Abstract
Multiple cell lines (estimated at 300–400) have been established from human small cell (SCLC) and non-small cell lung cancers (NSCLC). These cell lines have been widely dispersed to and used by the scientific community worldwide, with over 8000 citations resulting from their study. However, there remains considerable skepticism on the part of the scientific community as to the validity of research resulting from their use. These questions center around the genomic instability of cultured cells, lack of differentiation of cultured cells and absence of stromal–vascular–inflammatory cell compartments. In this report we discuss the advantages and disadvantages of the use of cell lines, address the issues of instability and lack of differentiation. Perhaps the most important finding is that every important, recurrent genetic and epigenetic change including gene mutations, deletions, amplifications, translocations and methylation-induced gene silencing found in tumors has been identified in cell lines and vice versa. These “driver mutations” represented in cell lines offer opportunities for biological characterization and application to translational research. Another potential shortcoming of cell lines is the difficulty of studying multistage pathogenesis in vitro.To overcome this problem, we have developed cultures from central and peripheral airways that serve as models for the multistage pathogenesis of tumors arising in these two very different compartments. Finally the issue of cell line contamination must be addressed and safeguarded against. A full understanding of the advantages and shortcomings of cell lines is required for the investigator to derive the maximum benefit from their use.
Keywords: Lung cancer, Cell lines, Preneoplasia, Oncogenes, Tumor suppressor genes, Genetic instability
1. Introduction
Lung cancer remains the commonest form of cancer deaths in the world. While most cases arise in smokers, lung cancer in lifetime never smokers is a major problem, and for reasons we have reviewed, may be considered a distinct entity [1]. Lung cancers represent several histologic types, and they may be divided into those arising from the central (mainly small cell, SCLC, or squamous cell carcinomas) or from the peripheral compartments (mainly ade-nocarcinomas) of the lung [1]. The mortality from lung cancer remains high, resulting in a great interest in studying this disease with the intention of developing a better understanding of its biology and translating these findings into improved therapeutic approaches.
Three major approaches are available for study of cancers: (1) fresh tumor tissue, (2) animal models and (3) cell cultures. A discussion of animal models is beyond the scope of this article. Tumor tissues and cell lines both have advantages and disadvantages. However, tumor tissues are limited in the amount available for any individual tumor, contain varying (and often unknown) amounts of non-malignant cells, and there are constraints about their acquisition, utilization and distribution. Inter-tumor variability, geographic and ethnic differences and differing pathologic criteria for classification add layers of complexity to their study and to the comparison of data from different investigators. For these reasons, cancer cell lines have been widely used for the study of lung cancer. However, there appears to be a high level of skepticism in the scientific community about the validity of this approach. For this reason we evaluate the pros and cons of lung cancer cell lines with emphasis on their relevance to lung cancer research. We also discuss in depth a novel in vitro approach for the study of multistage pathogenesis of lung cancer.
2. Establishment, availability and significance of lung cancer cell lines
We have reviewed the history of lung cancer culture, described methods for their propagation and media for their serum free culture [2,3], and these subjects will not be discussed in depth. Establishment of lung cancer cell lines began about 20 years after George Gey successfully cultured the first human cell line—HeLa, from a cervical cancer [4]. The initial emphasis was on SCLC, but later switched to non-small cell lung cancer (NSCLC). By the mid- 1980s lung cancer cell lines became widely used because of a number of factors:
Defined programs for tumor procurement and culture establishment.
Development of methods and media for culture.
Large number and variety of cell lines.
Ease of availability and widespread distribution to the scientific community.
An examination of the Wellcome Trust Sanger Institute public data base (http://www.sanger.ac.uk/) indicates that of the several hundred human tumor cell cultures they have studied (they have made an effort to collect all readily available lines), the number of lung cancer cell lines exceeds the combined total of lines from the other common epithelial malignancies. The disproportionately large number of lung lines is supported by an examination of the lines available from the American Type Culture Collection, a repository for cell cultures and other reagents (http://www.atcc.org/). While the figures from these sources are not strict scientific evidence, they indicate the very large number of in vitro reagents available to the scientific community. The widespread usage of human lung cancer cell lines has resulted in over 8000 citations in the PubMed data base of medical literature (http://www.ncbi.nlm.nih.gov/pubmed/).
2.1. Are lung cancer cell lines relevant?
Cancer cell lines offer certain advantages and disadvantages when compared to tumor materials. An excellent discussion on these issues is presented in a recent review [5], and the major points are enumerated in Table 1. Many of these points are self-explanatory, and they apply to all forms of cancer. Thus we do not discuss them in detail. However, to demonstrate the validity of the approach to use lung cancer cell lines for biomedical research, three points in particular needs elaboration, as discussed below.
Table 1.
Advantages and disadvantages of tumor cell lines.
| Advantages |
| Pure population of tumor cells |
| Possibility of wide distribution to investigators worldwide |
| Limitless replicative ability |
| Ability for clonal selection |
| Availability of in vivo and in vitro tests for the evaluation of invasiveness and tumorigenicity |
| Ability to utilize a single passage repeatedly |
| Identification of specific genetic, epigenetic and cytogenetic changes and confirmation of their importance to the origin or maintenance of the malignant state |
| Ability for phenotypic or genotypic selection or manipulation |
| Growth as substrate dependent or substrate independent cells |
| Determination of specific environmental conditions or growth factor requirements for optimal growth |
| Identification and testing of conventional and novel therapeutic approaches |
| Development of models to study multistage pathogenesis |
| Disadvantages |
| Possible selection of minor tumor subpopulations not characteristic of the original population |
| Possible acceleration of genomic instability |
| Absence of stromal, immune and inflammatory cells |
| Absence of vascularization |
| Difficulty of evaluating metastatic potential |
Are cell lines representative of tumors they were derived from? Cell lines probably arise from subpopulations of the original tumor that have inherent properties that allow them to grow as immortal cultures. Thus they may have specific genetic or epigenetic changes, activation of the immortality associated enzyme telomerase and contain a large fraction of cells with stem cell like features that differ from the major population in the original tumor. As a result of selection of the most robust, fast growing subpopulation, they may also be relatively undifferentiated and demonstrate epithelial to mesenchymal transformation. To some extent this is an inherent feature of the culture process. However, it can be minimized by special attention during the establishment process, including use of defined media that permits preferential growth of the cell type being cultured as well as patience, permitting relatively differentiated slow growing populations to be established in culture. Until a few years ago most breast cancer cell lines, derived from metastatic lesions, lacked estrogen receptor expression, unlike the majority of primary tumors. Successful cultures of primary cancers (some of which took 2 years to establish) resulted in cell lines that more closely resembled the primary tumor population, consisting of representative numbers of estrogen receptor positive, HER2 amplified and triple negative cultures [6–8]. SCLC, a tumor that has a rapid tumor doubling time in vivo, proved surprisingly difficult to culture, and its reproducible culture required defined media, considerable patience, and a realization that the cells lacked substrate adhesion. However, once established, they were found to express the entire program of neuroendocrine differentiation present in the tumors [9]. SCLC cell lines are of particular importance, because of the lack of easy access to fresh tumor material. A study of lung cancer lines has demonstrated major differences between the major signaling pathways in SCLC and NSCLC, and the importance of the hedgehog and notch pathways in SCLC [10–12]. These and related studies have identified potential novel targets for therapy.
Are cell lines so inherently unstable that they are irrelevant? One of the major criticisms of cell lines is their inherent instability, especially on long term culture. As discussed later, acquisition of the hallmarks of cancer by a tumor or premalignant cell leads to genomic instability. The more divisions such a cell undergoes, the greater the likelihood of accumulating multiple mutations. In fact detailed characterization of lung and other cancer cells has demonstrated the great complexity of tumor cells, with numerous cytogenetic, genetic and epigenetic changes [13,14]. Some of these changes are “driver” mutations or changes that are essential for the development or maintenance of the malignant phenotype, while many, probably most, are “passenger” mutations or changes without major contribution to the cancer phenotype. Because cell lines have short doubling times, they undergo more divisions, over a period of time, than do tumor cells. Thus cell lines are likely to undergo more molecular changes than their tumor counterparts. In addition, the culture process may have led to selective growth of rapidly growing cells having more molecular abnormalities than the tumor cells. For these reasons we compared some of the genomic properties of long cultured cells to their tumor counterparts, initially for breast [15], and later for lung [16] cancer.
2.2. Cell lines as experimental systems to study cancer biology and for translational research
Cell lines offer many advantages over tumors. Cell lines are populations of pure tumor cells without admixed stromal or inflammatory cells. The presence of large fractions of such cells in lung tumors (an average of 55%, author’s unpublished data) may make interpretation of profiling studies such as global expression profiling misleading. Contamination with non-malignant cells may mask the presence of mutations, and makes detection of gene deletions exceedingly difficult. The presence of high quality DNA, RNA and proteins from cell lines greatly aids testing and the interpretation of findings. Cell lines are capable of infinite replication, providing a limitless source of materials and permitting their dispersion to laboratories worldwide. Scientists can directly compare their results from identical materials. The absence of stromal cells has both advantages and disadvantages. Their absence results in a pure tumor cell population, greatly aiding tumor cell characterization. However stromal (and inflammatory) cells play crucial roles in tumor formation, growth and localized and metastatic spread. In addition, stroma is essential for angiogenesis.
For many decades a debate has raged as to the relevance of cell lines for the study of cancer biology and as in vitro models for translational biology. The answer is complex and multifaceted. An excellent review on this subject was published recently [5]. Before it can be addressed, the properties of cell lines and their respective tumors need to be examined. In general, cell lines maintain expression of the hallmarks with the exception of angiogenesis (which requires the presence of stromal tissues). The role of cell lines in understanding the molecular biology of lung cancer, and the ability to translate these findings to clinical applications would have been severely hampered and delayed without their availability (see below). Acquisition of the hallmarks of cancer results in genomic instability, with the appearance of numerous genetic and epigenetic changes which characterize the cancer genome [14]. They include driver mutations essential for the appearance and maintenance of the malignant phenotype, as well as many passenger mutations which contribute little or nothing. Cell lines have contributed greatly to sorting out the drivers from the passengers as tests for functionality and genetic manipulations are difficult if not impossible to perform in tumor tissues or animal models. Perhaps without exception, all of the important and recurring genetic and epigenetic changes present in lung cancers are represented in cell lines. While their frequencies may differ in tumors and cell lines, the latter provide essential models to study all of the important lung cancer genes. Some of the important contributions to our understanding of lung cancer pathogenesis are summarized in Table 1.
2.3. The crucial role of cell lines in elucidating the molecular and translational biology of lung cancer
The cancer genome is amazingly complex, as manifested by a detailed analysis of lung cancer cell lines [14]. Of the hundreds of genetic and epigenetic changes present in the typical cancer genome, most represent “passenger” mutations whose contribution to the cancer phenotype is negligible or absent, while a modest number represent “driver” mutations essential for the development and/or maintenance of malignant properties. Cell lines have played crucial roles in the identification and characterization of driver mutations. The large number and variety of lung cancer cell lines and their widespread availability to investigators throughout the world has resulted in much of our current understanding of lung cancer biology and of many translational applications. In some cases, the pure tumor cell populations of the cell lines has resulted in the discoveries being accelerated, while in other instances the discoveries would have been delayed considerably without the ready availability of cell lines. Every single important “driver” mutation present in lung cancer tumors is represented in the large bank of lung cancer cell lines available for investigation, providing crucial, and in some cases, essential, resources for the study of lung cancer pathogenesis.
The relevance of cell lines for the study of biomedical studies must reflect, to a great degree, how closely they resemble the tumors they were derived from. We have demonstrated that the genomic drift during culture life is not as great as commonly believed [16]. In preparation for this review, we compared the regions of frequent gain and loss in lung adenocarcinoma tumors and cell lines (Table 2) as determined by comparative genomic hybridization [17,18]. As shown in the table, many of the regions of frequent genomic gains and losses known to occur in tumors are represented in the cell lines. Many of these sites are the locations of genes known to be important in the pathogenesis of lung cancers. However, in general, the frequencies in cell lines are greater than in tumors. The higher frequencies in cell lines may reflect one or more of the following reasons: (a) preferential culture of tumors containing copy number changes at locations of crucial oncogenes and tumor suppressor genes (TSGs); (b) contamination of malignant cells by non-malignant stromal cells in tumors; or (c) enhanced frequencies of genomic instability reflecting the short doubling times of cultured cells. Perhaps the most important evidence that cell lines are useful models is the fact that all frequently recurring gene mutations, amplifications, deletions and translocations present in tumors are represented in tumor cell line at similar or greater frequencies. These findings present investigators with a wealth of materials to study tumor gene functions and interactions. These abilities allow cell lines to act as unique tools to determine whether the tumor findings reflect “driver” or “passenger” changes.
Table 2.
Frequent sites of gains and losses in lung adenocarcinomas: comparison of tumors versus cell lines.
| Amplifications | Frequency of gain in adenocarcinoma tumors (%) |
Frequency of gain in adenocarcinoma cell lines (%) |
Known oncogene |
|---|---|---|---|
| 1p11.2 | 46 | 46 | – |
| 1q21.2 | 51 | 72 | ARNT |
| 5p13.3 | 41 | 57 | TERT |
| 7p11.2 | 26 | 74 | EGFR |
| 7q31.2 | 20 | 44 | MET |
| 8q24.21 | 39 | 69 | MYC |
| 11q13.3 | 17 | 61 | CCND1 |
| 12p12.1 | 19 | 41 | K-RAS |
| 12q14.1 | 15 | 33 | CDK4 |
| 12q15 | 17 | 33 | MDM2 |
| 14q13.3 | 31 | 52 | TITF1 |
| 17q12 | 30 | 41 | ERBB2 |
| 20q13.2 | 29 | 62 | – |
| Deletions | Frequency of loss in adenocarcinoma tumors (%) |
Frequency of loss in adenocarcinoma cell lines (%) |
Known tumor suppressor gene |
|---|---|---|---|
| 3p14.2 | 36 | 62 | FHIT |
| 3p21.3 | 35 | 50 | RASSF1 |
| 5q22.2 | 22 | 25 | APC |
| 6q26 | 47 | 62 | PARK2 |
| 8p21.3 | 51 | 71 | – |
| 9p21.3 | 38 | 67 | CDKN2A/CDKN2B |
| 9q12 | 52 | 61 | – |
| 10q23.31 | 24 | 42 | PTEN |
| 13q14.12 | 37 | 74 | RB1 |
| 17p13.1 | 42 | 25 | TP53 |
| 19p13.3 | 29 | 18 | LKB1 |
Hanahan and Weinberg have defined six features (“hallmarks”) that are characteristic of the malignant phenotype [19]. These acquired capabilities result in acquisition of an enabling characteristic, namely genomic instability. The acquisition of these cancer hallmarks results in the development of genomic instability. Cell lines have made important, even crucial, contributions to the identification and characterization of oncogenes and TSGs, and lesser contributions to the other hallmarks. Perhaps the contributions of cell lines to the identification and characterization of oncogenes and TSGs (hallmarks 1 and 2) are greater than to the other hallmarks. We briefly discuss expression of the hallmarks of cancer, and of genomic instability, in lung cancer cell lines.
2.4. Hallmark 1: self-sufficiency in growth signals
Oncogenes may be activated by mutations (including point mutations, intragenic deletions or insertions),increased copy number or translocations. Not only are oncogenes activated in both tumors and cell lines, the mechanisms of activation are retained in cultures, as illustrated below.
The best characterized driver oncogenes in lung cancer are K-RAS and EGFR. K-RAS is often activated by point mutations limited to three codons, most frequently involving codon 12, in both tumors and cell lines [20,21]. A recent report, using short hairpin RNAs to deplete K-Ras in lung and pancreatic cancer cell lines harboring K-RAS mutations, two classes were identified—lines that do or do not require ras to maintain viability [22]. These findings, which could only have been performed in vitro, suggest that K-RAS oncogene “addicted” cancers represent candidate therapeutic targets.
EGFR mutations involving the kinase domain are characteristic of lung adenocarcinoma tumors and cell lines, and are rarely present in other tumor types [23]. These mutations target the first four exons of the kinase domain, and the vast majority are either a deletion of a conserved sequence in exon 19 or a single point mutation in exon 21 (L858R). These activating mutations usually confer an “addiction” to the oncogene, resulting in sensitivity to tyrosine kinase inhibitors (TKIs) [24,25]. However, secondary mutations in exon 20, especially T790M, are associated with resistance. EGFR and K-RAS mutations are almost always mutually exclusive [23]. The mutational spectrum in cell lines precisely follows those in tumors, and confers the same patterns of sensitivity and resistance [23,24]. In fact, in one of the two the original articles describing the finding of EGFR mutations, cell lines were used to confirm that the mutations functioned as “drivers” for lung cancer [26].
As shown in Table 2, virtually all the known oncogenes activated by copy number gains in lung cancers, are also amplified in cell lines. For instance, genome-wide approaches had identified TITF1 as being frequently amplified in lung adenocarcinomas and their cell lines [17,27], and the gene was identified as being responsible for lineage specific dependency [28]. The latter finding required the use of cell lines. Recently, the frequent finding of mutant allele specific imbalance (MASI) was described in cancer cells [24,29]. MASI for the EGFR and K-RAS genes was present in tumors and cell lines at similar frequencies, as was the mechanism of achieving MASI (usually by copy number gains in EGFR mutant cells, and by uniparental disomy in K-RAS mutant cells).
Relatively few examples of oncogene activation by translocations have been described in lung cancers. However, recently a small subset of lung cancers were found to harbor a small inversion within chromosome 2p, giving rise to a transforming fusion gene, EML4-ALK, which encodes an activated tyrosine kinase [30]. Most of these fusion proteins were present in adenocarcinomas arising in never smokers lacking EGFR or K-RAS mutations. We have identified activated ALK fusion proteins in three adenocarcinoma cell lines, all from never or light smokers and mutually exclusive of EGFR or K-RAS mutations (authors unpublished data).
2.5. Hallmark 2: insensitivity to growth-inhibitory (antigrowth) signals
Inactivation of TSGs is an integral part of the malignant process, and can be documented in every tumor cell. Common mechanisms of inactivation include inactivating mutations (missense or nonsense), methylation of the promoter region and/or 5′ end of the gene or via homozygous deletions. A study of cell lines has resulted in the initial identification of many TSGs or has elucidated their roles in lung cancer pathogenesis. Acquired inactivation of the RB gene in tumors was first identified in SCLC lines [31], and the role of TP53 and CDKN2 genes was confirmed and extended by this method [32,33]. A relatively crude identification (by today’s standards) of genome-wide regions of frequent loss in SCLC and NSCLC cell lines indicated novel regions of loss [34]. One of the novel regions of loss (chromosome 19p) identified in that study is now known to harbor two tumor suppressor genes frequently mutated and inactivated in NSCLC, namely LKB1 and BRG1 [35–37]. Cell lines also played crucial roles in the identification of these two TSGs, and also in elucidating their roles in lung cancer pathogenesis. Cell lines played a crucial role in the identification of RASSF1A as a major TSG inactivated in lung and breast [38,39] (and later in many other tumor types) and is located in the crucial 3p21.3 chromosomal region. Of interest, deletions of chromosome 3p, the first specific cytogenetic abnormality associated with lung cancer, was identified from a study of cell lines [40].
Many TSGs are inactivated by methylation (combined with loss of heterozygosity or other mechanism of inactivation of the second allele). Numerous genes (estimated in the hundreds per tumor) are methylated, and methylation of many of these putative TSGs results in loss of transcription [41,42]. Cell lines have played crucial roles in the identification of methylated TSGs including (as examples) RASSF1A, CDKN2A, RARβ and TCF21. In addition to identification, cell lines contribute to elucidating the role of methylated genes–gene silencing, pharmacologic reactivation after exposure to demethylating agents and loss of tumorigenic properties on reactivation.
2.6. Hallmark 3: evasion of programmed cell death (apoptosis)
Evasion of apoptotic death is a crucial and early event in tumor pathogenesis. While most of the major players (genes) and path-ways in the apoptotic cascade were identified without the use of lung cancer cell lines, the lines have played important roles in understanding the inter-relationships of the genes (including FLIP, survivin, death inducing signaling complex, TRAIL and the caspases), their roles in the cellular responses to cytotoxic therapies [43–47]. The finding that a SCLC cell line had a homozygous deletion at 2q33 encompassing the chromosomal location of the CASP8 gene led to the finding that the gene was frequently inactivated in SCLC and high grade neuroendocrine carcinomas [46].
2.7. Hallmark 4: limitless replicative potential
Most malignant tumors and cell cultures, unlike their non-malignant counterparts, have limitless replicative potential and avoid senescence [48]. Tumor cells may undergo a “crisis” during early culture life after which subpopulations emerge that evade normal checkpoint controls. These attributes are largely due to continuous expression of the enzyme telomerase which adds hexanucleotide repeats onto the ends of telomeric DNA. High telomerase expression is present in over 90% of NSCLC lines and all SCCL lines (authors’ unpublished data). However, the occasional NSCLC line lacking telomerase activity offers systems to study alternative models of immortalization.
2.8. Hallmark 5: sustained angiogenesis
Because tumor angiogenesis requires co-operation between malignant and non-malignant cells, cell cultures are less than ideal models for it study [49]. However, expression and modulation of angiogenic factors and their receptors have been studied in lung cancer cells both in vitro and as xenografts [50].
2.9. Hallmark 6: tissue invasion and metastasis
Tissue invasion and metastasis constitute another hallmark that is difficult to study using tumor cultures. As with angiogenesis, Three-dimensional extracellular matrix culture, on substrates such as Matrigel, restores many aspects of the differentiated state to non-malignant cells from a variety of tissues [51]. Use of such models has been applied to the study and inter-relationships of cancer related genes in tumor cells and models of multistage lung pathogenesis [52,53].
2.10. An enabling characteristic: genome instability
Acquisition of the six hallmarks discussed previously lead to genomic instability, as manifested by aneuploidy and cytogenetic aberrations. Spectral karyotyping (SKY) has indicated the complexity of the lung cancer genome of both tumors and cell lines [54–56]. These changes include amplifications (visible as double minutes or homogeneously staining regions), reciprocal and unbalanced translocations. Recurrent (non-random) changes may indicate the site of activation or loss of genes important in the pathogenesis of lung cancer.
Because tumors demonstrate, to varying degrees, genomic instability, tumor cell lines inherit this characteristic. The degree genomic instability is dependent, to some extent, on the number of cell divisions that have occurred since its onset. As cell lines have population short doubling times (hours or days) compared to tumors (months) it is to be expected that lines develop more genetic (and epigenetic changes) over the same time period. However, when we directly compared the changes in long cultured cell lines with their respective tumors, we came to the conclusion that “NSCLC cell lines in the large majority of instances retain the properties of their parental tumors for lengthy culture periods. NSCLC cell lines appear very representative of the lung cancer tumor from which they were derived and thus provide suitable model systems for biomedical studies of this important neoplasm” [16].
2.11. Other applications
One approach to identification of genes essential for tumor growth is to use genome-wide RNAi screens of cancer cell lines to identify multiple synthetic lethal interactions [57,58]. While initial attempts in lung cancer focused on identification of chemosensitizer loci [58], our collaborators have used such screens of multiple lung cancer lines to identify many new potential oncogenes whose knockdown results in growth cessation (Michael White, in collaboration with the authors).
Cell lines have proved advantageous in determining in vitro drug sensitivity to conventional and targeted therapies and for the understanding of mechanisms of drug resistance [24,59]. The advent of RNA-mediated interference (siRNA)-based functional genomics provides the opportunity to derive unbiased comprehensive collections of validated gene targets supporting critical biological systems outside the framework of preconceived notions of mechanistic relationships. We have combined a high-throughput cell-based one-well/one-gene screening platform with a genome-wide synthetic library of chemically synthesized small interfering RNAs for systematic interrogation of the molecular underpinnings of cancer cell chemoresponsiveness [58]. More recently a similar screen has been used to identify multiple putative oncogenes causing “oncogene addiction” in NSCLC cells (JDM and Michael White, unpublished data).
High-throughput biological assays such as microarrays permit us ask very detailed questions about how diseases operate, and promise to let us personalize therapy. However data processing is often inadequately described in the reports, leading to problems in interpretation and application. Methods for correcting the errors causing these problems have been described, permitting more widespread and beneficial applications of data analyses from these reports [60].
The concept that there are multi-potent, self-renewing and proliferative progenitor cell populations throughout the respiratory tree and also for lung cancers, has resulted in making pulmonary stem cell biology a growing field in biomedicine [61]. Cell lines have made major contributions to this still evolving field [62,63].
Cell lines have been used to study the relative radiosensitivity of classic or typical SCLC cells compared to NSCLC or the MYC amplified variant form of SCLC [64,65]. Cell lines have also demonstrated the complex inter-relationships between TKI therapy and radiotherapy, suggesting possible new therapeutic approaches for NSCLC [66].
A major new finding with major implications for cancer research has been the discovery of noncoding microRNAs [67]. These small RNAs (about 22 nucleotides long) regulate gene expression by hybridizing to complementary sequences in the 3′ untranslated region, and may influence the effects of oncogenes, tumor suppressor genes or methylation. Lung cancer cell lines have contributed to our understanding of microRNA regulation of the critical driver mutations K-RAS and EGFR [68,69]. Examination of a homozygously deleted region in a lung cancer cell line led to the identification of new microRNAs [70].
2.12. Pathway analysis for prediction of drug sensitivity
Utilization of gene expression signatures for pathway analysis has been suggested as an improved method for rational therapeutic drug selection (2). Recently we performed a detailed study of the epidermal growth factor receptor (EGFR) signaling pathway in a large panel of lung cancer cell lines and correlated the findings with response to tyrosine kinase inhibitors (TKIs) [24]. Mutations in seven pathway related genes were detected at frequencies similar to those described in resected tumors. The resistance or sensitivity of all cell lines, without exception, could be explained by their molecular signatures. This study confirmed and extended the clinical observations that (a) mutations of EGFR and copy number gains of EGFR and HER2 were independent factors related to TKI sensitivity, in descending order of importance and that (b) mutations of K-RAS were associated with increased in vitro resistance. Thus, in some cases after decades in culture, the cell lines retained the driver mutations, secondary resistance associated mutations and copy number gains, and their patterns of TKI response.
2.13. The use of lung cancer cell lines for global analyses
Lung cancer cell lines offer suitable models for the application of global approaches for the analysis of the genome (high-throughput DNA sequencing) [71], the transcriptome (microarray analyses) [72,73], the methylome (detection of genome-wide methylated sequences) [74], microRNA analyses and copy number changes [17,27,75]. As a result of these and similar studies on tumors and cell lines, the landscape of the cancer genome is rapidly evolving in complexity [14].
3. Immortalized lung epithelial cells as models for studying lung tumor transformation
Normal lung epithelial cell are valuable tools for studying the multistage pathogenesis of lung cancers. Two types of normal culture models are available for studies: primary cultured cells and immortalized cell lines. The major advantage of primary cell models is that it is more close to the lung tissue origin and thus more resemble the lung cell physiology. However, the inter-individual variability, the limited resource and more importantly, the finite life span that does not allow long term genetic manipulations, make the primary cell model less desirable. The major advantage of immortalized non-malignant epithelial lines is the cell lines can be genetically modified in an isogenic system to systematically study the genetic alteration in lung cancer. To this end, we have established more than 50 non-malignant lung epithelial cell lines including bronchial epithelial cells (HBEC) and small airway epithelial cells (HSAEC) [76] In addition, we have introduced several oncogenes and knocked down several tumor suppressor genes individually or in combination in these cells [77]. These genetically manipulated lines represent stages in the multistage pathogenesis of lung cancer. Thus we generated a spectrum of isogenic cell lines mimicking the common genetic alteration in lung tumor patients and provided the stable cells for further genetic modifications.
3.1. Comparing viral gene and non-viral gene immortalized cells
Normal lung epithelial cells stop proliferating in tissue cultures beyond 10–20 doublings. This is due to the stress related cell cycle arrest and to the telomere shortening-induced senescence. Thus, overcoming the cell cycle arrest and senescence is the key for achieving immortalization. In earlier work, SV40T antigens or E6/E7 genes from human papilloma virus 16 had been used for immortalizing bronchial epithelial cells [78–80]. These viral gene immortalized HBECs were aneuploid with numerical karyotypic abnormalities and some of the cells became tumorigenic at around passage 30 [78,80]. To avoid the usage of viral genes, we have developed a method of immortalizing HBECs with non-viral gene cyclin-dependent kinase 4 (Cdk4) from mouse in combination with the catalytic subunit of human telomerase (hTERT) [76]. To date, we have immortalized 40 human HBECs and over 10 HSAEC using Cdk4 and hTERT. We also immortalized several of the donor cells using both E6/E7/hTERT and Cdk4/hTERT methods as comparisons. We found many of these cells having near normal karyotype with duplications in chromosome 5 and 20 regions. The global gene expression pattern of the Cdk4/hTERT immortalized HBECs is more similar to the parental un-immortalized cells than the cells immortalized with HPV16 E6/E7/hTERT [76]. Cdk4/hTERT immortalized cells in many ways retain their normal phenotype including (1), no anchorage independent growth in soft agar and no tumor growth in nude or SCID mice (2), maintain intact p53 check point pathway (3). The growth is highly dependent on epidermal growth factor (EGF) and inhibited by EGF receptor (EGFR) inhibitors, a hall mark of epithelial cells (4), can be differentiated into ciliated columnar cells and mucin-producing cells in the presence of serum in three-dimensional organic culture [77,81]. One of the immortalized HBECs (HBEC3) has been cultured for more than 240 populations without losing these normal characteristics. Our results suggest that these cells have minimal disturbance in normal cellular pathways and are thus good models for studying multiple gene alteration in lung cancer. Recently, Fulcher et al. reported the immortalization of HBECs using polycomb ring finger oncogene Bmi-1 in combination with hTERT gene. It remains to be seen if these cells are fully immortalized and if the phenotype of the cells are stable at later passages [82]. Of interest, duplication of regions of chromosomes 5 and 20 have been found in many of the immortalized HBECs by us and by others suggesting the importance of genes in these regions in cellular immortalization [83]. Thus, it is a great challenge to minimize these chromosomal alterations by improving the current immortalization methods.
3.2. Malignant transformation effects by knocking-down p53 and introducing K-RASV12 mutation in HBECs
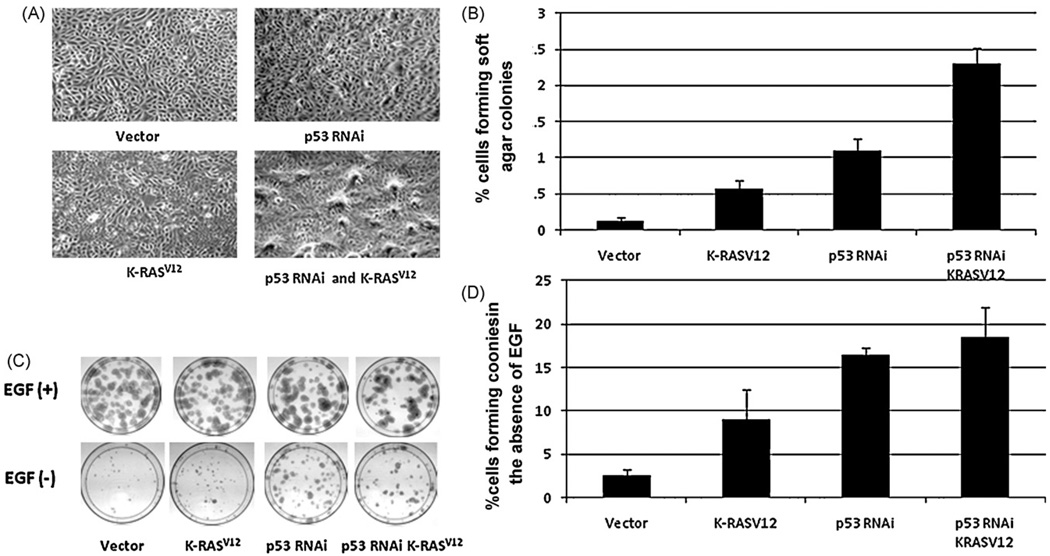
The most common gene alterations in lung cancer are p53 and K-RAS mutations. We knocked down the expression of p53 by RNAi and introduced K-RASV12 mutation into immortalized HBEC3 cells for studying the function of these genes in tumor transformation. We showed that knocking-down p53 (p53RNAi) and introducing K-RASV12 individually or in combination increased the transformation of these cells including the enhanced anchorage independent growth, reduction of EGF dependent growth and increased saturation density in culture, but these alteration are not sufficient for cells to grow tumors in nude or SCID mice (Fig. 1). Recently, we have introduced c-myc gene into the HBECs with p53 RNAi and K-RASV12 and found these cells are tumorigenic in SCID mice (Mitsuo Satoincollaboration with the authors, unpublished data). Thus, our results indicated that tumor transformation requires more than four genetic modifications such that inhibition of RB/p16 pathway, prevention of the telomere shortening, inactivation of p53 molecule and introduction of oncogenic K-RASV12 are not sufficient for full tumorigenic transformation.
Fig. 1.
Malignant transformation phenotype of p53 RNAi and K-RASV12 expressing HBEC3 cells. (Panel A) Contact inhibition is reduced in p53 RNAi and K-RASV12 expressing HBEC3 cells. Cells in vector control (top left) form smooth single layer cells at saturation density, p53 RNAi cells (top right) or K-RASV12 cells (bottom left) have partially lost contact inhibition and have overcrowded areas, and cells with p53 RNAi/K-RASV12 (bottom right) have lost contact inhibition and cells pile up forming many foci. (Panel B) Anchorage independent growth is increased considerably in p53 RNAi and K-RASV12 expressing HBEC3 cells. 1000 cells were grown in soft agar in the presence of EGF (5 ng/ml) and colonies were counted after 2 weeks. The numbers of colonies formed are increased 6, 11 and 23 fold in p53 RNAi, K-RASV12 and p53 RNAi/K-RASV12 expressing HBEC3 cells respectively compared with the vector control. (Panel C) Growth factor dependency is reduced in p53 RNAi and K-RASV12 expressing HBEC3 cells. 200 cells were cultured in KSFM medium in the presence (top, 5ng/ml)or absence of EGF (bottom) and colonies were stained with methylene blue after 2 weeks. Similar number of colonies formed in the presence of EGF in all cells (top), the colony number dramatically reduced in the absence of EGF in vector control (bottom left). (PanelD) Quantitation of colonies counted in bottom panel C. The number of colonies formed increased 4, 8 and 9-fold in p53 RNAi, K-RASV12 and p53 RNAi/K-RASV12 expressing HBEC3 cells respectively compared with the vector control. Figure modified from our previously published work [77].
3.3. Malignant transformation effects of wild-type and mutant EGFR in HBECs
One of the common mutations in lung adenocarcinoma occurs in the EGFR gene which confer an “addiction” to the oncogene [84]. As a result EGFR gene mutations (or increased copy number) offers great promise for targeted therapy [85]. Indeed, large scale clinical trials showed there is improved overall or progression free survival in patients treated with TKIs compared with patients in placebo group [85]. Interestingly, the survival benefit was not limited to patients with EGFR alterations. In addition, it was reported that patients with different EGFR mutation responded differently to TKIs [86]. These reports highlighted the current confusion regarding which lung cancer patient will respond and have survival benefit with TKIs. Thus, it is of great importance to study the biological process that involved in EGFR alterations. We introduced wild-type and mutated EGFR into the immortalized HBECs with or without oncogenic modification to dissect the pathways that altered by EGFR mutations and over-expression [77,87,88]. The major findings from our studies can be summarized as the following:
TKIs inhibited the proliferation and colony formation in HBECs with or without oncogene modification although the mechanism of the inhibition may be different.
Introducing either EGFR L858R mutation or exon 19 deletion (ΔE746–A750) enhanced the anchorage independent growth of HBEC cells. However, the enhancement is different with different mutations depending on the status of p53 gene.
Exogenously expressed wild-type or mutant EGFR in HBECs resulted in activation of EGFR which can be shown by phosphorylation of EGFR at four sites (Y845, Y992, Y1045, and Y1068). The extent of phosphorylation also depends on the p53 status in the cells.
Expression of either L858R or ΔE746–E750 but not wild-type EGFR resulted in dramatically increased sensitivity to ionizing radiation.
Recently, Guha et al. developed tyrosine phosphorylation profiles in HBECs exogenously expressing wild-type and mutant EGFR using stable isotope labeling and quantitative mass spectrometry [89]. The authors showed expressing either L858R or ΔE746–E750 resulted increased tyrosine phosphorylation of proteins in numerous signaling pathways compared with wild-type EGFR expressing cells. They confirmed the involvement of EGFR in previously known pathways but also discovered the involvement of unknown signaling molecules, for example polymerase transcript release factor, which had not been previously implicated in EGFR signaling.
3.4. Establishing lung tumor cell lines and matched normal bronchial cells lines
Matched tumor cell line and normal bronchial cell line from same individual are powerful tools for exploring the molecular and the functional difference between the normal and tumors. The paired cells are also ideal reagents for screening drugs that specifically kill tumor cells without damaging normal cells. We have recently established two matched normal and tumor cell lines from two NSCLC patients. Johnson et al. tested the biological activity of five lead compounds that reversing the methylation of potential tumor suppressor genes in these two matched lines and found two of the compounds selectively killed NSCLC while an other three compounds had no select activity against the tumor cell lines [90]. Thus this is a great example showing the utility of the paired cell lines.
3.5. Immortalized human small airway epithelial cells from the peripheral compartment of the lung
A majority of adenocarcinomas develop in the peripheral airways. Two recent findings:
Adenocarcinoma is the commonest form of lung cancer in never smokers.
EGFR mutations target the peripheral airway and give rise to lung adenocarcinomas, making it particular important to study the tumor transformation process in small airways.
There had been a few reports about immortalizing HSAECs but no detailed characterization of the cells available [91,92]. We have immortalized over 10 HSAECs using Cdk4 and hTERT. We showed these HSAECs share many characteristics of HBECs including being non-tumorigenic and lacking anchorage independent growth. In addition, these cells can be induced to express type 1 and type 2 pneumocyte markers in matrigel (BG unpublished results). We are currently exploring the difference between the immortalized HBEC and HSAECs.
3.6. Using fully transformed HBEC and HSAEC as models for studying lung cancer stem cells
Lung cancer kills more people than any other cancer in the world. In spite of the variety of new drug development, no major improvements in overall survival have been achieved. A new wave of treatment strategies focused on cancer stem cells. The hypothesis is a rare population of cancer cells, arise either from mutation of normal stem cells or gain stem cell properties, are responsible for tumor initiation, metastasis and drug resistant in cancer. The identification and characterization of cancer stem cells may be crucial for developing effective therapies. Cancer stem cells have been identified and characterized in leukemia and several solid tumors including breast, brain, prostate, colon and pancreatic tumors. However, the identification and characterization of lung cancer stem cells are lacking. We have shown the immortalized HBECs express basal cell markers such as p63 and keratin 14 and can differentiate in the presence of serum [76,81]. Thus immortalized HBECs may be enriched for cells with stem cell properties. It is possible the fully transformed HBEC cells, by introducing oncogenes or inactivation of tumor suppressor genes, are preferred models for studying lung cancer stem cells.
4. Concluding remarks
Cell lines, while not ideal model systems offer many advantages that complement the use of tumor tissues and animal models for the study of lung cancer. The very large number of lung cancer cells lines (more than for any other epithelial cancer) and their wide distribution to the scientific community have resulted in over 8000 citations in the medical literature. Clearly such a large, recent body of literature cannot be summarily dismissed as meaningless.
The major disadvantages of tumor cell lines that have emerged are:
Genetic instability or drift during long term passage.
Selective growth of subpopulations on initial culture or during long term passage.
Lack of interaction with other non-tumor components (stromal, vascular, inflammatory).
Genetic instability is a natural feature in the progression of tumors, caused by the development of the hallmarks of cancer. It is manifested both in vivo and in vitro. The rapid population doubling time of tumors raises the possibility instability may progress at a faster rate in vitro. Selection during successful culture of subpopulations having growth advantages may result in the cultured cells being less differentiated than the original tumor cells, and expressing greater epithelial to mesenchymal transition. While these concerns are real, they may be reduced by correct culture techniques. Cell cultures may retain the full program of neuroendocrine differentiation or expression of peripheral airway or squamous cell markers. Both tumors and cell lines contain multiple “passenger” mutations. However, because “driver” mutations are associated with the appearance or maintenance of the malignant phenotype, they are usually (if not always) maintained during cultured life. The ability to distinguish driver from passenger mutations is crucial in understanding the role of genetic changes important for carcino-genesis. Of great importance, all of the recurrent driver mutations, tumor suppressor genes and methylated genes demonstrated to play important roles in lung cancer pathogenesis are represented in lung cancer cell lines, providing invaluable reagents to investigate their roles in lung cancer. Interactions with stroma and other non-malignant cells is a major limitation of cultured tumor cells. Attempts at developing 3D models, models for invasiveness and differentiation etc have been only partially successful, and improvements in our ability to develop relevant models to study the interactions of tumor cells with their environment are needed.
Another limitation of tumor cell lines is the difficulty of studying multistage pathogenesis in vitro. In this report we describe an in vitro system using immortalized central or peripheral airway cells to study tumors arising in these two major compartments of the lung.
Contamination is a major problem. Mycoplasma and other microbial contaminations may skew results and cause contamination of other cultures. Perhaps more important is the widespread contamination by other cells, either of human or non-human species. Such contaminations have cast into doubt much of the in vitro studies with thyroid cancer [93]. However, contamination is largely an “iatrogenic disease” caused by the carelessness, ignorance or apathy on the part of the scientist. With proper and regular monitoring for provenance and contamination, this condition could be largely prevented or detected and corrected before it did major harm. Application of the appropriate measures require co-operation between scientists and editors of scientific journals who should require identification of cell line provenance as a prerequisite for publication [94,95].
Without cell lines our knowledge of lung cancer, its origins and treatment would be much less advanced. Cell lines have, and will continue to be invaluable tools for discovery. However, it remains important to know the model—how closely does it resemble the original tumor, what are the driver mutations, are differentiated properties maintained? Knowledge of these factors permits an evaluation of the in vitro model and the ability to interpret data from studies of the model.
Acknowledgments
We thank Drs. Will Lockwood and Wan Lam of the British Columbia Cancer Center, Vancouver, Canada, for their assistance with preparing Table 2. Supported by grants from the National Cancer Institute Bethesda, Maryland (Specialized Program of Research Excellence in Lung Cancer, P50CA70907, and Early Detection Research Network, U01CA084971) and from the Canary Foundation, Palo Alto, California.
Footnotes
Conflict of interest statement
The authors report no conflict of interests.
References
- 1.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 2.Gazdar AF, Minna JD. NCI series of cell lines: an historical perspective. J Cell Biochem. 1996 Suppl 24:1–11. doi: 10.1002/jcb.240630502. [DOI] [PubMed] [Google Scholar]
- 3.Oie HK, Russell EK, Carney DN, Gazdar AF. Cell culture methods for the establishment of the NCI series of lung cancer cell lines. J Cell Biochem. 1996 Supplement 24:24–31. doi: 10.1002/jcb.240630504. [DOI] [PubMed] [Google Scholar]
- 4.Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nat Rev Cancer. 2002;2:315–319. doi: 10.1038/nrc775. [DOI] [PubMed] [Google Scholar]
- 5.van Staveren WC, Solis DY, Hebrant A, Detours V, Dumont JE, Maenhaut C. Human cancer cell lines: experimental models for cancer cells in situ? For cancer stem cells? Biochim Biophys Acta. 2009;1795:92–103. doi: 10.1016/j.bbcan.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazdar AF, Kurvari V, Virmani A, Gollahon L, Sakaguchi M, Westerfield M, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78:766–774. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 9.Gazdar AF, Carney DN, Russell EK, Sims HL, Baylin SB, Bunn PA, Jr, et al. Establishment of continuous, clonable cultures of small-cell carcinoma of lung which have amine precursor uptake and decarboxylation cell properties. Cancer Res. 1980;40:3502–3507. [PubMed] [Google Scholar]
- 10.Coe BP, Lockwood WW, Girard L, Chari R, Macaulay C, Lam S, et al. Differential disruption of cell cycle pathways in small cell and non-small cell lung cancer. Br J Cancer. 2006;94:1927–1935. doi: 10.1038/sj.bjc.6603167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sriuranpong V, Borges MW, Ravi RK, Arnold DR, Nelkin BD, Baylin SB, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 2001;61:3200–3205. [PubMed] [Google Scholar]
- 12.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 13.Futreal PA. Backseat drivers take the wheel. Cancer Cell. 2007;12:493–494. doi: 10.1016/j.ccr.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wistuba II, Behrens C, Milchgrub S, Salahuddin S, Ahmadian M, Virmani AK, et al. Comparison of features of human breast cancer cell lines and their corresponding tumors. Clin Cancer Res. 1998;4:2931–2938. [PubMed] [Google Scholar]
- 16.Wistuba II, Bryant D, Behrens C, Milchgrub S, Virmani AK, Ashfaq R, et al. Comparison of features of human lung cancer cell lines and their corresponding tumors. Clin Cancer Res. 1999;5:991–1000. [PubMed] [Google Scholar]
- 17.Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garnis C, Lockwood WW, Vucic E, Ge Y, Girard L, Minna JD, et al. High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer. 2006;118:1556–1564. doi: 10.1002/ijc.21491. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 20.Mitsudomi T, Viallet J, Mulshine JL, Linnoila RI, Minna JD, Gazdar AF. Mutations of ras genes distinguish a subset of non-small-cell lung cancer cell lines from small-cell lung cancer cell lines. Oncogene. 1991;6:1353–1362. [PubMed] [Google Scholar]
- 21.Slebos RJ, Rodenhuis S. The molecular genetics of human lung cancer. Eur Respir J. 1989;2:461–469. [PubMed] [Google Scholar]
- 22.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shigematsu S, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba I, et al. Clinical and biological features associated with Epidermal Growth Factor Receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 24.Gandhi J, Zhang J, Xie Y, Soh J, Shigematsu H, Zhang W, et al. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS One. 2009;4:e4576. doi: 10.1371/journal.pone.0004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28 Suppl 1:24–31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 27.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 29.Soh J, Okumura N, Lockwood WW, Yamamoto H, Shigematsu H, Zhang W, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009;4:e4566. doi: 10.1371/journal.pone.0007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inamura K, Takeuchi K, Togashi Y, Hatano S, Ninomiya H, Motoi N, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol. 2009;22:508–515. doi: 10.1038/modpathol.2009.2. [DOI] [PubMed] [Google Scholar]
- 31.Harbour JW, Lai SL, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ. Abnormalities in structure and expression of the human retinoblastoma genein SCLC. Science. 1988;241:353–357. doi: 10.1126/science.2838909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, et al. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246:491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 33.Otterson GA, Kratzke RA, Coxon A, Kim YW, Kaye FJ. Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wildtype RB. Oncogene. 1994;9:3375–3378. [PubMed] [Google Scholar]
- 34.Virmani AK, Fong KM, Kodagoda D, McIntire D, Hung J, Tonk V, et al. Allelotyping demonstrates common and distinct patterns of chromosomal loss in human lung cancer types. Genes Chrom Cancer. 1998;21:308–319. doi: 10.1002/(sici)1098-2264(199804)21:4<308::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62:3659–3662. [PubMed] [Google Scholar]
- 36.Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Nieto S, Sanchez-Cespedes M. BRG1 and LKB1: tales of two tumor suppressor genes on chromosome 19p and lung cancer. Carcinogenesis. 2009;30:547–554. doi: 10.1093/carcin/bgp035. [DOI] [PubMed] [Google Scholar]
- 38.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 39.Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whang-Peng J, Kao-Shan CS, Lee EC, Bunn PA, Carney DN, Gazdar AF, et al. Specific chromosome defect associated with human small-cell lung cancer; deletion 3p(14–23) Science. 1982;215:181–182. doi: 10.1126/science.6274023. [DOI] [PubMed] [Google Scholar]
- 41.Shames DS, Minna JD, Gazdar AF. DNA methylation in health, disease, and cancer. Curr Mol Med. 2007;7:85–102. doi: 10.2174/156652407779940413. [DOI] [PubMed] [Google Scholar]
- 42.Wilson IM, Davies JJ, Weber M, Brown CJ, Alvarez CE, MacAulay C, et al. Epige-nomics: mapping the methylome. Cell Cycle. 2006;5:155–158. doi: 10.4161/cc.5.2.2367. [DOI] [PubMed] [Google Scholar]
- 43.Lin Y, Liu X, Yue P, Benbrook DM, Berlin KD, Khuri FR, et al. Involvement of c-FLIP and survivin down-regulation in flexible heteroarotinoid-induced apoptosis and enhancement of TRAIL-initiated apoptosis in lung cancer cells. Mol Cancer Ther. 2008;7:3556–3565. doi: 10.1158/1535-7163.MCT-08-0648. [DOI] [PubMed] [Google Scholar]
- 44.Lu B, Mu Y, Cao C, Zeng F, Schneider S, Tan J, et al. Survivin as a therapeutic target for radiation sensitization in lung cancer. Cancer Res. 2004;64:2840–2845. doi: 10.1158/0008-5472.can-03-3547. [DOI] [PubMed] [Google Scholar]
- 45.Cao C, Mu Y, Hallahan DE, Lu B. XIAP and survivin as therapeutic targets for radiation sensitization in preclinical models of lung cancer. Oncogene. 2004;23:7047–7052. doi: 10.1038/sj.onc.1207929. [DOI] [PubMed] [Google Scholar]
- 46.Shivapurkar N, Toyooka S, Eby MT, Huang CX, Sathyanarayana UG, Cunningham HT, et al. Differential inactivation of caspase-8 in lung cancers. Cancer Biol Ther. 2002;1:65–69. doi: 10.4161/cbt.1.1.45. [DOI] [PubMed] [Google Scholar]
- 47.Shivapurkar N, Reddy JL, Matta H, Sathyanarayana UG, Huang CX, Toyooka S, et al. Loss of expression of death-inducing signaling complex (DISC) components in lung cancer cell lines and the influence of MY Camplification. Oncogene. 2002;55:8510–8514. doi: 10.1038/sj.onc.1205941. [DOI] [PubMed] [Google Scholar]
- 48.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 49.Norrby K. In vivo models of angiogenesis. J Cell Mol Med. 2006;10:588–612. doi: 10.1111/j.1582-4934.2006.tb00423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu W, O’Reilly MS, Langley RR, Tsan RZ, Baker CH, Bekele N, et al. Expression of epidermal growth factor (EGF)/transforming growth factor-alpha by human lung cancer cells determines their response to EGF receptor tyrosine kinase inhibition in the lungs of mice. Mol Cancer Ther. 2007;6:2652–2663. doi: 10.1158/1535-7163.MCT-06-0759. [DOI] [PubMed] [Google Scholar]
- 51.Kenny PA. Three-dimensional extracellular matrix culture models of EGFR signalling and drug response. Biochem Soc Trans. 2007;35:665–668. doi: 10.1042/BST0350665. [DOI] [PubMed] [Google Scholar]
- 52.Upadhyay S, Liu C, Chatterjee A, Hoque MO, Kim MS, Engles J, et al. LKB1/STK11 suppresses cyclooxygenase-2 induction and cellular invasion through PEA3 in lung cancer. Cancer Res. 2006;66:7870–7879. doi: 10.1158/0008-5472.CAN-05-2902. [DOI] [PubMed] [Google Scholar]
- 53.Wang XQ, Li H, Van Putten V, Winn RA, Heasley LE, Nemenoff RA. Oncogenic K-Ras regulates proliferation and cell junctions in lung epithelial cells through induction of cyclooxygenase-2 and activation of metalloproteinase-9. Mol Biol Cell. 2009;20:791–800. doi: 10.1091/mbc.E08-07-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sy SM, Fan B, Lee TW, Mok TS, Pang E, Yim A, et al. Spectral karyotyping indicates complex rearrangements in lung adenocarcinoma of nonsmokers. Cancer Genet Cytogenet. 2004;153:57–59. doi: 10.1016/j.cancergencyto.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 55.Grigorova M, Lyman RC, Caldas C, Edwards PA. Chromosome abnormalities in 10 lung cancer cell lines of the NCI-H series analyzed with spectral karyotyping. Cancer Genet Cytogenet. 2005;162:1–9. doi: 10.1016/j.cancergencyto.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Ogiwara H, Kohno T, Nakanishi H, Nagayama K, Sato M, Yokota J. Unbalanced translocation, a major chromosome alteration causing loss of heterozygosity in human lung cancer. Oncogene. 2008;27:4788–4797. doi: 10.1038/onc.2008.113. [DOI] [PubMed] [Google Scholar]
- 57.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 59.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baggerly KA, Coombes KR. Deriving chemosensitivity from cell lines: forensic bioinformatics and reproducible reasearch in high throuput biology. Ann Appl Statist. 2009 in press. [Google Scholar]
- 61.Yagui-Beltran A, He B, Jablons DM. The role of cancer stem cells in neoplasia of the lung: past, present and future. Clin Transl Oncol. 2008;10:719–725. doi: 10.1007/s12094-008-0278-6. [DOI] [PubMed] [Google Scholar]
- 62.Seo DC, Sung JM, Cho HJ, Yi H, Seo KH, Choi IS, et al. Gene expression profiling of cancer stem cell in human lung adenocarcinoma A549 cells. Mol Cancer. 2007;6:75. doi: 10.1186/1476-4598-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meng X, Li M, Wang X, Wang Y, Ma D. Both CD133+ and CD133− subpop-ulations of A549 and H446 cells contain cancer-initiating cells. Cancer Sci. 2009;100:1040–1046. doi: 10.1111/j.1349-7006.2009.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan FJ, Carmichael J, Glatstein E, Mitchell JB. Radiation biology of lung cancer. J Cell Biochem Suppl. 1996;24:152–159. doi: 10.1002/jcb.240630510. [DOI] [PubMed] [Google Scholar]
- 65.Carmichael J, Degraff WG, Gamson J, Russo D, Gazdar AF, Levitt ML, et al. Radiation sensitivity of human lung cancer cell lines. Eur J Cancer Clin Oncol. 1989;25:527–534. doi: 10.1016/0277-5379(89)90266-6. [DOI] [PubMed] [Google Scholar]
- 66.Chen DJ, Nirodi CS. The epidermal growth factor receptor: a role in repair of radiation-induced DNA damage. Clin Cancer Res. 2007;13:6555–6560. doi: 10.1158/1078-0432.CCR-07-1610. [DOI] [PubMed] [Google Scholar]
- 67.Eder M, Scherr M. MicroRNA and lung cancer. N Engl J Med. 2005;352:2446–2448. doi: 10.1056/NEJMcibr051201. [DOI] [PubMed] [Google Scholar]
- 68.Weiss GJ, Bemis LT, Nakajima E, Sugita M, Birks DK, Robinson WA, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19:1053–1059. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]
- 69.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamada H, Yanagisawa K, Tokumaru S, Taguchi A, Nimura Y, Osada H, et al. Detailed characterization of a homozygously deleted region corresponding to a candidate tumor suppressor locus at 21q11–21 in human lung cancer. Genes Chrom Cancer. 2008;47:810–818. doi: 10.1002/gcc.20582. [DOI] [PubMed] [Google Scholar]
- 71.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 72.Zhao X, Li C, Paez JG, Chin K, Janne PA, Chen TH, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 73.Henderson LJ, Coe BP, Lee EH, Girard L, Gazdar AF, Minna JD, et al. Genomic and gene expression profiling of minute alterations of chromosome arm 1p in small-cell lung carcinoma cells. Br J Cancer. 2005;92:1553–1560. doi: 10.1038/sj.bjc.6602452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shames DS, Girard L, Gao B, Sato M, Lewis CM, Shivapurkar N, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006;3:e486. doi: 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lockwood WW, Chari R, Coe BP, Girard L, Macaulay C, Lam S, et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27:4615–4624. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 77.Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 78.Reddel RR, Salghetti SE, Willey JC, Ohnuki Y, Ke Y, Gerwin BI, et al. Development of tumorigenicity in simian virus 40-immortalized human bronchial epithelial cell lines. Cancer Res. 1993;53:985–991. [PubMed] [Google Scholar]
- 79.Viallet J, Liu C, Edmond J, Tsao M-S. Characterization of human bronchial epithelial cells immortalized by the E6 and E7 genes of human papillomavirus type 16. Exp Cell Res. 1994;212:36–41. doi: 10.1006/excr.1994.1115. [DOI] [PubMed] [Google Scholar]
- 80.Reddel RR, Ke Y, Gerwin BI, McMenamin MG, Lechner JF, Su RT, et al. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res. 1988;48:1904–1909. [PubMed] [Google Scholar]
- 81.Vaughan MB, Ramirez RD, Wright WE, Minna JD, Shay JW. A three-dimensional model of differentiation of immortalized human bronchial epithelial cells. Differentiation. 2006;74:141–148. doi: 10.1111/j.1432-0436.2006.00069.x. [DOI] [PubMed] [Google Scholar]
- 82.Fulcher ML, Gabriel SE, Olsen JC, Tatreau JR, Gentzsch M, Livanos E, et al. Novel human bronchial epithelial cell lines for cystic fibrosis research. Am J Physiol Lung Cell Mol Physiol. 2009;296:L82–L91. doi: 10.1152/ajplung.90314.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Farwell DG, Shera KA, Koop JI, Bonnet GA, Matthews CP, Reuther GW, et al. Genetic and epigenetic changes in human epithelial cells immortalized by telomerase. Am J Pathol. 2000;156:1537–1547. doi: 10.1016/S0002-9440(10)65025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 85.Gazdar AF. Personalized medicine and inhibition of EGFR signaling in lung cancer. N Engl J Med. 2009;361:1018–1020. doi: 10.1056/NEJMe0905763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jackman DM, Yeap BY, Sequist LV, Lindeman N, Holmes AJ, Joshi VA, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- 87.Das AK, Chen BP, Story MD, Sato M, Minna JD, Chen DJ, et al. Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res. 2007;67:5267–5274. doi: 10.1158/0008-5472.CAN-07-0242. [DOI] [PubMed] [Google Scholar]
- 88.Das AK, Sato M, Story MD, Peyton M, Graves R, Redpath S, et al. Non-small cell lung cancers with kinase domain mutations in the epidermal growth factor receptor are sensitive to ionizing radiation. Cancer Res. 2006;66:9601–9608. doi: 10.1158/0008-5472.CAN-06-2627. [DOI] [PubMed] [Google Scholar]
- 89.Guha U, Chaerkady R, Marimuthu A, Patterson AS, Kashyap MK, Harsha HC, et al. Comparisons of tyrosine phosphorylated proteins in cells expressing lung cancer-specific alleles of EGFR and KRAS. Proc Natl Acad Sci USA. 2008;105:14112–14117. doi: 10.1073/pnas.0806158105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson RL, Huang W, Jadhav A, Austin CP, Inglese J, Martinez ED. A quantitative high-throughput screen identifies potential epigenetic modulatorsofgene expression. Anal Biochem. 2008;375:237–248. doi: 10.1016/j.ab.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Piao CQ, Liu L, Zhao YL, Balajee AS, Suzuki M, Hei TK. Immortalization of human small airway epithelial cells by ectopic expression of telomerase. Carcinogenesis. 2005;26:725–731. doi: 10.1093/carcin/bgi016. [DOI] [PubMed] [Google Scholar]
- 92.Lundberg AS, Randell SH, Stewart SA, Elenbaas B, Hartwell KA, Brooks MW, et al. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 2002;21:4577–4586. doi: 10.1038/sj.onc.1205550. [DOI] [PubMed] [Google Scholar]
- 93.Schweppe RE, Klopper JP, Korch C, Pugazhenthi U, Benezra M, Knauf JA, et al. DNA profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J Clin Endocrinol Metab. 2008;93:4331–4341. doi: 10.1210/jc.2008-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nardone RM. Eradication of cross-contaminated cell lines: a call for action. Cell Biol Toxicol. 2007;23:367–372. doi: 10.1007/s10565-007-9019-9. [DOI] [PubMed] [Google Scholar]
- 95.Ringel MD. “Thyroid cancer” cell line misidentification: a time for proactive change. J Clin Endocrinol Metab. 2008;93:4226–4227. doi: 10.1210/jc.2008-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]