Abstract
In contrast to 5-methylcytosine (5-mC), which has been studied extensively1–3, little is known about 5-hydroxymethylcytosine (5-hmC), a recently identified epigenetic modification present in substantial amounts in certain mammalian cell types4,5. Here we present a method for determining the genome-wide distribution of 5-hmC. We use the T4 bacteriophage β-glucosyltransferase to transfer an engineered glucose moiety containing an azide group onto the hydroxyl group of 5-hmC. The azide group can be chemically modified with biotin for detection, affinity enrichment and sequencing of 5-hmC–containing DNA fragments in mammalian genomes. Using this method, we demonstrate that 5-hmC is present in human cell lines beyond those previously recognized4. We also find a gene expression level–dependent enrichment of intragenic 5-hmC in mouse cerebellum and an age-dependent acquisition of this modification in specific gene bodies linked to neurodegenerative disorders.
Parallel to the discovery of 5-hmC in mammalian genomes4,5, Tet proteins were shown to use dioxygen to oxidize 5-mC to 5-hmC in mammalian DNA5. Tet proteins are a group of iron(II)/α-ketoglutarate–dependent dioxygenases similar to the AlkB family proteins and hypoxia-inducible factor (HIF) prolyl-hydroxylases6,7. As Tet1 and Tet2 appear to affect embryonic stem (ES) cell maintenance and normal myelopoiesis, respectively8,9, these findings fostered speculation that this 5-hmC modification might also be an important epigenetic modification10.
To elucidate the biology of 5-hmC, the first step is to identify the locations of 5-hmC within genomic DNA, but so far it has remained challenging to distinguish 5-hmC from 5-mC and to enrich 5-hmC-containing genomic DNA fragments.
Widely used methods to probe 5-mC, such as bisulfite sequencing and methylation-sensitive restriction digestion, cannot discriminate between 5-hmC and 5-mC11,12. Anti-5-hmC antibodies have only recently become commercially available. However, attempts to use the antibodies to immuno-enrich 5-hmC-containing genomic DNA from complex genomes for sequencing have yet to be successful8. A single-molecule, real-time sequencing technology has been applied to distinguish between cytosine, 5-mC and 5-hmC, but further improvements are necessary to affinity-enrich 5-hmC–containing DNA and to achieve base-resolution sequencing13.
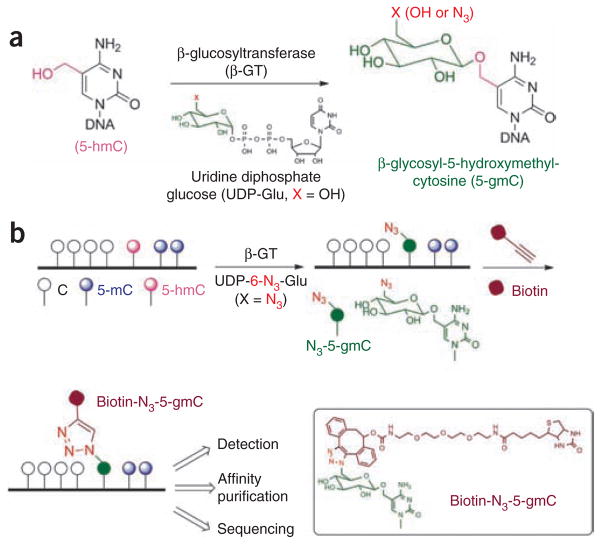
Here we present a chemical tagging technology to address both challenges. It has been shown that 5-hmC is present in the genome of the T-even bacteriophages. A viral enzyme, β-glucosyltransferase (β-GT), can catalyze the transfer of a glucose moiety from uridine diphosphoglucose (UDP-Glu) to the hydroxyl group of 5-hmC, yielding β-glucosyl-5-hydroxymethyl-cytosine (5-gmC) in duplex DNA14,15 (Fig. 1a). We took advantage of this enzymatic process and used β-GT to transfer a chemically modified glucose, 6-N3-glucose, onto 5-hmC for selective bio-orthogonal labeling of 5-hmC in genomic DNA (Fig. 1b). With an azide group present, a biotin tag or any other tag can be installed using Huisgen cycloaddition (click) chemistry for a variety of enrichment, detection and sequencing applications16–18.
Figure 1.
Selective labeling of 5-hmC in genomic DNA. (a) The hydroxyl group of 5-hmC in duplex DNA can be glucosylated by β-GT to form β-glucosyl-5-hydroxymethylcytosine (5-gmC) using UDP-Glu as a cofactor. (b) An azide group can be installed onto 5-hmC using chemically modified UDP-Glu (UDP-6-N3-Glu), which in turn can be labeled with a biotin moiety using click chemistry for subsequent detection, affinity purification and sequencing.
We used the biotin tag for high-affinity capture and/or enrichment of 5-hmC–containing DNA for sensitive detection and deep sequencing to reveal genomic locations of 5-hmC (Fig. 1b). The covalent chemical labeling coupled with biotin-based affinity purification provides considerable advantages over noncovalent, antibody-based immunoprecipitation as it ensures accurate and comprehensive capture of 5-hmC–containing DNA fragments, while still providing high selectivity.
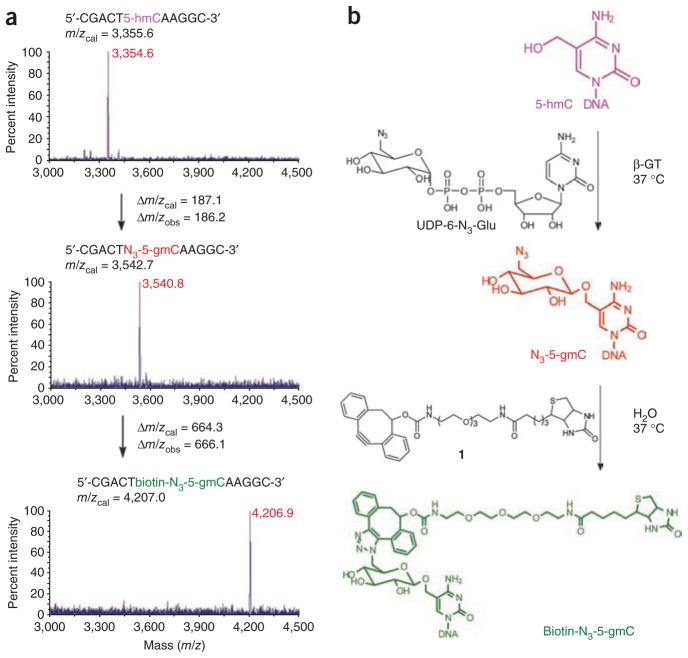
We chemically synthesized UDP-6-N3-Glu (Supplementary Fig. 1 and Supplementary Methods) and attempted the glycosylation reaction of an 11-mer duplex DNA containing a 5-hmC modification as a model system (Fig. 2). Wild-type β-GT worked efficiently using UDP-6-N3-Glu as the co-factor, showing only a sixfold decrease of the reaction rate compared to the native co-factor UDP-Glu (Supplementary Fig. 2). The 6-N3-glucose transfer reaction finished within 5 min with as low as 1% enzyme concentration. The identity of the resulting β-6-azide-glucosyl-5-hydroxymethyl-cytosine (N3-5-gmC) of the 11-mer DNA was confirmed by matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) analysis (Fig. 2). One can readily couple N3-5-gmC with dibenzocyclooctyne-modified biotin (compound 1) by copper-free click chemistry to introduce a biotin group (Fig. 2)19,20. Again, the identity of the 11-mer DNA with the biotin-N3-5-gmC label was confirmed by MALDI-TOF analysis (Fig. 2). High-performance liquid chromatography (HPLC) analysis indicated that the click chemistry is high yielding (~90%) (Supplementary Fig. 3). High-resolution mass spectroscopy (HRMS) analysis of the corresponding HPLC hydrolysates further verified that biotin-N3-5-gmC was formed (Supplementary Fig. 4).
Figure 2.
MS characterization of 5-hmC-, N3-5-gmC- and biotin-N3-5-gmC-containing 11-mer DNA in a model experiment. (a) MALDI-TOF of 5-hmC-, N3-5-gmC- and biotin-N3-5-gmC- containing 11-mer DNA, respectively, with the calculated molecular weight and observed molecular weight indicated. (b) Corresponding reactions of the β-GT–catalyzed formation of N3-5-gmC and the subsequent copper-free click chemistry to yield biotin-N3-5-gmC in duplex DNA. Reactions were performed in duplex DNA with the complementary strand; however, MS monitored the single-stranded DNA containing the modification.
The properties of 5-hmC in duplex DNA are quite similar to those of 5-mC in terms of its sensitivity toward enzymatic reactions such as restriction enzyme digestion and polymerization13–15. In an attempt to develop a method to differentiate these two bases in DNA, primer extension with a biotin-N3-5-gmC–modified DNA template was tested. Addition of streptavidin tetramer (binds biotin tightly) completely stops replication by Taq polymerase specifically at the modified position as well as one base before the modified position (Supplementary Fig. 5). Therefore, this method has the potential to provide single-base resolution of the location of 5-hmC in DNA loci of interest.
Next, we performed selective labeling of 5-hmC in genomic DNA from various cell lines and animal tissues. Genomic DNA from various sources was sonicated into small fragments (~100–500 base pairs), treated with β-GT in the presence of UDP-6-N3-Glu or regular UDP-Glu (control group) to yield N3-5-gmC or 5-gmC modifications and finally labeled with cyclooctyne-biotin (1) to install biotin. Because each step is efficient and bio-orthogonal, this protocol ensures selective labeling of most 5-hmC in genomic DNA. The presence of biotin-N3-5-gmC allows affinity enrichment of this modification and accurate quantification of the amount of 5-hmC in a genome using avidin–horseradish peroxidase (HRP).
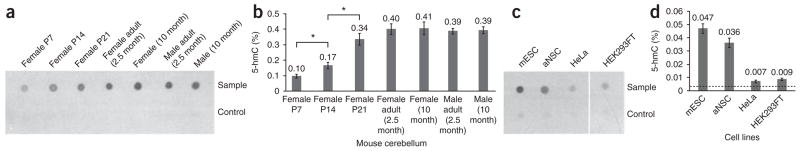
We determined the total amount of 5-hmC in mouse cerebellum at different stages of development (Fig. 3a,b). The control group showed almost no signal, demonstrating the high selectivity of this method. The amount of 5-hmC depends on the developmental stage of the mouse cerebellum (Fig. 3b). A gradual increase from post-natal day 7 (P7, 0.1% of total nucleotides in the genome) to adult stage (0.4% of total nucleotides) was observed21, which was further confirmed using antibody against 5-hmC through a dot-blot assay (Supplementary Fig. 6a). Our observation suggests that 5-hmC might play an important role in brain development. The 5-hmC level of mouse embryonic stem cells (mESC) was determined to be comparable to results reported previously (~0.05% of total nucleotides) (Fig. 3c,d)5. In addition, the amount of 5-hmC in mouse adult neural stem cells (aNSC) was tested, which proved comparable to that of mESC (~0.04% of total nucleotides) (Fig. 3c,d).
Figure 3.
Quantification of 5-hmC in various cell lines and tissues. (a) Dot-blot assay of avidin-HRP detection and quantification of mouse cerebellum genomic DNA containing biotin-N3-5-gmC. Top row: 40 ng of biotin-labeled samples using UDP-6-N3-Glu. Bottom row: 40 ng of control samples using regular UDP-Glu without biotin label. The exact same procedures were followed for experiments in both rows. P7, P14 and P21 represent postnatal day 7, 14 and 21, respectively. (b) Amounts of 5-hmC are shown in percentage of total nucleotides of mouse genome. *, P < 0.05, Student’s t-test; means ± s.e.m. for n = 4 experiments. (c) Dot-blot assay of avidin-HRP detection and quantification of genomic DNA samples from four cell lines (from same blot as in a), except that each dot contains 700 ng DNA. (d) Amounts of 5-hmC are shown in percentage of total nucleotides of the genome; means ± s.e.m. for n = 4 experiments. The dashed line indicates the limit of detection (~0.004%).
We also tested human cell lines (Fig. 3c,d). Notably, the presence of 5-hmC was detected in HeLa and HEK293FT cell lines, although in much lower abundance (~0.01% of total nucleotides) (Fig. 3d) than in other cells or tissues that have been previously reported to contain 5-hmC (previous studies did not show the presence of 5-hmC in HeLa cells due to the limited sensitivity of the methods employed4). These results suggest that this modification may be more widespread than previously anticipated. By contrast, no 5-hmC signal was detected in wild-type Drosophila melanogaster, consistent with a lack of DNA methylation in this organism22.
To further validate the utility of the method for biological samples we confirmed the presence of 5-hmC in the genomic DNA from HeLa cells. A monomeric avidin column was used to pull down the biotin-N3-5-gmC–containing DNA after genomic DNA labeling. These enriched DNA fragments were digested into single nucleotides, purified by HPLC and subjected to HRMS analysis. To our satisfaction, we obtained HRMS as well as MS/MS spectra of biotin-N3-5-gmC identical to the standard from synthetic DNA (Supplementary Fig. 4 and Fig. 6b,c). In addition, two 60-mer double-stranded (ds)DNAs, one with a single 5-hmC in its sequence and the other without the modification, were prepared. We spiked equal amounts of both samples into mouse genomic DNA and performed labeling and subsequent affinity purification of the biotinylated DNA. The pull-down sample was subjected to deep sequencing, and the result showed that the 5-hmC–containing DNA was >25-fold higher than the control sample (Supplementary Fig. 7).
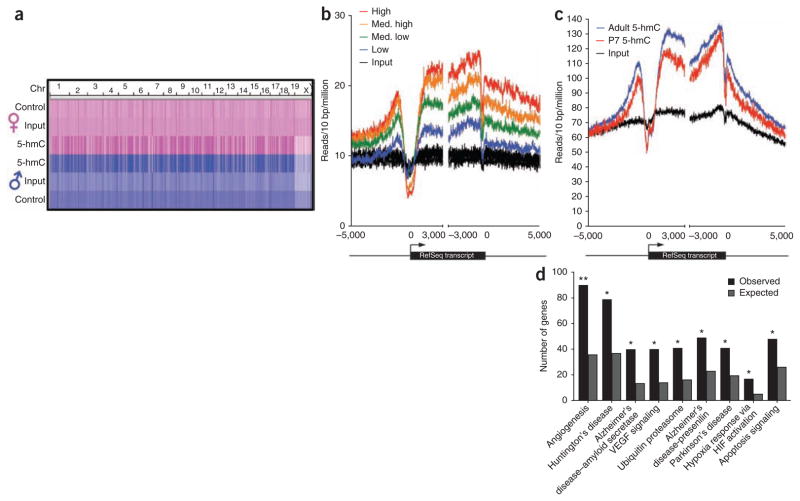
Next, we performed chemical labeling of genomic DNA from mouse cerebellum, subjecting the enriched fragments to deep sequencing such that 5-hmC–containing genomic regions could be identified. Initially, we compared male and female adult mice (2.5 months old), sequencing multiple independent biological samples and multiple libraries prepared from the same genomic DNA. Genome-scale density profiles are nearly identical between male and female and are clearly distinguishable from both input genomic DNA and control DNA labeled with regular glucose (no biotin) (Fig. 4a). Peak identification revealed a total of 39,011 high-confidence regions enriched consistently with 5-hmC in both male and female (Fig. 4a and Supplementary Table 1). All of the 13 selected, enriched regions were subsequently successfully verified in both adult female and male cerebellum by quantitative PCR (qPCR), whereas multiple control regions did not display enrichment (Supplementary Fig. 8).
Figure 4.
Genome-wide distribution of 5-hmC in adult mouse cerebellum and gene-specific acquisition of intragenic 5-hmC during mouse cerebellum development. (a) Genome-scale reproducibility of 5-hmC profiles and enrichment relative to genomic DNA and control-treated DNA in adult mouse cerebellum. Heatmap representations of read densities have been equally scaled and then normalized based on the total number of mapped reads per sample. Data are derived from a single lane of sequence from each condition. Control, UDP-Glu treated without biotin; Input, genomic DNA; 5-hmC, UDP-6-N3-Glu treated with biotin incorporated. (b) Metagene profiles of 5-hmC and input genomic DNA reads mapped relative to RefSeq transcripts expressed at different levels in adult mouse cerebellum. RefSeq transcripts were divided into four equally sized bins based on gene expression level and 5-hmC or input genomic DNA reads falling in 10-bp bins centered on transcription start sites or end sites. The reads were summed and normalized based on the total number of aligned reads (in millions). Input genomic DNA reads were mapped to each of the four gene expression level bins and are plotted here in black. The profiles completely overlap and so are collectively referred to as ‘Input’. (c) Proximal and intragenic enrichment of 5-hmC relative to surrounding regions in adult and P7 mouse cerebellum. Reads from 5-hmC-captured samples and input genomic DNA were summed in 10-bp intervals centered on either TSS or txEnds and normalized to the total number of aligned reads from each sample (in millions). (d) Enrichment of pathways associated with age-related neurodegenerative diseases in genes acquiring intragenic 5-hmC in adult mice relative to P7 mice. Shown are the number of genes that acquired 5-hmC in adult cerebellum and the number of genes expected based on the total number of genes associated with that pathway in mouse. **, P < 10−10; *, P < 10−5.
DNA methylation is widespread in mammalian genomes, with the exception of most transcription start sites (TSS)23–25. Previous studies have mostly assessed DNA methylation by bisulfite sequencing and methylation-sensitive restriction digests. It has since been appreciated that neither of these methods adequately distinguishes 5-mC from 5-hmC11,12. To determine the genome-wide distribution of 5-hmC, we generated metagene 5-hmC read density profiles for RefSeq transcripts. Normalized 5-hmC read densities differ by an average of 2.10 ± 0.04% (mean ± s.e.m.) in adult male and female cerebellum samples, indicating that the profiles are accurate and reproducible. We observed enrichment of 5-hmC in gene bodies as well as in proximal upstream and downstream regions relative to TSS, transcription termination sites (TTS) and distal regions (Fig. 4b). This is in contrast to previously generated methyl-binding domain–sequencing (MBD-Seq)26, as well as our own methylated DNA immunoprecipitation sequencing (MeDIP-Seq) from mouse cerebellum genomic DNA, in which the majority (~80%) of 5-mC–enriched DNA sequences were derived from satellite and/or repeat regions (Supplementary Fig. 9). Further analyses also reveal that both intragenic and proximal enrichment of 5-hmC is associated with more highly expressed genes, consistent with a role for 5-hmC in maintaining and/or promoting gene expression (Fig. 4b). Proximal enrichment of 5-hmC ~875 bp upstream of TSSs and ~160–200 bp downstream of the annotated TTSs further suggests a role for these regions in the regulation of gene expression through 5-hmC.
Quantification of bulk 5-hmC in the cerebellum of P7 and adult mice indicates genomic acquisition of 5-hmC during cerebellum maturation (Fig. 3a). We further explored this phenomenon by sequencing 5-hmC–enriched DNA from P7 cerebellum and compared these sequences to those derived from adult mice. Metagene profiles at RefSeq transcripts confirmed an increase in proximal and intragenic 5-hmC in adult relative to P7 cerebellum, although there was little to no difference and minimal enrichment over input genomic DNA in distal regions (Fig. 4c and Supplementary Table 2). Peak identification using P7 as background identified a total of 20,092 enriched regions that showed significant differences between P7 and adult tissues. Of those, 15,388 (76.6%) occurred within 5,425 genes acquiring intragenic 5-hmC in adult females (Supplementary Fig. 10 and Supplementary Table 3).
Gene ontology pathway analysis of the 5,425 genes acquiring 5-hmC during aging identified significant enrichment of pathways associated with age-related neurodegenerative disorders as well as angiogenesis and hypoxia response (Fig. 4d and Supplementary Table 4). This is of particular interest given that all these pathways have been linked to oxidation stress response and that the conversion of 5-mC to 5-hmC requires dioxygen5. Furthermore, an assessment of the gene list revealed that 15/23 genes previously identified as causing ataxia and disorders of Purkinje cell degeneration in mouse and human acquired intragenic 5-hmC in adult mice (Supplementary Fig. 11 and Supplementary Table 5)27. Together, these observations suggest that 5-hmC may play a role in age-related neurodegeneration.
Recently, β-GT was used to transfer a radiolabeled glucose for 5-hmC quantification28. (Our paper was under review when ref. 28 was published.) A major advantage of our technology is its ability to selectively label 5-hmC in genomic DNA with any tag. With a biotin tag attached to 5-hmC, DNA fragments containing 5-hmC can be affinity purified for deep sequencing to reveal distribution and/or location of 5-hmC in mammalian genomes. Because biotin is covalently linked to 5-hmC and biotin-avidin/streptavidin interaction is strong and highly specific, this technology promises high robustness as compared to potential anti-5-hmC, antibody-based, immune-purification methods8. Other fluorescent or affinity tags may be readily installed using the same approach for various other applications. For instance, imaging of 5-hmC in fixed cells or even live cells (if labeling can be performed in one step with a mutant enzyme) may be achieved with a fluorescent tag. In addition, the chemical labeling of 5-hmC with a bulky group could interfere with restriction enzyme digestion or ligation, which may be used to detect 5-hmC in specific genome regions. The attachment of biotin or other tags to 5-hmC also dramatically enhances the sensitivity and simplicity of the 5-hmC detection and/or quantification in various biological samples28. The detection limit of this method can reach ~0.004% (Fig. 3d) and the method can be readily applied to study a large number of biological samples.
With the technology presented here, we observed the developmental stage–dependent increase of 5-hmC in mouse cerebellum. Compared to postnatal day 7 at a time of massive cell proliferation in the mouse cerebellum, adult cerebellum has a significantly increased level of 5-hmC, suggesting that 5-hmC might be involved in neuronal development and maturation. Indeed, we also observed an increase of 5-hmC in aNSCs upon differentiation (unpublished data).
This technology enables us to selectively capture 5-hmC–enriched regions in the cerebellums from both P7 and adult mice, and determine the genome-wide distribution of 5-hmC by deep sequencing. Our analyses revealed general features of 5-hmC in mouse cerebellum. First, 5-hmC was enriched specifically in gene bodies as well as defined gene proximal regions relative to more distal regions. This differs from the distribution of 5-mC, where DNA methylation has been found both within gene bodies as well as in more distal regions23–25,29. Second, the enrichment of 5-hmC is higher in gene bodies that are more highly expressed, suggesting a potential role for 5-hmC in activating and/or maintaining gene expression. It is possible that conversion of 5-mC to 5-hmC is a pathway to offset the gene repression effect of 5-mC during this process without going through demethylation30. Third, we observed an enrichment of 5-hmC in genes linked to hypoxia and angiogenesis. The oxidation of 5-mC to 5-hmC by Tet proteins requires dioxygen5,8. A well-known oxygen sensor in mammalian systems that are involved in hypoxia and angiogenesis is the HIF protein, which belongs to the same mononuclear iron-containing dioxygenase superfamily as the active domain of the Tet proteins7. It is tempting to speculate that oxidation of 5-mC to 5-hmC by Tet proteins may constitute another oxygen-sensing and regulation pathway in mammalian cells. Lastly, the association of 5-hmC with genes that have been implicated in neurodegenerative disorders suggests that this base modification could potentially contribute to the pathogenesis of human neurological disorders. Should a connection between 5-hmC levels and human disease be established, the affinity purification approach shown in the current work could be used to purify and/or enrich 5-hmC–containing DNA fragments as a simple and sensitive method for disease prognosis and diagnosis.
In summary, we have developed an efficient and selective method to label and capture 5-hmC from genomic DNA. We have demonstrated the feasibility of using this approach to determine the genome-wide distribution of 5-hmC. Future application of this technology would enable us to understand the role(s) of the 5-hmC modification at molecular, cellular and physiological levels.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturebiotechnology/.
Supplementary Material
Acknowledgments
We would like to thank S. Warren for the helpful discussion and critical reading of the manuscript. This study was supported partly by the US National Institutes of Health (GM071440 to C.H. and NS051630/MH076090/MH078972 to P.J.) and the University of Chicago.
Footnotes
Accession codes. The sequencing data have been deposited in NCBI’s Gene Expression Omnibus with accession number GSE25398.
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
C.H., C.-X.S. and P.J. designed the experiments with help from Y.F. and B.T.L. Experiments were performed by C.-X.S., K.E.S., Y.F., C.Y. and Q.D. with the help of W.Z. and X.J.; Q.D. and J.W. carried out the chemical synthesis; K.E.S., X.L., Y.L. and P.J. provided the mouse cerebellum, mouse aNSC and fly samples, and performed deep sequencing; C.-H.C., L.Z., T.J.L. and L.A.G. helped with the mouse ESC, human HeLa, human HEK and related samples; B.Z. and L.M.H. performed the mass spectrometry analysis from HeLa cells. C.H., C.-X.S. and P.J. wrote the paper. All authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/.
Published online at http://www.nature.com/naturebiotechnology/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 3.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 4.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yi C, Yang CG, He C. A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc Chem Res. 2009;42:519–529. doi: 10.1021/ar800178j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 8.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010 Nov 7; doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loenarz C, Schofield CJ. Oxygenase catalyzed 5-methylcytosine hydroxylation. Chem Biol. 2009;16:580–583. doi: 10.1016/j.chembiol.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Josse J, Kornberg A. Glucosylation of deoxyribonucleic acid. III. alpha and beta-glucosyl transferases from T4-infected Escherichia coli. J Biol Chem. 1962;237:1968–1976. [PubMed] [Google Scholar]
- 15.Lariviere L, Morera S. Structural evidence of a passive base-flipping mechanism for beta-glucosyltransferase. J Biol Chem. 2004;279:34715–34720. doi: 10.1074/jbc.M404394200. [DOI] [PubMed] [Google Scholar]
- 16.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 17.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 18.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baskin JM, et al. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci USA. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ning X, Guo J, Wolfert MA, Boons GJ. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions. Angew Chem Int Ed. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munzel M, et al. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- 22.Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408:538–540. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- 23.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards JR, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim J, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 28.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.