The term, “epigenetics,” was first used to refer to the complex interactions between the genome and the environment that are involved in development and differentiation in higher organisms. Today, this term is used to refer to heritable alterations that are not due to changes in DNA sequence. Rather, epigenetic modifications, or “tags,” such as DNA methylation and histone modification, alter DNA accessibility and chromatin structure, thereby regulating patterns of gene expression. These processes are crucial to normal development and differentiation of distinct cell lineages in the adult organism. They can be modified by exogenous influences, and, as such, can contribute to or be the result of environmental alterations of phenotype or pathophenotype. Importantly, epigenetic programming has a crucial role in the regulation of pluripotency genes, which become inactivated during differentiation. Here, we review the major mechanisms in epigenetic regulation; highlight the role of stable, long-term epigenetic modifications that involve DNA methylation; and discuss those modifications that are more flexible (short-term) and involve histone modifications, such as methylation and acetylation. We will also discuss the role of nutritional and environmental challenges in generational inheritance and epigenetic modifications, concentrating on examples that relate to complex cardiovascular diseases, and specifically dissect the mechanisms by which homocysteine modifies epigenetic tags. Lastly, we will discuss the possibilities of modifying therapeutically acquired epigenetic tags, summarizing currently available agents and speculating on future directions.
Epigenetic Tags: acquisition, maintenance, and inheritance
Chromatin is the complex of chromosomal DNA associated with proteins in the nucleus (for review see 1). DNA in chromatin is packaged around histone proteins, in units referred to as nucleosomes. A nucleosome has 147 bp of DNA associated with an octomeric core of histone proteins, consisting of two H3-H4 histone dimers surrounded by two H2A-H2B dimers. N-terminal histone tails protrude from nucleosomes into the nuclear lumen. H1 histone associates with the linker DNA located between the nucleosomes. Nucleosome spacing determines chromatin structure which can be broadly divided into heterochromatin and euchromatin (Table 1) 1, 2. Chromatin structure and gene accessibility to transcriptional machinery are regulated by modifications to both DNA and histone tails (Figure 1).
Table 1.
Chromatin Domains
| Heterochromatin: transcriptional inactive, densely packed nucleosomes. |
| constitutive: highly repetitive DNA sequences, such as centromeric and telomeric domains, hypoacetylated nucleosomes, H3K9me* |
| facultative: includes silenced genes, such as inactive X chromosome or imprinted regions, or transcriptionally repressed genes, hypoacetylated nucleosomes, H3K27me |
| Euchromatin: transcriptional permissive chromatin, less densely packed. Accessible to nuclear factors and nuclear repressors, acetylated nucleosomes, H3K4me, H3L36me |
histone methylation sites are listed in abbreviated forms, for example H3K9me, histone lysine 9 methylation.
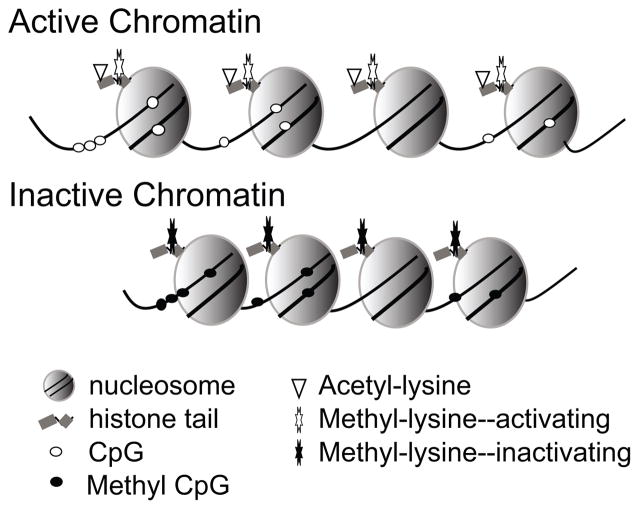
Figure 1. Epigenetic tags and chromatin structure.
Chromosomal DNA is packaged around histone cores to form nucleosomes. Nucleosome spacing in open structure that is accessible to nuclear factors is maintained, in part, by post-translational modification of histone tails, including lysine acetylation and specific lysine methylation. CpG dinucleotides are unequally distributed throughout chromosomal DNA, and may be concentrated in regions called CpG islands that can overlap gene promoters. Methylation of cytosines in CpG dinucleotides is overall associated with inactive, condensed states of the chromosome. Inactive chromatin is also maintained by specific histone lysine modifications.
DNA methylation
In differentiated mammalian cells, the principal epigenetic tag found in DNA is that of covalent attachment of a methyl group to the C5 position of cytosine residues in CpG dinucleotide sequences (referred to as CpG throughout this review)3. Recent findings suggest that in undifferentiated stem cells, cytosines, other than those in CpG, can be methylated, as well, 4 and that methylation of non-CpG cytosines is crucial for gene regulation in embryonic stem cells in particular. CpG methylation is, however, an important mechanism to ensure the repression of transcription of repeat elements and transposons, and also plays a crucial role in imprinting and X-chromosome inactivation 5. Transcriptional gene silencing by CpG methylation also restricts the expression of some tissue-specific genes during development and differentiation by repressing them in non-expressing cells.
During development, the pattern of CpG methylation changes in a predictable manner. In early embryogenesis, methylation is erased throughout the genome and then reestablished in all but CpG islands (regions of the genome with a concentration of CpG residues). CpG islands remain hypomethylated until later in development when some of them become methylated 6, 7. Subsequent methylation of cytosines in CpG islands and at other CpG dinucleotides is associated with transcriptional repression 6, 8, especially when these methylated sites involve promoter or other gene regulatory regions 3. DNA methylation may, however, be activating if it prevents binding or limits expression of transcriptional repressors. Recent studies defining the degree of methylation in mammalian promoters indicate that methylation occurs at only a small percentage of CpG dinucleotides and inhibits transcription of only a small subset of genes in differentiated cells. Many of these repressed genes are germline-specific 8, including pluripotency genes, suggesting that methylation is a crucial mechanism by which to suppress key genes during differentiation 8.
CpG methylation can supress transcription by several mechanisms. First, the presence of the methyl group at a specific CpG may directly block DNA recognition and binding by some transcription factors. For example, several studies have shown that transcriptional activation at GC-boxes is inhibited by methylation, which excludes binding of Sp1 and Sp3 transcription factors, at least in some promoter contexts 9, 10. Methylation has also been shown to block the ability of the nuclear factor, Hif1, from inducing erythropoietin transcription under hypoxic conditions 11. Alternatively, other factors may preferentially bind to methylated DNA, blocking transcription factor access. For example MeCP2 and other family members 12 bind to methyl CpG and contribute to transcriptional repression by the recruitment of histone-modifying proteins, such as histone deacetylases (HDAC). Subsequently, histone deacetylation promotes chromatin condensation, further repressing transcription 13, 14. This sequence of events illustrates how DNA methylation and certain histone modifications function together to contribute to the transcriptional on or off state of genes subject to epigenetic modification (Figure 1).
A family of DNA methyltransferase enzymes (DNMTs) is involved in de novo DNA methylation and its maintenance. During embryogenesis, de novo methylation is carried out by DNMT3A and DNMT3B 15. Although some studies suggest an ongoing role for DNMT3A and DNMT3B in maintaining methylation status in some cell types 16, 17, the ubiquitously expressed DNMT1 is predominantly responsible for maintaining cellular levels of CpG methylation. Interestingly, transcription from alternative promoters results in expression of a truncated oocyte-specific DNMT1 isoform, DNMT1o, that is essential for early embryogenesis 18. DNMT1 functions in a complex to recognize hemi-methylated DNA and to add methyl groups to the non-methylated daughter strand formed during replication 19. The base pairing of CpG allows for the reciprocal maintenance of methylation during subsequent replication cycles. In this manner, a non-genetic trait (DNA methylation) can be passed from cell to cell and, with it, the contextual effects on gene expression. Thus, methylation can be considered a long-term, relatively stable, epigenetic trait, the effects of which can contribute to maintaining the cellular phenotype.
Targeting DNA methylation: Role of histone modification, DNA-binding proteins and RNA
Owing to its heritability, DNA methylation is a powerful means by which to suppress the expression of unwanted or excess genes. Several basic questions remain unanswered, such as the mechanisms that promote targeting of specific CpG sites for methylation or prevent their modification. Many of the insights into the mechanistic aspects of targeting CpG methylation come from studies of imprinting and X-inactivation, where CpG methylation represses gene expression in chromosomal regions. X-inactivation occurs in somatic cells of females to limit the expression of most X-chromosome genes to those from one chromosome. Given the random nature of X-chromosome inactivation, female carriers may display a wide variation in phenotypic expression of X-linked disorders 20. Similarly, imprinting regulates autosomal gene expression to genes from only one parental allele in males and females. The importance of imprinting and gene dosage regulation in normal development can be appreciated by the consequences of imprinting disturbances that cause a number of human syndromic disorders, such as Prader-Willi, Angleman, Silver-Russell, and Beckwith–Wiedermann (reviewed in 21, 22).
In imprinting, clusters of genes in a chromosomal region are coordinately inhibited by methylation of an imprinting center; these centers are also referred to as differentially methylated regions (DMRs), and DMRs often overlap CpG islands. Recent studies suggest that transcription of DMRs in the oocyte may target them for subsequent CpG modification by maintaining an open chromatin structure accessible to de novo methylation 23. In both imprinting and X-inactivation, the expression of long non-coding RNAs 24, such as the Xist transcript in X-chromosome inactivation 25, may also play a regulatory role. Chromatin conformation is apparently essential for the imprinting of some maternally determined imprinted genes. In particular, histone H3-lysine 4 (H3K4) demethylase, LSD1, has been found to be essential for these processes, and deficiency of this enzyme results in embryonic stem cell lethality during early differentiation 26, 27. To date, the mechanisms involved in specifying methylation of DMRs during imprinting include recognition of specific DNA sequence motifs in imprinting centers, the expression of factors that are oocyte-specific to mark maternal chromosomes, chromatin remodeling, and/or combinations of these events. Similar mechanisms may also target methylation of specific gene promoters during differentiation.
CpG methylation is erased in a predictable manner during gametogeneis and following zygote fertilization. Recent findings suggest that these processes require the action of cytidine deaminases, such as AID, as well as DNA repair mechanisms 28, 29. Thus, enzymatic deamination of 5-methylcytosine leads to formation of thymine and T:G base-pair mismatches: base excision repair mechanisms subsequently delete thymine and restore C:G base pairing during epigenetic reprogramming. Spontaneous deamination of 5-methylcytosine also requires base excision repair mechanisms to repair base pairing mismatch, a process that is highly inefficient in most differentiated cells. Thus, spontaneous deamination of 5-methylcytosine has resulted in an overall depletion of CpG dinucleotide sequences in mammalian genomes (over evolutionary time)30. In differentiated, adult cells, CpG methylation is considered long-lasting and refractory to elimination, except by alterations in the expression or activity of DNTM1 (Figure 2) (or following spontaneous deamination and mismatch repair). Recently, however, a novel mechanism involving specific DNA demethylation in response to hormone stimulation has been discovered 31. In this system, the cytochrome p45027B1 (CYP27B1) gene is repressed by vitamin D-interacting repressor (VDIR)-mediated recruitment of DNA methylases and MBD4, a methyl DNA binding protein. Transcriptional suppression can be relieved by parathyroid hormone (PTH)-induced demethylation of promoter CpGs. Apparently, in the presence of PTH-induced phosphorylation, MBD4 can mediate demethylation of promoter CpGs through a base-excision repair mechanism that removes methylcytosine, apparently without deamination. This hormone-induced mechanism contrasts with excision-repair mechanisms described above in that it is targeted to a specific promoter by hormone action and it does not require deamination 32 (Figure 2). Other recent studies suggest a role for DNMT3 in active demethylation at estrogen receptor-α-responsive promoters that also exhibit hormone-induced alterations in CpG methylation status 33, 34. Although further studies are needed to confirm these mechanisms, the concept of hormone-induced methylation switching adds a new twist to epigenetic regulation. It remains to be seen whether other genes can be similarly regulated by methylation switching utilizing these or other, as yet, unknown mechanisms.
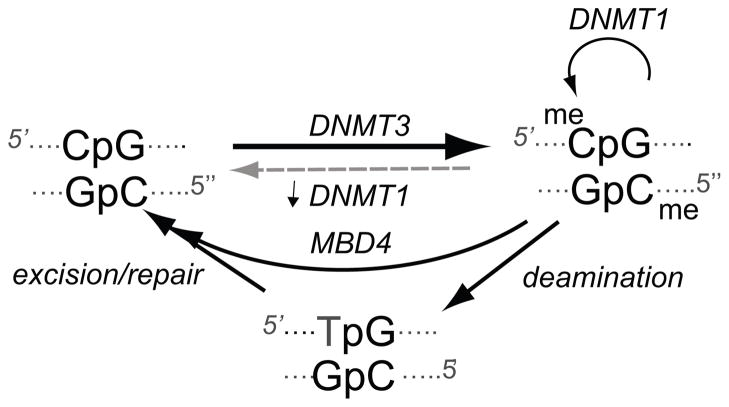
Figure 2. DNA methylaton and demethylation.
DNA methylation at the cytosine in CpG dinucleotides is initiated de novo by the DNA methyl transferases (DNMT) 3A and 3B. Following replication, DNMT1 plays a primary role in maintaining the methylation state in the daughter strands. Demethylation is thought to occur by reduction of DNMT1 activity or by excision repair mechanisms following deamidation of methyl cytosine (meC) to create a T:G mismatch. Recent findings (discussed further in the text) suggest that the methyl-DNA binding protein 4 (MBD4) may mediate demethylation by an hormonally regulated mechanism that does not involve deamination of meC, rather it involves the DNA glycosylase activity of MBD4 followed by a base-excision repair mechanism.
Histone regulation: readily reversible epigenetic changes
DNA methylation tags promote the persistence of certain histone states, such as deacetylation, thus providing a mechanism for perpetuating post-translational histone modifications. Histones can be post-translationally modified to restructure chromatin in many ways, including phosphorylation, ubiquitination, acetylation, and methylation35, 36. In fact, the ‘Histone Code Hypothesis’ suggests that different combinations of histone modifications may regulate chromatin structure and transcriptional status 37, 38. Details, such as the location of nucleosomes relative to the transcriptional start site of a gene, together with specific combinations of sites, types, and extents of histone modifications, add to the complexity of the histone code (reviewed in 1, 39). Nonetheless, efforts have been made to characterize patterns of histone modifications that contribute to cell-type specific regulation of genes in differentiated cells, such as those associated with smooth muscle-specific genes regulated by serum response factor binding to the CArG box 40.
Of the many described histone modifications, histone acetylation, at the ε-amino group of lysine residues in H3 and H4 tails, is most consistently associated with promoting transcription. However, this description oversimplifies a complex process, as acetylated, open-chromatin structure may also allow access of transcriptional repressors. For example, some bromodomain-containing factors, such as BRG1 and Brd4, target to acetylated histones where they can mediate the formation of repressor (or activator) complexes 41, 42. Acetylation is targeted to regions of chromatin by the recognition and binding of DNA sequence-specific transcription factors that recruit one of a growing family of histone acetyl transferase (HAT) cofactors such as CREB binding protein (CBP), and p300, MYST, and GNAT (see Table 2 for a list of histone modifying enzymes) 43. Disruption of the normal acetylation activity of CBP/p300 family members is associated with Rubenstein-Taybi syndrome, an autosomal dominant syndrome 44, 45, highlighting the importance of these cofactors in regulating the proper expression of gene combinations important in development and differentiation.
Table 2.
Histone Modifying Enzymes
| Category* | Properties |
|---|---|
| Histone acetyltranferase families | |
| GNAT MYST CBP/p300 |
Promotes open conformation of chromatin and gene activation, transcriptional-coactivators recruited by nuclear factors to specific genes |
|
| |
| Histone deacetylase families | Promotes condensation of chromatin and gene repression, recruited by transcriptional repressors to specific genes |
| Class I HDAC | Zn-dependent metallohydrolases |
| Class II HDAC | |
| Class IV HDAC |
|
| Class III HDAC (sirtuins) | NAD+-dependent deacetylases |
|
| |
| Histone lysine methyl-transferases | |
| Set domain proteins (partial list): | Histone specificity, transcriptional effect |
| Set1, MLL, Set7/9, SMYD3 | H3K4 specific, gene activation |
|
| |
| G9a; Suv91, SetB1, PRD14, CLL8, GLP, Suv39h1, Suv39h2 | H3K9 specific, mostly repressive |
|
| |
| EZH2 | H3K27, gene silencing |
|
| |
| NSD1 | H3K36, gene activation |
| H4K20, gene silencing | |
|
| |
| Suv4-20h1, Suv4-20h2, Set8/PR-SET7 | H4K20, gene silencing |
|
| |
| Histone lysine methyl-transferases | |
|
| |
| Dot1 domain protein | |
| Dot1L | H3K79, gene activation |
|
| |
| Histone lysine demethylases | |
| LS | amine oxidase superfamily; FAD-dependent |
| LSD1 | H3K4 me1/2, important in development |
| Histone lysine demethylases | |
| Jumonji (JmjC) domain proteins (partial list) | Hydroxylase activity requires iron and α-ketoglutarate cofactors |
| JMJD2A | H3K9me2/3; H3K36 me2/3 |
| JHDM1A | H3K36me1/2 |
| JAR1D1A | H3K4me2/3 |
subtypes of enzymes involved in histone lysine acetylation and methylation.
Deacetylation of histones correlates with CpG methylation and the inactive state of chromatin. There are 4 classes of histone deacetylase enzymes (HDACs), with members capable of deacetylation of histones and/or other protein targets 46. These regulatory proteins are themselves subject to regulation by acetylation, phosphorylation, and sumoylation 47, which can affect their function, subcellular distribution, and protein-protein associations 48. Several studies suggest that interactions with sequence-specific DNA binding proteins and co-repressor complexes can target certain HDAC proteins to histones in a gene-specific manner 49, 50.
Histone lysine methylation patterns and their effects on transcription are more complex than acetylation, in that some methylation sites are associated with transcriptionally permissive chromatin (euchromatin) and some are repressive, fostering heterochromatin formation. In addition, ε-amino groups of lysine residues can be mono-, di-, or tri-methylated. Overall, the H3K27me3 and H3K9me states are associated with silencing, whereas the H3K4me3 and H3K36me3 states are transcriptionally permissive modifications (see Table 3 for a list of histone methylation sites). Importantly, methylation marks recruit effector proteins that play essential roles in maintaining the transcriptional state of the chromatin, for example H3K9me recruits HP1, contributing to heterochromatin formation.
Table 3.
Histone methylation sites
| Histone | position | modification | Transcriptional effect/gene or chromatin location * |
|---|---|---|---|
| H3 | K4 | di-methyl | gene activation |
| tri-methyl | gene activation/5′ end transcriptionally active genes | ||
| K9 | mono-methyl | gene silencing/euchromatin | |
| di-methyl | gene silencing/euchromatin | ||
| tri-methyl | gene silencing/promoters & heterochromatin | ||
| tri-methyl | gene activation/gene coding region | ||
| K27 | mono-methyl | gene silencing/heterochromatin | |
| tri-methyl | gene silencing/inactive X-chromosome, imprinted regions & homeotic genes | ||
| K79 | di-methyl | gene activation | |
| tri-methyl | gene activation | ||
| H4 | K20 | di-methyl | gene silencing/heterochromatin |
| tri-methyl | gene silencing/heterochromatin |
most common effects and gene or chromatin region are listed, if known
Most histone lysine methyltransferases have a SET homology domain, a vast family of proteins that can be grouped into 7 subfamilies based on their structural similarities 39. SET1 family members specifically foster active chromatin by methylating H3K4. Other histone lysine methyltransferase families can methylate several histone targets. In addition, some of these methyl transferases have additional domains that specify binding to methylated DNA or to other proteins, such as CBP 39.
Until recently, histone methylation was considered a long-term epigenetic marker as the only mechanism for its removal was histone turnover; however, recent studies confirm the existence of multiple histone demethylases capable of demethylating histone lysine methyl groups. These enzymes include lysine-specific demethylase 1-(LSD1), which removes mono- or di-methyl groups from H3K4. The tri-methylated modification is targeted for removal by the Jumonji C-(JmjC) domain-containing demethylases 51 (see Table 2). Similar to histone methylases, LSD1 and JmjC family proteins may demethylate histones in a gene-specific manner, directed, in part, by interactions between demethylases and DNA sequence-specific nuclear factor complexes. In addition, recent studies show that specific histone demethylases may regulate androgen-mediated transcriptional responses and osteoblast differentiation 52–54.
Non-coding RNA, transcriptional gene silencing, and epigenetic modifications
As discussed above, long non-coding RNAs (lncRNAs) play an essential role in imprinting and X-chromosome inactivation. The gene silencing effects of these lncRNAs and other lncRNAs, such as HOTAIR in the human homeobox loci, are due, in part, to their recruitment of remodeling complexes such as the polycomb complex that foster histone methylation (notably the inhibitory H3K27me3) 55–57. In addition, lncRNAs can also suppress transcription by other mechanisms, such as the recruitment of RNA-binding proteins that interfere with histone deacetylation or exclude TFIID promoter association 58, 59.
There is also a growing literature on the role of small noncoding RNAs (sncRNAs) and their effects on transcriptional gene silencing. This group of RNAs includes the dicer-dependent microRNAs (miRNAs) and small inhibitory RNAs (siRNAs) formed by RNA-interference pathways, as well as the piRNAs, that are formed in a dicer-independent manner and specifically associate with the PIWI subfamily of argonaute proteins. Although each of these classes of sncRNAs have been shown to mediate epigenetic DNA and histone modifications, the piRNAs appear to have a distinct function of repressing transposon expression in germline cells by fostering de novo DNA methylation 60. A role for siRNA in RNA-mediating DNA methylation and transcriptional gene silencing was first discovered in plants 61 and has been found to exist in many species, including mammals 62. Recent findings in mammalian cells suggest that synthetic siRNAs and endogenous miRNAs that target gene promoters may direct transcriptional gene silencing by recruiting specific argonaute proteins and forming epigenetic remodeling complexes that suppress gene expression by fostering histone deacetylation, histone methylation (H3K9 and H3K27), and DNA methylation 63–65. In fact, although the mechanism has not been completely elucidated, dicer-deficient ES cells exhibit defects in differentiation that correlate with a loss of de novo DNA methylation and loss of miRNAs66, suggesting a role for endogenous miRNAs in regulating necessary epigenetic changes during differentiation. Other studies indicate that some siRNAs targeted to TATA-box sequences may block transcriptional inititation without causing epigenetic modifications to histones and DNA 67. Overall, these studies illustrate the important role of ncRNAs in modulating gene transcriptional silencing.
Role of Epigenetic Changes in Cardiovascular Diseases
It has been suggested that epigenetic changes may account for the missing heritability determinants of complex diseases, such as atherosclerosis, hypertension, metabolic syndrome, and diabetes, that, to date, have not been accounted for by genetic studies of sequence variation 68, 69. In a recent study, the influence of parental origin on disease association was examined by following the inheritance of single nucleotide polymorphisms (SNPs) near known imprinted genes. These results identified 6 SNPs in which parental origin of a gene alters risk 70. One of these SNPs that was associated with type 2 diabetes correlated directly with methylation status, as well. Thus, these findings suggest that additional, nonsequence-dependent variations may contribute to heritable traits. Below we review the relationships between epigenetics and genetics, epigenetics and nutrition, and how these relationships may influence cardiovascular disease.
DNA methylation and single nucleotide polymorphisms
Theoretically, SNPs that create CpG sites may be targets for epigenetic modifications, just as loss of these sites will prevent DNA methylation. The consequences of a polymorphism resulting in a CpG in the promoter region of the NDUFB6 gene illustrates this intersection between genetic and epigenetic regulation. NDUFB6 is a respiratory chain protein with diminished expression in type 2 diabetes. In muscle biopsies from elderly patients, NDUFB6 expression inversely correlates with the degree of DNA methylation, suggesting that the presence of a CpG site confers more risk for decreased expression (and potentially disease risk) than not having the site 71. Taken together, these findings support the concept that epigenetic modifications can influence risk in complex diseases.
Epigenetics, nutrition, and environment
Barker and colleagues hypothesized that environmental factors in crucial periods of early life (during fetal development, for instance) can influence risks for cardiovascular and metabolic diseases later in life. This concept is supported by a number of studies that have associated low birth weight in human populations with increased risk of cardiovascular disease (see for instance reviews 72, 73). For example, individuals prenatally exposed to famine during the Dutch Hunger Winter (1944–45) experienced higher prevalence of obesity and coronary heart disease as adults, when compared to adults born before or conceived after that period 74. In Barker’s seminal work, it was found that low birth-weight babies who survived infancy had an increased risk of coronary heart disease later in life, and that increasing birth weight was associated with a graded decrease in risk 75. In addition, in utero exposure to hypercholesterolemia has been associated with higher incidence and accelerated progression of lesions in humans, rabbits, and mice 76, 77. Exposure to different behavioral patterns during early postnatal life has also been shown to influence epigenetic modifications in experimental animal models 78. Thus, it has been suggested that these long-lasting changes arise, at least in part, from epigenetically mediated alterations in gene expression that occur very early in life 79. Applying these concepts to human populations, it has recently been proposed that social and environmental stresses during development may influence epigenetic processes that contribute to the adult race-based US health disparities in diseases like hypertension, diabetes, stroke, and coronary heart disease 80.
More recent studies in animal models have begun to characterize epigenetic modifications that are influenced by the intrauterine environment. For example, feeding a low-protein diet to pregnant rats causes low birth weight, hypertension, and endothelial dysfunction in the offspring. Studies have shown a role for the renin-angiotensin system in this phenotype as treatment of pregnant mothers with angiotensin converting enzyme inhibitors or angiotensin receptor (AT1R) antagonists 81 alleviates hypertension in the offspring. Consistent with these earlier findings, offspring of pregnant mothers fed low protein diets were found to have hypomethylated AT1bR gene promoters along with increased adrenal expression of AT1bR 82, suggesting a role for specific hypomethylation in regulating elevated blood pressure in this model. Other studies have reported that a low protein diet during pregnancy in rat results in overexpression of the hepatic glucocorticoid (GR) and peroxisomal proliferator-activated (PPARα) receptors in offspring 83, 84. Interestingly, it had been previously reported that DNMT1 contains a GR response element 85. Of importance, these studies established an underlying epigenetic mechanism that involves a decrease in the methylation patterns of GR and PPARα promoters, a decrease in DNMT1 expression, and an increase in the levels of transcription-permissive histone modifications. GR and PPARα receptors are key transcription factors whose altered expression may modulate the activity of a large number of metabolic pathways, and have been implicated in the pathology of numerous diseases, including obesity, diabetes, and atherosclerosis 86, 87.
As discussed further in the following section, the methyl-group responsible for DNA and histone methylation originates from S-adenosyl methionine (AdoMet), via Met biosynthesis through folate-dependent or -independent pathways of homocysteine (Hcy) remethylation (Figure 3). Of importance, supplementing a protein-restricted maternal diet in rats with methyl groups by addition of folate or glycine has been shown to decrease hypertension 88, improve endothelium-dependent vasodilation, and increase endothelial NO synthase mRNA levels 89, and to restore both the expression and promoter methylation status of the hepatic GR and PPARα receptors in offspring 83. These data support the hypothesis that folate and other methyl group donors can influence fetal development and the risk of cardiovascular disease in the next generation. Interestingly, supplying folate to the offspring, rather than the pregnant mothers, increased the methylation status of some, but not all, of the genes modified by maternal protein restriction 90, suggesting that some epigenetic modifications may not be reversible by nutritional interventions in the offspring.
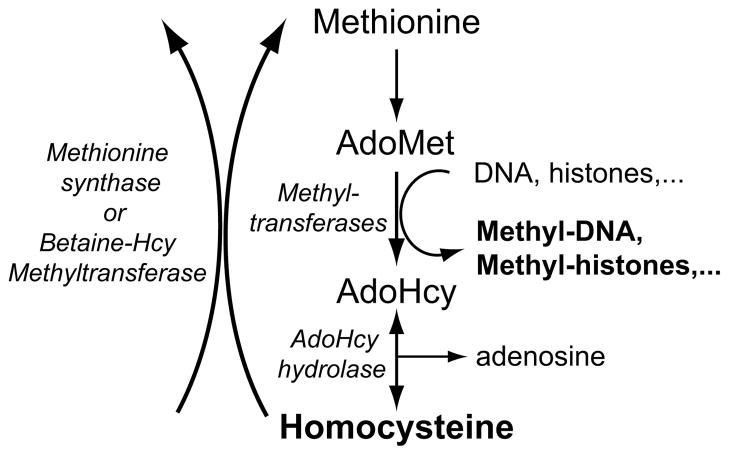
Figure 3. Homocysteine and methylation reactions.
S-adenosyl-methionine (AdoMet) is the primary source of methyl groups for hundreds of transmethylases that methylate DNA, RNA, histones, other proteins, and small biological molecules. Following transfer of methyl groups, S-adenosyl-homocysteine (AdoHcy) is formed. Accumulation of AdoHcy can inhibit methyltranferases. The hydrolysis of AdoHcy yields homocysteine (Hcy) and adenosine. Intracellular homocysteine can be removed from the cell, reform AdoHcy, become further metabolized in the transsulfuration pathways (not illustrated), or become methylated to form methionine by the folate-dependent methionine synthase or the folate-independent betaine-Hcy methyltransferase.
Other modifiers of methylation may also influence epigenetic tags. For instance, dietary status of choline (a betaine precursor that is involved in folate-independent pathways of methionine synthesis) was shown to affect DNA methylation 91. Moreover, in a study using apo-E deficient mice to assess the efficacy of dietary intervention in retarding atherogenesis, it was shown that betaine supplementation attenuated atherosclerotic lesion formation and growth 92. Treatments that alter epigenetic modifications may also have utility in human atherosclerosis where DNA methylation changes in target genes, such as the estrogen receptor α and β genes 93, 94, may contribute to the underlying pathogenic mechanisms (discussed further in 77).
Regulation of vascular nitric oxide (NO) production by endothelial nitric oxide synthase (eNOS) may influence atherogenesis as well as thrombosis. Interestingly, the restricted expression of eNOS to vascular endothelium is determined, in part, by DNA methylation and histone modifications 95, as treatment of non-expressing cell types with a DNMT inhibitor induced demethylation of the eNOS promoter and increased the levels of eNOS mRNA. Furthermore, local enrichment of acetylated H3 and H4 histones across the native eNOS locus was found in eNOS-expressing cells compared to non-endothelial cells, whose treatment with a HDAC inhibitor upregulated eNOS mRNA expression. Recent data suggests that NO itself may be an epigenetic factor, based on its regulatory function upon chromatin and gene expression, via modification (S-nitrosation and tyrosine-nitration) of nuclear factors, HDACs, and histones 96.
Homocysteine: a link between DNA methylation and vascular disease
Homocysteine (Hcy) is biochemically linked to the principal epigenetic tag found in DNA. Although increased circulating levels of Hcy are a risk factor for vascular disease, recent clinical trials that used folate and/or other vitamin B therapies to lower Hcy failed to reduce cardiovascular event rates, therefore casting doubt on homocysteine’s direct causative role in vascular disease97. Most studies, however, have not explored the consequences of elevated Hcy on methylation processes, even though Hcy plays a crucial role in methyl-donor biosynthesis 98. As depicted in Figure 3, the methyl group responsible for the establishment and maintenance of DNA methylation patterns originates from S-adenosyl methionine (AdoMet), an intermediate in Hcy metabolism. In addition to DNA methylation, AdoMet serves as the methyl donor for more than one hundred different cellular methyltransferase reactions, including histone methylation. Following the transfer of the methyl group, AdoMet is converted into S-adenosyl homocysteine (AdoHcy), which inhibits the majority of AdoMet-dependent methyltransferases. AdoHcy is further converted into Hcy and adenosine by AdoHcy hydrolase, which is widely distributed in mammalian tissues. This reaction is reversible and strongly favors AdoHcy synthesis rather than hydrolysis; however, both Hcy and adenosine are rapidly removed under physiological conditions, favoring the hydrolysis reaction. If Hcy accumulates, AdoHcy will accumulate as well, potentially inhibiting transmethylation reactions. Thus, increased Hcy may be regarded as a global DNA hypomethylation effector via AdoHcy accumulation.
There are many in vivo examples that suggest Hcy levels may modulate global DNA methylation. For example, in healthy humans, increased levels of plasma Hcy were associated with both increased AdoHcy concentrations and DNA hypomethylation in lymphocytes 99. This inverse relationship between Hcy plasma concentrations and DNA methylation patterns was further confirmed in other reports 100–102 (with one exception 103) and extended to several animal models 102. Several studies support the concept that DNA hypomethylation may be responsible, in part, for vascular complications associated with increased circulating levels of Hcy 104. For example, vascular disease patients manifested increased levels of both plasma Hcy and intracellular AdoHcy, together with decreased DNA methylation, supporting a role for hyperhomocysteinemia (HHcy) in modulating epigenetic mechanisms 105. This association has also been confirmed in several animal studies in which increased circulating levels of Hcy and AdoHcy were associated with endothelial dysfunction and aberrant DNA methylation patterns 106–108. [One human study, however, documented that coronary heart disease patients had increased global DNA methylation in the presence elevated serum Hcy levels 109.] In patients with renal functional impairment (and altered homocysteine clearance), which augments the risk for vascular disease, Ingrosso and colleagues reported increased levels of Hcy together with reduced global lymphocyte DNA methylation pattern 110. Of importance, Ingrosso and colleagues reported that global and specific DNA hypomethylation affected the expression of two genes (SYLB, a pseudoautosomal gene, and H19, an imprinted gene), and that both global DNA methylation patterns and allelic gene expression were normalized after lowering plasma Hcy levels with folate administration. Although two later reports failed to confirm these observations regarding global hypomethylation in patients with renal failure 111, 112, Ingrosso and colleagues work represents the first human study that causally linked Hcy with altered gene expression via DNA hypomethylation.
Aberrant global DNA methylation is only an index of the potential for epigenetic dysregulation. In addition to AdoHcy, a growing list of factors has been identified that can modify DNA methylation patterns. These include the rate of cell growth and DNA replication, chromatin accessibility, local availability of AdoMet, nutritional factors including folate supplementation, duration and degree of the hyperhomocysteinemic state, inflammation, dyslipidemias, oxidative stress, and aging 113. Thus, the relation between increased Hcy and DNA global hypomethylation may be masked in the clinical setting owing to the presence of these confounders, thereby possibly explaining some contradictory and counterintuitive findings reported to date. Another important aspect to consider is that DNA methylation is unequally distributed throughout chromosomes of differentiated cells 4. Thus, hyper- and hypomethylated regions can coexist in the genome, and global DNA methylation status need not correspond to the methylation status of specific genomic regions. For example, it has been recently shown that human cardiomyopathies of different etiologies display a unifying pattern of altered DNA methylation of three angiogenesis-related loci in which the differential (increased or decreased) methylation was correlated with the expression of the corresponding gene 114. It is likely that research on single gene methylation and expression may lead to a better understanding of the vascular effects of elevated Hcy 113, 115.
Several investigators have focused on target gene methylation patterns to explain some of the deleterious effects of Hcy. For example, in cultured endothelial cells, it was shown that Hcy, at physiological relevant concentrations, inhibits cell growth with a concomitant increase in intracellular AdoHcy concentration and a significant decrease in the AdoMet/AdoHcy ratio, suggesting that cellular hypomethylation could play a role in the observed phenotype 116. Subsequent studies suggested a role for transcriptional suppression of cyclin A in mediating Hcy-induced endothelial cell growth inhibition 117, 118, and found that Hcy triggers transcriptional inhibition of cyclin A through demethylation of a specific CpG site located in the core promoter, viz., on the cell-cycle dependent element (CDE). In addition, cyclin A promoter demethylation eliminates MeCP2 binding, which, in turn, impairs HDAC association, leading to an accumulation of acetylated H3 and H4. The authors concluded that the loss of DNA methylation in the CDE repressor site and the resulting chromatin remodeling increases chromatin accessibility to repressors, resulting in inhibition of cyclin A gene transcription. Of additional interest was the observation that a physiological concentration of plasma Hcy inhibits DNMT1 activity in this cell system, providing evidence that Hcy can directly modulate specific DNA methylation reactions. In other studies, Hcy was also shown to disrupt the growth of endothelial cells by downregulating fibroblast growth factor-2 (FGF2) via an epigenetic mechanism involving transcriptional repression 119. Apparently, the FGF2 gene promoter encompasses a CpG island and, in contrast with the cyclin A example, the FGF2 gene was heavily methylatedat cytosine residues despite significant AdoHcy accumulation. Normal levels of FGF2 transcription were restored when the cells were simultaneouslyexposed to a DNA demethylating agent and Hcy. Taken together with other examples in the literature 108, 120–122, these findings suggest that Hcy and AdoHcy accumulation can have complex effects on DNA methylation targets and their transcriptional potential.
Increasing evidence indicates that alterations in lipid metabolism may play a role in vascular pathology associated with hyperhomocysteinemia (HHcy) 123–126, and many studies suggest that epigenetics may play a role in these processes. For example, changes in DNA methylation have been suggested as a potential mechanism for altered apoA-I and apoA-IV gene expression in mice with HHcy 127, 128. In mice, both apoA-I and apoA-IV genes are contained within the apolipoprotein gene cluster on chromosome 9, and, notably, the cluster contains a CpG-rich regioncorresponding to the 3′ flanking region of the apoA-I gene andthe 5′ flanking region of the apoA-IV gene. Similarly, it has been shown that Hcy is significantly and inversely correlated with HDL-bound cholesterol and apoA-I in both human and murine models of HHcy 129. In murine primary hepatocyte cultures, cellular hypomethylation, induced by AdoHcy accumulation, was suggested as an explanation for the Hcy-induced inhibition of apoA-I protein synthesis 130; however, additional analysis indicated that Hcy regulation of apoA-I synthesis may also involve other, nonepigenetic mechanisms. Additional evidence for regulation of lipid biosynthesis by the epigenetic effects of Hcy was reported by Devlin and colleagues, who showed that a reduced methylation capacity (assessed by AdoMet/AdoHcy ratio) in liver from mice with mild or moderate HHcy was associated with hepatic changes in phospholipid species and impaired long-chain polyunsaturated fatty acid metabolism that could be attributed, in part, to differential CpG methylation of genes involved in these pathways 131, 132. Taken together, these findings indicate that Hcy may influence gene expression by modulating epigenetic pathways. As discussed above, the list of potential modifiers of methylation is long, suggesting that there are still many questions yet unresolved about Hcy’s effects on methylation, gene expression, and cardiovascular disease risk.
Future Directions and Therapeutic interventions
The plasticity of certain epigenetic modifications can be followed throughout development and differentiation and in response to environmental stimuli. The fact that modifications can accumulate in aging is supported by studies in genetically identical monozygotic twins: younger twins were far more concordant in terms of the patterns of DNA methylation and histone acetylation than older twins, suggesting that these tags are acquired or modified over time 133. Thus, it seems possible that epigenetic modifications may be amenable to pharmacological interventions. For example, 5-azacytidine, a DNMT inhibitor, has been used to increase fetal hemoglobin production by causing hypomethylation of γ-globin genes in sickle cell disease patients 134, and, as discussed above, methyl donor treatments (folate, glycine and betaine) can increase DNA methylation, altering gene expression. Recent studies have also reported a microRNA (microRNA-29b) that can cause global hypomethylation, by reducing the expression of DNMT enzymes 135. These approaches, however, are not specific and may have undesirable consequences on the expression of genes distinct from those of primary interest.
Additional studies are necessary to unravel the mechanisms that select specific genes for epigenetic regulation prior to developing targeted therapeutic approaches to reprogram these modifications. To date, most epigenetic therapies have focused on modulating chromatin structure. For example, there has been a surge in the development of many class and isoform-selective HDAC-inhibitors 136, some of which may have utility in cancer, Huntingtons disease, sickle cell disease, or cardiovascular diseases 137, 138. The usefulness of these approaches, again, may depend on the ability of a target HDAC to modulate subsets of genes, rather than cause global changes. This goal may not be that unrealistic: sirt6, a sirtuin deacetylase, apparently coordinately regulates the expression of multiple glycolytic genes 139, suggesting a possible target through which to regulate cellular metabolism. In addition, the specific methylation/demethylation of CYP27B1 31 discussed above suggests novel mechanisms of regulating epigenetic tags at the DNA level by modulating proteins that mediate methylation switching. In addition siRNA-based methods may provide a targeted means to transcriptionally silence genes. Recent findings also suggest that this method may be adapted to provide long term (with epigenetic changes) or short term (without epigenetic changes) regulation of gene transcription, depending on the targeting site in the promoter; this type of flexibility may have many therapeutic advantages 62.
Since Waddington made his initial observations about the environmental influences in development 140, much progress has been made to uncover the molecular mechanisms involved in epigenetic regulation. At the present time, additional studies are needed to define the human epigenome, its role in development and disease, and the processes that regulate its formation and dynamic modulation throughout the life of an individual.
Acknowledgments
The authors thank Stephanie Tribuna for her assistance with the preparation of this manuscript.
Funding Sources
This work was supported by NIH grants HL61795, HV28178, HL81587, and HL70819 (JL and DH) and by Fundação para a Ciência e Tecnologia (Portugal) and Fundação Calouste Gulbenkian (Portugal) grants Serviço de Formação de Recursos Humanos/Bolsa Sabática (SFRH/BSAB)/921/2009 and 105783 (RC).
Footnotes
Disclosures
The authors have no conflicts of interest to disclose.
References
- 1.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 2.Fedorova E, Zink D. Nuclear architecture and gene regulation. Biochim Biophys Acta. 2008;1783:2174–2184. doi: 10.1016/j.bbamcr.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 3.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 4.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 6.Illingworth RS, Bird AP. CpG islands--‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 9.Nomura J, Hisatsune A, Miyata T, Isohama Y. The role of CpG methylation in cell type-specific expression of the aquaporin-5 gene. Biochem Biophys Res Commun. 2007;353:1017–1022. doi: 10.1016/j.bbrc.2006.12.126. [DOI] [PubMed] [Google Scholar]
- 10.Aoyama T, Okamoto T, Nagayama S, Nishijo K, Ishibe T, Yasura K, Nakayama T, Nakamura T, Toguchida J. Methylation in the core-promoter region of the chondromodulin-I gene determines the cell-specific expression by regulating the binding of transcriptional activator Sp3. J Biol Chem. 2004;279:28789–28797. doi: 10.1074/jbc.M401273200. [DOI] [PubMed] [Google Scholar]
- 11.Rossler J, Stolze I, Frede S, Freitag P, Schweigerer L, Havers W, Fandrey J. Hypoxia-induced erythropoietin expression in human neuroblastoma requires a methylation free HIF-1 binding site. J Cell Biochem. 2004;93:153–161. doi: 10.1002/jcb.20133. [DOI] [PubMed] [Google Scholar]
- 12.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 14.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 15.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 16.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirio MC, Martel J, Mann M, Toppings M, Bartolomei M, Trasler J, Chaillet JR. DNA methyltransferase 1o functions during preimplantation development to preclude a profound level of epigenetic variation. Dev Biol. 2008;324:139–150. doi: 10.1016/j.ydbio.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 20.Orstavik KH. X chromosome inactivation in clinical practice. Hum Genet. 2009;126:363–373. doi: 10.1007/s00439-009-0670-5. [DOI] [PubMed] [Google Scholar]
- 21.Butler MG. Genomic imprinting disorders in humans: a mini-review. J Assist Reprod Genet. 2009;26:477–486. doi: 10.1007/s10815-009-9353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 23.Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23:105–117. doi: 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green K, Lewis A, Dawson C, Dean W, Reinhart B, Chaillet JR, Reik W. A developmental window of opportunity for imprinted gene silencing mediated by DNA methylation and the Kcnq1ot1 noncoding RNA. Mamm Genome. 2007;18:32–42. doi: 10.1007/s00335-006-0092-9. [DOI] [PubMed] [Google Scholar]
- 25.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 26.Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, Gaudet F, Li E, Chen T. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 28.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wossidlo M, Arand J, Sebastiano V, Lepikhov K, Boiani M, Reinhardt R, Scholer H, Walter J. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. Embo J. 2010;29:1877–1888. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, Kitagawa H, Takeyama K, Shibuya H, Ohtake F, Kato S. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 32.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 33.Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 34.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 35.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 37.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 38.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 39.Volkel P, Angrand PO. The control of histone lysine methylation in epigenetic regulation. Biochimie. 2007;89:1–20. doi: 10.1016/j.biochi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 40.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circ Res. 2007;100:1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 41.Ooi L, Belyaev ND, Miyake K, Wood IC, Buckley NJ. BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. J Biol Chem. 2006;281:38974–38980. doi: 10.1074/jbc.M605370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 43.Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol. 2008;18:682–689. doi: 10.1016/j.sbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zimmermann N, Acosta AM, Kohlhase J, Bartsch O. Confirmation of EP300 gene mutations as a rare cause of Rubinstein-Taybi syndrome. Eur J Hum Genet. 2007;15:837–842. doi: 10.1038/sj.ejhg.5201791. [DOI] [PubMed] [Google Scholar]
- 45.Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ, Breuning MH. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 46.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellert HS, McMahon SB. Biochemical pathways that regulate acetyltransferase and deacetylase activity in mammalian cells. Trends Biochem Sci. 2009;34:571–578. doi: 10.1016/j.tibs.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Codd R, Braich N, Liu J, Soe CZ, Pakchung AA. Zn(II)-dependent histone deacetylase inhibitors: suberoylanilide hydroxamic acid and trichostatin A. Int J Biochem Cell Biol. 2009;41:736–739. doi: 10.1016/j.biocel.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 49.Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. Embo J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 51.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 52.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 53.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 54.Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. Embo J. 29:68–79. doi: 10.1038/emboj.2009.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagschal A, Sutherland HG, Woodfine K, Henckel A, Chebli K, Schulz R, Oakey RJ, Bickmore WA, Feil R. G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol Cell Biol. 2008;28:1104–1113. doi: 10.1128/MCB.01111-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 58.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 59.Nagano T, Fraser P. Emerging similarities in epigenetic gene silencing by long noncoding RNAs. Mamm Genome. 2009;20:557–562. doi: 10.1007/s00335-009-9218-1. [DOI] [PubMed] [Google Scholar]
- 60.Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, Sakaki Y, Sasaki H, Hohjoh H. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Res. 2010 Apr 12; doi: 10.1093/nar/gkq229. (in press) Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matzke MA, Primig M, Trnovsky J, Matzke AJ. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. Embo J. 1989;8:643–649. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morris KV. RNA-directed transcriptional gene silencing and activation in human cells. Oligonucleotides. 2009;19:299–306. doi: 10.1089/oli.2009.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 64.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 67.Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. Embo J. 2009;28:1708–1719. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182:845–850. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P, Besenbacher S, Jonasdottir A, Sigurdsson A, Kristinsson KT, Jonasdottir A, Frigge ML, Gylfason A, Olason PI, Gudjonsson SA, Sverrisson S, Stacey SN, Sigurgeirsson B, Benediktsdottir KR, Sigurdsson H, Jonsson T, Benediktsson R, Olafsson JH, Johannsson OT, Hreidarsson AB, Sigurdsson G, Ferguson-Smith AC, Gudbjartsson DF, Thorsteinsdottir U, Stefansson K. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ling C, Poulsen P, Simonsson S, Ronn T, Holmkvist J, Almgren P, Hagert P, Nilsson E, Mabey AG, Nilsson P, Vaag A, Groop L. Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J Clin Invest. 2007;117:3427–3435. doi: 10.1172/JCI30938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barker DJ. The developmental origins of well-being. Philos Trans R Soc Lond B Biol Sci. 2004;359:1359–1366. doi: 10.1098/rstb.2004.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 74.Painter RC, Roseboom TJ, Bleker OP. Prenatal exposure to the Dutch famine and disease in later life: an overview. Reprod Toxicol. 2005;20:345–352. doi: 10.1016/j.reprotox.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 75.Godfrey KM, Barker DJ. Fetal programming and adult health. Public Health Nutr. 2001;4:611–624. doi: 10.1079/phn2001145. [DOI] [PubMed] [Google Scholar]
- 76.Palinski W, Napoli C. The fetal origins of atherosclerosis: maternal hypercholesterolemia, and cholesterol-lowering or antioxidant treatment during pregnancy influence in utero programming and postnatal susceptibility to atherogenesis. Faseb J. 2002;16:1348–1360. doi: 10.1096/fj.02-0226rev. [DOI] [PubMed] [Google Scholar]
- 77.Turunen MP, Aavik E, Yla-Herttuala S. Epigenetics and atherosclerosis. Biochim Biophys Acta. 2009;1790:886–891. doi: 10.1016/j.bbagen.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 78.Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 79.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol. 2009;5:401–408. doi: 10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- 80.Kuzawa CW, Sweet E. Epigenetics and the embodiment of race: developmental origins of US racial disparities in cardiovascular health. Am J Hum Biol. 2009;21:2–15. doi: 10.1002/ajhb.20822. [DOI] [PubMed] [Google Scholar]
- 81.Sherman RC, Langley-Evans SC. Early administration of angiotensin-converting enzyme inhibitor captopril, prevents the development of hypertension programmed by intrauterine exposure to a maternal low-protein diet in the rat. Clin Sci (Lond) 1998;94:373–381. doi: 10.1042/cs0940373. [DOI] [PubMed] [Google Scholar]
- 82.Bogdarina I, Welham S, King PJ, Burns SP, Clark AJ. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ Res. 2007;100:520–526. doi: 10.1161/01.RES.0000258855.60637.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 84.Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr. 2007;97:1064–1073. doi: 10.1017/S000711450769196X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rouleau J, Tanigawa G, Szyf M. The mouse DNA methyltransferase 5′-region. A unique housekeeping gene promoter. J Biol Chem. 1992;267:7368–7377. [PubMed] [Google Scholar]
- 86.Das SK, Chakrabarti R. Role of PPAR in cardiovascular diseases. Recent Pat Cardiovasc Drug Discov. 2006;1:193–209. doi: 10.2174/157489006777442441. [DOI] [PubMed] [Google Scholar]
- 87.Walker BR. Glucocorticoids and cardiovascular disease. Eur J Endocrinol. 2007;157:545–559. doi: 10.1530/EJE-07-0455. [DOI] [PubMed] [Google Scholar]
- 88.Jackson AA, Dunn RL, Marchand MC, Langley-Evans SC. Increased systolic blood pressure in rats induced by a maternal low-protein diet is reversed by dietary supplementation with glycine. Clin Sci (Lond) 2002;103:633–639. doi: 10.1042/cs1030633. [DOI] [PubMed] [Google Scholar]
- 89.Torrens C, Brawley L, Anthony FW, Dance CS, Dunn R, Jackson AA, Poston L, Hanson MA. Folate supplementation during pregnancy improves offspring cardiovascular dysfunction induced by protein restriction. Hypertension. 2006;47:982–987. doi: 10.1161/01.HYP.0000215580.43711.d1. [DOI] [PubMed] [Google Scholar]
- 90.Burdge GC, Lillycrop KA, Phillips ES, Slater-Jefferies JL, Jackson AA, Hanson MA. Folic acid supplementation during the juvenile-pubertal period in rats modifies the phenotype and epigenotype induced by prenatal nutrition. J Nutr. 2009;139:1054–1060. doi: 10.3945/jn.109.104653. [DOI] [PubMed] [Google Scholar]
- 91.Niculescu MD, Craciunescu CN, Zeisel SH. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. Faseb J. 2006;20:43–49. doi: 10.1096/fj.05-4707com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lv S, Fan R, Du Y, Hou M, Tang Z, Ling W, Zhu H. Betaine supplementation attenuates atherosclerotic lesion in apolipoprotein E-deficient mice. Eur J Nutr. 2009;48:205–212. doi: 10.1007/s00394-009-0003-4. [DOI] [PubMed] [Google Scholar]
- 93.Post WS, Goldschmidt-Clermont PJ, Wilhide CC, Heldman AW, Sussman MS, Ouyang P, Milliken EE, Issa JP. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res. 1999;43:985–991. doi: 10.1016/s0008-6363(99)00153-4. [DOI] [PubMed] [Google Scholar]
- 94.Kim J, Kim JY, Song KS, Lee YH, Seo JS, Jelinek J, Goldschmidt-Clermont PJ, Issa JP. Epigenetic changes in estrogen receptor beta gene in atherosclerotic cardiovascular tissues and in-vitro vascular senescence. Biochim Biophys Acta. 2007;1772:72–80. doi: 10.1016/j.bbadis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 95.Matouk CC, Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circ Res. 2008;102:873–887. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- 96.Illi B, Colussi C, Grasselli A, Farsetti A, Capogrossi MC, Gaetano C. NO sparks off chromatin: tales of a multifaceted epigenetic regulator. Pharmacol Ther. 2009;123:344–352. doi: 10.1016/j.pharmthera.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 97.Antoniades C, Antonopoulos AS, Tousoulis D, Marinou K, Stefanadis C. Homocysteine and coronary atherosclerosis: from folate fortification to the recent clinical trials. Eur Heart J. 2009;30:6–15. doi: 10.1093/eurheartj/ehn515. [DOI] [PubMed] [Google Scholar]
- 98.Joseph J, Handy DE, Loscalzo J. Quo vadis: whither homocysteine research? Cardiovasc Toxicol. 2009;9:53–63. doi: 10.1007/s12012-009-9042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem. 2000;275:29318–29323. doi: 10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]
- 100.Stern LL, Mason JB, Selhub J, Choi SW. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol Biomarkers Prev. 2000;9:849–853. [PubMed] [Google Scholar]
- 101.Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, Selhub J. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Castro R, Rivera I, Blom HJ, Jakobs C, Tavares de Almeida I. Homocysteine metabolism, hyperhomocysteinaemia and vascular disease: an overview. J Inherit Metab Dis. 2006;29:3–20. doi: 10.1007/s10545-006-0106-5. [DOI] [PubMed] [Google Scholar]
- 103.Kok RM, Smith DE, Barto R, Spijkerman AM, Teerlink T, Gellekink HJ, Jakobs C, Smulders YM. Global DNA methylation measured by liquid chromatography-tandem mass spectrometry: analytical technique, reference values and determinants in healthy subjects. Clin Chem Lab Med. 2007;45:903–911. doi: 10.1515/CCLM.2007.137. [DOI] [PubMed] [Google Scholar]
- 104.Pogribny IP, Beland FA. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell Mol Life Sci. 2009;66:2249–2261. doi: 10.1007/s00018-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Castro R, Rivera I, Struys EA, Jansen EE, Ravasco P, Camilo ME, Blom HJ, Jakobs C, Tavares de Almeida I. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003;49:1292–1296. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- 106.Lund G, Andersson L, Lauria M, Lindholm M, Fraga MF, Villar-Garea A, Ballestar E, Esteller M, Zaina S. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem. 2004;279:29147–29154. doi: 10.1074/jbc.M403618200. [DOI] [PubMed] [Google Scholar]
- 107.Devlin AM, Arning E, Bottiglieri T, Faraci FM, Rozen R, Lentz SR. Effect of Mthfr genotype on diet-induced hyperhomocysteinemia and vascular function in mice. Blood. 2004;103:2624–2629. doi: 10.1182/blood-2003-09-3078. [DOI] [PubMed] [Google Scholar]
- 108.Devlin AM, Bottiglieri T, Domann FE, Lentz SR. Tissue-specific changes in H19 methylation and expression in mice with hyperhomocysteinemia. J Biol Chem. 2005;280:25506–25511. doi: 10.1074/jbc.M504815200. [DOI] [PubMed] [Google Scholar]
- 109.Sharma P, Kumar J, Garg G, Kumar A, Patowary A, Karthikeyan G, Ramakrishnan L, Brahmachari V, Sengupta S. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol. 2008;27:357–365. doi: 10.1089/dna.2007.0694. [DOI] [PubMed] [Google Scholar]
- 110.Ingrosso D, Cimmino A, Perna AF, Masella L, De Santo NG, De Bonis ML, Vacca M, D’Esposito M, D’Urso M, Galletti P, Zappia V. Folate treatment and unbalanced methylation and changes of allelic expression induced by hyperhomocysteinaemia in patients with uraemia. Lancet. 2003;361:1693–1699. doi: 10.1016/S0140-6736(03)13372-7. [DOI] [PubMed] [Google Scholar]
- 111.Nanayakkara PW, Kiefte-de Jong JC, Stehouwer CD, van Ittersum FJ, Olthof MR, Kok RM, Blom HJ, van Guldener C, ter Wee PM, Smulders YM. Association between global leukocyte DNA methylation, renal function, carotid intima-media thickness and plasma homocysteine in patients with stage 2–4 chronic kidney disease. Nephrol Dial Transplant. 2008;23:2586–2592. doi: 10.1093/ndt/gfn040. [DOI] [PubMed] [Google Scholar]
- 112.Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, Heimburger O, Barany P, Alvestrand A, Nordfors L, Qureshi AR, Ekstrom TJ, Schalling M. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease? J Intern Med. 2007;261:488–499. doi: 10.1111/j.1365-2796.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- 113.Ingrosso D, Perna AF. Epigenetics in hyperhomocysteinemic states. A special focus on uremia. Biochim Biophys Acta. 2009;1790:892–899. doi: 10.1016/j.bbagen.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 114.Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS One. 2010;5:e8564. doi: 10.1371/journal.pone.0008564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Perna AF, Ingrosso D, Violetti E, Luciano MG, Sepe I, Lanza D, Capasso R, Ascione E, Raiola I, Lombardi C, Stenvinkel P, Massy Z, De Santo NG. Hyperhomocysteinemia in uremia--a red flag in a disrupted circuit. Semin Dial. 2009;22:351–356. doi: 10.1111/j.1525-139X.2009.00579.x. [DOI] [PubMed] [Google Scholar]
- 116.Wang H, Yoshizumi M, Lai K, Tsai JC, Perrella MA, Haber E, Lee ME. Inhibition of growth and p21ras methylation in vascular endothelial cells by homocysteine but not cysteine. J Biol Chem. 1997;272:25380–25385. doi: 10.1074/jbc.272.40.25380. [DOI] [PubMed] [Google Scholar]
- 117.Wang H, Jiang X, Yang F, Chapman GB, Durante W, Sibinga NE, Schafer AI. Cyclin A transcriptional suppression is the major mechanism mediating homocysteine-induced endothelial cell growth inhibition. Blood. 2002;99:939–945. [PMC free article] [PubMed] [Google Scholar]
- 118.Jamaluddin MD, Chen I, Yang F, Jiang X, Jan M, Liu X, Schafer AI, Durante W, Yang X, Wang H. Homocysteine inhibits endothelial cell growth via DNA hypomethylation of the cyclin A gene. Blood. 2007;110:3648–3655. doi: 10.1182/blood-2007-06-096701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chang PY, Lu SC, Lee CM, Chen YJ, Dugan TA, Huang WH, Chang SF, Liao WS, Chen CH, Lee YT. Homocysteine inhibits arterial endothelial cell growth through transcriptional downregulation of fibroblast growth factor-2 involving G protein and DNA methylation. Circ Res. 2008;102:933–941. doi: 10.1161/CIRCRESAHA.108.171082. [DOI] [PubMed] [Google Scholar]
- 120.Yideng J, Jianzhong Z, Ying H, Juan S, Jinge Z, Shenglan W, Xiaoqun H, Shuren W. Homocysteine-mediated expression of SAHH, DNMTs, MBD2, and DNA hypomethylation potential pathogenic mechanism in VSMCs. DNA Cell Biol. 2007;26:603–611. doi: 10.1089/dna.2007.0584. [DOI] [PubMed] [Google Scholar]
- 121.Lenz B, Bleich S, Beutler S, Schlierf B, Schwager K, Reulbach U, Kornhuber J, Bonsch D. Homocysteine regulates expression of Herp by DNA methylation involving the AARE and CREB binding sites. Exp Cell Res. 2006;312:4049–4055. doi: 10.1016/j.yexcr.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 122.Huang YS, Zhi YF, Wang SR. Hypermethylation of estrogen receptor-alpha gene in atheromatosis patients and its correlation with homocysteine. Pathophysiology. 2009;16:259–265. doi: 10.1016/j.pathophys.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 123.Werstuck GH, Lentz SR, Dayal S, Hossain GS, Sood SK, Shi YY, Zhou J, Maeda N, Krisans SK, Malinow MR, Austin RC. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J Clin Invest. 2001;107:1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Namekata K, Enokido Y, Ishii I, Nagai Y, Harada T, Kimura H. Abnormal lipid metabolism in cystathionine beta-synthase-deficient mice, an animal model for hyperhomocysteinemia. J Biol Chem. 2004;279:52961–52969. doi: 10.1074/jbc.M406820200. [DOI] [PubMed] [Google Scholar]
- 125.Jacobson TA, Miller M, Schaefer EJ. Hypertriglyceridemia and cardiovascular risk reduction. Clin Ther. 2007;29:763–777. doi: 10.1016/j.clinthera.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 126.Mikael LG, Wang XL, Wu Q, Jiang H, Maclean KN, Rozen R. Hyperhomocysteinemia is associated with hypertriglyceridemia in mice with methylenetetrahydrofolate reductase deficiency. Mol Genet Metab. 2009;98:187–194. doi: 10.1016/j.ymgme.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 127.Devlin AM, Lentz SR. ApoA-I: a missing link between homocysteine and lipid metabolism? Circ Res. 2006;98:431–433. doi: 10.1161/01.RES.0000214406.87060.e0. [DOI] [PubMed] [Google Scholar]
- 128.Mikael LG, Genest J, Jr, Rozen R. Elevated homocysteine reduces apolipoprotein A-I expression in hyperhomocysteinemic mice and in males with coronary artery disease. Circ Res. 2006;98:564–571. doi: 10.1161/01.RES.0000204825.66410.0b. [DOI] [PubMed] [Google Scholar]
- 129.Liao D, Tan H, Hui R, Li Z, Jiang X, Gaubatz J, Yang F, Durante W, Chan L, Schafer AI, Pownall HJ, Yang X, Wang H. Hyperhomocysteinemia decreases circulating high-density lipoprotein by inhibiting apolipoprotein A-I Protein synthesis and enhancing HDL cholesterol clearance. Circ Res. 2006;99:598–606. doi: 10.1161/01.RES.0000242559.42077.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liao D, Yang X, Wang H. Hyperhomocysteinemia and high-density lipoprotein metabolism in cardiovascular disease. Clin Chem Lab Med. 2007;45:1652–1659. doi: 10.1515/CCLM.2007.358. [DOI] [PubMed] [Google Scholar]
- 131.Devlin AM, Singh R, Wade RE, Innis SM, Bottiglieri T, Lentz SR. Hypermethylation of Fads2 and altered hepatic fatty acid and phospholipid metabolism in mice with hyperhomocysteinemia. J Biol Chem. 2007;282:37082–37090. doi: 10.1074/jbc.M704256200. [DOI] [PubMed] [Google Scholar]
- 132.Devlin AM, Singh R, Bottiglieri T, Innis SM, Green TJ. Hepatic acyl-coenzyme a:cholesterol acyltransferase-2 expression is decreased in mice with hyperhomocysteinemia. J Nutr. 140:231–237. doi: 10.3945/jn.109.112920. [DOI] [PubMed] [Google Scholar]
- 133.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saunthararajah Y, Hillery CA, Lavelle D, Molokie R, Dorn L, Bressler L, Gavazova S, Chen YH, Hoffman R, DeSimone J. Effects of 5-aza-2′-deoxycytidine on fetal hemoglobin levels, red cell adhesion, and hematopoietic differentiation in patients with sickle cell disease. Blood. 2003;102:3865–3870. doi: 10.1182/blood-2003-05-1738. [DOI] [PubMed] [Google Scholar]
- 135.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, Havelange V, Volinia S, Blum W, Rush LJ, Perrotti D, Andreeff M, Bloomfield CD, Byrd JC, Chan K, Wu LC, Croce CM, Marcucci G. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bieliauskas AV, Pflum MK. Isoform-selective histone deacetylase inhibitors. Chem Soc Rev. 2008;37:1402–1413. doi: 10.1039/b703830p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pons D, de Vries FR, van den Elsen PJ, Heijmans BT, Quax PH, Jukema JW. Epigenetic histone acetylation modifiers in vascular remodelling: new targets for therapy in cardiovascular disease. Eur Heart J. 2009;30:266–277. doi: 10.1093/eurheartj/ehn603. [DOI] [PubMed] [Google Scholar]
- 138.Willyard C. The saving switch. Nat Med. 16:18–21. doi: 10.1038/nm0110-18. [DOI] [PubMed] [Google Scholar]
- 139.Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Holliday R. Epigenetics: a historical overview. Epigenetics. 2006;1:76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]