Reeder and Hawley (1996) and tested an 18-nucleotide DNA oligonucleotide that could anneal to RNA but not DNA because it contained Gs opposite the 8 Us between nucleotides −33 and −16 (UG-15, Fig. 2B; see Materials and Methods). This oligonucleotide reduced the pause half-life by a factor of 5, consistent with the weaker pairing expected relative to the A-containing −15 oligonucleotide and supportive of the conclusion that antisense oligonucleotides reduce pausing by disrupting the pause RNA hairpin.
Oligonucleotides whose pairing extended past the pause hairpin to within the 7- to 9-nucleotide 3′-proximal region characteristic of ρ-independent terminators still reduced pausing, but did not terminate transcription (e. g., −7 oligonucleotide; Fig. 3A), even at concentrations of KCl (⩾0.5 m) that allow weak termination when the pause hairpin stem is extended to within this region (the ‘perfect hairpin’ pause site; Chan and Landick 1997). However, we could not conclude from this result that direct hairpin RNAP interaction is also required for termination because pausing could be an obligatory step in the termination pathway. Thus, the strong inhibition of pausing caused by antisense oligonucleotides itself could prevent termination.
Figure 3.
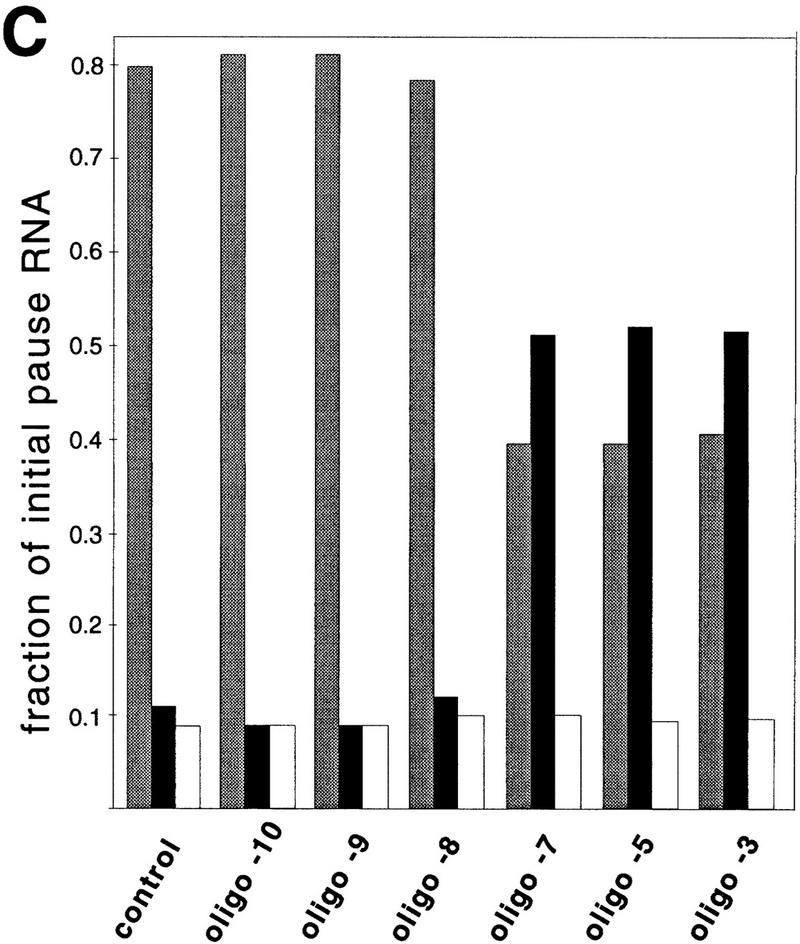
Transcript release by antisense oligonucleotides. (A) A29 complexes or preformed paused complexes were eluted from the beads (see Materials and Methods) and incubated for 5 min in the presence of either the −10 or −7 oligonucleotide. Elongation was allowed to resume in the presence of 1 m KCl by addition of NTPs and samples were taken at the times indicated above each lane. (B) Preformed paused complexes were left on beads, incubated with 500-fold excess of each oligonucleotide in 1 m KCl, and chased with NTPs. The supernatants (released RNA) and the beads (RNA remaining in TCs) from samples before and after the chase were collected and electrophoresed separately. (C) Relative concentrations of pause RNA that was extended (shaded bars) released from TCs (solid bars), or retained on beads (open bars) after addition of NTPs to the preformed paused complexes were determined from PhosphorImager scans and are plotted as a fraction of the total pause RNA.
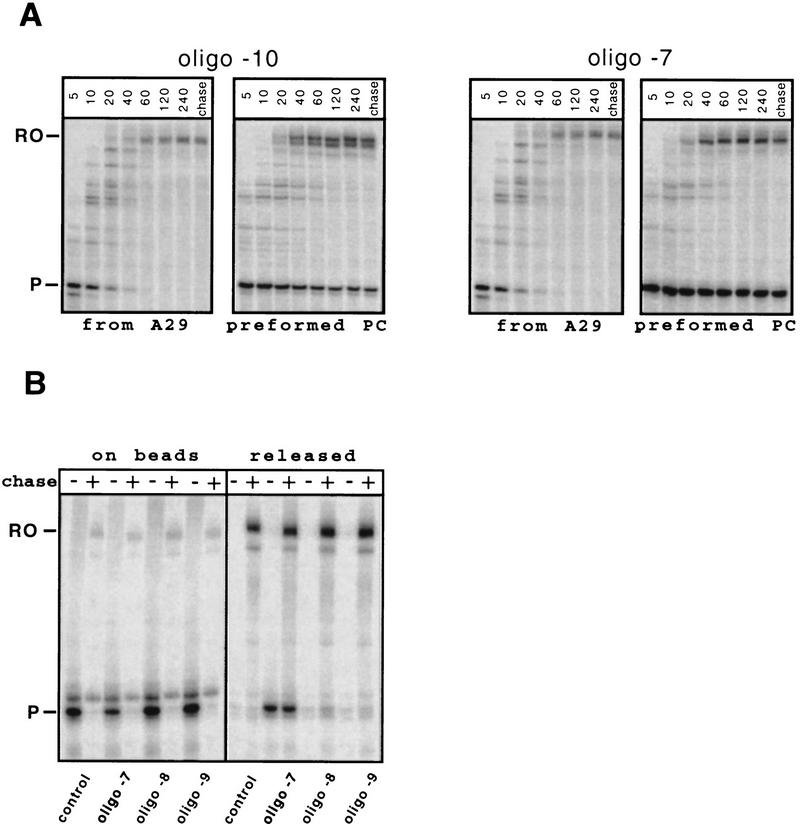
Therefore, we tested the effects of oligonucleotides on preformed paused TCs. We formed paused TCs from A29 complexes immobilized on beads by repeated rounds of stepwise transcription (Wang et al. 1995; Kashlev et al. 1996), and then tested whether oligonucleotides that could invade the 7- to 9-nucleotide 3′-proximal region could terminate transcription at 1 m KCl. Initially we tested for extention of the pause RNA after incubation of paused TCs with the oligonucleotides. Oligonucleotides that annealed ⩾9 nucleotides from the RNA 3′ end reduced the pause half-life by the same factor of ∼10 observed when they were present as TCs elongating through the pause site (e.g., oligonucleotide −10; Fig. 3A; note that incubation of the −11 oligonucleotide, not shown, with paused TCs still did not stimulate pausing). The small amount (∼20%) of pause RNA that failed to extend when oligonucleotides annealing ⩾9 nucleotides from the RNA 3′ end were added was attributable to partial inactivation of the paused TC during its preparation, as the same fraction also failed to extend after addition of a noncomplementary oligonucleotide or of no oligonucleotide. However, oligonucleotides that could hybridize to within 8 nucleotides or less of the pause RNA 3′ end prevented extension of ∼65% of the pause RNA when they were added either 5 min before or together with 150 μm NTPs (e.g., −7 oligonucleotide; Fig. 3A).
To determine whether the pause RNA that failed to extend in the presence of the −7, −5, and −3 oligonucleotides was trapped in arrested TCs or was actually released, we separated and analyzed the supernatant and bead fractions of the transcription reactions (Fig. 3B). Oligonucleotides −7, −5, and −3 released about fivefold more pause RNA (50% of the starting sample) than oligonucleotides −8, −9, or the control oligonucleotide (∼10% of the starting sample). Thus, all three antisense oligonucleotides that could pair to the base 8 nucleotides from the pause RNA 3′ end stimulated transcript release from preformed paused complexes to the same extent. The need to preform paused complexes to observe release is unlikely to reflect an inability of the oligonucleotides to hybridize rapidly enough to affect a moving RNAP, as oligonucleotides were able to reduce the pause half-life while RNAP is moving. Furthermore, we did not observe RNA release at the pause site when the complexes were walked to positions −7 or −2, incubated with −7 oligonucleotide to allow time for hybridization, and then chased (data not shown). These results suggest both that oligonucleotide-stimulated release requires preexistence of the paused complex and that release does not occur at template positions (−7 and −2) that do not form paused complexes, even when oligonucleotides can hybridize to the base 8 nucleotides from the transcript 3′ end.
From these experiments with antisense oligonucleotides we concluded tentatively that pausing requires direct interaction of a nascent hairpin structure with RNAP, but that once a paused TC is formed, transcript release requires only indirect disruption of the −8 nucleotide’s interactions with RNAP or with the DNA template. Importantly, the −8 nucleotide appears to form the upstream most base-pair in the TC RNA–DNA heteroduplex (Lee and Landick 1992; Nudler et al. 1997; see Discussion).
The structure and geometry of the pause hairpin in the nascent RNA is essential for its function
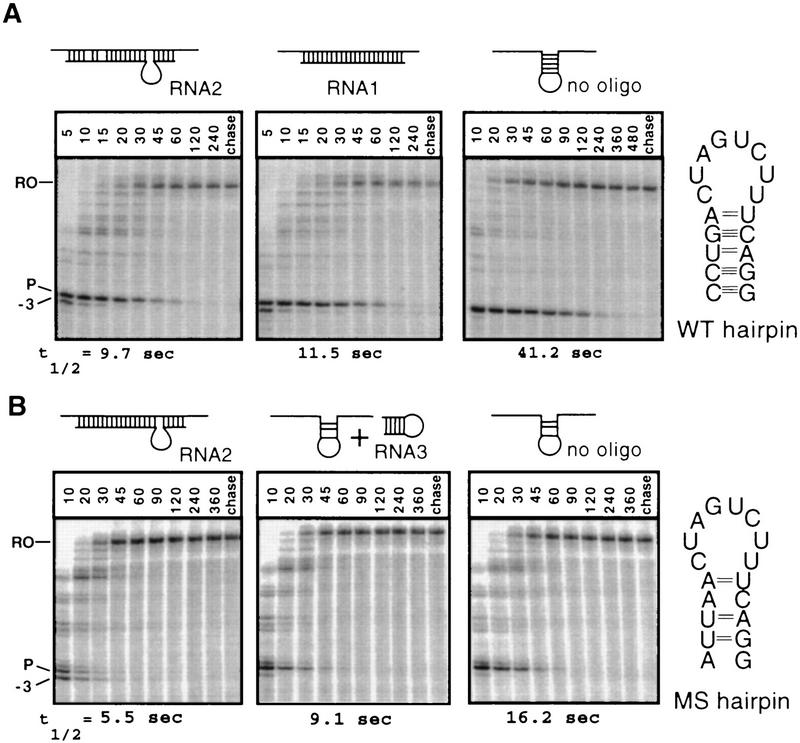
Although the results described so far suggested that the nascent pause RNA hairpin makes relatively specific and functionally important contact to RNAP, it remained possible that indirect effects of the hairpin could not be recapitulated by less stable DNA-to-RNA base-pairing, that oligonucleotide pairing to the transcript could stimulate pausing if it also presented a suitably located single-stranded loop to interact with RNAP, or that the hairpin might not need to be anchored to the nascent RNA at all. To test these possibilities, we used RNA oligonucleotides that could form an RNA–RNA duplex in place of the wild-type pause hairpin (RNA1), that could form an RNA–RNA duplex with a single-stranded loop where the pause hairpin loop normally would occur (RNA2), or that could form the pause hairpin structure free in solution (RNA3). We tested RNA oligonucleotides on both a wild-type template (WT, Fig. 4A) and a template that specified a mutant pause hairpin that contains three mismatches in the hairpin stem (MS hairpin, Fig. 4B; Chan and Landick 1993; Wang et al. 1995). The MS hairpin appears to be stabilized partially by interaction with RNAP; therefore, it reduces but does not eliminate pausing (Wang and Landick 1997). Thus, it allowed us to test for either inhibition or stimulation of pausing by RNA3, which could hypothetically either compete for or mimic the effect of a nascent RNA hairpin (stimulation of pausing could be indistinguishable from no effect of the oligonucleotide on the wild-type template).
Figure 4.
Effect of RNA oligonucleotides on pausing on the wild-type (WT; A) and multisubstituted (MS; B) hairpin templates. A29 complexes were walked to the −7 (U64) position, incubated in the presence or absence of oligonucleotide, and then chased (see Materials and Methods). RNA1 is complementary to the WT pause hairpin (CCUGAAAGACUAGUCAGGAUGA), RNA2 oligonucleotide is complementary to the MS hairpin and contains the 7-nucleotide insertion of the trp hairpin loop sequence (underlined; CCUGACUAAUGAAAGACUAGUUAAUAUGA); for both RNA1 and RNA2 oligonucleotides the 5′ end is positioned at −11. RNA3 oligonucleotide corresponds to the his pause hairpin (CCUGACUAGUCUUUCAGG). Structures predicted to form with annealing of oligonucleotides to the RNA transcript are shown schematically above each lane. Calculated pause half-lives are indicated below each panel.
Ternary complexes were walked by successive rounds of elongation with NTP subsets to the −7 position of both templates, incubated with 500-fold molar excess of RNA oligonucleotides to allow formation of RNA–RNA hybrids, and then chased at limiting GTP (10 μm). On both templates, addition of RNA1 or RNA2 reduced the half-life by a factor of 3 to 4 (Fig. 4A,B). The effects of RNA oligonucleotides on pausing are less than those of the DNA oligonucleotides, most likely because the formation of hairpin structures in the RNA oligonucleotides competes with antisense oligonucleotide–nascent RNA pairing. The addition of the pause RNA hairpin as a separate molecule (RNA3) reduced pausing slightly on the MS hairpin template (Fig. 4B), but not on the WT template (data not shown). The slight inhibition on the MS template may reflect competition by RNA3 for the interaction of the MS hairpin with RNAP, but it might also be explained by direct disruption of the weak MS hairpin structure by pairing of complementary bases in RNA3.
From these results we conclude that neither 20 μm RNA hairpin in trans nor an RNA–RNA duplex that creates the proper 3′-proximal spacing in conjunction with a properly positioned loop sequence is sufficient to stimulate pausing in the manner observed for a nascent RNA hairpin. Thus, the pause hairpin structure must be present in the nascent RNA and makes contacts that depend on its precise location and structure.
Antisense oligonucleotides mimic disruption of pause hairpin interaction with RNAP
If antisense oligonucleotides affect pausing solely by pairing to the nascent RNA (rather than to the nontemplate DNA or through interactions with RNAP), their effects should be reduced under conditions that compromise the pause hairpin interaction because previous results have established that the effect of the pause hairpin on half-life is additive with other interactions (Chan et al. 1997). Thus, to test our conclusions further, we analyzed the effects of antisense oligonucleotides on pausing when IMP was substituted for GMP in the pause hairpin and in the presence of elevated [Cl−].
Substitution of IMP for GMP in the hairpin stem should destabilize it by ∼4 Kcal/mole by reducing the number of hydrogen bonds (∼1.3 Kcal/mole per substitution; SantaLuca et al. 1992) and replacing GTP with ITP in transcription reactions is known to inhibit some pauses (Reisbig and Hearst 1981; Levin and Chamberlin 1987). By stepwise transcription, we substituted IMP residues only for the three Gs in the his pause hairpin and then measured the pause half-life in the absence and presence of an antisense DNA oligonucleotide (Fig. 5A,B). Substitution of IMP in the hairpin stem reduced the half-life by a factor of ∼4 (from 97 to 27 sec) in the absence of DNA oligonucleotide. Addition of oligonucleotide further decreased the half-life to 9 sec on both templates, therefore destabilization of the hairpin with IMP reduced the oligonucleotide effect to a factor of 3 rather than by a factor of 12. These results and those with the MS hairpin (Fig. 4; see above) show that although either IMP substitution or base mismatches in the hairpin stem reduce pausing by destabilizing the structure, neither eliminates completely the hairpin’s effect on pausing. Importantly, in both cases, antisense oligonucleotides reduce the pause half-life only to the same half-life they produce on a wild-type template, rather than by the same factor as would be expected if they affected pause escape through interactions other than with the nascent RNA.
Figure 5.
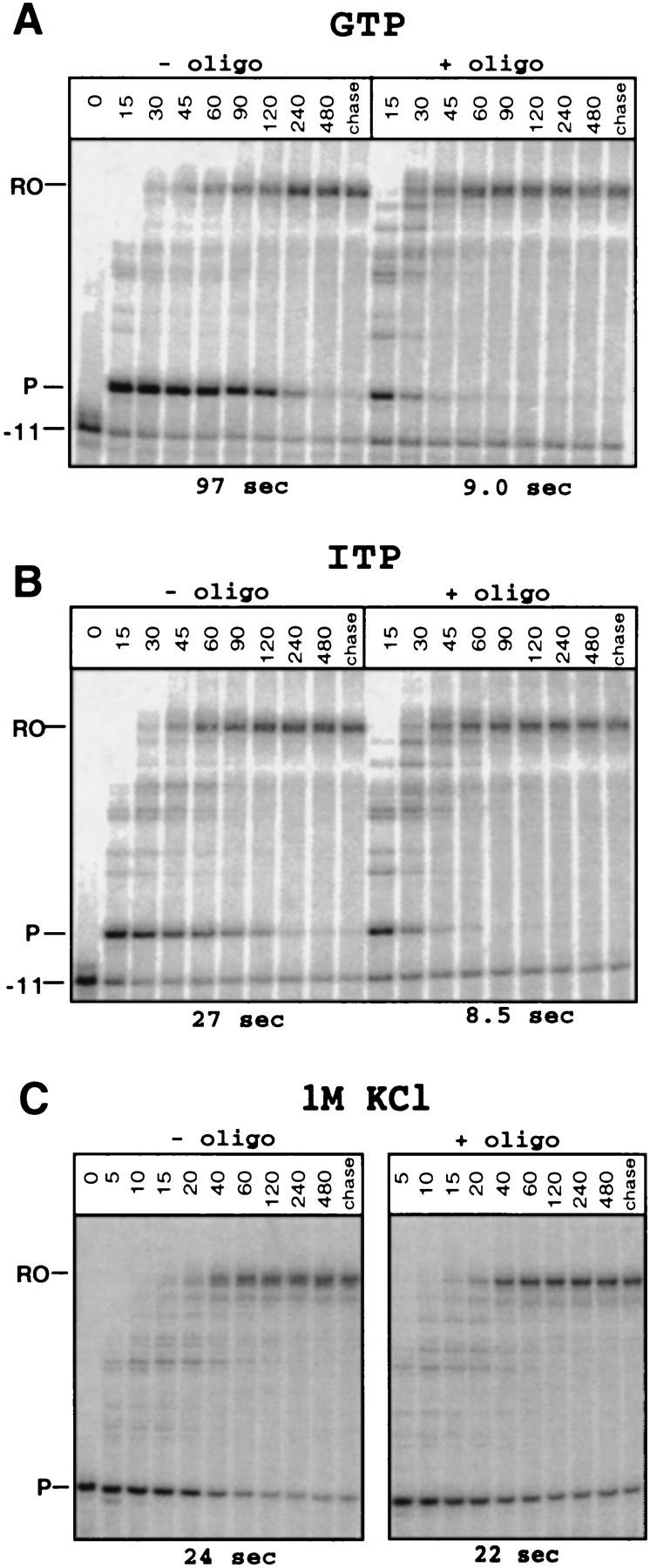
Effect of DNA oligonucleotides on pausing on templates with hairpins destabilized by ITP substitution within the stem or KCl addition. A29 complexes were walked to the −11 (G60) position (A,B) or to the pause site (U71, C). Either GTP (A) or ITP (B) was incorporated at three positions within the hairpin stem (see Fig. 1). Complexes were eluted from the beads and incubated at 37°C for 5 min in the absence or presence of oligonucleotide −15, and, in C, at 1 m KCl. Elongation was resumed and samples were analyzed as above. Half-lives determined as described in Materials and Methods are shown below each panel.
Elevated concentrations of KCl decrease the rate of elongation at most template positions, but increase the rate of escape from the his pause site apparently by blocking hairpin interaction, leading to a net decrease in the pause half-life by a factor of 3 at 1 m KCl (Chan and Landick 1997). If this idea is correct, elevating KCl concentration and adding an oligonucleotide complementary to the hairpin should not produce synergistic effects on pause half-life, as both are predicted to eliminate pause hairpin–RNAP interaction. We tested this prediction by measuring the half-life of the paused complex in the presence of DNA oligonucleotide at low salt or at 1 m KCl (Fig. 5C). As expected, the combination of oligonucleotide and Cl− did not reduce pause half-life any further than Cl− alone, even though elevated KCl should promote antisense oligonucleotide pairing to the nascent RNA. Importantly, this result confirms that elevated Cl− alone can eliminate the effect of the pause hairpin and that its 3-fold rather than the >10-fold effect reflects the general slowing of RNA chain elongation at elevated Cl−.
The pause hairpin inhibits nucleotide addition by disengaging the RNA 3′ end from the active site
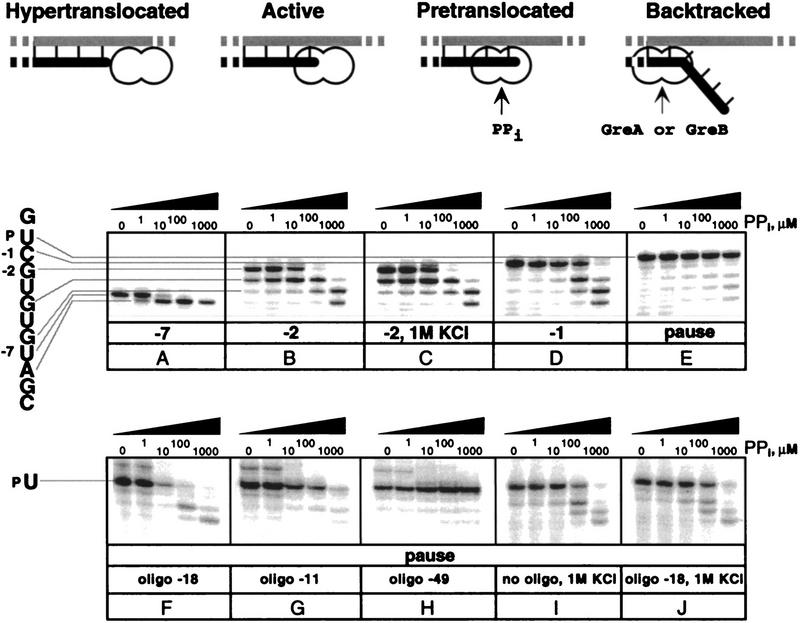
Finally, we wished to use antisense oligonucleotides to test the idea that the pause hairpin functions by disengaging the RNA 3′ end from the active site (e.g., by hypertranslocation; see Fig. 6 and Chan et al. 1997) rather than by causing backtracking of RNAP along the template as has been proposed for some pause sites (Guajardo and Sousa 1997; Komissarova and Kashlev 1997b; Nudler et al. 1997). Backtracking of RNAP may precede both ρ-independent termination and formation of a hairpin-stabilized paused TC (Nudler et al. 1995; Wang et al. 1995; Landick 1997); extensive backtracking can lead to transcription arrest (Reeder and Hawley 1996; Komissarova and Kashlev 1997a). However, the resistance of the his paused complex to GreA-stimulated transcript cleavage (Feng et al. 1994) argues against backtracking in the hairpin-stabilized paused TC because GreA stimulates cleavage of 2- to 3-nucleotide fragments from the 3′ end when RNAP is backtracked (Borukhov et al. 1993). Nonetheless, it remained formally possible that RNAP is backtracked by only 1 nucleotide at the his pause site and therefore, does not serve as a substrate for cleavage. If this were the case, then the pause RNA should be susceptible to pyrophosphorolysis. Furthermore, if backtracking occurred, antisense oligonucleotide pairing might inhibit pyrophosphorolysis at the pause as it does transcript cleavage at an arrest site (Reeder and Hawley 1996; Komissarova and Kashlev 1997a). In contrast, if hypertranslocation occurred, antisense oligonucleotide pairing might actually promote pyrophosphorolysis.
Figure 6.
Transcript sensitivity to pyrophosphorolysis. (Top) Different conformations of TC can be distinguished by their sensitivity to transcript cleavage and pyrophosphorolysis. (Bottom) Immobilized TCs were halted along the template encoding the WT his pause signal at the positions indicated below each panel and treated with increasing concentrations of PPi (indicated above each panel; see Materials and Methods). Oligonucleotides, 1 m KCl, or both were added when indicated.
We tested complexes halted at the pause and at several positions before the pause, −7, −2, and −1, for their sensitivity to pyrophosphate (PPi; Fig. 6). The paused complex was resistant to PPi at all concentrations tested (up to 1mm; Fig. 6E), whereas complexes halted at positions −7U, −2G, and −1C were all sensitive, although to different extents (Fig. 6A,B,D). Complexes with a U at the transcript 3′ end were unusually sensitive to pyrophosphorolysis: the −7 RNA was shortened by 1 nucleotide at 1 μm PPi (Fig. 6A), and RNAs corresponding to −3U, −5U were cleaved too rapidly to be detected during degradation of −2C and −1G complexes (Fig. 6B,C). Therefore, the conformation of the PPi-resistant, his paused complex must be different from other complexes with a 3′-terminal UMP. We interpret this PPi resistance to mean that the pause hairpin prevents backtracking even by 1 nucleotide to the PPi senstitive, pretranslocated conformation and most likely favors the hypertranslocated conformation (Fig. 6). In agreement with this conclusion, addition of the −18 antisense oligonucleotide that destroys the pause hairpin stimulated pyrophosphorolysis (Fig. 6F), whereas an oligonucleotide that pairs upstream from the pause hairpin had no effect (Fig. 6H). The −11 antisense oligonucleotide, which can create the same 3′-proximal spacing as the pause hairpin but could not stimulate pausing (see Fig. 2), also could not recapitulate the PPi resistance caused by the wild-type pause hairpin (Fig. 6G). Thus, direct hairpin interaction with RNAP appears to be required to prevent pyrophosphorolysis as well as to stimulate transcriptional pausing.
Finally, if our hypothesis that Cl− ion inhibits pause hairpin–RNAP interaction is correct and this interaction is required to disengage the 3′ OH from the active site, then ⩾0.5 m KCl should stimulate pyrophosphorolysis in the his paused TC (see Fig. 5; Chan and Landick 1997). In confirmation of this prediction, we found that 1 m KCl stimulated pyrophosphorolysis of the his pause RNA (Fig. 6I), but had no effect at the −2 position (Fig. 6C) and slightly inhibited pyrophosphorolysis caused by addition of the −18 oligonucleotide to the paused TC (cf. Fig. 6F,J). Similar effects were observed with 0.5 m KCl (data not shown). We conclude that, in the his paused TC, direct interaction of the pause hairpin with RNAP inhibits nucleotide addition and pyrophosphorolysis by moving the RNA 3′ OH away from the NTP-binding subsite, most likely to a hypertranslocated state, rather than by favoring backtracking of RNAP.
Discussion
The principal conclusion of our study is that nascent RNA structures stimulate transcriptional pausing or transcript release through different types of interactions with the TC. At the pause site, a nascent RNA hairpin inhibits nucleotide addition through a direct interaction with RNAP that cannot be mimicked by structures formed with antisense RNA or DNA oligonucleotides. In contrast, antisense oligonucleotides can stimulate transcript release, provided that RNAP is not locked in a cycle of rapid chain extension. This suggests that terminator hairpins could act indirectly simply by disrupting interaction of ssRNA with RNAP or template DNA. In addition, our results argue that interaction of the his pause RNA hairpin with RNAP blocks nucleotide addition by disrupting contact between the active site and the nascent RNA. We will discuss these conclusions in the context of a current model for RNA chain elongation and termination.
Antisense oligonucleotides may affect different steps in transcript elongation
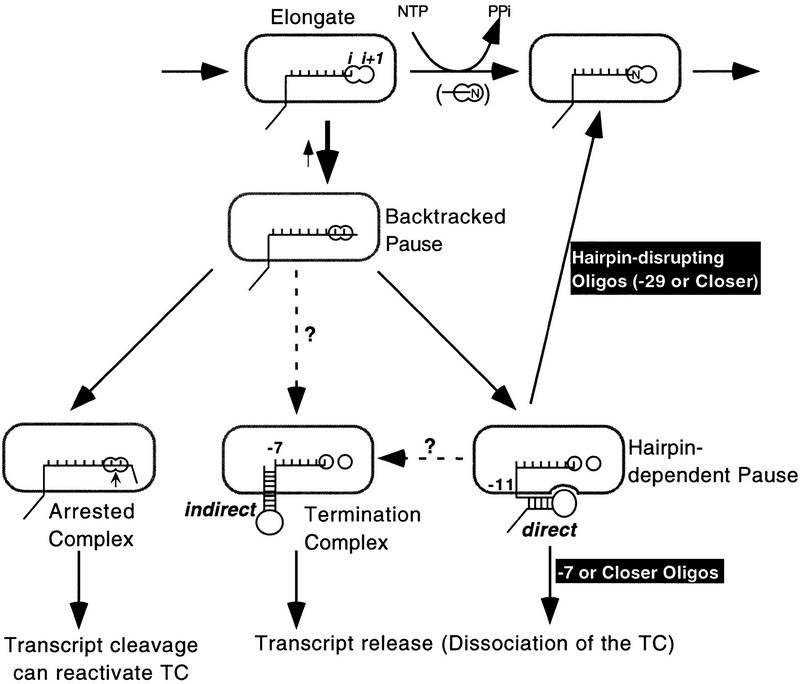
Rapid chain elongation occurs in the bipartite active site of RNAP in a cycle of at least four steps: (1) NTP binding in subsite i + 1; (2) nucleophilic displacement of the NTP’s pyrophosphate by the transcript’s 3′ OH, which is positioned in subsite i; (3) pyrophosphate release; and (4) translocation of the new 3′-terminal nucleotide back to subsite i (Fig. 7, DNA omitted for clarity; see Erie et al. 1992). Transcriptional pausing, termination, or arrest, all events that interrupt chain elongation, appear to begin with transient backtracking of RNAP along the RNA and DNA chains, which removes RNAP from the rapid cycle of chain extension (Nudler et al. 1995, 1997; Wang et al. 1995; Reeder and Hawley 1996; Komissarova and Kashlev 1997a,b; Landick 1997). Backtracking, also called reverse translocation, means that both the RNA–DNA hybrid and RNAP contacts to RNA and DNA slide upstream by a few nucleotides so that the active site becomes occluded by the RNA chain. This prevents nucleotide addition, but allows transcript cleavage. Backtracking is thought to be triggered at sequences for which the RNA–DNA hybrid and RNAP–DNA contacts in the active conformation (with site i + 1 open) are less stable than those formed when RNAP shifts upstream. This transient movement may slow nucleotide addition for a length of time that depends on the ‘positional equilibrium’ among the active conformation and the nearby backtracked states (Guajardo and Sousa 1997; Komissarova and Kashlev 1997b; Landick 1997; Nudler et al. 1997); thus unstable RNA–DNA and RNAP–nucleic acid contacts may generate one type of transcriptional pausing.
Figure 7.
Model of events that interrupt RNA chain elongation and the effects of antisense oligonucleotides on them. The steps in rapid chain elongation (see text) are represented horizontally at the top. RNAP in an elongating TC is represented by the oval; the DNA strands were omitted for clarity. RNAP’s bipartite active site is represented by the double circle (modified from Erie et al. 1992 to use i for the binding site of the 3′-terminal nucleotide and i + 1 for the binding site of the incoming NTP). Transcript bases that are paired to the DNA template are shown by vertical lines; other transcript bases are not shown (the RNA-to-DNA hybrid is thought to be ∼8 bp; see Lee and Landick 1992; Landick 1997; Nudler et al. 1997, and references therein). This model of hairpin-dependent pausing and ρ-independent termination differs superficially from other versions we have published recently (Chan et al. 1997; Landick 1997; Mooney et al. 1998) because it shows the backtracked pause as a separate intermediate to illustrate the possibility that, at least for some ρ-independent terminators, RNAP–RNA hairpin interaction may not be essential for transcript release.
Once backtracking has disrupted the cycle of rapid NTP addition and PPi release, a second rearrangement may lead to transcriptional arrest, to hairpin-dependent pausing, or to termination depending on RNA and DNA sequences and structures that surround the 10- to 12-nucleotide 3′-proximal region (Fig. 7). If the transcript lacks structures that block further backtracking, then further retreat of the TC may extrude enough of the 3′ region of transcript to form a stable structure or an association with RNAP that arrests transcription until it can be reactivated by transcript cleavage. If a hairpin can form ∼10–12 nucleotides from the RNA 3′ end, then it may stabilize a hairpin-dependent paused conformation. Finally, if the hairpin can reach to within 7–9 nucleotides of the 3′ end, then the TC is destabilized and, if the remaining 3′-proximal RNA is U-rich, may dissociate even at low salt (intrinsic termination).
Direct contact between the pause hairpin and RNAP disrupts association of the transcript with the active site at the his pause site
Previous results, including cross-linking of the pause hairpin to β subunit residues 904–950 (Wang and Landick 1997) and inhibition by Cl− ion of only the hairpin-dependent component of transcriptional pausing (Chan and Landick 1997), suggested that the contribution of the pause hairpin to inhibition of nucleotide addition at the his pause site was mediated through a direct interaction with RNAP. However, it is difficult to rule out the possibility that hairpin cross-linking to RNAP simply reflects its proximity to the enzyme, rather than a functional interaction. Furthermore, the Cl− effect is potentially complex. Although the threefold acceleration of nucleotide addition at the pause that is caused by Cl− almost certainly involves the pause hairpin (Chan and Landick 1997), as only substitutions in the pause hairpin and not the other pause signal components reverse the Cl− effect, it is possible that binding of Cl− supplants interactions of single-stranded transcript with RNAP that facilitate chain elongation and that are disrupted by the pause hairpin. Furthermore, Cl− is a competitor of many protein–DNA interactions (Leirmo et al. 1987) and thus may also compete for stabilizing interactions of the nontemplate or template DNA strands with RNAP (see below).
Our finding that antisense RNA or DNA oligonucleotides are unable to substitute for the effect of the pause hairpin provides important confirmation for the ideas that pause hairpin function requires a specific interaction with RNAP and that this interaction can be disrupted by Cl− binding to RNAP. The following key points establish these conclusions. First, all antisense oligonucleotides that can disrupt the pause hairpin reduce the pause half-life by a factor of ∼15, even when they should recapitulate the same 3′-proximal spacing as the pause hairpin (Fig. 2B). Thus, the increase in pause half-life caused by the pause hairpin must be ∼15-fold, higher than the 6- to 10-fold estimated from base substitutions (Chan and Landick 1993). Second, neither addition of the pause hairpin RNA as a separate molecule at high concentration, nor pairing to the transcript of antisense oligonucleotides that should recreate the pause hairpin loop were able to substitute for the wild-type pause hairpin in the nascent RNA (Fig. 4). Third, oligonucleotide effects were reduced when the pause hairpin contained natural or inosine base substitutions, but not when the antisense oligonucleotides contained Gs that should pair to RNA but not DNA (Figs. 2, 4, and 5). Thus, the effects of oligonucleotides clearly were through pairing to the nascent RNA and not the nontemplate DNA. Fourth, the ability of Cl− to accelerate escape from the pause was eliminated by antisense oligonucleotides (Fig. 5). This argues strongly that both Cl− and antisense oligonucleotides accelerate nucleotide addition at the pause by competing for RNA hairpin–RNAP interaction. Finally, both antisense oligonucleotides and Cl− ion are able to ameliorate the unusual resistance of the paused complex to pyrophosphorolysis (Fig. 6). Thus, interaction of the pause hairpin with RNAP, which can be disrupted by either antisense oligonucleotides or Cl−, must disengage the nascent RNA 3′ OH from reactive alignment in the active site. Backtracking by one or morenucleotide as an explanation for hairpin-dependent pausing is ruled out by the paused complex’s resistance to both pyrophosphorolysis (Fig. 6) and transcript cleavage (Feng et al. 1994).
Therefore, we propose that direct RNA hairpin–RNAP interaction is an integral component of the mechanism of transcriptional pausing at the his pause site and that it stabilizes a conformation of the TC that is different than a backtracked TC. This conformation most likely is hypertranslocated, with the RNA 3′ end displaced upstream from its normal position, whereas interaction of RNAP with the downstream DNA, which also is required for strong pausing (Lee and Landick 1992; Wang et al. 1995), may anchor the front end of the enzyme on the template and prevent the NTP-binding subsite from moving with the pause RNA 3′ end. Most nascent RNA contacts in the active site could be retained in the paused conformation, however, as separating the 3′ OH− and NTP-binding subsites i and i + 1 by only a few tenths of an angstrom should be sufficient to inhibit nucleotide addition.
An important implication of these findings is the existence of at least two types of paused transcription complexes. Hairpin-dependent pauses disrupt the transcript-active site association and require hairpin–RNAP interaction. Backtracked pauses, which set up formation of the hairpin-dependent pause, may in some cases create strong delays in transcript elongation by occluding the i + 1 site with nascent RNA (for instance, a strong backtracked pause occurs in the early transcribed region HIV-1; see Palangat et al. 1998). Whether pause hairpin–RNAP interaction simply provides the free energy required to pull the RNA 3′ end out of the active site, causes a direct allosteric change in RNAP that alters the active site, or possibly creates a physical barrier to binding of NTP or PPi substrates into the active site remain questions for future study.
Role of RNA hairpin in transcription termination
Our results also have important implications for the mechanism of ρ-independent termination. First, because we were able to observe transcript release only when antisense oligonucleotides were added to preformed paused TCs, and not when they were present during elongation and thus counteracted pausing (Fig. 3), our results support the generally held view that pausing of some type must precede transcript release (Farnham and Platt 1980; von Hippel and Yager 1991; McDowell et al. 1994). However, although transcript release clearly can occur from a hairpin-dependent paused complex, we cannot exclude the possibility that formation of a terminator hairpin could release the transcript from a backtracked pause and not require the hairpin–RNAP interaction present in the hairpin-dependent paused complex (Fig. 7). Because intrinsic terminators are typically U-rich after the hairpin, they encode strong signals for backtracking, which has been observed at least for λ tR2 (Nudler et al. 1995). The backtracked pause formed at the his pause site may not delay RNAP sufficiently for oligonucleotide-mediated transcript release. Although one might argue that the requirement for 1 m KCl to observe oligonucleotide-mediated release in our experiments as well as transcription termination when the pause hairpin stem is extended to within 9 nucleotides of the RNA end reflects a need to disrupt a stabilizing hairpin interaction with RNAP, both antisense oligonucleotides and the perfect hairpin already disrupt this interaction at low salt. Thus, the requirement for high salt to observe transcript release at the pause more likely reflects Cl− displacement of an RNAP–RNA or RNAP–DNA interaction, such as with the nontemplate strand, that is compromised by the U-rich 3′-proximal region at a ρ-independent terminator (see below).
The second important implication of our results for the mechanism of termination derives from the remarkably narrow window of spacing from the RNA 3′ end within which antisense oligonucleotide pairing stimulates transcript release. Only when an antisense oligonucleotide pairs to the −8 nucleotide did we observe release, and the efficiency of release was unchanged when pairing closer to the 3′ end was possible (Fig. 3; cf. effects of −8, −7, −5, and −3 oligonucleotides). Presumably pairing to the -8 nucleotide reflects disruption of an interaction critical for TC stability, which is accomplished at an intrinsic terminator by the terminator hairpin. There are at least three such interactions whose loss could be a critical step in intrinsic termination. The first is pairing of the nascent RNA to the DNA template, whose thermodynamic contribution to TC stability near a termination site may depend on backtracking of RNAP. Because backtracking appears to compensate energetically for the unstable rU · dA base-pairs produced at a terminator, the possible oscillation of the TC among different positional conformers may keep the TC stable until backtracking is prevented by formation of the terminator hairpin. Restricting the positional equilibrium of the TC to an unstable hybrid might then cause transcript release. Antisense oligonucleotides have been shown to prevent backtracking of RNAP (Reeder and Hawley 1996; Komissarova and Kashlev 1997a), therefore they also could produce this effect.
The second possibility is a related idea originally postulated by Yager and von Hippel (1991), who argued that pairing of nascent RNA to the DNA template compensated energetically for formation of the single-stranded transcription bubble and that transcript release occurs when a terminator hairpin shortens the RNA–DNA heteroduplex (postulated then to be 12 bp) because the remaining rU · dA base-pairs are too weak to prevent collapse of the transcription bubble. Thus, antisense oligonucleotide pairing to the −8 nucleotide might allow nontemplate–template strand pairing at a critical position that would lead to complete bubble collapse because the remaining bubble is too short to be stabilized by RNAP.
The third possibility, originally suggested by Chamberlin and co-workers (Reynolds et al. 1992; Chamberlin 1995), is that ssRNA interaction with RNAP stabilizes the TC, which becomes unstable when a terminator hairpin disrupts the ssRNA–RNAP interaction. This idea was elaborated further by Nudler et al. (1996, 1997) who suggested that terminator hairpins may disrupt critical RNAP–ssRNA interactions in the transcript exit channel that could help keep RNAP wrapped tightly around the downstream DNA duplex. Antisense oligonucleotide pairing similarly could disrupt these interactions and produce transcript release.
These three possibilities are not mutually exclusive. Antisense oligonucleotides may destabilize the TC by causing more than one of these effects and our results do not distinguish among them unambiguously. However, the critical role of antisense oligonucleotide pairing to the −8 nucleotide leads us to suggest that competition between RNA–DNA base pairing and transcripition bubble collapse deserves more careful consideration than it has received recently. Both the Chamberlin and Goldfarb groups have argued that RNA–DNA base pairing cannot be responsible for the resistance of the TC to dissociation (Chamberlin 1995; Nudler et al. 1996; Uptain and Chamberlin 1997). However, some evidence central to this conclusion relies on studies of TCs that lack a nontemplate strand in the critical region 7–10 nucleotides upstream from the RNA 3′ end. TC formed on ssDNA (Uptain and Chamberlin 1997) or by template hopping to short DNAs that only occupy the downstream half of RNAP’s DNA-binding surfaces (Nudler et al. 1996) cannot recapitulate a normal TC structure in which maintenance of an unpaired transcription bubble necessarily imposes a destabilizing contribution to the overall free energy of the TC (Yager and von Hippel 1991).
If antisense oligonucleotides merely acted to prevent backtracking, it seems more likely that pairing to −12 would be sufficient to trigger transcript release at a terminator, as the pause hairpin blocks effectively backtracking even by 1 nucleotide to a conformation able to undergo pyrophosphorolysis. Likewise, if disruption of protein–RNA interactions alone were sufficient to destabilize the TC, then antisense oligonucleotide pairing in the −11 to −8 region should exert some effect as these are the bases that appear to reside in the RNA exit channel (Nudler et al. 1997) and thus to stabilize the downstream DNA clamp. On the other hand, it seems reasonable that the −8 RNA–DNA base pair could play a critical role in maintaining an unpaired transcription bubble. If the transcription bubble normally is maintained by a combination of RNAP–DNA interaction and RNA–DNA base-pairing, then exposure of the −8 template base to pairing with the nontemplate strand might reduce the transcription bubble below some critical size that can be stabilized by RNAP. We favor the idea that a minimal size of the hybrid is critical, rather than only its stability, for two reasons. First, if reducing hybrid stability per se were critical for transcript release, then oligonucleotides that disrupted more of the hybrid than the −7 oligonucleotide (e.g., −5 and −3 oligonucleotides) should have produced a higher efficiency of release, rather than the sharp boundary in destabilization that we observed (see Fig. 3). Second, substitution of iodo-UMP at three positions within the 3′-proximal region, which is predicted to increase the stability of RNA–DNA hybrid (Ferrer et al. 1997), does not reduce the efficiency of −7 oligonucleotide-stimulated transcript release (I. Artsimovitch and R. Landick, unpubl.). However, because transcript release at the pause requires high salt, even with the −7 antisense oligonucleotide, there must be an additional destabilizing force at a ρ-independent terminator. It is tempting to speculate that this is provided by the three U residues invariably present immediately after essentially all ρ-independent terminator hairpins (d’Aubenton Carafa et al. 1990). If terminator hairpin disruption of the −8 RNA–DNA base pair can nucleate bubble collapse, three immediately adjacent, weak rU· dA base pairs may cause it to become favorable even without the contribution of Cl− competition for RNAP–nontemplate strand interaction. Because the actual 3′ end of a terminated RNA can be shifted slightly downstream by accelerating nucleotide addition (McDowell et al. 1994), the normal heterogeneity in terminated RNA 3′ ends may arise because chain extension continues until bubble collapse is complete.
Further study will be required to test these ideas about transcript release, which we are led to by consideration of ability of some antisense oligonucleotides to trigger by disrupting a critical interaction of the −8 nascent RNA base. However, our results clearly demonstrate that some type of paused TC must be formed before these events become possible.
Materials and methods
Sources of proteins, DNAs, and oligonucleotides
RNA polymerase was purified from Escherichia coli strain MRE600 (Midwest Grain Processing Co.) using the method of Burgess and Jendrisak (1975) with modifications (Hager et al. 1990). His-tagged RNA polymerase was prepared as described previously (Wang et al. 1995). Transcription templates were prepared by PCR amplification of pCL102b (wild-type; Chan and Landick 1997) or pDW308 (MS hairpin; Wang et al. 1995) with primers 947 and 645. All DNA oligonucleotides were obtained from Operon Technologies. Oligonucleotides −7 to −49 and the noncomplementary control oligonucleotide were 22 nucleotide in length. Oligonucleotides −5 and −3 were 24 and 26 nucleotides, respectively, with 3′ ends identical to the −7 oligonucleotide. Oligonucleotide UG-15 (GGGGGCTGGTCGGGGTGG; Fig. 2B) was designed to form U–G base pairs with nascent RNA; it is predicted not to form a stable hybrid with the nontemplate DNA strand (8 mismatches/18 nucleotides). For in vitro transcription reactions all oligonucleotides were used at 500-fold molar excess relative to the ternary complex.
In vitro transcription reactions
A29 TCs were formed at 40 nm in transcription buffer (20 mm Tris HCl, 20 mm NaCl, 14 mm MgCl2, 14 mm β-mercaptoethanol, 0.1 mm Na2EDTA) with 32P derived from [α32P]CTP (New England Nuclear; 3000 Ci/mmole) as described previously (Landick et al. 1996). Immobilized A29 TCs were formed with 3 pmoles of His-tagged RNA polymerase immobilized on Ni2+–NTA agarose beads (Qiagen), moved stepwise to the desired template positions by successive rounds of incubation with subsets of NTPs (Wang et al. 1995), and then kept on ice before use. Where indicated, preformed paused complexes were eluted from the beads with 150 mm imidazole and then diluted to 30 mm imidazole; control experiments demonstrated that this concentration of imidazole did not affect pausing (data not shown). When used, KCl and DNA or RNA oligonucleotides were added to 1 m and at 500-fold molar excess, respectively, to the halted complexes, followed by 5 min of incubation at room temperature and equilibration at 37°C. Elongation was allowed to resume by addition of 10 μm GTP, 150 μm each of ATP, CTP, UTP (Pharmacia), and 100 μg heparin/ml. Samples were taken at the desired time points and mixed with the equal volume of 2× stop solution (Landick et al. 1996). After completion of the time course, the reactions were incubated for an additional 5 min with 250 μm each NTP (chase).
Pyrophosphorolysis and transcript release assays
Immobilized on Ni2+–NTA beads complexes were walked to the desired positions on the template, preincubated for 5 min at room temperature in the presence of 500-fold molar excess of oligonucleotides, 1 m KCl, or both, and then equilibrated at 37°C. Halted complexes were treated with increasing concentrations of PPi (1 μm, 10 μm, 100 μm, and 1 mm) in transcription buffer for 15 min at 37°C, washed with transcription buffer five times and mixed with equal volume of 2× stop solution. Alternatively, immobilized complexes were chased for 5 min at 37°C with 250 μm NTPs while still on beads. The supernatants (released RNA) were mixed with equal volume of 2× stop solution; the beads (arrested or paused RNA) were washed four times with transcription buffer and resuspended in 1× stop solution. Samples were analyzed on 10% denaturing gels.
Sample analysis and quantitation
Samples were denatured for 2 min at 90°C and electrophoresed through 9% denaturing gels (19:1 (wt/wt) acrylamide to bis-acrylamide, 7 m urea) in 1× TBE [44 mm Tris-borate ( pH 8.3), 7.5 mm EDTA]. Relative concentrations of RNA species were determined using a PhosphorImager and ImageQuaNT software from Molecular Dynamics. Pause half-lives (the time period during which half of the complexes enter the elongation pathway) and efficiencies (fraction of transcribing RNAP molecules that recognize the pause) were determined by nonlinear regression analysis (Microsoft Excel) and kinetic simulation (KINSIM and FITSIM programs) as described previously (Landick et al. 1996).
Acknowledgments
We thank members of the Landick laboratory for many productive discussions and for suggesting revisions during preparation of the manuscript. This work was supported by National Institutes of Health grant GM38660.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL landick@macc.wisc.edu; FAX (608) 262-9865.
References
- Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Burgess RR, Jendrisak JJ. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Chamberlin MJ. The Harvey lectures. New York, NY: Wiley-Liss; 1995. New models for the mechanism of transcription elongation and its regulation; pp. 1–21. [PubMed] [Google Scholar]
- Chan C, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- ————— Effects of neutral salts of transcript elongation and pausing suggest the his leader pause RNA hairpin interacts with an easily disordered region of RNA polymerase. J Mol Biol. 1997;268:37–53. doi: 10.1006/jmbi.1997.0934. [DOI] [PubMed] [Google Scholar]
- Chan C, Wang D, Landick R. Spacing from the transcript 3′ end determines whether a nascent RNA hairpin interacts with RNA polymerase to prolong pausing or triggers termination. J Mol Biol. 1997;268:54–68. [Google Scholar]
- Cheng S-W, Lynch EC, Leason KR, Court DL, Shapiro BA, Friedman DI. Functional importance of sequence in the stem-loop of a transcription terminator. Science. 1992;254:1205–1207. doi: 10.1126/science.1835546. [DOI] [PubMed] [Google Scholar]
- d’Aubenton Carafa Y, Brody E, Thermes C. Prediction of Rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- Erie DA, Yager TD, von Hippel PH. The single-nucleotide addition cycle in transcription: A biophysical and biochemical perspective. Annu Rev Biophys Biomol Struct. 1992;21:379–415. doi: 10.1146/annurev.bb.21.060192.002115. [DOI] [PubMed] [Google Scholar]
- Farnham PJ, Platt T. A model for transcription termination suggested by studies on the trp attenuator in vitro using base analogs. Cell. 1980;20:739–748. doi: 10.1016/0092-8674(80)90320-7. [DOI] [PubMed] [Google Scholar]
- Feng G, Lee DN, Wang D, Chan CL, Landick R. GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- Ferrer E, Wiersma M, Kazimierczak B, Muller CW, Eritja R. Preparation and properties of oligodeoxynucleotides containing 5-iodouracil and 5-bromo- and 5-iodocytosine. Bioconjug Chem. 1997;8:757–761. doi: 10.1021/bc970042l. [DOI] [PubMed] [Google Scholar]
- Fisher R, Yanofsky C. A complementary DNA oligomer releases a transcription pause complex. J Biol Chem. 1983;258:9208–9212. [PubMed] [Google Scholar]
- ————— Use of complementary DNA oligomers to probe trp leader transcript secondary structures involved in transcription pausing and termination. Nucleic Acids Res. 1984;12:3295–3302. doi: 10.1093/nar/12.7.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper HB, Hearst JE. A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell. 1982;29:81–90. doi: 10.1016/0092-8674(82)90092-7. [DOI] [PubMed] [Google Scholar]
- Guajardo R, Sousa R. A model for the mechanism of polymerase translocation. J Mol Biol. 1997;265:8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- Hager DA, Jin DJ, Burgess RR. Use of Mono Q high-resolution ion-exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry. 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- Kashlev M, Nudler E, Severinov K, Borukhov S, Kommisarova N, Goldfarb A. Histidine-tagged RNA polymerase of Escherichia coli and transcription in solid phase. Methods Enzymol. 1996;274:326–334. doi: 10.1016/s0076-6879(96)74028-4. [DOI] [PubMed] [Google Scholar]
- Komissarova N, Kashlev M. Arrest of transcription: E. coli RNA polymerase translocates backward leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci. 1997a;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J Biol Chem. 1997b;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- Landick R. RNA polymerase slides home: Pause and termination site recognition. Cell. 1997;88:741–744. doi: 10.1016/s0092-8674(00)81919-4. [DOI] [PubMed] [Google Scholar]
- Landick R, Wang D, Chan C. Quantitative analysis of transcriptional pausing by RNA polymerase: The his leader pause site as a paradigm. Methods Enzymol. 1996;274:334–352. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- Lee DN, Landick R. Structure of RNA and DNA chains in paused transcription complexes containing Escherichia coli RNA polymerase. J Mol Biol. 1992;228:759–777. doi: 10.1016/0022-2836(92)90862-e. [DOI] [PubMed] [Google Scholar]
- Leirmo S, Harrison C, Cayley DS, Burgess RR, Record MT., Jr Replacement of potassium chloride by potassium glutamate dramatically enhances protein–DNA interactions in vitro. Biochemistry. 1987;26:2095–2101. doi: 10.1021/bi00382a006. [DOI] [PubMed] [Google Scholar]
- Levin JR, Chamberlin MJ. Mapping and characterization of transcriptional pause sites in early genetic region of bacteriophage T7. J Mol Biol. 1987;196:61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- McDowell JC, Roberts JW, Jin DJ, Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- Mooney R, Artsimovitch I, Landick R. Information processing by RNA polymerase: Recognition of regulatory signals during RNA chain elongation. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Kashlev M, Nikiforov V, Goldfarb A. Coupling between transcription termination and RNA polymerase inchworming. Cell. 1995;81:351–357. doi: 10.1016/0092-8674(95)90388-7. [DOI] [PubMed] [Google Scholar]
- Nudler E, Avetissova E, Markovstov V, Goldfarb A. Transcription processivity: RNA polymerase–DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA:DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- Palangat M, Meier T, Keene R, Landick R. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol Cell. 1998;1:1033–1042. doi: 10.1016/s1097-2765(00)80103-3. [DOI] [PubMed] [Google Scholar]
- Reeder T, Hawley D. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- Reisbig RR, Hearst JE. Escherichia coli deoxyribonucleic acid dependent ribonucleic acid polymerase transcriptional pause sites on SV40 DNA F1. Biochemistry. 1981;20:1907–1918. doi: 10.1021/bi00510a029. [DOI] [PubMed] [Google Scholar]
- Reynolds R, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA polymerase. II. Construction and analysis of hybrid terminators. J Mol Biol. 1992;224:53–63. doi: 10.1016/0022-2836(92)90575-5. [DOI] [PubMed] [Google Scholar]
- Reynolds R, Bermúdez-Cruz RM, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA polymerase. Analysis of 13 rho-independent terminators. J Mol Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- Richardson J, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhardt F, Curtiss III R, Ingraham J, Lin E, Low K, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger H, editors. Escherichia coli and Salmonella : Cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 822–848. [Google Scholar]
- SantaLuca J, Jr, Kierzek R, Turner D. Context dependence of hydrogen bond free energgy revealed by substitutions in an RNA hairpin. Science. 1992;256:217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- Telesnitsky A, Chamberlin M. Terminator-distal sequences determine the in vitro efficiency of the early terminators of bacteriophages T3 and T7. Biochemistry. 1989;28:5210–5218. doi: 10.1021/bi00438a044. [DOI] [PubMed] [Google Scholar]
- Uptain S, Chamberlin M. Escherichia coli RNA polymerase terminates transcription efficiently at rho-independent terminators on single-stranded DNA templates. Proc Natl Acad Sci. 1997;94:13548–13553. doi: 10.1073/pnas.94.25.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain S, Kane C, Chamberlin M. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- von Hippel PH, Yager TD. Transcript elongation and termination are competitive kinetic processes. Proc Natl Acad Sci. 1991;88:2307–2311. doi: 10.1073/pnas.88.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Landick R. Preferential interaction of the his pause RNA hairpin with RNA polymerase β subunit residues 904-950 correlates with strong transcriptional pausing. Proc Natl Acad Sci. 1997;94:8433–8438. doi: 10.1073/pnas.94.16.8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Meier T, Chan C, Feng G, Lee D, Landick R. Discontinuous movements of DNA and RNA in E. coli RNA polymerase accompany formation of a paused transcription complex. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- Wilson K, von Hippel P. Transcription termination at intrinsic terminators: The role of the RNA hairpin. Proc Natl Acad Sci. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M, Mullis K, Barnett J, Stroynowski I, Yanofsky C. Transcription termination at the tryptophan operon attenuator is decreased in vitro by an oligomer complementary to a segment of the leader transcript. Proc Natl Acad Sci. 1982;79:2181–2185. doi: 10.1073/pnas.79.7.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager TD, von Hippel PH. A thermodynamic analysis of RNA transcript elongation and termination in Escherichia coli. Biochemistry. 1991;30:1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- Zaychikov E, Denissova L, Heumann H. Translocation of the Escherichia coli transcription complex observed in the registers 11 to 20: ‘jumping’ of RNA polymerase and asymmetric expansion and contraction of the transcription bubble. Proc Natl Acad Sci. 1995;92:1739–1743. doi: 10.1073/pnas.92.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]