Abstract
smc of Bacillus subtilis encodes a homolog of eukaryotic SMC proteins involved in chromosome condensation, pairing, and partitioning. A null mutation in B. subtilis smc caused a temperature-sensitive–lethal phenotype in rich medium. Under permissive conditions, the mutant had abnormal nucleoids, ∼10% of the cells were anucleate, and assembly of foci of the chromosome partitioning protein Spo0J was altered. In combination with a null mutation in spo0J, the smc mutation caused a synthetic phenotype; cell growth was slower and ∼25% of the cells were anucleate. Our results demonstrate that the B. subtilis Smc protein, like its eukaryotic counterpart, plays an important role in chromosome structure and partitioning.
Keywords: Bacillus subtilis, chromosome condensation, chromosome partitioning, green fluorescent protein, Smc, sporulation
Efficient partitioning of chromosomes into dividing cells is important for cell survival. In Escherichia coli and Bacillus subtilis, chromosomes partition to daughter cells with high fidelity (Hiraga et al. 1989; Ireton et al. 1994). Although proteins, sites, and mechanisms involved in physical separation (decatenation) of bacterial chromosomes have been characterized, much less is known about the mechanisms governing efficient partitioning to daughter cells (for review, see Hiraga 1992; Wake and Errington 1995). Recent work has shown that the chromosomal region around the origin of replication (oriC) is in a defined orientation for most of the bacterial (B. subtilis and E. coli) cell cycle and that newly replicated oriC regions are rapidly separated from each other (Glaser et al. 1997; Gordon et al. 1997; Lin et al. 1997; Webb et al. 1997). The origin regions are found toward the poles of the highly condensed nucleoid body, oriented toward the ends of the cell. The rapid separation and localization of oriC regions indicate the function of a mitotic-like apparatus in prokaryotes (Glaser et al. 1997; Gordon et al. 1997; Lin et al. 1997; Webb et al. 1997).
Cellular proteins contributing to efficient chromosome partitioning have recently been characterized. Spo0J from B. subtilis and ParB from Caulobacter crescentus are required for efficient chromosome partitioning and are similar to a family of plasmid-encoded proteins required for plasmid partitioning in E. coli (e.g., ParB for P1 and SopB for F). ParB of C. crescentus is essential for growth, and overexpression causes a defect in chromosome partitioning (Mohl and Gober 1997). Deletion of spo0J in B. subtilis causes an ∼100-fold increase in the number of anucleate cells, resulting in accumulation of 1%–2% anucleate cells in a growing culture (Ireton et al. 1994). Spo0J binds to at least eight sites located in the origin proximal 20% of the chromosome (Lin and Grossman 1998). Spo0J is found in the cell in single discrete foci located near the poles of the nucleoid body (Glaser et al. 1997; Lin et al. 1997), in a pattern similar to that observed for the region around the origin of replication (Lewis and Errington 1997; Webb et al. 1997). Visualization of the foci of Spo0J by immunofluorescence microscopy or by use of a Spo0J–green fluorescent protein (GFP) fusion indicates the assembly of a large nucleoprotein complex containing Spo0J. The function of Spo0J and other proteins of this family is still unknown, though they are thought to be involved in pairing and/or positioning sister chromosomes (Nordström and Austin 1989; Niki and Hiraga 1997; Lin and Grossman 1998).
The muk genes of E. coli were identified in an elegant screen for mutants that produce anucleate cells (Hiraga et al. 1989). The mukB gene product has features of a myosin-like motor protein and is involved in chromosome condensation and/or movement [(Niki et al. 1991; Hiraga 1992; Wake and Errington 1995; Hu et al. 1996), and references therein]. mukE and mukF, which are in an operon with mukB, are also required for efficient partitioning and their products are thought to interact with MukB (Yamanaka et al. 1996). Of the ∼12 bacterial genomes that have been sequenced, mukB, mukE, and mukF are found only in E. coli and Haemophilus influenzae.
B. subtilis, along with many other bacterial species (but not E. coli or H. influenzae), contains a homolog of the eukaryotic Smc (structural maintenance of chromosomes) proteins (Oguro et al. 1996). Several eukaryotes have multiple smc genes, and eukaryotic Smc proteins play a role in chromosome condensation, pairing, and/or segregation (for review, see Hirano et al. 1995; Koshland and Strunnikov 1996; Heck 1997). For example, mutations in the SMC genes of Saccharomyces cerevisiae cause defects in chromosome condensation, segregation, and sister chromatid cohesion (Guacci et al. 1997; Michaelis et al. 1997). DNA condensation by the 13S condensin of Xenopus laevis requires two Smc proteins, XCAP-C and XCAP-E (Hirano et al. 1997). Dosage compensation in Caenorhabditis elegans involves specific interaction of an Smc homolog, Dpy-27, and other proteins, with the X chromosome (Chuang et al. 1994, 1996). Although the precise biochemical function of the Smc proteins is not known, recent work has shown that Smc proteins, or complexes containing Smc proteins, can affect DNA topology in vitro (Kimura and Hirano 1997; Sutani and Yanagida 1997).
The smc gene of B. subtilis encodes a 135-kD protein that is homologous to eukaryotic Smc proteins (Oguro et al. 1996). B. subtilis Smc is ∼24% identical and ∼46% similar to SMC1 and SMC2 (S. cerevisiae), XCAP-C and XCAP-E (X. laevis), Dpy-27 (C. elegans), and Cut3 and Cut14 (Schizosaccaromyces pombe). It contains all of the domains associated with the Smc family; an amino-terminal NTP-binding domain, two internal coiled–coil regions separated by a hinge, and the carboxy-terminal signature “DA-box” motif (Hirano et al. 1995; Koshland and Strunnikov 1996). Whereas the existence of Smc proteins in eukaryotes is well documented, their prevalence in bacteria and archaebacteria is only beginning to be appreciated. A search of GenBank and individual sequence databases (both completed and in progress) revealed that at least 11 bacteria and 2 archaebacteria contain genes encoding homologs of Smc. In addition to B. subtilis, the list includes Streptococcus pyogenes, Streptococcus pneumoniae, Mycobacterium tuberculosis, several Mycoplasma species (M. genitalium, M. pneumoniae, and M. hyorhinis), Borrelia burgdorferi, Treponema pallidum, Synechocystis sp., Neisseria gonorrhoeae, and the archaebacteria Methanococcus jannaschii and Archaeoglobus fulgidus.
We report the characterization of the B. subtilis smc gene. Null mutations in smc caused a conditional lethal phenotype, alterations in nucleoid appearance, a defect in chromosome partitioning, and a synthetic phenotype with a null mutation in spo0J. These findings indicate that the function of Smc proteins is highly conserved.
Results and Discussion
Defects in cell growth and sporulation in an smc null mutant
We constructed a null mutation in the B. subtilis smc gene by replacing most of the coding region with a gene whose product confers resistance to kanamycin (Materials and Methods). In rich medium (LB) the smc null mutant was temperature sensitive for growth; it formed colonies at 24°C but not at 37°C or higher temperatures. Cells grown in LB at 24°C had a plating efficiency of ∼0.05% at 37°C and 43°C. Many of the colonies that grew at high temperatures appeared to contain compensatory mutations. Shift of a culture grown in LB medium at 24°C to 45°C resulted in the rapid loss of viability.
The smc null mutant also had a growth defect in minimal medium, but the defect was less severe than that in rich medium. In defined minimal glucose medium, the smc null mutant was viable at all temperatures tested, up to 45°C, although the generation time (∼125 min at 30°C) was significantly longer than that of wild type (∼85 min at 30°C) (Table 1). Accumulation of compensatory mutations was much less of a problem in minimal medium.
Table 1.
Growth and chromosome partitioning defects of smc mutants
| Strain
|
Relevant genotype
|
Doubling time (min)a
|
Percent anucleateb
|
|---|---|---|---|
| JH642 | wild type | 85 | <0.03 (0/3685) |
| RB35 | Δsmc | 126 | 10.4 (158/1514) |
| AG1468 | Δspo0J | 85 | 0.6 (39/6437) |
| RB41 | Δsmc Δspo0J | 178 | 26.4 (120/454) |
Generation times were determined for cells grown at 30°C in defined minimal glucose medium.
The percentage of anucleate cells was determined during exponential growth in minimal glucose medium at 30°C. The number of anucleate cells observed out of the total number of cells counted is indicated in parentheses. Previous results with cells grown at 37°C in rich medium (2 × SG medium) indicated that the percent anucleate cells in an exponentially growing culture is 0.017% (4 anucleate cells of 23,250 total cells) for wild type and 1.4% (322/22,945) for Δspo0J (Ireton et al. 1994).
The smc null mutation also caused a defect in sporulation. In nutrient sporulation medium at 24°C, the sporulation frequency of the smc mutant was ∼10−3–10−4 that of wild type when assayed ∼24 hr after the end of exponential growth. The defects in growth and sporulation are probably due to alterations in nucleoid structure that are described below.
Defects in chromosome partitioning and nucleoid structure
Similar to its eukaryotic counterparts, B. subtilis smc is required for efficient chromosome partitioning. Under permissive growth conditions in defined minimal glucose medium at 30°C, approximately 10% of the smc null mutant cells were anucleate, as judged by DAPI staining and combined fluorescence–Nomarski microscopy of fixed cells (Fig. 1A,B; Table 1). This frequency is ∼200- to 1000-fold greater than that of wild-type cells (Ireton et al. 1994). In minimal medium, the frequency of anucleate cells in the smc mutant did not change significantly at 37°C.
Figure 1.
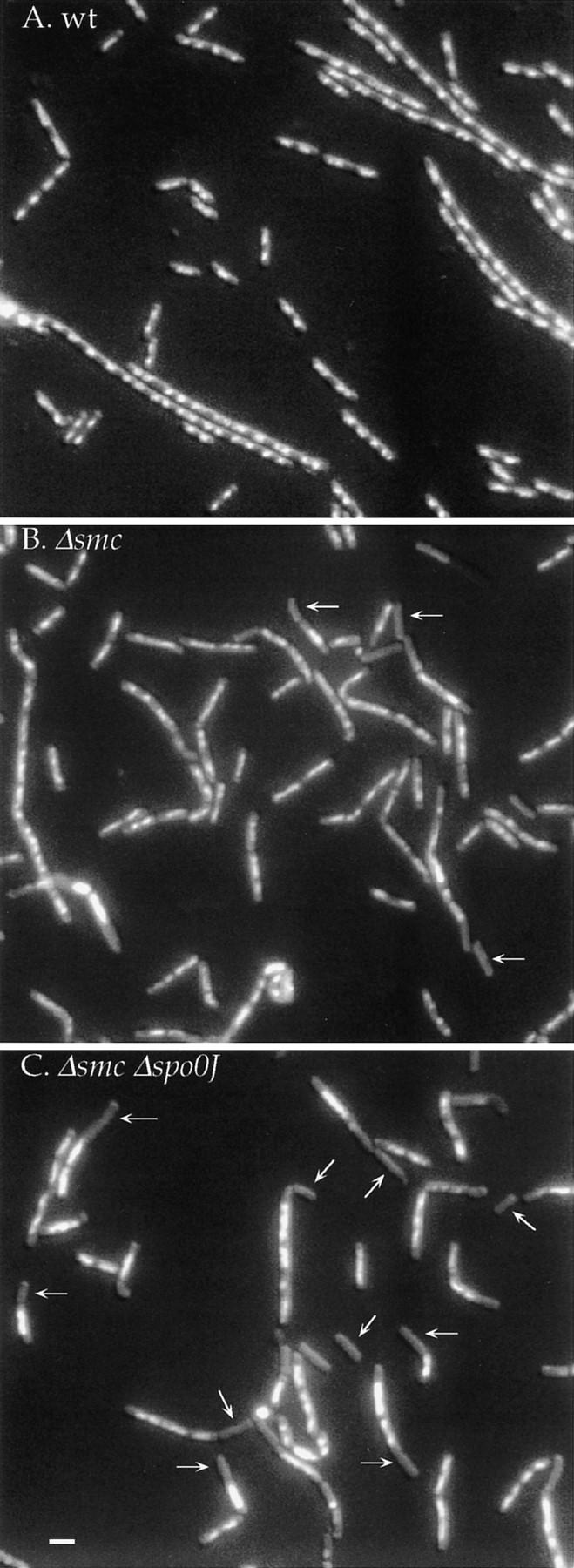
smc mutants produce anculeate cells and have aberrant nucleoids. Cells were grown for several generations in defined minimal medium at 30°C, and samples were taken during exponential growth, fixed, and stained with DAPI. Photographs are combined Nomarski–fluorescence images. Arrows indicate some of the anucleate cells. (A) Wild-type (JH642); (B) Δsmc (RB35); (C) Δsmc Δspo0J (RB41). Note that the nucleoid bodies in many of the cells from the smc and smc spo0J mutants are irregular and some cells appear to have increased DNA content. Scale bar, ∼1 μm.
In addition to the defect in chromosome partitioning, the smc mutant had a defect in nucleoid structure. The normally compact, condensed, regular nucleoid bodies seen in fixed wild-type cells (Fig. 1A) often appeared decondensed, stretched, and/or elongated in the smc mutant (Fig. 1B). These results indicate that Smc of B. subtilis has profound effects on both chromosome structure and partitioning. These effects were more exaggerated in LB medium at nonpermissive temperatures. After 4 hr at 45°C the mutant had a much higher fraction of anucleate cells and many cells appeared to have incomplete nucleoids. In the cells with DNA, the nucleoid structures were highly irregular. We suspect that the temperature-sensitive–lethal phenotype of the smc mutant in rich medium is due to these effects on the nucleoid.
Similar results on the growth, partitioning, and condensation phenotypes of a B. subtilis smc mutant have been obtained independently (S. Moriya, E. Tsujikawa, A.K.M. Hassen, K. Asai, T. Kodama, and N. Ogasawara, pers. comm.). The smc phenotypes are remarkably similar to those caused by mukB mutations in E. coli (Niki et al. 1991; Hu et al. 1996). MukB and Smc proteins have some similarities; both have an amino-terminal domain containing a nucleotide binding motif and a central domain containing two large coiled–coil regions. The major difference is in the carboxy-terminal domain where the signature DA-box motif found in Smc proteins is not found in MukB. These similarities in phenotype and domains indicate that Smc and MukB have similar functions.
We suspect that the primary defect in the smc mutant is in chromosome condensation (folding) and that the other phenotypes (growth, sporulation, partitioning) are secondary. It is formally possible that the defects in partitioning and nucleoid structure result from a defect in DNA replication. However, we think that this is unlikely. In addition to the anucleate cells produced by the smc mutant, we observe some mutant cells with increased DNA content (Fig. 1B). Furthermore, the DNA replication mutants that we have examined (dnaB, dnaI, dnaA) do not cause such dramatic defects in nucleoid structure (K.P. Lemon, P.A. Levin, and A.D. Grossman, unpubl.). If there are alterations in DNA replication in the smc mutant, we suspect that they would be caused by the defects in chromosome condensation.
Spo0J localization in the smc null mutant
The chromosome partitioning protein Spo0J localizes in discrete foci near the poles of the nucleoid body, as visualized by immunofluorescence microscopy with anti-Spo0J antibodies or by use of a Spo0J–GFP fusion (Glaser et al. 1997; Lin et al. 1997). Each focus represents a large nucleoprotein complex of Spo0J bound to multiple sites. There are at least eight Spo0J binding sites, all in the origin proximal 20% of the chromosome (Lin and Grossman 1998). The two most distal sites are ∼775 kb apart, and there is a cluster of five sites that extend over ∼115 kb that are likely to be the major sites in vivo (Lin and Grossman 1998). Assembly of the Spo0J–DNA complex most likely requires considerable bending and compaction of the DNA.
We used a Spo0J–GFP fusion to monitor assembly and location of the Spo0J foci (Fig. 2A–D). Each visible focus was also an indication of the location of the origin region (Glaser et al. 1997; Lewis and Errington 1997; Lin et al. 1997; Lin and Grossman 1998). In wild-type cells grown at 30°C in defined minimal glucose medium, ∼70% of the cells had two distinct foci of Spo0J–GFP (Table 2), and ∼90% of these had one focus at each pole of the nucleoid body (Fig. 2A). These are cells with partly replicated chromosomes that have separated the origin regions to opposite ends of the nucleoid (Lin et al. 1997). Approximately 20% of the cells had either three or four foci of Spo0J–GFP (Table 2). These cells were farther along in the division cycle and often had two apparently separated nucleoids. These results are similar to those described previously (Glaser et al. 1997; Lin et al. 1997).
Figure 2.
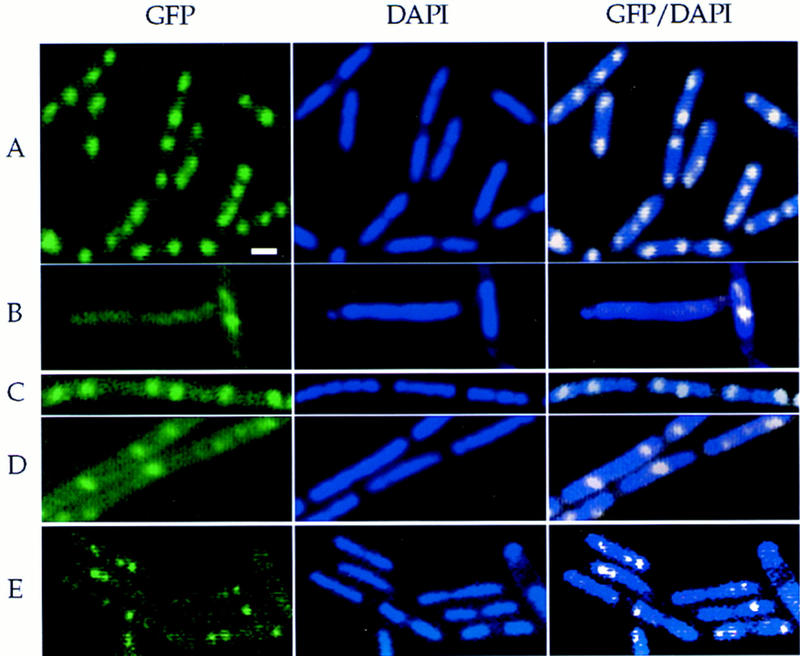
Spo0J–GFP and Smc–GFP localization in live cells. Cells were grown in defined minimal medium at 30°C and samples taken during exponential growth. Three panels are shown for each field of cells: endogenous fluorescence from the GFP fusion (left); nucleoids stained with DAPI (middle); and overlays of the GFP and DAPI images (right). In the DAPI/GFP overlay, the color of the GFP signal was changed to white to better contrast with the DAPI image. Scale bar, ∼1 μm. (A) Spo0J–GFP in wild-type cells (PL657); (B–D) Spo0J–GFP in smc mutant cells (RB40). (B) Example of a cell with no discrete focus of Spo0J–GFP; (C) two nucleoids (left and middle), each with two foci of Spo0J–GFP, one polar and one medial; (D) example of a nucleoid (center of panel) with a single medial spot of Spo0J–GFP; (E) localization of Smc–GFP. The fluorescence signal from Smc–GFP was much more faint than that from Spo0J–GFP, and discrete foci were visible on ∼50% of the nucleoids.
Table 2.
Spo0J–GFP foci in smc mutant and wild-type cells
| Spo0J–GFP foci/cell
|
Percentage of cells with indicated number of Spo0J–Gfp focia
|
|||||
|---|---|---|---|---|---|---|
| 0
|
1
|
2
|
3
|
4
|
⩾5
|
|
| Wild type | <0.3% (0) | 7% (24) | 71% (242) | 12% (40) | 8% (26) | 2% (6) |
| Δsmc | 11% (45) | 25% (104) | 34% (141) | 20% (83) | 8.6% (36) | 2.6% (11) |
A total of 338 wild-type cells (PL657) and 420 Δsmc mutant cells (RB40) were analyzed for the number and localization of Spo0J–GFP foci. The percentage of cells with a given number of foci is indicated with the total numbers in parentheses. Due to rounding, the percentages do not always add up to 100.
The smc mutant was defective in assembly of the large Spo0J–DNA complexes. A much greater fraction of mutant cells had no visible Spo0J–GFP foci (Fig. 2B)—∼10% compared to ≤0.3% in wild type (Table 2). Only those cells with a nucleoid were counted. Furthermore, the fraction of cells with one focus of Spo0J–GFP was ∼25% in the mutant compared to only 7% in the wild type. The increase in cells with zero and one focus was at the expense of those with two foci of Spo0J–GFP; only ∼35% of the mutant cells had two discrete foci, compared to ∼70% of wild type (Table 2). These results indicate that proper assembly and function of Spo0J is partly dependent on Smc, and we postulate that this effect on Spo0J assembly is due to defects in chromosome condensation and folding in the smc mutant.
The position of Spo0J–GFP on the nucleoid appeared to be altered in many of the smc mutant cells (Fig. 2B–D). In wild-type cells with two foci, ∼90% had a Spo0J–GFP focus near each pole of the nucleoid (Fig. 2A), consistent with previous findings (Glaser et al. 1997; Lin et al. 1997). In contrast, in smc mutant cells with two foci, ∼57% had a Spo0J–GFP focus near each pole of the nucleoid and ∼40% had one focus near a pole and the other near the middle of the nucleoid (Fig. 2C). These results might indicate that Spo0J, and hence oriC, is mislocalized in the smc mutant.
Alternatively, the apparent mislocalization might be a consequence of the decrease in the number of Spo0J foci. For example, we suspect that some of the mutant cells with two foci may contain four separated origins. In this case, based on that seen in wild-type cells, the expected pattern of localization of four foci is two polar and two medial (Fig. 2A) (Lin et al. 1997). If, in the smc mutant, one or two of the foci did not assemble, then the relative position of the remaining foci with respect to the nucleoid would appear altered.
The straightforward prediction from this interpretation would be that the fraction of smc mutant cells with four foci of Spo0J should be reduced. This does not appear to be the case (Table 2); 8.6% of the mutant cells have four foci compared to 8% of the wild type. However, for each anucleate cell produced in the mutant, there should be a cell with an extra chromosome (assuming the chromosome is not degraded). Several of the mutant cells appear to have increased DNA content (Fig. 1B,C). Therefore, we expect that the smc mutant should actually have extra cells with four or more Spo0J foci. As there is a defect in assembly of the foci, the fraction of mutant cells with ⩾4 foci is reduced and appears similar to that of wild type. There is an increase in the fraction of mutant cells with three foci, consistent with this interpretation.
Clearly, Spo0J localization is disrupted in the smc mutant. Our data do not permit us to conclude whether this is due to mislocalization of the oriC region and/or defects in assembly of Spo0J foci. If localization of the assembled foci is relatively normal, then the implication is that Smc is not directly involved in origin movement.
An smc spo0J double mutant has a synthetic phenotype
Because both smc and spo0J mutants have defects in chromosome partitioning, we tested the phenotype of a double mutant. The smc spo0J double mutant grew much more slowly than either single mutant (Table 1). The spo0J single mutant has a generation time similar to that of wild type (∼85 min at 30°C in defined minimal glucose medium), and the smc single mutant doubles in ∼125 min. In contrast, the double mutant has a generation time of ∼180 min. The smc spo0J double mutant also had a more severe partitioning defect than either single mutant; ∼25% of the cells were anucleate (Table 1; Fig. 1C). In addition, many more of the cells of the double mutant had aberrant nucleoid structures. Together, these results suggest that Smc and Spo0J functions are partially redundant and that both proteins contribute to nucleoid organization.
Subcellular localization of Smc
We used an Smc–GFP fusion protein to determine the subcellular localization of Smc in live cells. The smc–gfp fusion was integrated in single copy in the B. subtilis chromosome, and the fusion protein was functional (Materials and Methods). Smc–GFP appeared in discrete foci, apparently associated with the nucleoid. There were typically one to two foci per nucleoid body. Approximately 70% of the cells with Smc–GFP foci had at least one focus of Smc–GFP located near a pole of the nucleoid (Fig. 2E), and ∼30% of the cells had a focus associated with the nucleoid but not obviously near a pole. If the foci of Smc represent the active protein, as opposed to perhaps a reservoir of inactive protein, then the implication is that Smc functions in a discrete location, possibly a site for catalyzing chromosome condensation.
Models for Smc function in prokaryotes
In eukaryotes, Smc proteins function in chromosome condensation, pairing, and segregation (Hirano et al. 1995; Koshland and Strunnikov 1996; Heck 1997). The effects of an smc null mutation in B. subtilis, the accumulation of anucleate cells, disruption of nucleoid structure, misassembly of Spo0J foci, and synthetic phenotype with spo0J, demonstrate the importance of Smc in chromosome dynamics in at least one prokaryote. We suspect that Smc will have similar functions in other prokaryotes.
We consider two possible models, not mutually exclusive, for the function of Smc in chromosome dynamics. Smc, like its counterparts in eukaryotes, may be involved in chromosome condensation, contributing to the compaction of the nucleoid mass. Alternatively, Smc may be a motor protein involved in movement of chromosomes, or parts of chromosomes, during partitioning. It seems more likely that the multiple phenotypes of the smc mutant (anucleate cells, aberrant nucleoids, decreased foci of Spo0J) are due to a primary defect in chromosome condensation and folding rather than a primary defect in oriC movement. Therefore, we favor the condensation model and suspect that other, as yet unidentified, proteins constitute the actual partitioning machinery.
Combining spo0J and smc mutations causes a partitioning and growth defect more severe than that caused by either mutation alone, probably because both proteins affect chromosome folding and organization. We propose that Spo0J, bound to multiple sites, forms a complex that helps organize the oriC region of the chromosome. Upon replication, sister origin regions pair in a Spo0J-mediated process. Smc facilitates assembly of the Spo0J complex and pairing via its role in chromosome condensation. In a spo0J null mutant, in which pairing of the origins is weakened or disrupted, the condensed state of the chromosome could keep the sister origins in relatively close proximity, allowing them to be presented to the segregation machinery. In the absence of Smc, pairing of the origins, mediated by Spo0J, could provide a small but significant level of structure to the oriC region. When both functions are lost, structure and orientation of the origin region are further disrupted, causing a severe defect in cell growth and chromosome partitioning.
Based on database searches, many of the bacterial species that have an Smc protein appear to have a Spo0J homolog. The use of genetic and cell biological approaches in B. subtilis, combined with biochemical analysis, should continue to provide insights into the function of these highly conserved proteins.
Materials and methods
Plasmids and strains
The smc gene was amplified by PCR (sequences of all primers are available on request) and cloned into the vector pGEMcat (Youngman et al. 1989) to give pDL54. pDL59 contains the Δsmc::kan mutation that was introduced into the chromosome by transformation. The kanamycin-resistance cassette was cloned between the SalI and SnaBI sites in smc in pDL54, deleting from bp 723 to 2714 of the 3561-bp smc gene to give pDL59.
pDL62 contains the 3′ end of smc fused in-frame to gfp in the vector pGEMcat. Briefly, a fragment of smc was amplified such that the stop codon was changed to contain an XhoI site, and the fragment was used to replace the fragment of spo0J in the spo0J–gfp fusion in pDL50B (Lin et al. 1997). Integration of pDL62 into the B. subtilis chromosome by single crossover at smc results in fusion of full length smc to gfp. The gfp allele contained the S65T and V163A mutations (Kahana and Silver 1996).
Several additional plasmids were used to disrupt smc by plasmid integration. Each plasmid contains a fragment internal to smc cloned into the vector pGEMcat. pRB7 contains a SalI – PstI fragment (bp 723-1274); pRB9 contains a PstI–BsmI fragment (bp 1274–1790), and pDL55 contains a PstI–KpnI fragment (bp 1274–1932) of smc. Integration of each of these into the chromosome caused a null phenotype (data not shown).
To confirm that the observed phenotypes were due to disruption of smc and not to possible polar effects on the downstream srb gene (Oguro et al. 1996), we used a plasmid, pDL53, that contains a fragment extending from bp 1274 of smc to bp 58 of srb. Integration of this plasmid into the chromosome should affect expression of srb exactly the same as integration of pRB9 and pDL55 but should not disrupt smc. Integration of pDL53 had no noticeable effect on chromosome partitioning and did not cause the conditional lethal phenotype observed in the smc mutants, indicating that the phenotypes of the smc mutants were not due to a polar effect on srb.
All B. subtilis strains are derived from JH642 (aka AG174; trp phe) and were constructed by standard methods (Harwood and Cutting 1990). The spo0J–gfp fusion was subcloned from pDL50B (Lin et al. 1997) into the spectinomycin-resistant vector pUS19 (Benson and Haldenwang 1993) to give pPL52. pPL52 was integrated into the B. subtilis chromosome by a single recombination at spo0J to generate the spo0J–gfp fusion. B. subtilis strains include AG1468, Δspo0J::spc (Ireton et al. 1994); RB35, Δsmc::kan; PL657, spo0J–gfp spc; RB40, Δsmc::kan spo0J–gfp spc; RB41, Δsmc::kan Δspo0J::spc; DCL305, smc–gfp cat. Cells containing the smc–gfp fusion appeared to have a wild-type phenotype at 30°C, indicating that the Smc–GFP fusion protein is functional.
Microscopy
Nucleoid morphology and distribution were analyzed by combined Nomarski/fluorescence microscopy. Cells were grown at 30°C in S750 minimal medium with glucose (1%), glutamate (0.1%), and required amino acids, essentially as described (Vasantha and Freese 1980; Jaacks et al. 1989). Cells (10 μl) from exponential growth were added to a well of a microscope slide that had been treated with poly-l-lysine. After 5 min, the fluid was removed and the slide allowed to dry. Methanol was added for 5 min to fix the cells. Slides were washed five times in tap water and allowed to dry. DAPI (2 μg/ml) was added to the slides to visualize the DNA. Microscopy was performed with a Zeiss Axioplan 2. Images were captured with a cooled CCD camera (Optronics Engineering) and a CG-7 framegrabber (Scion) using Scion Image 1.62 software, and merged in Adobe Photoshop 3.0.
Spo0J–GFP and Smc–GFP were visualized in live cells, essentially as described (Glaser et al. 1997). Briefly, 2 μl of 0.5% agarose was applied to a well of a slide and allowed to cool for ∼1 min. Ten microliters of culture was added to the well. After 5 min, excess liquid was removed and DAPI (3 μl, 2 μg/ml) was added to stain DNA. Fluorescence microscopy and image processing were as described above.
Acknowledgments
We thank Dr. S. Moriya for communicating results prior to publication, and S. Sanders for advice and use of her fluorescence microscope. We are grateful to S. Bell for comments on the manuscript and members of our laboratory, particularly K.P. Lemon and P.A. Levin, for useful discussions and comments on the manuscript. R.A.B. was supported, in part, by a postdoctoral fellowship from the National Institutes of Health. D.C.H.L. was supported, in part, by a predoctoral fellowship from the National Science Foundation. This work was supported, in part, by U.S. Public Health Service grant (GM41934) to A.D.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL adg@mit.edu; FAX (617) 253-2643.
References
- Benson AK, Haldenwang WG. Regulation of σB levels and activity in Bacillus subtilis. J Bacteriol. 1993;175:2347–2356. doi: 10.1128/jb.175.8.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang P-T, Albertson DG, Meyer BJ. DPY-27: A chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–474. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- Chuang P-T, Lieb JD, Meyer BJ. Sex-specific assembly of a dosage compensation complex on the nematode X chromosome. Science. 1996;274:1736–1739. doi: 10.1126/science.274.5293.1736. [DOI] [PubMed] [Google Scholar]
- Glaser P, Sharpe ME, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes & Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- Gordon GS, Sitnikov D, Webb CD, Teleman A, Straight A, Losick R, Murray AW, Wright A. Chromosome and low copy plasmid segregation in E. coli: Visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular biological methods for Bacillus. Chichester, UK: John Wiley & Sons; 1990. [Google Scholar]
- Heck M. Condensins, cohesins, and chromosome architecture: How to make and break a mitotic chromosome. Cell. 1997;91:5–8. doi: 10.1016/s0092-8674(01)80002-7. [DOI] [PubMed] [Google Scholar]
- Hiraga S. Chromosome and plasmid partition in Escherichia coli. Annu Rev Biochem. 1992;61:283–306. doi: 10.1146/annurev.bi.61.070192.001435. [DOI] [PubMed] [Google Scholar]
- Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffe A. Chromosome partitioning in Escherichia coli: Novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ, Swedlow JR. The SMC family: From chromosome condensation to dosage compensation. Curr Opin Cell Biol. 1995;7:329–336. doi: 10.1016/0955-0674(95)80087-5. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Hu KH, Liu E, Dean K, Gingras M, DeGraff W, Trun NJ. Overproduction of three genes leads to camphor resistance and chromosome condensation in Escherichia coli. Genetics. 1996;143:1521–1532. doi: 10.1093/genetics/143.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Gunther IV NW, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaacks KJ, Healy J, Losick R, Grossman AD. Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana JA, Silver PA. Supplement 34: Use of the A. victoria green fluorescent protein to study protein dynamics in vivo. In: Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, NY: John Wiley & Sons; 1996. pp. 9.7.22–9.7.28. [Google Scholar]
- Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: A biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Lewis PJ, Errington J. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol Microbiol. 1997;25:945–954. doi: 10.1111/j.1365-2958.1997.mmi530.x. [DOI] [PubMed] [Google Scholar]
- Lin DC-H, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;32:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Lin DC-H, Levin PA, Grossman AD. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc Natl Acad Sci. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Gober JW. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- Niki H, Hiraga S. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K, Austin SJ. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- Oguro A, Kakeshita H, Takamatsu H, Nakamura K, Yamane K. The effect of Srb, a homologue of the mammalian SRP receptor α-subunit, on Bacillus subtilis growth and protein translocation. Gene. 1996;172:17–24. doi: 10.1016/0378-1119(96)00181-3. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yanagida M. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature. 1997;388:798–801. doi: 10.1038/42062. [DOI] [PubMed] [Google Scholar]
- Vasantha N, Freese E. Enzyme changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J Bacteriol. 1980;144:1119–1125. doi: 10.1128/jb.144.3.1119-1125.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake RG, Errington J. Chromosome partitioning in bacteria. Annu Rev Genet. 1995;29:41–67. doi: 10.1146/annurev.ge.29.120195.000353. [DOI] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC-H, Grossman AD, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Ogura T, Niki H, Hiraga S. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol & Gen Genet. 1996;250:241–251. doi: 10.1007/BF02174381. [DOI] [PubMed] [Google Scholar]
- Youngman P, Poth H, Green B, York K, Olmedo G, Smith K. Methods for genetic manipulation, cloning, and functional analysis of sporulation genes in Bacillus subtilis. In: Smith I, Slepecky R, Setlow P, editors. Regulation of procaryotic development. Washington, D.C.: American Society for Microbiology; 1989. pp. 65–87. [Google Scholar]


