Abstract
While the three-letter genetic code that maps nucleotide sequence to protein sequence is well known, there must exist other codes that are embedded in the human genome. Recent work points to sequence-dependent variation in DNA shape as one mechanism by which regulatory and other information could be encoded in DNA. Recent advances include the discovery of shape-dependent recognition of DNA that depends on minor groove width and electrostatics, the existence of overlapping codes in protein-coding regions of the genome, and evolutionary selection for compensatory changes in nucleotide composition that facilitate nucleosome occupancy. It is becoming clear that DNA shape is important to biological function, and therefore will be subject to evolutionary constraint.
Introduction
Elucidation of the genetic code undoubtedly is one of the most important discoveries of modern biology. The ubiquity, across all known life forms, of using a triplet DNA sequence lookup table to encode amino acids underscores the elegance of this finding.
Although the triplet genetic code is the most common lookup table used to decode genomic information, the possibility exists of additional codes in the genome, both within and outside coding regions (Figure 1). The flexibility in DNA sequence afforded by the degeneracy of the genetic code allows for additional information to be encoded within protein-coding sequences. Motivation for searching for codes outside protein coding regions of the genome stems from the prescient suggestion that regulatory mutations would have a larger biological impact than would mutations in coding sequences [1].
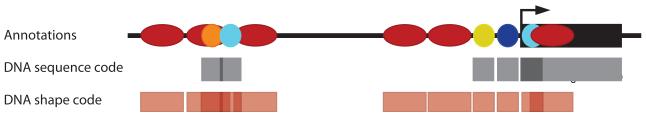
Figure 1.
Different genomic codes are subject to selective pressures. Shown is a hypothetical genomic region (top) with a gene (black rectangle), nucleosomes (red ellipses), and various transcription factors (colored circles). Gray rectangles (middle) represent genomic regions in which the primary order of the DNA nucleotide sequence encodes functional information. Pink rectangles (bottom) represent regions where genomic information is encoded in DNA shape. These various encodings can be dispersed, overlapping, redundant, and, importantly, can represent different types of biological function. Considering the molecular structure of DNA in addition to the primary nucleotide sequence can help in understanding how biological function is encoded in genomes. Note that symbols and spacing are not drawn to scale.
In this review we discuss progress over the past two years in elucidating some of the many codes embodied in a genome, and how evolution may be sculpting these codes. Our approach to this question has its basis in the structural biology of DNA. While the elegance of the three-letter genetic code often has focused analysis of the human genome on the sequence of nucleotides, this approach neglects the molecular nature of DNA, the physical embodiment of the information that is encoded in the genome. Recent work has reminded us that subtle variations in DNA shape can be exploited by biological systems.
Since evolution is the guiding principle of biology [2], one might ask whether structural features of DNA might be under evolutionary constraint. Recent work has demonstrated that substantially more territory in the human genome is under selection for maintaining DNA shape than for the exact sequence of nucleotides [3]••. This work showed that segments in the human genome that are DNA shape-constrained encompass a substantial fraction of experimentally determined functional regions (enhancers, deoxyribonuclease I hypersensitive sites, promoters, etc.) [4], evidence that maintaining DNA shape is important to at least some aspects of genomic function.
Evolution and genetic codes
Protein-coding sequences
Of the evolutionarily constrained nucleotides that have been identified in the human genome, about one-third occur in coding sequences [4]. A large fraction of these constrained coding bases are likely the result of selective pressure at the level of protein structure and function. But a recent study shows that even in protein-coding sequences, overlapping codes exist [5]••, which implies that selection within these regions could operate through other means. In an impressive effort, Itzkovitz et al. analyzed over 600 different genomes from diverse phyla including viruses, bacteria, fungi, plants, and vertebrates. They found that coding regions encode additional information, including known alternate codes like bacterial translation initiation sites, as well as codes that are yet unknown. A striking example of the existence of overlapping genomic codes is the presence of enhancer regions that have recently been identified within coding sequences [6-10].
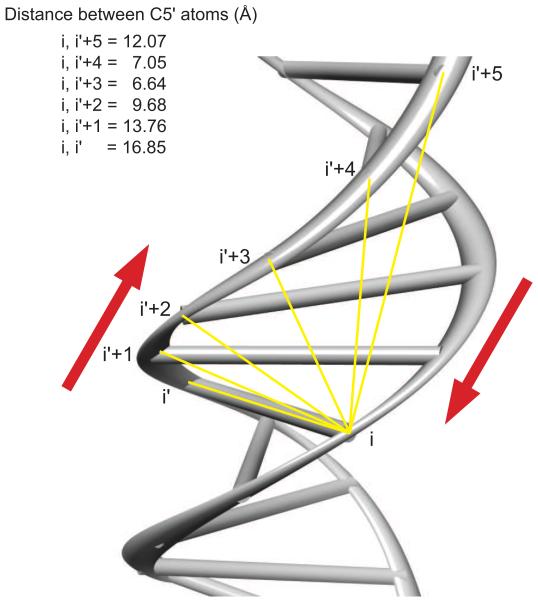
An earlier study had found that the organization of the genetic code allows for superimposition of a DNA structural signal onto a protein-coding sequence via amino acid substitution [11]. How might this occur? In the DNA double helix, backbone atoms that are in closest proximity across the minor groove, and therefore influence DNA shape [12], are separated by three nucleotides on the complementary strands (Figure 2). Accordingly, positions offset by three nucleotides in adjacent codons are ideal places for selective constraint to act on DNA shape in coding regions.
Figure 2.
Three-dimensional structure of the DNA minor groove. The minor groove of a B-form DNA molecule (PDB code: 355D) is shown in this Chimera rendering. The backbone is represented as a ribbon and the bases as a ladder. The numbering system corresponds to nucleotides that are either complementary (i and i’), or offset by different numbers of nucleotides across the strands, in the 3′ direction. The red arrows point toward the 3′ end of each strand. Distances between C5′ atoms on each strand are depicted with yellow lines. Note that the C5′ atoms closest to each other across the two DNA strands belong to nucleotides offset by +3 and +4. (This figure was adapted from Jason A Greenbaum, PhD thesis, Boston University, 2006, and used with permission.)
Non-coding sequences
The majority of evolutionarily constrained bases in the human genome, totaling two-thirds of all constrained positions [4], reside outside of coding sequences. Selective pressures imposed upon non-coding regions likely differ from selection that operates on coding sequences, which are subject to the strict rules of the genetic code. In support of this idea, a recent study found that broadly expressed genes have highly constrained protein sequences, but relatively plastic regulatory sequences [13]. Another study found differential constraint patterns—including DNA shape-based constraint—operating on the non-coding and coding regions of a recently duplicated gene pair [14]. Additional evidence that different mechanisms of evolutionary selection operate in non-coding regions comes from observations that substitution biases depend on local nucleotide context and proximity to genes [15], and that positively selected human-specific insertions/deletions (indels) are enriched in non-coding regions nearby genes [16].
Because protein-DNA interactions are so diverse and use a wide variety of recognition mechanisms [17], we are not likely to find a universal code that explains functional non-coding sequences. Further, functional sequences tend to turn over frequently [18], rendering identification across species using comparative methods a difficult task. In fact, enhancers can evolve beyond recognizable sequence similarity and still retain function [19]. Given that different DNA sequences can encode similar shapes [3,20,21] (Figure 3), DNA shape-based functional equivalence becomes an interesting concept to investigate, in parallel with traditional investigations of functional equivalence based on nucleotide sequence identity.
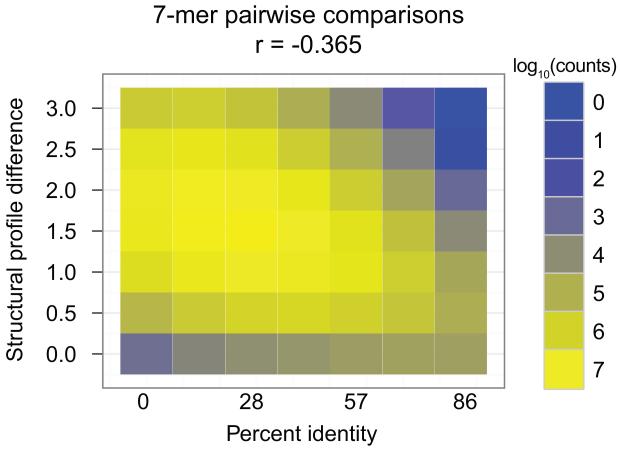
Figure 3.
The relationship between sequence identity and similarity in structural profile in a pairwise comparison of all 7-mer DNA oligonucleotide duplexes. DNA structural profiles were predicted using the ORChID database [21], and similarity in structure was measured using Euclidean distance. The color scale represents log10(counts) for each cell within the matrix, representing the number of sequences having a particular nucleotide sequence identity and a particular structural similarity. The cell in the lower left corner, which corresponds to sequences with 0% identity but very similar structural profiles, contains 1,365 distinct 7-mer pairs. The overall correlation (Pearson’s r) between sequence identity and structural profile change is −0.365.
DNA shape as a mechanism for genomic encoding
How might the molecular nature of DNA be used to encode information in a genome? Proteins are known to exploit nuances in DNA shape for recognition. A recent review covered how advances in computational methods, particularly molecular simulation, have advanced our understanding of how DNA shape depends on sequence [12]. A new and remarkably widespread mechanism for shape-specific recognition of DNA was discovered by comprehensive data-mining of three-dimensional protein-DNA structures [22]••. Many DNA-binding proteins (including the histones that make up the nucleosome core particle) were found to insert positively-charged arginine side chains into especially narrow segments of the DNA minor groove, thereby exploiting the enhanced negative electrostatic potential of a narrow minor groove for shape-dependent recognition. Another study found an alternative mechanism for creating a narrow minor groove through the use of Hoogsteen base pairing within a canonical B-DNA helix, and demonstrated the importance of DNA shape for p53 binding [23]••. The concepts presented in these recent findings reveal a clearer structural picture of how DNA shape can specify important genomic signals.
Although the bacterial gene architectural protein Fis is considered to be a protein that binds to DNA with little sequence preference, there are some sequences to which it binds with sub-nanomolar affinity. The authors of a recent study investigated the basis for Fis binding selectivity by solving X-ray crystal structures of Fis complexed with11 different DNA sequences [24]•. The authors concluded that Fis initially selects binding targets that have a narrow DNA minor groove at the center of the binding site, which allows the helix-turn-helix units of Fis to bind to the adjacent segments of the major groove. The ultimate stability of the Fis complex is governed by the ability of a DNA binding site to bend, which depends on the location of pyrimidine-purine (YR) steps in the site.
A recent paper compared the structures of the DNA in three protein-DNA complexes with the X-ray structures of the naked DNA molecules [25]••. The authors found that structural nuances observed in protein-bound DNA also existed in the unbound DNA target, suggesting how recognition might occur. This paper demonstrates the power of having detailed structural information for naked DNA molecules to enable the elucidation of the pathway for recognition by a DNA binding protein.
Nucleosome positioning and DNA physical properties
Wrapping eukaryotic genomic DNA around histone octamers to form an array of nucleosome core particles influences the functions encoded in the underlying sequence. The proteins that comprise the nucleosome core particle are among the most highly constrained [26], an observation that underscores their biological significance. Recent improvements in high-throughput genome profiling methods have yielded high-resolution nucleosome occupancy maps for a number of species [27-29]. Examination of nucleosome-bound sequences has led to the conclusion that nucleosome positioning is sequence-directed [30-32], yet the histone octamer makes no base-specific contacts with DNA [17]. This scenario leads to the intriguing hypothesis that DNA structure, and not sequence per se, can direct nucleosome positioning.
Properties of DNA that influence nucleosome positioning can be segregated into two general categories—those that are conducive to nucleosome formation, and those that exclude nucleosomes. A recent study performed a statistical analysis of sequence features that are predictive of nucleosome occupancy, and concluded that G+C content is the most dominant [33]. While this is an informative conclusion, the DNA structural property of minor groove width also was found to be important and, unsurprisingly, G+C content generally correlates with many other DNA structural features [34]. Consistent with this finding is extensive evidence that long A-tracts—stretches of consecutive deoxyadenosine nucleotides on one strand of the double helix—strongly influence nucleosome organization [35]. A-tracts are enriched in eukaryotic genomes, and have unique structural and mechanical properties that likely resist the DNA structural deformation required for nucleosome formation. Systematic mutagenesis and subsequent functional analysis of short A/T-rich sequences found that they can act as core promoter elements [36]. To explain this finding, the authors proposed complementary and redundant mechanisms of nucleosome exclusion by A/T-rich sequences, and binding site recognition by TFIID.
Other efforts to explain nucleosome positioning focused more on physical properties of DNA. For example, nucleosome occupancy in yeast and fly can be predicted using only DNA flexibility and curvature [37]. Another group developed the Repositioned Mutation (RM) test, which is an elegant algorithm designed to detect evolutionary selection for nucleosome positioning by comparing patterns at orthologous loci (originally proposed in [38]). Implementation of the RM test on the yeast genome revealed that the biophysical property of nucleosomal deformation energy is preserved across species so as to maintain chromatin organization in non-coding regions [39]. Together, these results suggest that physical properties of DNA are crucial for chromatin organization.
Two recent studies that used different methods to carefully analyze nucleotide substitution patterns in the yeast genome are particularly insightful about nucleosome positioning codes and the selective pressures that can act upon them. In the first study, the authors performed a thorough analysis of substitution patterns overlaid on high resolution nucleosome positioning data [40]••. Knowing that G+C content and A-tracts influence nucleosome positioning (see above), the authors focused on substitution patterns that would affect these signals in regions that positioning data show are important (for example, well-defined nucleosomes or nucleosome-depleted regions). Remarkably, they found regionally linked compensatory substitutions that serve to maintain nucleosome-positioning dynamics. They conclude that local sequence composition is influenced by nucleosome organization. The other study also looked at substitution patterns, but took a different approach in which the authors measured the effects of substitutions on a structurally based model of nucleosomal deformation energy [41]••. They observed a strong anti-correlation between substitution frequency and the DNA structure-based energetics of nucleosome formation. Together, these studies demonstrate the functional conservation of chromatin organization through natural selection operating on DNA shape-based signals.
Selection for DNA structural features that maintain nucleosomal positioning signals could result in a large fraction of the genome being under structural constraint. This could be considered a kind of low-level and pervasive form of DNA structural selection, whereas DNA structural selection acting on transcription factor binding sites would likely be less pervasive and, possibly, more intense. It is interesting to note that a DNA structure-based code for nucleosome positioning has been found in protein coding regions [42]•, indicating the compatible superimposition of the genetic code and a nucleosome positioning code. An intriguing possibility is that variations in local DNA shape that are encoded along genomic sequences could have a profound impact on chromatin organization, and therefore the evolution of regulatory systems.
Disease-associated SNPs and DNA shape
In the rapidly approaching age of personalized genomics and genomic medicine, the ability to interpret non-coding variation will be critical [43,44]. This is made clear by a recent meta-analysis of 465 unique human trait-associated single nucleotide polymorphisms (SNPs) that were identified across a series of genome wide association studies (GWAS), which showed that 89% of the variants occur in non-coding regions [45]. This work suggests that sequence differences in regulatory regions of the genome may capture more trait-associated variation than do differences in coding sequences. As a specific example, a recent study discovered differences in allelic enhancers at several type 2 diabetes-associated loci through comprehensive identification of regulatory regions in human pancreatic islet cells [46]••. Another study used DNA shape as a guide to find a functional non-coding variant [47]••. Based on these early results, it is clear that analyses of DNA shape will contribute to the pressing endeavor of interpreting non-coding variations and assessing their affect on disease.
Conclusions and prospects
In this brief review we have discussed how the shape and physical properties of DNA can influence biological function. The existence of such phenomena suggests that functional genomic codes can utilize DNA shape in addition to nucleotide sequence. The implication is that DNA shape can be a substrate for selective evolutionary pressure.
A recent example points the way to new DNA structure-based biological phenomena. High throughput sequencing was used to determine the spectrum of mutations that occurred after treatment of DNA with a mutagen [48]••. This experimental approach allows for exhaustive characterization of mutation frequency, in contrast to standard methods that rely on phenotypic change. The frequency of mutation at a given position was found to vary depending on the identities of the nucleotides neighboring the site of mutation, beyond nearest-neighbors, an unexpected result. The authors used their results to advance the idea that a genotype itself has a phenotype, which is expressed only when the genotype is embodied as a molecular entity, DNA.
The flood of genome-wide non-coding functional data that are emerging from the ENCODE [4] and modENCODE [49,50] Projects only whets our appetite for similar data from other organisms in the tree of life. Such data will allow us to directly test the correspondence of nucleotide sequence conservation with conservation of function [51], and so perhaps detect the presence of new kinds of genomic signals. A pioneering effort in this realm used chromatin immunoprecipitation combined with high-throughput sequencing to compare maps of the genome-wide occupancy of two transcription factors in the livers of five vertebrates [52].
Multi-species genome alignments are the foundation of comparative genomics, but current methods align genomes based strictly on nucleotide sequence. New methods are emerging [53,54] that can incorporate other information besides sequence (including DNA shape) to drive multi-species alignments. This, and other, advances will give us new ways to interpret the complex and overlapping codes that are hidden in a genome.
Acknowledgements
We thank Adam Woolfe for help in identifying papers reporting exonic enhancers, and Jason A Greenbaum for permission to use Figure 2. This work was supported by a grant to TDT from the National Human Genome Research Institute of the National Institutes of Health (R01 HG003541). SCJP was supported by the Intramural Research Program of the NHGRI, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended reading
- 1.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 2.Dobzhansky T. Nothing in biology makes sense except in the light of evolution. American Biol Teacher. 1973;35:125–129. [Google Scholar]
- 3.Parker SCJ, Hansen L, Abaan HO, Tullius TD, Margulies EH. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009;324:389–392. doi: 10.1126/science.1169050. ••This paper introduced a new method for assessing evolutionary constraint based on DNA shape, and showed that more than 10% of the human genome is under selection for structure. Of particular significance is the finding that a high proportion of functional genomic elements occur in regions that are under structural constraint.
- 4.ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itzkovitz S, Hodis E, Segal E. Overlapping codes within protein-coding sequences. Genome Res. 2010;20:1582–1589. doi: 10.1101/gr.105072.110. •• This study presents a clever method to measure information content in coding sequences. The authors found that across diverse phyla coding sequences encode information beyond the standard genetic code.
- 6.Lampe X, Samad OA, Guiguen A, Matis C, Remacle S, Picard JJ, Rijli FM, Rezsohazy R. An ultraconserved Hox-Pbx responsive element resides in the coding sequence of Hoxa2 and is active in rhombomere 4. Nucleic Acids Res. 2008;36:3214–3225. doi: 10.1093/nar/gkn148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong X, Navratilova P, Fredman D, Drivenes Ø , Becker TS, Lenhard B. Exonic remnants of whole-genome duplication reveal cis-regulatory function of coding exons. Nucleic Acids Res. 2010;38:1071–1085. doi: 10.1093/nar/gkp1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barthel KKB, Liu X. A transcriptional enhancer from the coding region of ADAMTS5. PLoS ONE. 2008;3:e2184. doi: 10.1371/journal.pone.0002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tümpel S, Cambronero F, Sims C, Krumlauf R, Wiedemann LM. A regulatory module embedded in the coding region of Hoxa2 controls expression in rhombomere 2. Proc Natl Acad Sci U S A. 2008;105:20077–20082. doi: 10.1073/pnas.0806360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen HP, Lin A, Bloom JS, Khan AH, Park CC, Smith DJ. Screening reveals conserved and nonconserved transcriptional regulatory elements including an E3/E4 allele-dependent APOE coding region enhancer. Genomics. 2008;92:292–300. doi: 10.1016/j.ygeno.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Baisnée PF, Baldi P, Brunak S, Pedersen AG. Flexibility of the genetic code with respect to DNA structure. Bioinformatics. 2001;17:237–248. doi: 10.1093/bioinformatics/17.3.237. [DOI] [PubMed] [Google Scholar]
- 12.Rohs R, West SM, Liu P, Honig B. Nuance in the double-helix and its role in protein-DNA recognition. Curr Opin Struct Biol. 2009;19:171–177. doi: 10.1016/j.sbi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaffney DJ, Blekhman R, Majewski J. Selective constraints in experimentally defined primate regulatory regions. PLoS Genet. 2008;4:e1000157. doi: 10.1371/journal.pgen.1000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sánchez-Gracia A, Romero-Pozuelo J, Ferrús A. Two frequenins in Drosophila: unveiling the evolutionary history of an unusual neuronal calcium sensor (NCS) duplication. BMC Evol Biol. 2010;10:54. doi: 10.1186/1471-2148-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nevarez PA, DeBoever CM, Freeland BJ, Quitt MA, Bush EC. Context dependent substitution biases vary within the human genome. BMC Bioinformatics. 2010;11:462. doi: 10.1186/1471-2105-11-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C-H, Chuang T-J, Liao B-Y, Chen F-C. Scanning for the signatures of positive selection for human-specific insertions and deletions. Genome Biol Evol. 2009;1:415–419. doi: 10.1093/gbe/evp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sathyapriya R, Vijayabaskar MS, Vishveshwara S. Insights into protein-DNA interactions through structure network analysis. PLoS Comput Biol. 2008;4:e1000170. doi: 10.1371/journal.pcbi.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meader S, Ponting CP, Lunter G. Massive turnover of functional sequence in human and other mammalian genomes. Genome Res. 2010;20:1335–1343. doi: 10.1101/gr.108795.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meireles-Filho ACA, Stark A. Comparative genomics of gene regulation-conservation and divergence of cis-regulatory information. Curr Opin Genet Dev. 2009;19:565–570. doi: 10.1016/j.gde.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Gardiner EJ, Hunter C, Lu X, Willett P. A structural similarity analysis of double-helical DNA. J Mol Biol. 2004;343:879–889. doi: 10.1016/j.jmb.2004.08.092. [DOI] [PubMed] [Google Scholar]
- 21.Greenbaum JA, Pang B, Tullius T. Construction of a genome-scale structural map at single-nucleotide resolution. Genome Res. 2007;17:947–953. doi: 10.1101/gr.6073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. •• This important paper established a widespread mechanism for shape-selective recognition of DNA. A key feature of this recognition mechanism is that it relies on the interplay of DNA shape and minor groove electrostatic potential, and so represents a DNA shape-encoded genomic signal.
- 23.Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, Honig B, Shakked Z. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nature Struct Mol Biol. 2010;17:423–429. doi: 10.1038/nsmb.1800. ••This paper reports the remarkable discovery of Hoogsteen base pairs in a p53-DNA complex. The narrow minor groove and concomitant negative electrostatic potential that result from a Hoogsteen base pair are recognized by an essential arginine residue of p53. This structure reveals a new way that DNA shape can be sculpted to form a recognition site.
- 24.Stella S, Cascio D, Johnson RC. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Devel. 2010;24:814–826. doi: 10.1101/gad.1900610. • This paper takes advantage of a very large set of X-ray structures of Fis-DNA complexes to work out the details of how the protein recognizes a high-affinity binding site. Having this remarkable catalog of structures of a DNA-protein complex makes it possible to deduce the mechanism of DNA shape-dependent recognition.
- 25.Locasale JW, Napoli AA, Chen S, Berman HM, Lawson CL. Signatures of protein-DNA recognition in free DNA binding sites. J Mol Biol. 2009;386:1054–1065. doi: 10.1016/j.jmb.2009.01.007. •• This paper presents a strong argument for solving three-dimensional structures of naked DNA molecules that also associate with protein in a protein-DNA complex. Unfortunately, very few examples currently exist, but it is clear from the results presented in this paper that important insights into protein-DNA recognition will be learned from such structural comparisons.
- 26.Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 27.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nature Rev Genet. 2010;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radman-Livaja M, Rando O. Nucleosome positioning: How is it established, and why does it matter? Dev Biol. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ioshikhes IP, Albert I, Zanton SJ, Pugh BF. Nucleosome positions predicted through comparative genomics. Nature Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett A, Tillo D, Field Y, Leproust E, Hughes T, Lieb J, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tolstorukov MY, Colasanti AV, McCandlish D, Olson WK, Zhurkin VB. A novel ‘roll-and-slide’ mechanism of DNA folding in chromatin. Implications for nucleosome positioning. J Mol Biol. 2007;371:725–738. doi: 10.1016/j.jmb.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tillo D, Hughes TR. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes A, Rando OJ. Chromatin ‘programming’ by sequence - is there more to the nucleosome code than %GC? J Biol. 2009;8:96. doi: 10.1186/jbiol207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugihara F, Kasahara K, Kokubo T. Highly redundant function of multiple AT-rich sequences as core promoter elements in the TATA-less RPS5 promoter of Saccharomyces cerevisiae. Nucleic Acids Res. 2011;39:59–75. doi: 10.1093/nar/gkq741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miele V, Vaillant C, d’Aubenton-Carafa Y, Thermes C, Grange T. DNA physical properties determine nucleosome occupancy from yeast to fly. Nucleic Acids Res. 2008;36:3746–3756. doi: 10.1093/nar/gkn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babbitt GA, Kim Y. Inferring natural selection on fine-scale chromatin organization in yeast. Mol Biol Evol. 2008;25:1714–1727. doi: 10.1093/molbev/msn127. [DOI] [PubMed] [Google Scholar]
- 39.Babbitt GA, Tolstorukov MY, Kim Y. The molecular evolution of nucleosome positioning through sequence-dependent deformation of the DNA polymer. J Biomol Struct Dynam. 2010;27:765–780. doi: 10.1080/07391102.2010.10508584. [DOI] [PubMed] [Google Scholar]
- 40.Kenigsberg E, Bar A, Segal E, Tanay A. Widespread compensatory evolution conserves DNA-encoded nucleosome organization in yeast. PLoS Comput Biol. 2010;6:e1001039. doi: 10.1371/journal.pcbi.1001039. •• Careful analysis of substitution patterns in yeast coupled with high-resolution nucleosome positioning data showed that there is a spatial coupling of compensatory mutations. These mutation patterns serve to maintain nucleosome positioning, demonstrating a direct link between chromatin organization and sequence evolution.
- 41.Babbitt GA, Cotter CR. Functional conservation of nucleosome formation selectively biases presumably neutral molecular variation in yeast genomes. Genome Biol Evol. 2011;3:15–22. doi: 10.1093/gbe/evq081. •• This study reports a strong anti-correlation between substitution patterns in yeast and an energetics-based model of nucleosomal formation. This anti-correlation is highest at points on the DNA backbone that interface with the nucleosome surface. Thus, there is a clear linkage between the evolution of DNA structural properties and nucleosome positioning.
- 42.Cohanim AB, Haran TE. The coexistence of the nucleosome positioning code with the genetic code on eukaryotic genomes. Nucleic Acids Res. 2009;37:6466–6476. doi: 10.1093/nar/gkp689. • This study measured A- and G-tract occurrences in exons from yeast, worm, arabidopsis, zebrafish, and human. Comparison to nucleosome positioning data revealed that A-tract avoidance is specific to exon regions with well-positioned nucleosomes, and not in linker regions. These results show that eukaryotic genomes use known nucleosome positioning signals in coding regions.
- 43.Cooper DN, Chen J-M, Ball EV, Howells K, Mort M, Phillips AD, Chuzhanova N, Krawczak M, Kehrer-Sawatzki H, Stenson PD. Genes, mutations, and human inherited disease at the dawn of the age of personalized genomics. Hum Mut. 2010;31:631–655. doi: 10.1002/humu.21260. [DOI] [PubMed] [Google Scholar]
- 44.Feero WG, Guttmacher AE, Collins FS. Genomic medicine--an updated primer. NEJM. 2010;362:2001–2011. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- 45.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stitzel ML, Sethupathy P, Pearson DS, Chines PS, Song L, Erdos MR, Welch R, Parker SCJ, Boyle AP, Scott LJ, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. •• In this thorough study, the authors used a variety of methods to identify regulatory regions human pancreatic islet cells. Some regions occur in known type 2 diabetes-associated loci, and a subset display allele-specific enhancer activity. This study highlights the importance of comprehensively understanding how non-coding variations affect biological function.
- 47.Sommer WH, Lidström J, Sun H, Passer D, Eskay R, Parker SCJ, Witt SH, Zimmermann US, Nieratschker V, Rietschel M, et al. Human NPY promoter variation rs16147:T>C as a moderator of prefrontal NPY gene expression and negative affect. Hum Mut. 2010;31:E1594–1608. doi: 10.1002/humu.21299. •• Changes to DNA shape induced by a single nucleotide polymorphism in a non-coding genomic region linked to stress response were used to discover potentially functional non-coding variants. Subsequent analyses revealed measurable biochemical changes to protein binding and gene regulation due to this alteration.
- 48.Petrie KL, Joyce GF. Deep sequencing analysis of mutations resulting from the incorporation of dNTP analogs. Nucleic Acids Res. 2010;38:8095–8104. doi: 10.1093/nar/gkq685. •• This paper provides a wonderful illustration of how high-throughput DNA sequencing can give new insights into a problem that has been well-studied in the past using a less direct experimental readout. The authors’ conclusion that “a genotype ... has considerable phenotype” is provocative and potentially far-reaching in its implications.
- 49.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE Project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weirauch MT, Hughes TR. Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends Genet. 2010;26:66–74. doi: 10.1016/j.tig.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grabherr MG, Russell P, Meyer M, Mauceli E, Alföldi J, Di Palma F, Lindblad-Toh K. Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics. 2010;26:1145–1151. doi: 10.1093/bioinformatics/btq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kemena C, Notredame C. Upcoming challenges for multiple sequence alignment methods in the high-throughput era. Bioinformatics. 2009;25:2455–2465. doi: 10.1093/bioinformatics/btp452. [DOI] [PMC free article] [PubMed] [Google Scholar]