Abstract
There is little information as to the location of early tRNA biosynthesis. Using fluorescent in situ hybridization in the budding yeast, Saccharomyces cerevisiae, examples of nuclear pre-tRNAs are shown to reside primarily in the nucleoli. We also probed the RNA subunit of RNase P. The majority of the signal from RNase P probes was nucleolar, with less intense signals in the nucleoplasm. These results demonstrate that a major portion of the tRNA processing pathway is compartmentalized in nucleoli with rRNA synthesis and ribosomal assembly. The spatial juxtaposition suggests the possibility of direct coordination between tRNA and ribosome biosynthesis.
Keywords: RNase P, nucleolus, tRNA processing
The physical organization of transcription and RNA processing events in eukaryotic nuclei has been studied extensively in the cases of pre-rRNAs and pre-mRNAs. Different stages in the expression of these RNAs are often associated with distinctive subnuclear structures, and these associations are integral to the synthesis and processing events. There is no direct information on the sites of tRNA gene transcription, although it is generally assumed that tRNA synthesis will be distributed throughout the nucleus; unlike the tandemly repeated ribosomal genes, the tRNA genes are not usually clustered in the chromosomes. In the budding yeast Saccharomyces cerevisiae, the 275 tRNA genes are scattered throughout the linear genome map (Cherry et al. 1997). Information on the sites of the numerous pre-tRNA processing events in the nucleus is also limited and has not provided a clear model for localization of the pathway.
Maturation of eukaryotic pre-tRNA primary transcripts is often an ordered process, with 5′- and 3′-terminal processing preceding splicing (Tobian et al. 1985; Lee et al. 1991) and intron removal in turn preceding exit from nucleus to cytoplasm. In addition, multiple nucleotide modifications occur in the nucleus (Etcheverry et al. 1979; Hopper and Martin 1992). Although the order of processing events is not uniform for all pre-tRNAs and is not absolutely obligatory even for individual pre-tRNAs, the ordering suggests possible spatial organization of the maturation pathway. In yeast, current information on the sites of pre-tRNA processing could be consistent with either a dispersed pathway or one that is highly localized, especially to some portion of the nuclear periphery. Immunoelectron microscopy showed at least some of the tRNA splicing ligase to be near nuclear pores, leading to the speculation that the late splicing events in the nuclear pathway occur near the time that intron-containing pre-tRNAs exit to the cytoplasm (Clark and Abelson 1987). For intron-containing pre-tRNAs, it is therefore assumed that most of the nuclear precursor will retain the introns and that earlier processing events, such as removal of the termini, will need to take the introns into account (Leontis et al. 1988). Immunofluorescence microscopy has also localized a base modification enzyme, N2,N2-dimethylguanosine-specific tRNA methyltransferase, to the interior of the nuclear periphery (Li et al. 1989).
There is limited information about the possible location of other tRNA processing events. RNase P, which cleaves pre-tRNAs early in the pathway to produce mature 5′ termini, has been found at different sites in multiple studies of mammalian cells. Using fluorescent oligonucleotide probes complementary to the RNA subunit of human RNase P, one set of reports found at least one small, bright signal near the surface of a nucleolus in HeLa cells, although the majority of the RNase P signal appeared to distribute in the cytoplasm for unknown reasons (Matera et al. 1995; Lee et al. 1996). A more recent report using similar methods to localize endogenous RNase P in rat kidney epithelial cells and HeLa cells, as well as fluorescent RNase P RNA injections, showed transient association of RNase P RNA to the nucleolus but steady-state localization in the nucleoplasm (Jacobson et al. 1997). In yeast there are no previous reports of direct microscopic localizations of RNase P, but recent characterizations of the RNase P subunit composition suggest possible nucleolar localization of the yeast enzyme (Lygerou et al. 1994; Chamberlain et al. 1996; Chu et al. 1997; Dichtl and Tollervery 1997; Stolc and Altman 1997; Chamberlain et al. 1998). Unlike the simple, bacterial RNase P, the yeast nuclear holoenzyme has both a large RNA subunit and nine integral protein subunits that are essential for function (Chamberlain et al. 1998). Eight of nine protein subunits, comprising 94% of the protein mass, are also used for RNase MRP (mitochondrial RNA processing), a nucleolar rRNA processing enzyme previously suspected of being related to RNase P because of the similarity of its RNA subunit to that of RNase P. Only two of the protein subunits common to the human RNase P and RNase MRP have been characterized, but this partial information suggests that the complex subunit structures of the two enzymes might be widely maintained in eukaryotes (Lygerou et al. 1996; Stolc and Altman 1997).
The high degree of shared protein content in the two yeast enzymes prompted us to examine the location of RNase P and other aspects of the pre-tRNA processing pathway in yeast. Using a variety of probes to pre-tRNAs and RNase P RNA subunits, we find that a great deal of the early pre-tRNA processing pathway is likely to reside in the nucleolus in yeast.
Results
To examine the location of nuclear pre-tRNA processing intermediates, probes were used to either intron-containing or intronless precursors. Two fluorescent deoxyoligonucleotides were prepared that spanned the introns of two different pre-tRNAs (Leu-3 and Trp) but had minimal complementarity to the mature tRNA domains. The position of these probes relative to the pre-tRNA primary transcripts is given in Figure 1. Each probe will detect intermediates both before and after 5′- and 3′-end processing, of which the end-matured intermediates are more abundant (Lee et al. 1991). Fluorescein signal from the combined pre-tRNA intron probes (green in Fig. 2a) colocalizes almost exclusively in the single crescent-shaped nucleolus of each cell, along with the CY3-labeled probe used to detect the nucleolar U14 small nucleolar RNA (U14 snoRNA, red in Fig. 2b). Only small amounts of nucleoplasmic staining with the pre-tRNA intron probes are observed. The nucleoplasm is shown in blue (DAPI staining). Overlap between pre-tRNA and U14 signals is shown as yellow in Figure 2c to highlight the nucleoloar localization of the pre-tRNA intron signal.
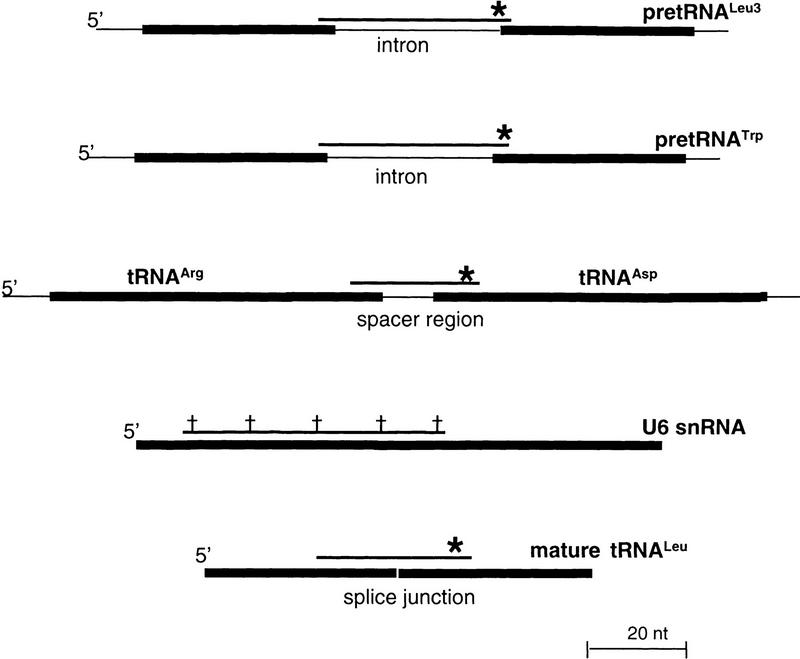
Figure 1.
Schematic depiction of fluorescent probes. Oligodeoxynucleotide probes that anneal to several small RNAs are used in the experiments shown in Fig. 2. The positions at which the fluorescent probes bind to their target RNAs are shown relative to the full-length primary transcripts. 5′-Fluorescein labels are denoted by asterisks, with internal CY3 labels denoted by daggers. The fluorescent antisense RNA probes used to detect U14 snoRNA have been described previously (Samarsky et al. 1998).
Figure 2.
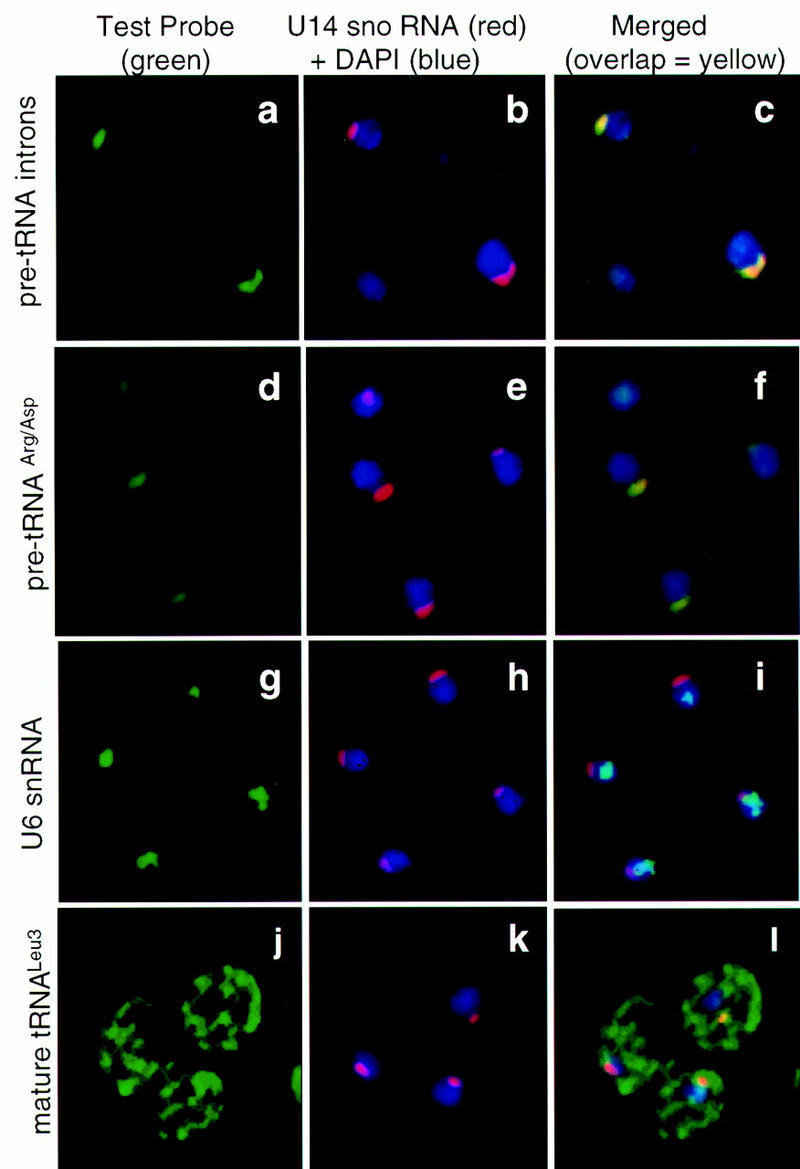
Nuclear tRNA precursors are found predominantly in the nucleolus of S. cerevisiae. In situ hybridization of fluorescently labeled oligonucleotide probes to endogenous RNAs was carried out as described in the text. The left panel of each triplet shows staining with the probe specific for the RNA named at the left of the panels. The center panels show staining of the same cells as at left but with a probe directed against U14 snoRNA (red) and DNA detection with DAPI (blue). Merging of the left and center panels is shown at right, with overlap between the green and red signals shown in yellow. Overlap between green and blue appears as blue–green. (a–c) Introns to pre-tRNALeu3 and pre-tRNATrp are probed simultaneously (a,c). (d–f) The intergenic spacer of pre-tRNAArg/tRNAAsp dimeric transcripts is probed (d,f). (g–i) U6 snRNA is probed (g,i). (j–l) Mature tRNALeu3 is probed (j,l).
Because it is theoretically possible that only pre-tRNAs with introns are selectively localized in the nucleolus, probes were tested that annealed to the 5′ leader sequences of different pre-tRNAs prior to cleavage by RNase P. Obtaining significant signal above the low autofluorescence background proved difficult, as RNase P cleavage and 3′ processing occur soon after synthesis. Although it might be possible to use artificial pre-tRNA variants with slow terminal processing (Lee et al. 1997) or very high-copy-number plasmids containing a tRNA gene to boost pre-tRNA signal, either of these strategies could lead to inappropriate localization of pre-tRNAs. The probe that gave the best signal to endogenous transcripts was directed against the transcribed spacer region in tRNAArg/tRNAAsp dimeric transcripts (Fig. 1). Neither of these tRNAs has an intron, but a dimeric processing intermediate accumulates prior to separation into tRNAArg and tRNAAsp by RNase P (Schmidt et al. 1980; Engelke et al. 1985). The probe to the spacer of this Arg/Asp intermediate gave clearly detectable nucleolar signal (green in Fig. 2d) that colocalized with U14 snoRNA (Fig. 2e and f). As controls to demonstrate that these methods do not routinely stain nucleoli, we probed both nucleoplasmic U6 small nuclear RNA (U6 snRNA, Fig. 2g) and the cytoplasmic mature domain of tRNALeu3 (Fig. 2j). As predicted by its involvement in pre-mRNA splicing and the location of the mammalian counterpart, the U6 snRNA was found to be nucleoplasmic (green in Fig. 2g and blue–green in Fig. 2i, relative to red U14 snoRNA nucleolar marker and blue DAPI nucleoplasm in Fig. 2, h and i). The mature tRNALeu3 probe, which spanned the tRNA splice junction of the spliced tRNA, was found primarily in the cytoplasm as expected (green in Fig. 2, j and l, relative to red U14 snoRNA nucleolar marker and blue nucleoplasm in Fig. 2, k and l). The small amount of overlap between signal from the mature tRNA probe and the nucleolar signal is not consistently present, and it is not clear whether it is a result of insufficient resolution or suggests that some spliced tRNA builds up in the nucleolus before export.
If pre-tRNAs that await both splicing and terminal processing accumulate in the nucleolus, then RNase P, the endonuclease that removes the 5′ leader sequences early in the processing pathway, would also be expected to be present in the nucleolus. Two different types of fluorescent oligodeoxynucleotide probes were used to demonstrate that most RNase P is nucleolar in S. cerevisiae. Signals from probes to RNase P RNA were expected to be weak, as there are estimated to be <400 RNase P molecules per yeast cell (Pagán-Ramos et al. 1996). Different oligonucleotide probes to the wild-type RNase P RNA were tested, and the one that gave the best signal is indicated in Figure 3. A second probe is also shown in Figure 3 that was developed to guard against artifactual signal. A strain was constructed in which a unique 20-nucleotide sequence was inserted at position 135 in the mature RNA domain of the single-copy, essential RPR1 gene. The insertion was made in the chromosomal copy of the gene to eliminate any possible effects from expressing this RNA subunit from an unusual locus. The artificial insertion is in a dispensable region of the RNA subunit that protrudes into solution from the holoenzyme (Fig. 3; Tranguch et al. 1994), leaving the RNase P functional. Cells containing the modified RNase P RNA grow normally, have no pre-tRNA processing defects, and make normal amounts of the slightly larger RNase P RNA subunit (Pagán-Ramos et al. 1994; F. Houser-Scott, A. Kendall, and D.R. Engelke, unpubl.).
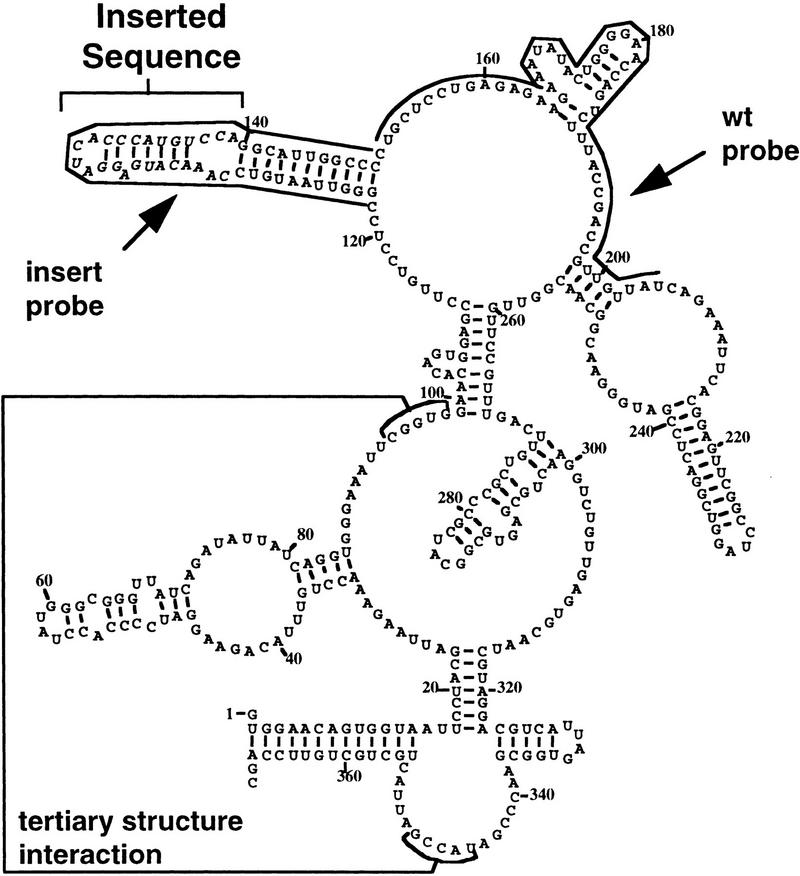
Figure 3.
Fluorescent probes for wild-type RNase P RNA and an inserted sequence. The proposed secondary structure of the S. cerevisiae RNase P RNA (RPR1 RNA) subunit is shown, including the long-distance base-pairing that contributes to tertiary structure. The positions of the artificial 20-nucleotide insertion are italicized and indicated by a bracket. CY3-labeled complementary oligonucleotide probe positions to either the wild-type RPR1 sequence or the inserted sequence are indicated by lines along the sequence.
The fluorescent probe to the wild-type RNA subunit of RNase P, RPR1 RNA, showed clear localization of most of the signal to the nucleolus (Fig. 4a–c). A fluorescent probe complementary to the artificial insertion does not give a signal with cells containing wild-type RNase P (Fig. 4d) but gives a nucleolar signal in the strain containing the insertion in RPR1 (f and h) indistinguishable from the normal RPR1 RNA probe. Thus, the RNase P RNA signal is likely to be a true representation of the position of the enzyme, rather than hybridization to ribosomal sequences or binding to nucleolar proteins.
Figure 4.
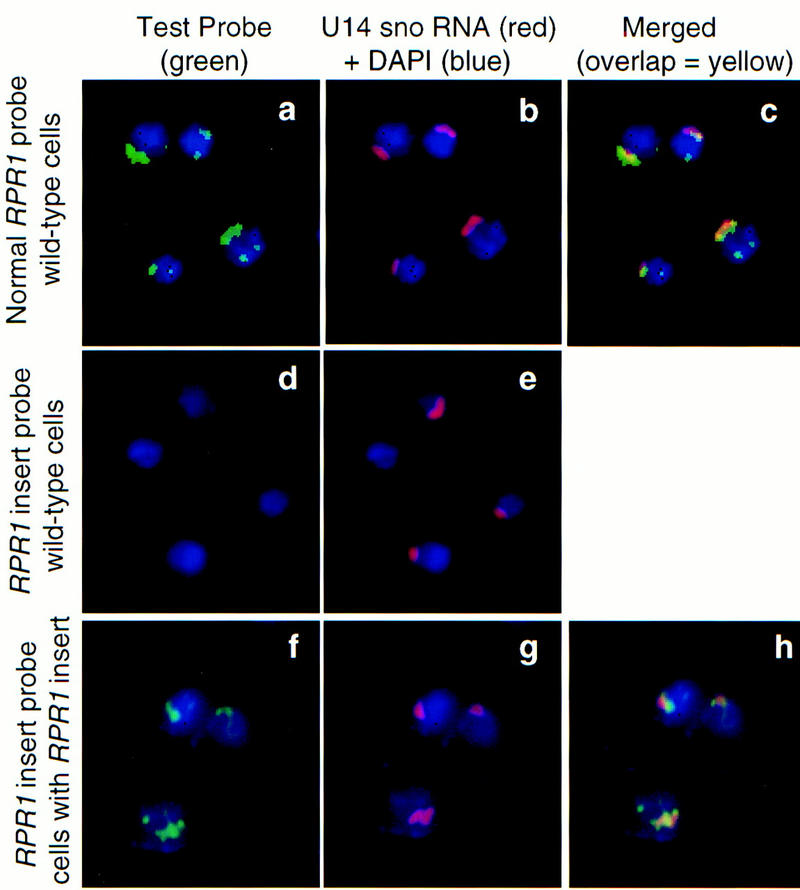
RNase P is located primarily in the nucleolus in S. cerevisiae. All panels are stained for nuclear DNA with DAPI (blue). Probes for RNase P RNA or an artificial insert in RNase P RNA (Fig. 3) are shown in green in the left panels. The center panels show probes of the same cells for U14 snoRNA (red). The right panels show the left and center panels merged, with overlap between the green and red signals in yellow. (No merged panel is given for the center set, as there is no RNA signal in the left panel.) (a–c) The mature domain of wild-type RNase P RNA is probed (RPR1 RNA, a,c). (d–e) The probe to an artificial insert in RPR1 RNA is applied to wild-type cells, in which, as expected, it gives no signal above background. (f–h) The artificial insert probe is used (f,h) with cells having the modified RPR1 allele containing inserted sequences complementary to the probe.
Both the RNase P probe and the artificial insertion probe reproducibly show secondary nucleoplasmic sites in addition to the main signal at the nucleolus. The relative distribution of signal is 66% ± 7% to the nucleolus versus 34% to the nucleoplasm when data are collected through multiple optical sections of individual cells. This is more pronounced nucleoplasmic signal than is seen with the faint secondary staining from pre-tRNA intron probes. It is possible that the secondary signals, which usually manifest as one or more punctate signals well separated from the nucleolus, represent secondary biosynthetic loci outside a main organizational site at the nucleolus. With >200 tRNA genes likely to be active in S. cerevisiae, it is conceivable that some classes of transcripts or genes in different chromosomal environments might have different organizational fates.
Discussion
In retrospect it makes a great deal of sense that much, if not all, of the pre-tRNA processing pathway should be localized to nucleoli. The subcompartmentalization of the precursors might facilitate moving the intermediates through the required series of nucleolytic cleavages and nucleotide modifications. In addition, the nucleolus is the site of synthesis and processing for rRNAs, the other class of heavily modified, abundant RNA involved in translation of mRNAs. Colocalization of the two pathways might well provide the opportunity to share aspects of regulation, including transport to the cytoplasm. The resolution of the fluorescence data used here is not sufficient to tell whether RNase P and the pre-tRNAs are restricted to subcompartments of the nucleolus that are distinct or coincident with various aspects of rRNA biosynthesis. Further studies using electron microscopy and comparative markers from the ribosomal processing cascade might shed more light on this question.
It is not yet clear what percentage of nuclear tRNA precursors are nucleolar, and it is possible that some classes of pre-tRNAs might undergo alternative nucleoplasmic pathways. This appears to be true in at least one case. Similar in situ localizations with probes to the unusually large intron of pre-tRNAIle show primarily nucleoplasmic localization (S. Sarkar and A.K. Hopper, in prep.). Although the bulk of the RNase P is at the nucleolus, the secondary nucleoplasmic signals would be consistent with such alternative pathways. It is also not known how many other pre-tRNA processing activities might be nucleolar. Previous data have not indicated a strong nucleolar link for splicing ligase or one modification enzyme (Clark and Abelson 1987; Li et al. 1989), but the locations of most enzymes in the pathway remain to be explored.
The experiments presented here do not address whether the pre-tRNA transcripts originate at the nucleolus, as is the case for rRNA gene transcription. The tandemly repeated rRNA gene clusters are nucleolar, and synthesis of the primary transcripts by RNA polymerase I coincides with RNA processing and assembly of ribosomal subunits (Raué and Planta 1991; Eichler and Craig 1994). The 5S rRNA genes transcribed by RNA polymerase III (Pol III) are also nucleolar in Xenopus (Narayanswami and Hamkalo 1990) and likely to be nucleolar in S. cerevisiae, as they are part of the large rRNA repeating unit. Nucleolar localization of the tRNA genes would be counterintuitive, as they are scattered in the linear genome map, but there is currently no evidence as to where the tRNA genes reside. It is not impossible that unique features of the tRNA transcription complex with Pol III might be used to organize a majority of the genes to the vicinity of the nucleolus. Earlier work examining suppressors of defects in Pol III transcription components have identified both Pol III transcription factor subunits and nucleolar antigens, suggesting a link between Pol III transcription units and the nucleolus (Lalo et al. 1993; Lefebvre et al. 1993). Possible candidates for interactions with nucleolar components include the subunits common to Pol I and Pol III, but not Pol II, and the multisubunit transcription factors TFIIIB and TFIIIC that are used for both tRNA gene and 5S rRNA gene transcription.
The data presented here suggest that the pre-tRNAs are nucleolar from before RNase P cleavage to at least near the time when their introns are removed. Further study of the pathways by which they enter and exit the nucleolus might shed light on the extent to which tRNA biosynthesis is coordinated with that of ribosomes.
Materials and methods
Fluorescent in situ hybridization
Yeast cells in logarithmic growth phase in liquid culture were fixed, probed with fluorescently labeled oligodeoxynucleotides, stained with DAPI, and imaged as described previously (Long et al. 1995; Samarsky et al. 1998) except that in the case of the Arg/Asp pre-tRNA probe only 10% formamide was used in buffers. Imaging was performed essentially as described (Carter et al. 1993; Samarsky et al. 1998). Images were captured using CellSCAN software (Scanalytics, Fairfax, VA) on an Optiplex GXpro computer (Dell, Austin, TX) with a CH-250 16-bit, cooled CCD camera (Photometrics, Tucson, AZ) mounted on a Provis AX70 fluorescence microscope (Olympus, Melville, NY) with a PlanApo 60×, 1.4 NA objective (Olympus) and HiQ bandpass filters (Chroma Technology, Brattleboro, VT). The fluorescence illumination was controlled by the software using a Uniblitz VS25 shutter (Vincent Associates, Rochester, NY). When images were restored, a three-dimensional data set, composed of 20–25 images separated by 200 nm in the axial direction, was acquired and deconvolved with an acquired point spread function (PSF) using EPR software (Scanalytics). The software controlled the axial position of the objective using a PZ54 E piezoelectric translator (Physik Instrumente, Costa Mesa, CA). The PSF is a data set, composed of 40–50 images separated by 200 nm in the axial direction, of a fluorescent microsphere (Molecular Probes, Eugene, OR) that was 200 nm in diameter. A single median plane was recorded for blue filtered images. Experiments in Figure 2 used the diploid strain MH2 (ade2-101/ade2-101 trp1D/trp1D ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3/his3 lys4/lys4). Oligonucleotide probes for the introns of pre-tRNATrp and pre-tRNALeu3 were labeled with a single fluorescein attached to the 5′ end and had the sequences 5′-GATTGCAATCTTATTCCGTGGAATTTCCAAGATTTAATTG and 5′-TGAGTATTCCCACAGTTAACTGCGGTCAAGATATTTCTTG. Both intron probes gave weak nucleolar signals individually and are shown simultaneousy to boost the signal-to-noise ratio. The dimeric Arg/Asp intergenic probe was 5′-fluorescein-TCACGGAAGAAACAAAGCACTCAC. The sequence of the U6 snRNA probe was 5′T*GATCATCTCTGT*ATTGTTTCAAAT*TGACCAAATGT*CCACGAAGGGT*T, with CY3 incorporated at the T sites followed by asterisks. The sequence of the mature tRNALeu3 probe across the processed splice junction was 5′-fluorescein-ACGATACCTGAGCTTGAATCAGGC. The RPR1 RNA hybridization probes were (wild-type) 5′-T*AACAACGGTCGGT*AAAGACTGGTT*CCCCAGTATATTTCT*TCTCTCAGGAGCAT*G and (with insert) 5′-T*GCCAATGCCT*GGACATGGGT*GATCCTCATGTT*TGGACATTAAT*C, with asterisks indicating CY3-modified T positions. The derivatized antisense RNA probe for U14 snoRNA was prepared as described previously (Long et al. 1995; Samarsky et al. 1998). The U14 snoRNA probe was postlabeled either with Oregon Green 488 if the other RNA probe was labeled with CY3 or with CY3 if the other RNA probe was labeled with fluorescein. For consistency, U14 is always shown in red pseudocolor, and the other probed RNA is shown in green.
Strain containing insertion-tagged RPR1
For RNase P panels, the haploid yeast strain W3031A was used (MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100). For integrating the 20-nucleotide insertion in RPR1, starting strain JYL1 was used (W3031A with chromosomal RPR1 disrupted by HIS3, surviving because of a wild-type RPR1 copy on URA3-marked plasmid). DNA fragments were used to transform the strain that spanned the coding region and contained the 20-nucleotide insertion at position 135 of the mature domain (5′-gucCAAACAUGAGGAUCACCCAUGUCCAggc), with lowercase letters being old sequences at insertion site and uppercase letters being newly inserted sequence (see Fig. 3). Uptake of the altered RPR1 gene by reciprocal recombination was selected by ability to lose the plasmid-borne copy in medium containing 5-fluoro-orotic acid. Correct insertion at the genomic RPR1 locus was subsequently established by screening for loss of the HIS3 allele that previously disrupted the chromosomal RPR1 gene and then by the size of the genomic PCR product within the RPR1 locus across the insertion site. Confirmation that the altered, larger RNA was expressed at normal levels and that the wild-type RNA was absent was obtained by RNA blot analysis as described (Tranguch and Engelke 1993).
Acknowledgments
We thank Norman Pace and Dennis Thiele for comments on the manuscript and Anita Hopper for helpful discussions and communicating results prior to publication. We also thank Pascal Chartrand for conveying information and materials. The study was supported by grants from the National Institutes of Health to D.R.E (GM34869) and R.H.S. (GM54887 and GM57071).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL engelke@umich.edu; FAX (734) 763-7799.
References
- Carter KC, Bowman D, Carrington W, Fogarty K, McNeil JA, Fay FS, Lawrence JB. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- Chamberlain JR, Pagán-Ramos E, Kindelberger DW, Engelke DR. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 1996;24:3158–3166. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes & Dev. 1998;12:1679–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J.M., C. Ball, S. Weng, G. Juvik, R. Schmidt, C. Adler, B. Dunn, S. Dwight, L. Riles, R.K. Mortimer, and D. Botstein. 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature (Suppl. 6632) 387: 67–73. [PMC free article] [PubMed]
- Chu S, Zengel JM, Lindahl L. A novel protein shared by RNase MRP and RNase P. RNA. 1997;3:382–391. [PMC free article] [PubMed] [Google Scholar]
- Clark MW, Abelson J. The subnuclear localization of tRNA ligase in yeast. J Cell Biol. 1987;105:1515–1526. doi: 10.1083/jcb.105.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Tollervey D. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 1997;16:417–429. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler DC, Craig N. Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1994;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- Engelke DR, Gegenheimer P, Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem. 1985;260:1271–1279. [PubMed] [Google Scholar]
- Etcheverry T, Colby D, Guthrie C. A precursor to a minor species of tRNASer contains an intervening sequence. Cell. 1979;18:11–26. doi: 10.1016/0092-8674(79)90349-0. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Martin NC. Processing of yeast cytoplasmic and mitochondrial precursor tRNAs. In: Jones EW, Pringle JR, Broach JR, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae. Vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 99–141. [Google Scholar]
- Jacobson MR, Cao L-G, Taneja K, Singer RH, Wang Y-l, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Knapp G, Beckmann JS, Johnson PF, Fuhrman SA, Abelson J. Transcription and processing of intervening sequences in yeast tRNA genes. Cell. 1978;14:221–236. doi: 10.1016/0092-8674(78)90109-5. [DOI] [PubMed] [Google Scholar]
- Lalo D, Carles C, Sentenac A, Thuriaux P. Interactions between three common subunits of yeast RNA polymerases I and III. Proc Natl Acad Sci. 1993;90:5524–5528. doi: 10.1073/pnas.90.12.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Matera AG, Ward DC, Craft J. Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolus: A possible coordinate role in ribosome biogenesis. Proc Natl Acad Sci. 1996;93:11471–11476. doi: 10.1073/pnas.93.21.11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Rohlman CE, Molony LA, Engelke DR. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol Cell Biol. 1991;11:721–730. doi: 10.1128/mcb.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kindelberger DW, Lee J-Y, McClennen S, Chamberlain J, Engelke DR. Nuclear pre-tRNA terminal structure and RNase P recognition. RNA. 1997;3:175–185. [PMC free article] [PubMed] [Google Scholar]
- Lefebvre O, Rüth J, Sentenac A. A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5S RNA synthesis. Identification of two classes of suppressors. J Biol Chem. 1993;269:23374–23381. [PubMed] [Google Scholar]
- Leontis N, DaLio A, Strobel M, Engelke D. Effects of tRNA-intron structure on cleavage of precursor tRNAs by RNase P from Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:2537–2552. doi: 10.1093/nar/16.6.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-M, Hopper AK, Martin NC. N2,N2-Dimethylguanosine-specific tRNA methyltransferase contains both nuclear and mitochondrial targeting signals in Saccharomyces cerevisiae. J Cell Biol. 1989;109:1411–1419. doi: 10.1083/jcb.109.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Elliott DJ, Stutz F, Roshbash M, Singer RH. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z, Mitchell P, Petfalski E, Séraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes & Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Pluk H, van Venrooij WJ, Séraphin B. hPop1: An autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J. 1996;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding proteins, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanswami S, Hamkalo BA. High resolution mapping of Xenopus laevis 5S and ribosomal RNA genes by EM in situ hybridization. Cytometry. 1990;11:144–152. doi: 10.1002/cyto.990110117. [DOI] [PubMed] [Google Scholar]
- Pagán-Ramos E, Tranguch AJ, Kindelberger DW, Engelke DR. Replacement of the Saccharomyces cerevisiae RPR1 gene with heterologous RNase P RNA genes. Nucleic Acids Res. 1994;22:200–207. doi: 10.1093/nar/22.2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán-Ramos E, Lee Y, Engelke DR. Mutational analysis of Saccharomyces cerevisiae nuclear RNase P: Randomization of universally conserved positions in the RNA subunit. RNA. 1996;2:441–451. [PMC free article] [PubMed] [Google Scholar]
- Raué HA, Planta RJ. Ribosome biogenesis in yeast. Prog Nucleic Acid Res Mol Biol. 1991;41:89–129. doi: 10.1016/s0079-6603(08)60007-0. [DOI] [PubMed] [Google Scholar]
- Samarsky, D., M.J. Fournier, R.H. Singer, and E. Bertrand. 1998. The snoRNA box C/D motif is necessary and sufficient for nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. (in press). [DOI] [PMC free article] [PubMed]
- Schmidt O, Mao J, Ogden R, Beckmann J, Sakano H, Abelson J, Söll D. Dimeric tRNA precursors in yeast. Nature. 1980;287:750–752. doi: 10.1038/287750a0. [DOI] [PubMed] [Google Scholar]
- Stolc V, Altman S. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes & Dev. 1997;11:2414–2425. doi: 10.1101/gad.11.18.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobian JA, Drinkard L, Zasloff M. tRNA Nuclear transport: Defining the critical regions of human tRNAmeti by point mutagenesis. Cell. 1985;43:415–422. doi: 10.1016/0092-8674(85)90171-0. [DOI] [PubMed] [Google Scholar]
- Tranguch AJ, Engelke DR. Comparative structural analysis of nuclear RNase P RNAs from yeast. J Biol Chem. 1993;268:14045–14053. [PubMed] [Google Scholar]
- Tranguch AJ, Kindelberger DW, Rohlman CE, Lee J-Y, Engelke DR. Structure-sensitive RNA footprinting of yeast nuclear ribonuclease P. Biochemistry. 1994;33:1778–1787. doi: 10.1021/bi00173a022. [DOI] [PubMed] [Google Scholar]