Abstract
Here we report an in vitro model system for studying the molecular and cellular mechanisms that underlie the neurodegenerative disease amyotrophic lateral sclerosis (ALS). Embryonic stem cells (ESCs) derived from mice carrying normal or mutant transgenic alleles of the human SOD1 gene were used to generate motor neurons by in vitro differentiation. These motor neurons could be maintained in long-term coculture either with additional cells that arose during differentiation or with primary glial cells. Motor neurons carrying either the nonpathological human SOD1 transgene or the mutant SOD1G93A allele showed neurodegenerative properties when cocultured with SOD1G93A glial cells. Thus, our studies demonstrate that glial cells carrying a human SOD1G93A mutation have a direct, non–cell autonomous effect on motor neuron survival. More generally, our results show that ESC-based models of disease provide a powerful tool for studying the mechanisms of neural degeneration. These phenotypes displayed in culture could provide cell-based assays for the identification of new ALS drugs.
ALS is a progressive neurodegenerative disease characterized by the loss of upper and lower motor neurons, culminating in muscle wasting and death from respiratory failure1. The majority of ALS cases are apparently sporadic, with 90% of patients presenting disease symptoms without a family history of disease. The remaining 10% of patients are diagnosed with familial ALS1–3, and approximately 25% of these familial cases are caused by dominant mutations in the gene encoding super oxide dismutase (SOD1)4. Identification of pathogenic alleles of SOD1 has led to the generation of transgenic mouse and rat models for the study of ALS5–8. Overproduction of pathogenic human SOD1 protein in these models leads to late-onset, progressive neuro-degenerative disease5,7,8. Studies with these animals have led to the identification of intrinsic pathogenic characteristics of ALS motor neurons, including the formation of protein aggregates, cytoskeletal abnormalities, proteosome dysfunction and increased sensitivity to cell death signals1,9.
In contrast, studies of chimeric mice suggest that non–cell autonomous processes contribute to motor neuron death in ALS10. In animals bearing both wild-type cells and cells harboring the SOD1G93A transgene, wild-type neurons surrounded by transgenic non-neuronal cells acquired phenotypes characteristic of ALS10. Conversely, transgenic neurons associated with wild-type cells were increasingly spared. Unfortunately, as a result of the complex cellular milieu of both the spinal cord and the muscle, it was not possible in these animal studies to identify the specific cells involved in pathological interactions with motor neurons. Conditional mutagenesis experiments also suggest that the SOD1 protein can have both cell autonomous and non–cell autonomous effects in the disease. When multicopy SOD1 transgenes were specifically removed from motor neurons and microglial cells, an increase in animal lifespan was observed11. However, these experiments did not directly address the effect of the SOD1G93A allele in microglia on motor neuron survival, as mortality was used as the primary endpoint of the study.
Here we report the establishment of an in vitro model system for studying ALS in which both the cell types and factors that directly influence motor neuron survival can be investigated. This system is based on the differentiation of ESCs derived from mice bearing the human SOD1G93A transgene. Transgenic ESCs were differentiated into motor neurons in large numbers12 and cocultured either with ESC-derived cells that were also generated during the differentiation of motor neurons, or with primary mouse glial cells from mutant and wild-type SOD1 mice. Notably, motor neurons in these cultures showed pathogenic properties observed in ALS patients and in transgenic animals. In addition, coculture experiments with glial cells from wild-type and mutant SOD1 mice identified mutant glia as a source of adverse non–cell autonomous effects on motor neuron survival.
RESULTS
Derivation of ESC lines from SOD1G93A ALS mice
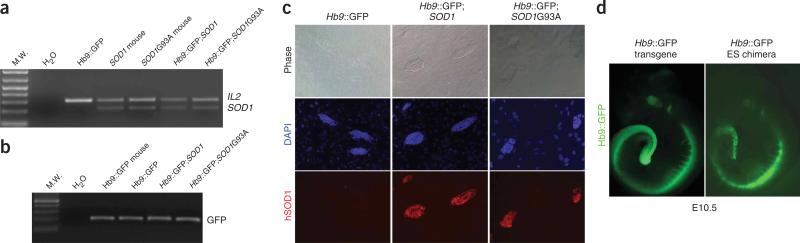
ESC lines were derived from embryos generated by crossing hemizygous mice carrying either the pathogenic (mutant) SOD1G93A transgene or the non-pathogenic (wild-type) SOD1 transgene5 with hemizygous mice carrying a transgenic reporter gene in which expression of the green fluorescent protein (GFP) is driven by promoter elements from the Hb9 gene (Hb9::GFP)12. The Hb9 gene encodes a homeodomain transcription factor that is expressed in postmitotic motor neurons13,14. Thus, the Hb9::GFP transgene provides both a marker for the differentiation of ESCs into motor neurons12 and a means of identifying motor neurons from other cell types. Blastocyst-stage embryos were retrieved from these mouse crosses and used to derive ESC lines that were genotyped by PCR (Fig. 1). These analyses identified ESC lines that carried only the Hb9::GFP transgene (Hb9GFP), lines that carried both Hb9::GFP and the wild-type SOD1 transgene (SOD1), and a line that carried both Hb9::GFP and the mutant SOD1G93A transgene (SOD1G93A).
Figure 1.
Derivation of Hb9::GFP;SOD1 mouse ESC lines. (a,b) PCR for human SOD1 and the internal control Il2 (a), and for GFP in Hb9::GFP, Hb9::GFP;SOD1 and Hb9::GFP;SOD1G93A ESC lines (b). (c) Expression of human SOD1 protein in Hb9::GFP, Hb9::GFP;SOD1 and Hb9::GFP;SOD1G93A ESC lines. (d) A control Hb9::GFP transgenic embryo and an E10.5 chimera generated using the Hb9::GFP ESC line.
To determine whether the expression of the Hb9::GFP transgene in these ESCs accurately reflects that of the endogenous Hb9 gene, we examined GFP fluorescence in the undifferentiated ESCs and in chimeras created by injecting these cells into nontransgenic blastocysts. Each of the undifferentiated cell lines lacked obvious GFP expression (Fig. 2). However, in embryonic day 10.5 (E10.5) chimeras created with these cells, specific GFP expression was observed in the developing eye, hindbrain and spinal cord, where Hb9 is known to be expressed (Fig. 1d)12,14. Immunostaining with antibodies that preferentially recognize the human SOD1 protein confirmed the PCR genotyping of the cell lines and showed that both the SOD1 and SOD1G93A transgenes are expressed in the undifferentiated ESCs (Fig. 1c).
Figure 2.
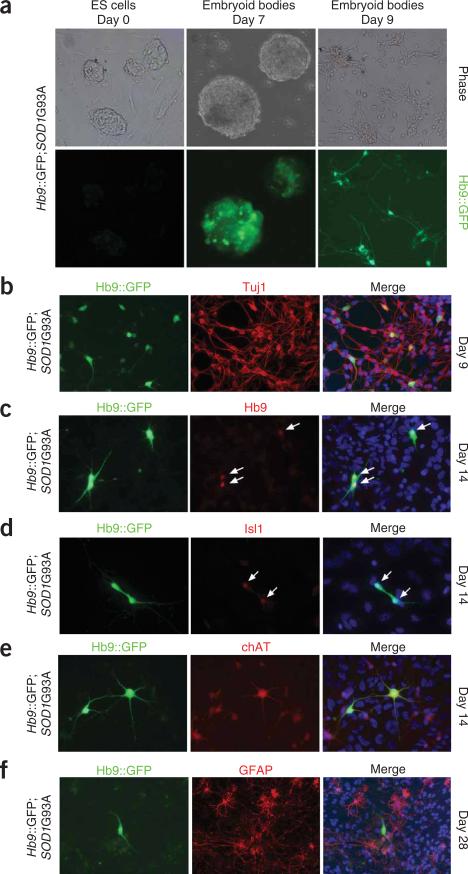
Differentiation of the SOD1G93A mouse ESC lines into motor neurons. (a) GFP expression of the ESC during different phases of the differentiation into motor neurons. (b–e) Expression of neuronal markers Tuj1 (b), Hb9 (c), Isl1 (d) and choline acetyltransferase (ChAT) (e) in motor neurons derived from SOD1G93A ESC lines at 9 and 14 d after the beginning of the differentiation process. (f) Presence of GFAP-positive cells surrounding a GFP motor neuron after 28 d. The white arrows indicate the nuclei of motor neurons immunoreactive for Hb9 or Isl1 proteins.
Motor neurons generated by differentiation of ESCs
To determine whether pathogenic properties associated with ALS can be recapitulated in vitro, we generated motor neurons by differentiating the transgenic ESC lines as previously described12. Briefly, ESC were dissociated into a single-cell suspension, allowed to spontaneously aggregate into embryoid bodies over 48 h and then treated with retinoic acid (RAc) and soluble sonic hedgehog (Shh) protein for 5 d (ref. 12).
We found that the SOD1G93A genotype did not interfere with the initial specification or differentiation of motor neurons, as no significant qualitative or quantitative differences were observed in the differentiation of the three cell lines. GFP expression in embryoid bodies derived from the different cell lines, including SOD1G93A, first appeared 5 d after treatment with Shh and RAc (Fig. 2a). Two days later, when embryoid bodies were dissociated with papain and plated, GFP-positive cells with an obvious neuronal morphology could be observed (Fig. 2a). We used fluorescence-activated cell sorting (FACS) to determine the percentage of differentiating ESCs that expressed GFP and found no statistically significant differences between the cell lines (Hb9GFP 33% ± 4%, SOD1 33% ± 9%, SOD1G93A 26% ± 6%) (P > 0.05, Supplementary Fig. 1 online).
To confirm that cells expressing GFP in embryoid bodies had differentiated into bona fide motor neurons, we dissociated the embryoid bodies and performed immunostaining with antibodies specific for proteins known to be expressed in motor neurons (Fig. 2b–f). As was previously observed with this differentiation protocol12, we found that GFP-positive cells derived from the SOD1G93A cell line expressed a neuronal form of tubulin (Tuj1, Fig. 2b), the transcription factors Hb9 and Isl1/2 (Fig. 2c,d and Supplementary Fig. 2 online) and the enzymatic machinery required to generate acetylcholine (Chat, Fig. 2e).
Survival of SOD1G93A motor neurons in culture
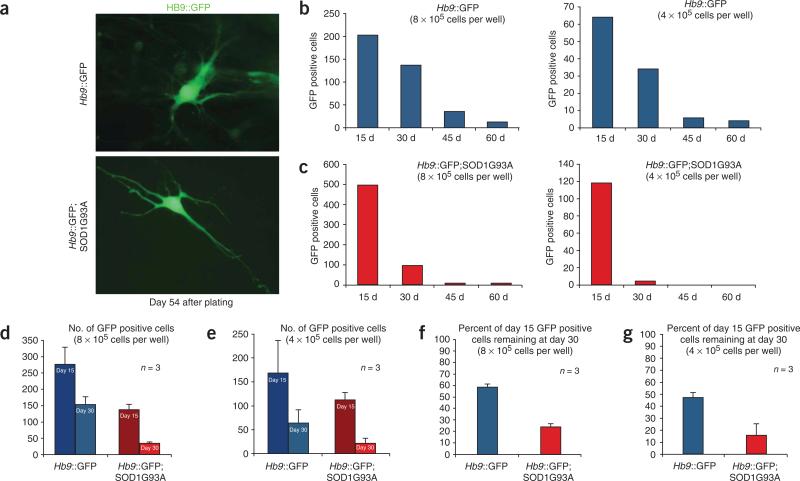
ALS is a late-onset neurodegenerative disease, and mice carrying the human SOD1G93A transgene develop symptoms as a consequence of motor neuron loss after several weeks. Therefore, it seemed possible that motor neurons derived from ESCs might show neurodegenerative properties only after they have been maintained in culture for a prolonged length of time. To determine the period of time that ESC-derived motor neurons can survive in culture, we dissociated Hb9GFP and SOD1G93A embryoid bodies 7 d after differentiation and plated the resulting mixture of GFP-positive and negative cells at two different densities in the presence of neurotrophic factors12 (Fig. 3). We observed that the number of GFP-positive cells decreased precipitously during the first 2 weeks after plating and continued to decrease over the following weeks. However, GFP-positive cells could still be detected in both Hb9GFP- and SOD1G93A-derived cultures 54 d after plating (Fig. 3a). Although GFP-positive cells were present in both cultures at later time points, the number of cells expressing GFP decreased more rapidly in the SOD1G93A (Fig. 3c) cultures than in Hb9GFP control cultures (Fig. 3b).
Figure 3.
Effect of SOD1 genotype on motor neuron survival. (a) GFP-positive motor neurons derived from SOD1G93A and Hb9::GFP cell lines 61 d after differentiation. (b,c) Number of GFP-positive cells derived from Hb9::GFP (b) and SOD1G93A (c) ESC lines present 15, 30, 45 and 60 d after dissociation of embryoid bodies plated at two different concentrations (8 × 105 and 4 × 105 cells per well). (d,e) Number of GFP-positive motor neurons derived from Hb9::GFP and SOD1G93A 15 and 30 d after embryoid bodies dissociation plated at concentrations of 8 × 105 (d) and 4 × 105 (e) per well. (f,g) Same experiments analyzed as percentage of GFP-positive motor neurons derived from Hb9::GFP and SOD1G93A cell lines present at 15 d still remaining at 30 d after plating at 8 × 105 (f) and 4 × 105 (g) cells per well. Error bars represent s.d.
To confirm the effect of the SOD1G93A genotype on the number of GFP-positive cells, we differentiated both the SOD1G93A and Hb9GFP ESCs into motor neurons, plated equal numbers of cells at two different concentrations (8 × 105 (n = 3) and 4 × 105 (n = 3) cells per well) and counted the number of GFP-positive neurons in the cultures at 2 and 4 weeks (Fig. 3d–g). Under both plating conditions, significantly fewer GFP-positive cells were observed in the SOD1G93A cultures at both 2 and 4 weeks (P < 0.005, Fig. 3d–g).
ESC-derived motor neurons show ALS pathology
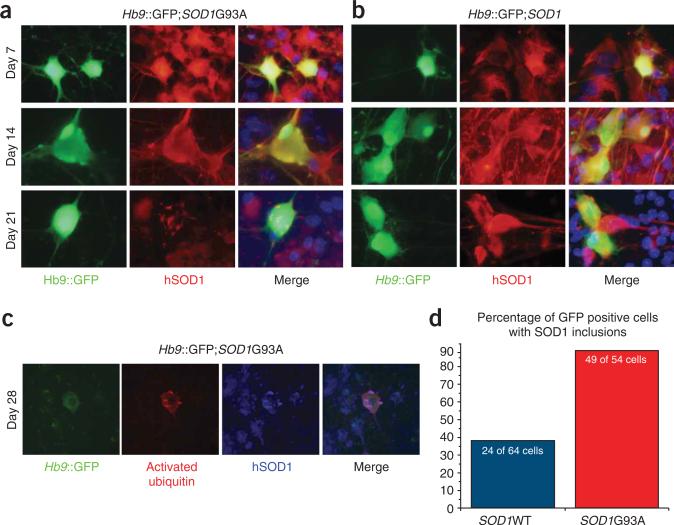
Hb9GFP and SOD1G93A ESCs differentiated into motor neurons with similar efficiencies, but the cultures showed differences in the number of GFP-positive cells over time. Thus, pathological processes may have underlain the preferential loss of GFP-positive cells in the SOD1G93A cultures. To investigate these processes and to determine whether they mirror events that occur during the progression of ALS, we examined motor neurons in culture for the presence of histopathological hallmarks of the disease. Motor neurons in ALS patients and transgenic mice carrying the SOD1G93A allele accumulate protein inclusions that are recognized by antibodies specific for the SOD1 protein1,7. We therefore determined whether aggregation of the mutant SOD1 protein accompanies the loss of GFP-positive motor neurons in SOD1G93A cultures by staining with antibodies specific for the human SOD1 protein at 7, 14 and 21 d after dissociation of embryoid bodies (Fig. 4). At 7 and 14 d after dissociation, both the wild-type SOD1 protein and the mutant SOD1G93A protein were localized broadly and evenly in the cytoplasm of GFP-positive motor neurons (Fig. 4a,b). However, at 14 d punctate structures stained with the SOD1 antibody could be observed in a small proportion of motor neurons expressing the SOD1G93A protein (Fig. 4a).
Figure 4.
Intracellular aggregation of SOD1 protein in cultured motor neurons. (a,b) Expression of human SOD1 protein in motor neurons derived from SOD1G93A (a) and SOD1 (b) cell lines at 7, 14 and 21 d after embryoid body dissociation. (c) Co-expression of ubiquitin and SOD1 protein in cells derived from the SOD1G93A cell line, 28 d after differentiation. (d) Percentage of GFP-positive motor neurons with SOD1 inclusions present after 21 d in culture.
When cultures were examined 21 d after dissociation, a shift in protein localization was observed in the SOD1G93A motor neurons (Fig. 4a,d). In 43 of 54 (79.75 ± 7.75%) of the motor neurons selected at random for analysis by GFP expression, the SOD1G93A protein localized to inclusions in the perinuclear space, the cell body and also the neural processes (Fig. 4a,d and Supplementary Fig. 3 online). When control motor neurons expressing wild-type SOD1 were examined 21 d after differentiation (Fig. 4b,d), we observed inclusions in a smaller proportion of cells, 23 of 64 (35.93 ± 0.43%). The inclusions in SOD1G93A motor neurons were also significantly larger in area, were significantly longer and had a higher optical density than those in control neurons, suggesting that they contained more SOD1 protein at a higher concentration (P < 0.005, Supplementary Fig. 3).
The levels of ubquitinated proteins are substantially elevated in motor neurons of ALS patients and SOD1G93A animals during neural degeneration7,15–17. Examination of the ESC-derived motor neurons showed an increase in staining with antibodies specific for ubiquitin, relative to other cells in the culture. This staining often colocalized with the SOD1 protein inclusions (Fig. 4c).
The death of motor neurons in ALS patients and transgenic mice carrying mutant SOD1 genes occurs, at least in part, through activation of programmed cell death pathways. It has been proposed that apoptosis in these cells is mediated through the release of cytochrome c and the activation of caspase-3 (refs. 18,19). Examination of the SOD1G93A motor neurons 14 d after dissociation of embryoid bodies showed that some neurons expressed activated caspase-3 and contained diffuse cytoplasmic staining with cytochrome c–specific antibodies (Supplementary Fig. 4 online). Thus, the SOD1G93A motor neurons can initiate cell death pathways in vitro that are similar to those activated in vivo during the course of disease18.
SOD1G93A glial cells adversely effect motor neuron survival
Both autonomous defects in motor neurons and toxic non–cell autonomous interactions with other cell types in the spinal cord have been implicated in ALS pathology9–11,20. Only a subset of cells in embryoid bodies became motor neurons under our differentiation conditions. We therefore considered the possibility that cells within the embryoid bodies might also develop into other cell types that normally associate with neurons in the spinal cord. These additional cells might, as suggested by chimeric experiments, contribute non–cell autonomously to the loss of neurons in SOD1G93A cultures. Glial cells are closely associated with motor neurons and both are derived from a common progenitor in vivo21. We therefore addressed the possibility that glial cells were present in our cultures. To determine whether these cells were also produced by our differentiation protocol, we stained SOD1G93A cultures with antibodies specific for glial fibrillary acidic protein (GFAP)22. Indeed, GFAP-positive cells were found in close association with GFP-positive motor neurons (Fig. 2f). We found that approximately 30% of the cells at 28 d were GFAP-positive (Hb9GFP, 31%, SOD1, 32%, SOD1G93A, 36%). Thus, it seemed possible that the SOD1G93A glial cells in these cultures might be adversely affecting motor neuron survival.
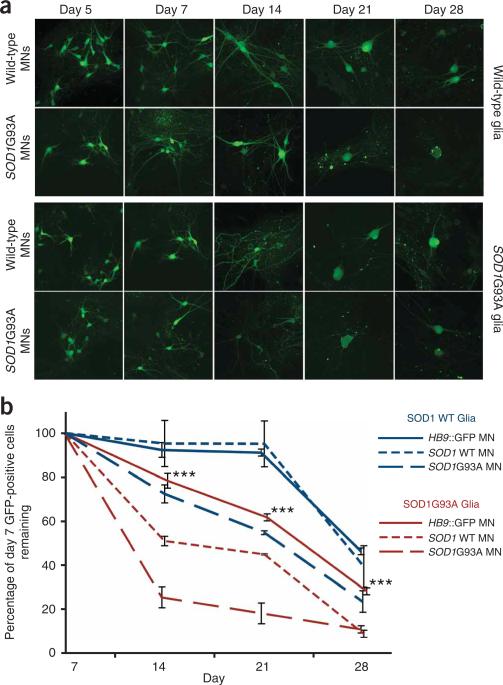
To examine this possibility, we differentiated the three ESC lines (Hb9GFP, SOD1 and SOD1G93A) into motor neurons and plated the neurons on established monolayers of primary glia isolated from the cortex of neonatal mice with differing SOD1 genotypes (wild-type SOD1 and the mutant SOD1G93A)23. During the first 7 d after plating on either glial monolayer, motor neurons of all genotypes increased in size and took on a more mature morphology (Fig. 5a). However, by 14 d there was a 50% decrease in the number of wild-type SOD1– derived motor neurons in cocultures with SOD1G93A glia compared with the same preparation of neurons plated on wild-type SOD1 glia. Similarly, we did not see a significant reduction (P > 0.05) in the number of Hb9GFP motor neurons when plated on SOD1 glia, but we did see a reduction of 30% if the same neurons were cocultured with SOD1G93A glia (Fig. 5b). These data suggest that wild-type SOD1 glia provide a permissive environment for motor neuron growth and differentiation, comparable to those of other well-established in vitro systems for motor neuron culture24–26. In contrast, when we cocultured wild-type motor neurons (SOD1 or Hb9GFP) with glia from SOD1G93A mice, we observed a marked reduction in motor neuron survival (Fig. 5a,b).
Figure 5.
Glial cell genotype directly affects motor neuron survival in culture. (a) GFP- positive motor neurons at 5, 7, 14, 21 and 28 d in culture after embryoid body dissociation. (b) Graph shows percentage of Hb9::GFP– positive cells over time in all the conditions studied. Experiments were done in triplicate and results were normalized to the number of cells found at 7 d in vitro. Error bars represent s.e.m.
We next investigated the effect of the mutant SOD1 transgene in motor neurons in the coculture system. When mutant SOD1G93A motor neurons were plated on wild-type SOD1 glial cells, we observed a 27% decrease in the number of neurons between 7 and 14 d. The loss of motor neurons increased to 75% when the mutant SOD1G93A motor neurons were cultured with mutant SOD1G93A glia (Fig. 5a,b).
Together, these results show that the SOD1G93A genotype in glial cells has a negative effect on motor neuron survival, regardless of the motor neuron genotype. However, a greater negative effect is observed with the SOD1G93A motor neurons. Thus, glial cells have a non–cell autonomous effect on motor neuron survival and mutant motor neurons are more sensitive to the effect.
To determine whether the differing influences of the wild-type SOD1 and the mutant SOD1G93A glial cell cultures could be explained by the presence of different proportions of distinct glial cell types, we characterized the two populations by immunostaining with established glial markers. We did not observe a significant difference (P > 0.05) in the wild-type SOD1 and the mutant SOD1G93A glial populations over time (Supplementary Fig. 5 online).
DISCUSSION
The capacity of ESCs to self renew in culture while retaining their developmental potential allows the production of unlimited numbers of differentiated cells to replenish those lost during disease27,28. An alternative use of ESCs is to provide insights into disease mechanisms29,30. ESCs carrying the genes responsible for a particular disease can be induced to differentiate into the cell types affected in that disease. Studies of these differentiated cells in culture could provide important information regarding the molecular and cellular nature of events leading to pathology.
We have used this approach to develop an in vitro model of amyotrophic lateral sclerosis (ALS). ESC lines were derived from normal mice, and from mice that overexpress the wild-type human SOD1 transgene or the mutant SOD1G93A transgene, the latter of which is responsible for one type of familial ALS. Using previously established methods12, the ESC lines were differentiated into motor neurons in culture. The wild-type SOD1 and the mutant SOD1G93A motor neurons produced high levels of the corresponding human SOD1 proteins, and they both showed properties of bona fide motor neurons. These motor neurons could be maintained in long-term culture, providing the opportunity to detect differences between the mutant SOD1G93A motor neurons and those derived from control cell lines.
A number of changes characteristic of neurodegeneration in ALS were observed in the mutant SOD1G93A motor neurons between 14 and 28 d. First, the intracellular localization of the SOD1G93A protein changed, forming inclusions that increased in size and density. Second, the amount of ubiquitin increased. Third, some motor neurons expressed activated caspase-3 and showed cytoplasmic staining with cytochrome c antibodies. Finally, a significant difference in survival was observed between mutant SOD1G93A motor neurons and the controls. Thus, many of the late-onset pathologies observed in both human ALS and SOD1G93A mice are recapitulated in this in vitro model, including the loss of motor neurons, which is the ultimate cause of symptoms in humans.
Several studies have suggested that cells in the spinal cord may have pathological, non–cell autonomous effects on motor neurons or on the rate of disease progression10,11. However, these studies either were unable to identify the cell types that caused these effects10 or could not determine whether they acted directly to affect motor neuron survival11. We found that cultures of ESC-derived motor neurons contain other cell types, including astroglia, and thus considered the possibility that these ESC-derived cells have a non–cell autonomous effect on motor neuron survival in vitro. We therefore systematically examined the effects of coculturing motor neurons with primary glia from SOD1G93A mice and from mice expressing the wild-type SOD1 protein. We found that mutant SOD1G93A glia reduced the survival of both wild-type and mutant motor neurons. However, the effect was significantly greater on mutant SOD1G93A motor neurons. Therefore, our studies show for the first time that an ALS genotype in glial cells directly and negatively affects the survival of motor neurons, and confirm that there is a non-cell autonomous component to motor neuron degeneration.
Consistent with the results reported here, another study described in this issue has shown that primary astrocyte cultures expressing ALS-associated mutant SOD1 proteins contain diffusible factor(s) that are toxic to primary and ESC-derived motor neurons31. In this study, motor neurons were the only cell types affected by these mutant glial cells and only SOD1G93A glial cells, not muscle cells or fibroblasts, adversely affected motor neuron survival. Although mutant primary neurons showed morphometric alterations, their survival up to 14 d in culture was indistinguishable from that of their wild-type counterparts. In our studies, differences in survival between wild-type SOD1 and mutant SOD1G93A ESC-derived motor neurons were observed at 14 and 28 d in culture. The differences between the two studies may therefore originate in the source (embryo- or ESC-derived) or number of the motor neurons used, and the timeframe of the investigations.
The results reported here and elsewhere in this issue31 provide the basis for detailed mechanistic studies of the interactions between SOD1 mutant motor neurons and glia, and they provide an assay for diffusible factor(s) that are either toxic or beneficial for motor neuron survival. The model system described here may also provide a high-throughput cell-based assay for small molecules that promote survival of mutant SOD1 motor neurons. Finally, these studies validate the use of ESCs carrying disease-causing genes to define disease mechanisms. In this regard, our work suggests that the future development of human ESC lines from patients via somatic cell nuclear transplantation could provide the opportunity to study the nature of sporadic ALS, which affects the majority of ALS patients.
METHODS
Derivation of mouse ESCs
ESC lines were derived from crosses between mice transgenic for Hb9::GFP (Jackson lab, stock number 005029) and mice transgenic for SOD1G93A (Jackson lab, stock number 004435) or SOD1WT (Jackson lab, stock number 002297). Transgenic Hb9::GFP females were injected IP with 7.5 units of pregnant mares’ serum (Calbiochem), followed 46–50 h later with 7.5 units of human chorionic gonadotropin (Calbiochem). After administration of human chorionic gonadotropin, females were mated with SOD1G93A or SOD1 transgenic males. Females were killed 3 d later and blastocysts flushed from the uterine horn with mES cell medium (knockout-DMEM (GIBCO), 15% Hyclone fetal bovine serum (Hyclone), 100 unit ml–1 penicillin and 0.1 mg ml–1 streptomycin (GIBCO), 2 mM glutamine (GIBCO), 100 mM nonessential amino acids (GIBCO), 55 mM 2-mercaptoethanol (GIBCO) and 1,000 units ml–1 leukocyte inhibiting factor (Chemicon)). Blastocysts were plated individually into one 10-mm well of a tissue culture dish containing mitotically inactivated mouse embryonic fibroblasts and mouse ESC medium supplemented with the MEK kinase inhibitor PD98059 (Cell Signaling). At 48 h after plating, one half volume of fresh medium was added to each culture well. Medium was changed daily after 3 d. At 4–5 d after plating, inner cell mass-derived outgrowth were dislodged with a Pasteur pipette, washed in a drop of PBS and incubated for 10 min in 0.25% trypsin at 37 °C. ESC clumps were gently dissociated with a Pasteur pipette and transferred onto a fresh layer of fibroblasts. For routine culture, the mouse ESCs were generally split 1:6 with a solution of 0.25% trypsin (GIBCO) every 2–3 d.
Generation of chimeric embryos
Chimeric embryos were generated as previously described32. Recipient mothers were killed at 10.5 d postcoitum, and then embryos were removed from the uterus and placed at 4 °C in PBS for whole mount analysis of GFP fluorescence.
Differentiation of mES cells into motor neurons
Mouse ESCs were differentiated into motor neurons according to methods previously described11. The ESCs were grown to 70–80% confluence in 10-cm plates (Falcon) in mouse ESC medium. To form embryoid bodies, cells were washed once with PBS to eliminate ESC medium and then incubated with 1 ml of 0.25% trypsin (GIBCO) for 5–10 min at room temperature (21-25 °C). Cells were then resuspended in 10 ml of DM1 medium (DMEM-F12 (GIBCO), 10% knockout serum (GIBCO), penicillin, streptomycin, glutamine (GIBCO) and 2-mercaptoethanol (GIBCO)), counted and plated at a concentration of 200,000 cells per ml in Petri dishes (Falcon). Two days later, embryoid bodies were split from one dish into four Petri dishes containing DM1 medium supplemented with RAc (100 nM; stock: 1 mM in DMSO, Sigma) and Shh (300 nM, R&D Systems). Medium was changed after 3–4 d.
On day 7 the embryoid bodies were dissociated in single-cell suspensions. The embryoid bodies were pelleted in a 15-ml Falcon tube, washed once with PBS and incubated in Earle's balanced salt solution with 20 units of papain and 1,000 units of DNase I (Worthington Biochemical) for 30–60 min at 37 °C. The mixture was then triturated with a 10-ml pipette and centrifuged for 5 min at 300 g. The resulting cell pellet was washed with PBS and resuspended in F12 medium (F12 medium (GIBCO) with 5% horse serum (GIBCO), B-27 supplement (GIBCO), N2 supplement (GIBCO)) with neurotrophic factors (GDNF, CNTF, NT3 and BDNF (10 ng ml–1, R&D Systems)). The cells were counted and plated on poly-d-lysine/laminin culture slides (BD Biosciences) or on a layer of primary glia cells. For the motor neuron survival experiments, GFP-positive cells with visible axons and dendrites were counted at different time points after plating (7, 14, 21 and 28 d).
Polymerase chain reaction
PCR reactions were performed using an MJ Research Thermal Cycler, and TaKaRa Ex Taq HS (Takara) enzymes. For SOD1 genotyping, the forward primer CAT CAG CCC TAATCC ATC TGA and reverse primer CGC GAC TAA CAA TCA AAG TGA amplified a 236-bp fragment in the fourth exon of the gene. As an internal control, a set of primers that amplified a 324-bp fragment of the IL-2 gene (forward: CTA GGC CAC AGA ATT GAA AGA TCT; reverse: CAT CAG CCC TAA TCC ATC TGA) were used. The annealing temperature for this reaction was 60 °C for 35 cycles. For GFP, a set of primers (forward: AAG TTC ATC TGC AAC ACC; reverse: TCC TTG AAG AAG ATG GTG CG) that amplified a fragment of 173 bp of the gene were used, with an annealing temperature of 60 °C for 35 cycles.
FACS
For FACS, a BD Biosciences LSRII flow cytometer was used. Embryoid bodies were dissociated with papain and resuspended in cold PBS with 2% FBS. Calcein blue (Invitrogen) was used to assay cell viability. The FACS Diva software package (BD Biosciences) was used for data analysis.
Glia cultures
Glia monolayers were obtained from P2 mice. Tissue was isolated in calcium- and magnesium-free Hanks’ BSS (HBSS). Under a dissecting microscope the cortex was isolated and carefully striped of the meninges. The tissue was split into small pieces and transferred to a 50-ml centrifuge tube in a final volume of 12 ml of HBSS. Tissue digestion was performed using trypsin-EDTA (GIBCO BRL no.25200) and 1% DNAse (Sigma no. DN-25) at 37 °C for 15 min, swirling the mixture periodically. Dissociated tissue was triturated using a fire-polished Pasteur glass pipette and filtered through a 72-μm nylon mesh (NITEX 100% polyamide nylon fiber, TETKO) to remove any undissociated tissue. Filtered material was pelleted at 300 g for 5 min and resuspended in 2 ml of glia medium (minimum essential medium with Earl's salts, GIBCO BRL no. 11095-080, 20% glucose, penicillin-streptomycin, GIBCO BRL no. 15145-014, and 10% horse serum, GIBCO BRL no. 26050-070) and the cell number was determined. One brain generally yielded enough glia to plate one T75 flask (Falcon no. 3084). Once monolayers were confluent (generally in 10–14 d), cells were replated on 12- or 24-well multiwell dishes over glass cover slips that had been coated with poly-d-lysine (0.5 mg ml–1 for 30 min at 21-25 °C).
Immunocytochemistry analysis
Cells were fixed with 4% paraformaldehyde-PBS, blocked and permeabilized with BSA (1%)–Triton X-100 (0.1%). They were then incubated overnight with the following antibodies: mouse monoclonal anti-Tuj1 (Covance), Islet 1 and RC2 (Developmental Studies Hybridoma Bank University of Iowa), SOD1 (SIGMA), S100 (Chemicon) and CNPase (Abcam); rabbit anti-Hb9 (T. Jessell, Columbia University), GFAP (Chemicon) and anti-ubiquitin (DAKO); goat anti-vimentin (Chemicon) and rat anti-CD11b (Abcam) (Supplementary Fig. 6 online). The cells were then incubated with donkey antibodies specific for rabbit, conjugated to Cy3 (1:100, 2 h), and/or donkey antibodies specific for mouse, conjugated to Cy5 antibodies (1:100, 2 h; Jackson ImmunoResearch). After the samples were mounted in Vectashield (Vector Labs), confocal or epifluorescence microscopy was performed using Olympus FV 1000, 40× and 60× oil immersion objective, 1.45 NA, or an Olympus IX70. Image acquisition was performed using FLUOVIEW software 4.0 (Olympus). For relative fluorescence analysis, all settings such as exposure time, magnification and gain were maintained constant for all samples. Offline analysis of relative intensities for all the samples was done using Metamorph 4.5 (Universal Imaging). Only cells having morphological features of neurons (that is, phase bright soma and several neurites) were considered for subsequent analysis.
Neuronal density
Hb9-GFP–positive motor neuorns were counted for each condition studied (Zeiss microscope, 40×, 1.3 NA, oil immersion objective). The density of neurons was normalized as a percent of initial numbers counted at 7 or 14 d and was established for all conditions studied. These experiments were carried out three times as independent experiments.
Data analysis
Statistical analyses were performed using Student's t-test or ANOVA and are expressed as arithmetic mean ± s.e.m.; t-test values of *P < 0.05, **P < 0.01, ***P < 0.005 were considered statistically significant. Each set of data presented represents experiments that were performed in sister cultures to reduce variability. Similar significances were found expressing the data in cumulative distributions plots. Therefore, we chose to present the data as mean ± s.e.m. to simplify its presentation.
Supplementary Material
ACKNOWLEDGMENTS
We thank J.W. Lichtman and J.R. Sanes for helpful discussions and use of confocal microscopy facilities at The Center for Brain Science, Harvard University. We are grateful to H. Akutsu for assistance in the derivation of mouse ESC lines, G. Birkhoff for preparing figures and B. Tilton for assistance with flow cytometry. We thank S. Przedborski and other members of the Columbia University research groups for communicating their results to us prior to publication, and for their criticals comments in our manuscripts. We thank Project ALS for supporting contact and cooperation between the research groups at Columbia and Harvard and for donating transgenic SOD1G93A mice. We thank T.M. Jessell and H. Wichterle for providing Hb9 antisera. This work was supported by the Stowers Medical Institute and Harvard Stem Cell Institute and US National Institutes of Health R01 HD046732-01A1 to K. Eggan and by the ALS Association to T. Maniatis. M.A. Carrasco is a Milton Safenowitz Post-Doctoral Fellow of the ALS Association and K. Eggan is a MacArthur Fellow of the John D. and Catherine T. MacArthur Foundation.
Footnotes
Note: Supplementary information is available on the Nature Neuroscience website.
COMPETING INTERESTS STATEMENT The authors declare no competing financial interests.
References
- 1.Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Brown RH., Jr. Amyotrophic lateral sclerosis. Insights from genetics. Arch. Neurol. 1997;54:1246–1250. doi: 10.1001/archneur.1997.00550220050013. [DOI] [PubMed] [Google Scholar]
- 3.Cole N, Siddique T. Genetic disorders of motor neurons. Semin. Neurol. 1999;19:407–418. doi: 10.1055/s-2008-1040855. [DOI] [PubMed] [Google Scholar]
- 4.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 5.Gurney ME, et al. Motor neuron degeneration in mice that express a human Cu/Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 6.Nagai M, et al. Rats expressing human cytosolic copper-zinc superoxide dismutase transgenes with amyotrophic lateral sclerosis: associated mutations develop motor neuron disease. J. Neurosci. 2001;21:9246–9254. doi: 10.1523/JNEUROSCI.21-23-09246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruijn LI, et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 8.Wong PC, et al. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 9.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 10.Clement AM, et al. Wild-type non-neuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- 11.Boillee S, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 12.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 13.Arber S, et al. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 14.Thaler J, et al. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23:675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- 15.Ince PG, Tomkins J, Slade JY, Thatcher NM, Shaw PJ. Amyotrophic lateral sclerosis associated with genetic abnormalities in the gene encoding Cu/Zn superoxide dismutase: molecular pathology of five new cases, and comparison with previous reports and 73 sporadic cases of ALS. J. Neuropathol. Exp. Neurol. 1998;57:895–904. doi: 10.1097/00005072-199810000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. Copper-binding–site null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum. Mol. Genet. 2003;12:2753–2764. doi: 10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe M, et al. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol. Dis. 2001;8:933–941. doi: 10.1006/nbdi.2001.0443. [DOI] [PubMed] [Google Scholar]
- 18.Pasinelli P, Houseweart MK, Brown RH, Jr., Cleveland DW. Caspase-1 and 3 are sequentially activated in motor neuron death in Cu/Zn superoxide dismutase–mediated familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA. 2000;97:13901–13906. doi: 10.1073/pnas.240305897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raoul C, et al. Motoneuron death triggered by a specific pathway downstream of Fas potentiation by ALS-linked SOD1 mutations. Neuron. 2002;35:1067–1083. doi: 10.1016/s0896-6273(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 20.Beers DR, et al. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA. 2006;103:16021–16026. doi: 10.1073/pnas.0607423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 22.Bignami A, Dahl D. Astrocyte-specific protein and neuroglial differentiation. An immunofluorescence study with antibodies to the glial fibrillary acidic protein. J. Comp. Neurol. 1974;153:27–38. doi: 10.1002/cne.901530104. [DOI] [PubMed] [Google Scholar]
- 23.Banker G, Goslin K. Culturing Nerve Cells. 2nd ed. MIT Press; Cambridge, Massachusetts: 1998. [Google Scholar]
- 24.Ullian EM, Harris BT, Wu A, Chan JR, Barres BA. Schwann cells and astrocytes induce synapse formation by spinal motor neurons in culture. Mol. Cell. Neurosci. 2004;25:241–251. doi: 10.1016/j.mcn.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Allen NJ, Barres BA. Signaling between glia and neurons: focus on synaptic plasticity. Curr. Opin. Neurobiol. 2005;15:542–548. doi: 10.1016/j.conb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Ullian EM, Christopherson KS, Barres BA. Role for glia in synaptogenesis. Glia. 2004;47:209–216. doi: 10.1002/glia.20082. [DOI] [PubMed] [Google Scholar]
- 27.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 28.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lerou PH, Daley GQ. Therapeutic potential of embryonic stem cells. Blood Rev. 2005;19:321–331. doi: 10.1016/j.blre.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Nun IF, Benvenisty N. Human embryonic stem cells as a cellular model for human disorders. Mol. Cell. Endocrinol. 2006;252:154–159. doi: 10.1016/j.mce.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 31.Nagai M, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. doi: 10.1038/nn1876. advance online publication, 15 April 2007 (doi:10.1038/nn1876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hogan B, Beddington R, Costantini F, Lacey E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.