Abstract
Motivation: Transcription factor (TF) ChIP-seq datasets have particular characteristics that provide unique challenges and opportunities for motif discovery. Most existing motif discovery algorithms do not scale well to such large datasets, or fail to report many motifs associated with cofactors of the ChIP-ed TF.
Results: We present DREME, a motif discovery algorithm specifically designed to find the short, core DNA-binding motifs of eukaryotic TFs, and optimized to analyze very large ChIP-seq datasets in minutes. Using DREME, we discover the binding motifs of the the ChIP-ed TF and many cofactors in mouse ES cell (mESC), mouse erythrocyte and human cell line ChIP-seq datasets. For example, in mESC ChIP-seq data for the TF Esrrb, we discover the binding motifs for eight cofactor TFs important in the maintenance of pluripotency. Several other commonly used algorithms find at most two cofactor motifs in this same dataset. DREME can also perform discriminative motif discovery, and we use this feature to provide evidence that Sox2 and Oct4 do not bind in mES cells as an obligate heterodimer. DREME is much faster than many commonly used algorithms, scales linearly in dataset size, finds multiple, non-redundant motifs and reports a reliable measure of statistical significance for each motif found. DREME is available as part of the MEME Suite of motif-based sequence analysis tools (http://meme.nbcr.net).
Contact: t.bailey@uq.edu.au
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
It is estimated that the human genome contains 1500 transcription factors (TFs) that play a key role in regulating transcription by binding to the genome alone or in protein complexes (Vaquerizas et al., 2009). Chromatin immunoprecipitation (ChIP) followed by high-throughput sequencing (ChIP-seq) is the current method of choice for determining the genomic binding locations of a TF. The resolution of ChIP-seq location data is such that the actual binding location is typically within ~50 bp of the predicted location (Wilbanks and Facciotti, 2010). A TF ChIP-seq experiment generally yields hundreds to tens of thousands of predicted locations (often called ‘ChIP-seq peaks’). In addition to providing evidence of direct regulation by the TF proximity to individual genes, the ChIP-seq data provide a rich resource for exploring the in vivo DNA-binding affinity of the ChIP-ed TF and cofactor TFs that bind DNA in complex or nearby, and for discovering the identities of these cofactors.
Ab initio motif discovery algorithms applied to ChIP-seq peak regions can usually discover the DNA-binding motif of the ChIP-ed TF. However, most existing algorithms have limitations that make them less than ideal for discovering all the cofactor motifs in a ChIP-seq dataset. One limitation of many popular algorithms is that they do not scale well to inputs containing thousands of sequences. As a result, many studies to date have used only a fraction (say 500 sequences) of the available ChIP-seq peak regions for motif discovery, greatly decreasing the chances of discovering motifs for infrequent cofactors. A second limitation is that many algorithms do not provide reliable motif statistics to enable the biologist to discern functional motifs from statistical artifacts. This limitation can cause valid motifs to be ignored, or time to be wasted investigating random motifs. A third limitation is that most algorithms do not allow two ChIP-seq datasets to be directly compared, finding motifs that are relatively enriched. This limitation makes it difficult to determine whether two TFs always bind as a heterodimer or even if the motif for the ChIP-ed TF has been found at all. Other limitations possessed by some algorithms that make them unsatisfactory for ChIP-seq data analysis include only finding a single motif or being ‘hard-wired’ to use a particular type of sequence (e.g. promoters).
Here, we describe a novel motif discovery algorithm lacking the above limitations, and demonstrate how to use it to mine ChIP-seq datasets for primary and cofactor motifs and for insights into cooperative or indirect binding by TFs. We show that the new algorithm is significantly faster than several algorithms that are commonly used for analyzing ChIP-seq datasets, yet is able to discover substantially more cofactor motifs. The algorithm searches for motifs expressed as simplified ‘regular expressions’ (consensus sequences allowing wildcards but not variable length gaps). It achieves speed partly by limiting its search to short motifs (up to 8 bp wide), which turns out to be ideal for identifying and studying the DNA-binding motifs of monomeric eukaryotic TFs, but may cause it to miss some wider motifs due to binding by large TF complexes. Our new algorithm has some similarities with a few existing, word-based algorithms such as Trawler (Ettwiller et al., 2007) and Amadeus (Linhart et al., 2008)), but our approach is simpler and we demonstrate that it finds substantially more cofactor motifs. Consequently, our algorithm complements rather than replaces existing motif discovery tools for the analysis of ChIP-seq data.
2 METHODS
2.1 DREME: discriminative regular expression motif elicitation
Our design objective for a motif discovery algorithm tailored to eukaryotic ChIP-seq data is the ability to quickly discover multiple, short, non-redundant, statistically significant, discriminative motifs in extremely large sets of short sequences. The focus on short (from 4 to 8 bp) motifs is motivated by the observation that this range encompasses the DNA-binding region of most eukaryotic monomeric TFs, and existing algorithms can easily find the larger, more information-rich motifs characteristic of binding by TF protein complexes. In the interest of computational speed, we restrict the search for motifs to a simplified form of ‘regular expression’: words over the IUPAC alphabet, which adds eleven wildcard characters to the standard DNA alphabet, ACGT. For example, the binding motif of the Klf family of TFs is well represented by the IUPAC regular expression (RE) motif CMCRCCC (called a ‘CACC-box’), where M matches A or C and R matches the purines A or G.
Our search for motifs is exhaustive for exact words (no wildcards), but heuristic for words with wildcards, again in the interest of speed. To identify statistically significant, discriminative motifs, we compute the significance of the relative enrichment of each motif in two sets of sequences using the Fisher's Exact Test. This test computes the probability that the fraction of sequences in the first set matching the motif would be as great as observed (or greater), given the fraction of matching sequences in the second set. In a typical application of our approach, one set of sequences would be a set of ChIP-seq peak regions from a TF ChIP-seq experiment, and the other would either be similar data from a different ChIP-seq experiment or shuffled versions of the first sequences. To avoid problems with self-overlapping motifs, we only count the number of sequences containing a motif (not the number of occurrences of the motif) in each of the two datasets. However, once the motif with the highest significance has been found, all of its non-overlapping occurrences in the first set of sequences are aligned to create a position-specific probability matrix (Stormo, 2000). Finally, to find multiple, non-redundant motifs in a set of sequences, we simply ‘erase’ the best motif found by setting all its occurrences to a special letter that cannot match any motif, and then repeat the search for motifs.
DREME implements this motif discovery approach. The input to DREME is two sets of DNA sequences and a significance threshold. The algorithm's outer loop performs a heuristic search of RE motifs, determines the most significant motif, reports it and erases all its occurrences in the input datasets. The outer loop is repeated until no new motif has E-value less than the given significance threshold.
The central task of the DREME algorithm is searching the space of RE motifs. Our approach uses a beam search that starts with a set of highly significant ‘seed’ RE motifs and attempts to find more significant generalizations of them. To initialize the beam search, DREME computes the significance of each word (no wildcards) of length three to eight that occurs in the positive sequences. DREME does this by counting how many positive and negative sequences contain each word, and computing the (uncorrected) p-value of the Fisher's Exact Test. This is done in time linear in the size of the datasets. The 100 most significant words are then passed as ‘seed’ REs to an inner loop that performs the beam search.
At each iteration of the inner loop, DREME considers all generalizations of each seed RE that differ from the seed RE by containing exactly one additional wildcard. If all the words that match a generalization of the seed RE are significant (uncorrected p ≤ 0.01), DREME estimates the generalized RE's statistical significance, otherwise, the generalization is discarded. To save computation time, DREME estimates the significance of candidate REs without scanning the input sequences. When all seed REs have been generalized, DREME sorts the new, more general REs by estimated significance, and then computes the exact significance of the top 100 to use as seed REs in the next iteration. The inner loop stops when no seed RE can be generalized to include an additional wildcard, typically after three or four iterations.
To estimate the number of sequences matching a generalized RE, DREME uses the fact that this is always equal to the size of the union of the matching sets of two more specific REs, as shown in Supplementary Table S1. DREME assumes that these two matching sets (RE1 and RE2) are independent samples from a set of N sequences, so the size of the union (RE3) is given by
| (1) |
where |RE| is the size of the set of sequences matching a given RE. For example, if RE1 = ATGCG is a seed RE, and RE2 = ATGCT, which differs from it only in the last position, is significant, then DREME estimates the number of positive (or negative) sequences matching the generalization RE3 = ATGCK using Equation (1). DREME applies the Fisher's Exact Test to the estimated numbers of positive and negative sequences matching ATGCK and caches the result for later use in estimating the numbers of sequences matching ATGCD and ATGCH. By performing generalizations in the order shown in Supplementary Table S1, DREME efficiently estimates the numbers of sequences matching a generalized RE.
2.2 Evaluating motif discovery
We evaluate DREME and compare it with a number of existing motif discovery algorithms using several TF ChIP-seq datasets from mouse embryonic stem cells (mES cells), mouse erythrocytes and a human lymphoblastoid cell line. In our evaluation, we consider speed, ability to identify the primary motif, identification of secondary motifs and illustrate the usefulness of being able to do discriminative motif discovery in ChIP-seq datasets. To determine what motifs an algorithm discovers, we compare its output motifs to a large panel of known motifs using the TOMTOM (Gupta et al., 2007) algorithm and use gene expression data from the Gene Expression Atlas (Kapushesky et al., 2010) to determine which TF among TFs with similar DNA-binding affinity may bind to secondary motifs.
2.2.1 Running motif discovery algorithms
We run DREME using its default settings. When only one set of sequences is input, DREME creates a dinucleotide-shuffled version of the input sequence set as the negative sequence set. For discriminative motif discovery, we provide two sets of input sequences to DREME. In both cases, the significance threshold for motifs is E = 0.05.
We compare the speed and accuracy of several popular motif discovery algorithms with that of DREME using the 13 mESC ChIP-seq datasets. Two of these algorithms [MEME (Bailey and Elkan, 1995) and nestedMICA (Down and Hubbard, 2005)] are extremely slow when run on more than 500 sequences, so we randomly choose 500 sequences from each ChIP-seq dataset to create datasets of reduced size. We run the other three algorithms [WEEDER (Pavesi et al., 2004), Trawler (Ettwiller et al., 2007) and Amadeus (Linhart et al., 2008)] on the same datasets that were used above with DREME. Where possible, we parameterize the programs to search for motifs of 4–7 bp. The exceptions are WEEDER up to 8 bp; Trawler minimum 4 bp; Amadeus exactly 7 bp. The number of motifs to search for is a required parameter for nestedMICA, and we set it to 10 to keep running times reasonably short. (It does not finish running after 134 h on the full-size E2f1 dataset, which contains 20 699 sequences.) We set the maximum number of motifs to find to 20 for MEME; WEEDER, Trawler and Amadeus have no such parameter. With MEME, we use a 0-order background model and with nestedMICA we use a 0-order four class background model. As background sequences for DREME, Amadeus and Trawler, we use 1, 5 and 10 dinucleotide-shuffled copies, respectively, of the foreground sequences. With WEEDER, we use its pre-computed background model for mouse, which contains the frequencies of all 8mers in regions 1000 bp upstream of genes. Exact command lines and more details are given in the Supplementary Material.
2.2.2 ChIP-seq datasets
We use 13 mESC ChIP-seq datasets (Chen et al., 2008), 3 mouse erythrocyte datasets (Cheng et al., 2009; Kassouf et al., 2010; Tallack et al., 2010) and 1 human lymphoblastoid cell line dataset (Ramagopalan et al., 2010). The mESC datasets are for 12 TFs that are key to the maintenance of pluripotency, plus CTCF. The mouse erythrocyte datasets are for Gata1, Klf1 and Tal1, key regulators of erythropoiesis. The human lymphoblastoid cell line data is for the vitamin D receptor (VDR) and includes separate ChIP-seq data for cells before and after stimulation with calcitriol. To prepare the datasets for use with motif discovery algorithms, we map the (centers of the) ChIP-seq peaks declared by the respective authors to the genome, and extract the 100 bp of genomic sequence centered on each peak. [The one exception is Gata1, where we used the peaks declared by Tallack et al. (2010) in the Cheng et al. (2009) data.]
2.2.3 Identifying discovered motifs
To determine the success of motif discovery, we compare the motifs to a database of known TF motifs using TOMTOM (Gupta et al., 2007). The motif database comprises all vertebrate motifs in the JASPAR database (Sandelin et al., 2004) (146 motifs) and all mouse motifs in the UniProbe database (Berger and Bulyk, 2009) (386 motifs). Because the two motif reference datasets lack a motif for the key pluripotency TF Nanog, we also include an in vitro motif for Nanog taken from the literature (Jauch et al., 2008), for a total of 533 motifs. We only consider statistically significant TOMTOM predictions (E ≤ 0.05, corrected for 533 tests).
Because many TFs have very similar DNA-binding affinities, many of the known motifs in this database are similar to each other, so some discovered motifs have significant matches to multiple motifs in the database. Therefore, in order to determine to which TF a discovered motif may correspond, we use prior knowledge from the literature and evidence of increased expression in the ChIP-ed tissue type. To resolve multiple matches, we take the most significant TOMTOM prediction among matching TFs that are either upregulated in, or known to be central regulators of, the ChIP-ed cell type. We consider a TF as upregulated in mouse ES cells if it was so marked for at least one experiment in cell type EF0_000462 in the Gene Expression Atlas (Kapushesky et al., 2010) (http://www.ebi.ac.uk/gxa/). For motifs discovered in mouse erythrocytes and human lymphoblastoid cell lines, we restrict matching candidate motifs to TFs previously shown to be important to transcriptional regulation in those cell types.
3 RESULTS
3.1 Discovering motifs in a single ChIP-seq dataset using DREME
3.1.1 Discovering motifs in mouse ES cell ChIP-seq data
DREME reports between 10 and 33 significant motifs (P < 0.05) for each of the mESC datasets (Table 1, column 3), and the number of significant motifs it finds is highly correlated with the number of ChIP-seq peaks in the input dataset. It discovers the ChIP-ed (primary) TF motif in 10 of the 13 mESC ChIP-seq datasets (Table 1, column 4), and it is the most significant motif found by DREME in those 10 cases. In two of the remaining three cases (Nanog and E2f1 ChIP-seq datasets), DREME finds a highly significant motif that is similar to the known motif, but the TOMTOM p-value does not meet our 0.05 threshold due to the large number of multiple tests (533 reference motifs). However, the uncorrected p-values are quite low (Nanog, p = 0.0021; E2f1, p = 0.0016). Since DREME reports 24 motifs in the Nanog dataset, we could reject a slightly different null hypothesis that stated that the known Nanog motif was not found (Nanog, p = 0.05, corrected for 24 tests) and similarly for E2f1 (E2f1, p = 0.04, corrected for 25 tests). In the final case (Smad1 ChIP-seq dataset), DREME does not find a motif that matches the only available in vitro Smad-family motif (UniProbe Smad3_primary), nor the motif reported by Chen et al. (The complete DREME and TOMTOM output on the mESC datasets is given in the Supplementary Material.)
Table 1.
Primary and cofactor motifs found by DREME in 13 mES cell ChIP-seq datasets
| TF | Peaks | m | r | Cofactor motifs |
|---|---|---|---|---|
| CTCF | 39609 | 29 | 1 | Myc, STAT3, GABPA |
| cMyc | 3422 | 12 | 1 | STAT3, Egr1 |
| E2f1 | 20699 | 25 | 2a | STAT3, Myc, Klf4, Fox, |
| CREB/ATF | ||||
| Esrrb | 21647 | 29 | 1 | Klf4, Sox2, STAT3, Oct4, Myc, |
| Rxra, Zic3, Ewsr1 | ||||
| Klf4 | 10875 | 26 | 1 | STAT3, Esrrb, Sox2, Oct4, Sp1, |
| Gata3, Myc, Zfp161 | ||||
| Nanog | 10343 | 24 | 4a | Sox2, Oct4, Zic3, Klf4, Elf5, |
| Esrrb, Tead1 | ||||
| nMyc | 7182 | 21 | 1 | STAT3, Smad1, CREB/ATF, |
| Sfpib | ||||
| Oct4 | 3761 | 17 | 1 | Sox2, Klf4, CREB/ATF, Esrrb |
| STAT3 | 2546 | 13 | 1 | Klf4, Esrrb, Sox2, Oct4, Myc, |
| Sp1, Irf4 | ||||
| Smad1 | 1126 | 10 | No | Sox2, Oct4, Esrrb, Zic3, Klf4, |
| Zfp740 | ||||
| Sox2 | 4526 | 19 | 1 | Oct4, Klf4, STAT3, Zic3, Esrrb |
| Tcfcp2l1 | 26910 | 33 | 1 | STAT3, Klf4, Esrrb, Egr1, Sox2, |
| Oct4, Fox, Myc, Sp1, Tead1, | ||||
| CREB/ATF | ||||
| Zfx | 10338 | 20 | 1 | STAT3, Myc, Esrrb |
Columns show the name of the ChIP-ed TF; the number of ChIP-seq peaks; the number of significant motifs (m) found by DREME (E < 0.05); the rank (r) of the ChIP-ed TF's motif; and cofactor motifs found. Cofactor TFs are listed in the order of DREME significance and in bold font if they are 1 of the 12 pluripotency TFs. Only the cofactor TF family name is given when several family members match the DREME motif (e.g. ‘Myc’).
aSee text for discussion of E2f1 and Nanog motifs.
In addition to finding the primary motif, DREME discovers binding motifs for up to 12 cofactors in each of the mESC ChIP-seq datasets (Table 1, column 4). Many of the motifs found by DREME in each mES cell dataset correspond to 1 of 12 key pluripotency TFs (shown in bold font in Table 1), and these predictions correspond well to the actual overlap of ChIP-seq peak regions reported by 0 (). For example, in addition to finding the motif for the ChIP-ed factor in the Tcfcp2l1 dataset, DREME also discovers the motifs for pluripotency factors STAT3, Klf4, Esrrb, Sox2, Oct4 and c-Myc/n-Myc (Myc-family motifs are essentially indistinguishable). In the CTCF dataset, DREME identifies the CTCF motif as well an unknown motif that was recently found to frequently occur upstream of the CTCF motif (consensus GCTGCAGT) (Boyle et al., 2010) in human cells.
Many of the other motifs found by DREME can be assigned to TFs that are known to be upregulated in mES cells, but whose roles in the maintenance of pluripotency are unknown (shown in normal font in Table 1, column 4). Several of these cofactor TF motifs are found in more than one mESC ChIP-seq dataset. These cofactor motifs include Sp1, CREB/ATF and Zic3, which was recently suggested as being required for the maintenance of pluripotency (Lim et al., 2010). On the other hand, we are currently unable to assign many of the motifs discovered by DREME to any known motif. Although some of these unassigned motifs may be artifacts, others may simply correspond to TFs whose motifs are not contained in the two motif databases we searched.
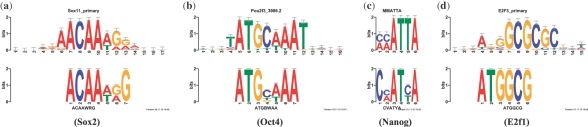
Several of the motifs discovered by DREME in the mES cell datasets differ substantially from those reported by Chen et al. in their original data analysis (0, ). Of particular note, DREME discovers the monomer motifs for Oct4 and Sox2 in their respective ChIP-seq datasets, rather than the Oct-Sox heterodimer motif. The in vivo binding motifs discovered by DREME for Oct4 and Sox2 closely match the in vitro motifs derived using protein binding microarray (PBM) technology (Berger and Bulyk, 2009), suggesting that the PBM motifs accurately capture DNA binding of these factors in vivo (Fig. 1a and b). This illustrates a particular advantage of focusing on short, core motifs and we discuss this issue in detail below. We also explore the hypothesis (suggested by Chen et al.) that Oct4 and Sox2 bind DNA exclusively as a homodimer. (The fact that the most significant motif found by DREME in each of the two ChIP-seq datasets is the primary factor's suggests this hypothesis may be false.)
Fig. 1.
Comparison of DREME mESC TF ChIP-seq motifs with in vitro motifs. Each panel shows the logo of the in vivo binding motif discovered by DREME in the designated TF ChIP-seq dataset (lower logo) aligned with the logo of the best available in vitro motif (upper logo). Since no in vitro motifs are available for Sox2, Oct4 and E2f1, UniProbe motifs for closely related TF family members Sox11, Pou2f3 and E2f3 are used. The in vitro motif for Nanog is taken from Jauch et al. (2008).
DREME finds a motif similar to the known E2f1 motif in the E2f1 ChIP-seq dataset. This is in contrast to Chen et al., who report not finding a motif for E2f1 using either the WEEDER (Pavesi et al., 2004) or nestedMICA (Down and Hubbard, 2005) motif discovery algorithms. A previous study of E2f1 ChIP-chip data also failed to find a convincing E2f1 motif, concluding that E2f1 is mainly recruited to promoters via a mechanism distinct from recognition and binding to the consensus site (Bieda et al., 2006). Nonetheless, in the E2f1 dataset, the second most significant motif discovered by DREME is the word ATGGCG, which occurs in 1274 of the 20 699 ChIP-seq peak regions (E = 10 − 98) and resembles the in vitro motif for E2f-family member E2f3 (Fig. 1d). The strong enrichment of this word in the E2f1 ChIP-seq data and its resemblance to the in vitro E2f3 motif suggests that it may represent at least a subset of the in vivo binding sites of E2f1 in mES cells. The DREME motif also is similar to the motif for YY1 (JASPAR MA0095.1, consensus ATGG), suggesting the hypothesis that the sites discovered by DREME might bind both (but not simultaneously) E2f1 or YY1 in mES cells.
3.1.2 Discovering motifs in mouse erythrocyte ChIP-seq data
When we apply DREME individually to three ChIP-seq datasets from mouse erythroid cells [TFs Gata1 (Cheng et al., 2009), Klf1 (Tallack et al., 2010) and Tal1 (Kassouf et al., 2010)], the top motif is the ChIP-ed motif in the first two cases, but not in the last case. With the Gata1 dataset, DREME finds the Gata1 motif (E = 10 − 892) followed by the motif of SPI1 (E = 10 − 79, a key myeloid developmental regulator (Zakrzewska et al., 2010). It also finds the Klf1 motif (E = 10 − 37) and and E-box motif (E = 10 − 19) that might be due to Tal1 binding. DREME also discovers the motif for Runx1 (E = 10 − 2), which may also be involved in erythropoeisis (Yokomizo et al., 2008). In all, DREME finds 21 significant motifs in the Gata1 dataset.
DREME finds a total of six significant motifs in the Klf1 dataset. It identifies the Klf1 motif as the most significant (E = 10 − 49), followed by Gata1 (E = 10 − 42). Interestingly, the third most significant motif is the ‘secondary’ Klf motif discovered from in vitro binding data (Berger and Bulyk, 2009) (E = 10 − 11).
In the Tal1 ChIP-seq dataset, the most significant motif found by DREME (among 10 motifs found) is Gata1 (E = 10 − 262), whereas the actual Tal1 binding motif (E-box) is the third most significantly found motif (E = 10 − 26). The lower enrichment of the Tal1 motif compared with Gata1 may be due to the fact that Tal1 often binds DNA as a complex with Gata, Lmo2, Ldb1 and E47 (Wadman et al., 1997). In this complex, Tal1 has reduced contact with the DNA, binding to a motif consisting of one-half of an E-box followed ~10 bp later by a Gata1 site (Kassouf et al., 2010). This ‘semi-direct’ binding would reduce the prevalence of the full Tal1 motif in the Tal1 ChIP-seq dataset. DREME also finds motifs for Klf1 (E = 10 − 23) and SPI1 (E = 10 − 16) in this dataset.
3.1.3 Human lymphoblastoid cell line motifs
We apply DREME to two vitamin D receptor (VDR) ChIP-seq datasets from lymphoblastoid cells before and after stimulation with calcitriol (Ramagopalan et al., 2010). DREME finds 19 significant motifs in the unstimulated cell data, and only 11 in the stimulated cell data (E < 0.05). VDR binds DNA as a heterodimer with retinoid X receptor (RXR) and the left and right halves of the binding motif both have the consensus sequence (A/G)(A/G)G(T/G)TCA. DREME finds a single motif matching the consensus in the ChIP-seq data from the calcitriol-stimulated cells (fourth most significant motif, E = 10 − 11). The top-scoring motif in both datasets is an ETS-factor motif, possibly due to binding of Ets-1, which has been shown to cause VDR to function as a constitutive activator (Tolón et al., 2000). The second most significant motif in the stimulated cell data is an E-box, which has been proposed as being involved in negative regulation by VDR (Kato et al., 2007). The third most significant motif found in stimulated cell data strongly resembles STAT1, which is known to interact with VDR (Vidal et al., 2002). The fifth motif found is another Ets motif, and the sixth motif is a Klf motif. The seventh and eighth motifs significantly resemble Runx2 and Ap1/Fos/Jundm2 (TOMTOM E < 0.05), respectively, both of which are known to interact with VDR (Marcellini et al., 2010). In all, six of the eight most significant motifs found in the stimulated cell data appear to represent the ChIP-ed factor (VDR) or likely cofactors.
3.2 Discriminative motif discovery
3.2.1 Determining if binding is solely via a heterodimer
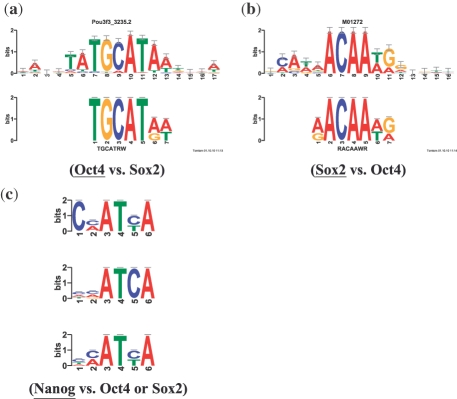
It has been suggested that Sox2 and Oct4 bind their targets exclusively as a heterodimer in mES cells (0, ). If this is so, we should not observe enrichment of either motif when we use the ChIP-seq datasets for these two TFs as the positive and negative inputs to DREME. We performed this experiment (using default values for all DREME parameters), and found that the Oct4 dataset is enriched in Oct4 binding sites compared with the Sox2 dataset, and vice versa.
Using Oct4 as the positive and Sox2 as the negative input, DREME discovers the Oct4 motif (Fig. 2a, E = 10 − 29), but not the Sox2 motif (E > 0.05). DREME reports that this motif occurs in 18% of the Oct4 ChIP-seq peaks, but in only 9% of the Sox2 peaks. The Oct4 motif is thus significantly enriched in Oct4 ChiP-seq peaks compared with Sox2 peaks. Reversing the roles of the two ChIP-seq datasets, DREME finds the Sox2 motif (Fig. 2a, E = 10 − 158), but not the Oct4 motif. The Sox2 motif found occurs in 55% of Sox2 peaks, but in only 28% of the Oct4 peaks. These two results strongly suggest that both Oct4 and Sox2 often bind DNA in mES cells alone or with other partners, rather than exclusively with each other as a heterodimer.
Fig. 2.
Discriminative motif discovery in mESC ChIP-seq datasets. Panels (a) and (b) show the logo of the binding motif discovered by DREME in the two designated TF ChIP-seq datasets (lower logo) aligned with the logo of a known motif for the ChIP-ed TF (upper logo). (a) Upper logo is known Oct4 motif (Pou-family member Pou3f3, UniProbe Pou3f3_3235.2). (b) Upper logo is known Sox2 motif (TRANSFAC M01272). (c) Shows the most significant motif found by DREME in the Nanog dataset using (top to bottom) the shuffled Nanog dataset, the Oct4 dataset or the Sox2 dataset as the negative set.
3.2.2 Finding context-dependent motifs
It was recently reported that Oct4 may preferentially bind to a different motif with consensus ATGCGCAT when not binding near Sox2 (Mason et al., 2010). They used Oct4 and Sox2 ChIP-seq data from Marson et al. (2008) to create two Oct4 sequence datasets. The first dataset, Oct4woSox2, contains only Oct4 regions where there is no Sox2 peak within 5000 bp. The second dataset, Oct4wSox2, contains Oct4 regions that are within 50 bp of a Sox2 peak. Using the first and second datasets as positive and negative inputs, respectively, to their discriminative motif discovery algorithm CMF, they discovered the ATGCGCAT ‘Oct-only’ motif. They also noted that the ChIP-seq peaks containing this motif are enriched for a different set of transcription factors (nMyc, cMyc, E2f1 and Zfx1) than the Oct4 peaks containing a nearby Sox2 binding site (Nanog, Sox2 and Smad1), suggesting that the preferred in vivo binding motif of Oct4 is context dependent.
We applied DREME in an analogous way and found that it also discovers the ‘Oct-only’ motif when applied discriminatively to the Oct4woSox2 and Oct4wSox2 datasets. On these datasets, the ‘Oct-only’ motif is the most significant one found by DREME (E = 1e-47), and DREME runs about 5.4 times faster than CMF (74 versus 378 s). Interestingly, the ‘Oct-only’ motif is also among the 17 motifs found by DREME in the Chen et al. Oct4 ChIP-seq dataset, raising the total number of identifiable motifs in that dataset to five (Table 1, column 5).
3.2.3 Refining the search for the ChIP-ed factor's motif
Sometimes the most significant motif found by DREME is not that of the ChIP-ed TF, and it may not even be clear whether its motif has been found at all. For example, the most significant motifs found by DREME in the Nanog dataset are Oct4 and Sox2. This suggests that Nanog might sometimes be binding indirectly via one or both of these TFs. We wondered if a discriminative approach—looking for motifs enriched in the Nanog dataset relative to the Oct4 or Sox2 dataset—might increase our confidence in the motif we proposed as the Nanog DNA-binding motif (Fig. 1c). We, therefore, ran DREME using the Nanog mESC ChIP-seq dataset as the positive input, and each of the other two TFs' datasets (individually) as the negative input. The most significant motif in each of these two cases (Nanog versus Oct4, Nanog versuss Sox2) is indeed highly similar to the DREME motif we suggest above as the putative Nanog motif (Fig. 2c), and both are more similar to the previously published in vitro motif for Nanog [(C/A)(C/A)ATTA] than the motif found by DREME in non-discriminative mode. When we use TOMTOM to compare these two new putative Nanog motifs to our two reference motif databases plus the TRANSFAC motif database (Matys et al., 2006), each is most similar to a motif for Pbx1 (data not shown). So, as noted above, determining whether this motif is indeed the binding motif for Nanog, Pbx1 or a combination thereof, may require ChIP-seq data for Pbx1 in mES cells. However, the fact that this is the most highly enriched motif in the Nanog dataset relative to both the Oct4 and Sox2 datasets suggests that the motif is not merely due to the presence of Pbx1 binding sites in the Nanog dataset.
3.3 Comparison with other motif discovery algorithms
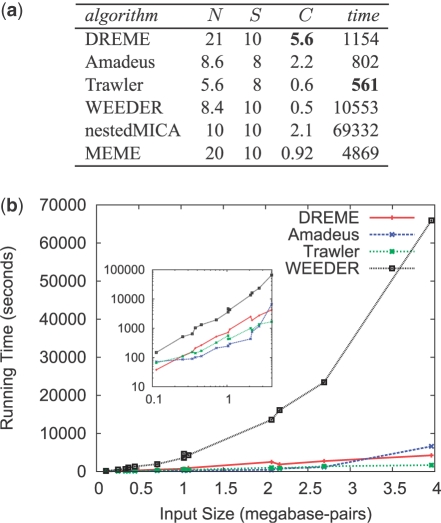
We compare the speed and accuracy of several popular motif discovery algorithms with that of DREME using the 13 mESC ChIP-seq datasets. On this particular task, DREME finds substantially more cofactor motifs than the other five algorithms (Fig. 3a, column 4). Although generally somewhat slower than Amadeus and Trawler, DREME discovers more than twice as many identifiable cofactor motifs on average compared with Amadeus, and almost 10 times as many as Trawler. The later two algorithms only find 8 of the 13 ChIP-ed motifs, whereas DREME, WEEDER, nestedMICA and MEME find 10 (Fig. 3a, column 3).
Fig. 3.
Comparison of motif discovery algorithms. (a) The table shows the average number of motifs discovered (N), number of datasets in which the ChIP-ed motif was found (S), the average number of identifiable co-factor motifs found (C), and the average running time in seconds of the algorithm on the mESC ChIP-seq datasets. Bold font indicates best performance. Note: Times for nestedMICA and MEME are for the reduced size datasets (0.5 megabase-pairs). (b) The plot shows the running times for DREME, Amadeus, Trawler and WEEDER on the full-size mESC ChIP-seq datasets. Inset plot is the same data plotted with log scales on both axes.
WEEDER is much slower than DREME, and its running time scales non-linearly with dataset size (Fig. 3b), as do MEME and nestedMICA (data not shown). The full-size datasets contain on average about 10 000 sequences of length 100 bp, and DREME completes its analysis in 19 min on average, whereas WEEDER requires almost 10 times as long. Both MEME and nestedMICA are really too slow to handle ChIP-seq datasets containing thousands of sequences, failing to finish running after many days. For this reason, results in Figure 3a for nestedMICA and MEME are for the reduced-size datasets containing 500 000 bp.
4 DISCUSSION
Focusing on short, core motifs in TF ChIP-seq datasets appears to be a very profitable approach to understanding patterns of DNA binding by TFs. Additional insight can also be gained by searching for motifs that are relatively enriched in one ChIP-seq dataset compared with another. The ab initio discovered motifs can often be associated with the probable TF that binds them by comparison to existing compendia of TF motifs, using expression in the ChIP-ed tissue and prior knowledge as a filter. The large numbers of motifs discovered by DREME that can be reliably assigned to TFs suggests that unassigned motifs may represent binding by other TFs whose motifs are not yet known. Of course, because it is only designed to find short, core motifs, DREME is intended only to complement existing motif finders (such as those tested here). A complete analysis of a TF ChIP-seq dataset should also include a search for longer, more complex motifs.
Our motif discovery algorithm incorporates ideas from several existing algorithms (Barash et al., 2001; Sharov and Ko, 2009; Sinha and Tompa, 2003). Barash et al. (2001) uses the Fisher's Exact Test to measure the significance of enrichment of motifs in two sets of sequences. Their motifs are not regular expressions but ‘δ-balls’: the set of words that are within a set Hamming distance from a given word. This motif definition treats variations from the consensus word the same, regardless of position within the motif. Real TF motifs are less tolerant of variation in certain positions (Matys et al., 2006), and this is better captured by regular expressions, which explicitly list all the allowed variations (if any) at each motif position. Several algorithms including YMF (Sinha and Tompa, 2003) use regular expressions at some stage in the search for motifs, but differ from our algorithm in other respects. For example, YMF only allows a subset of the IUPAC wildcards and scores motifs using a different statistical test (Z-score). Counting the number of sequences (not occurrences) is equivalent to the ‘Zero or One Occurrence Per Sequence’ (ZOOPS) model used by numerous motif discovery algorithms, and finding multiple motifs by erasing is reminiscent of MEME (Bailey and Elkan, 1995). CisFinder (Sharov and Ko, 2009) also uses the idea of counting words and computing relative enrichment in two sets of sequences, is extremely fast, and can find many cofactor motifs in ChIP-seq datasets. Unlike DREME, however, it requires a motif clustering step to (partially) remove redundant motifs, and cannot be restricted to finding short, core motifs. Also, in contrast with DREME, CisFinder reports a p-value for each motif that is not a measure of the significance of the motif, but only of a single word (without wildcards) matching the motif.
ACKNOWLEDGEMENTS
We wish to acknowledge Peter Clote for allowing to use his dinucleotide shuffle algorithm in the MEME suite.
Funding: NIH R0-1 grant (RR021692-05).
Conflict of Interest: none declared.
REFERENCES
- Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- Barash Y, et al. A simple hyper-geometric approach for discovering putative transcription factor binding sites. In: Gascuel O, Moret BME, editors. Algorithms in Bioinformatics: Proceedings of the First International Workshop. 2001. Vol. 2149 in Lecture Notes in Computer Science, Springer, pp. 278–293. [Google Scholar]
- Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the dna-binding specificities of transcription factors. Nat. Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieda M, et al. Unbiased location analysis of E2F1-binding sites suggests a widespread role for e2f1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2010;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cheng Y, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, Hubbard TJP. NestedMICA: sensitive inference of over-represented motifs in nucleic acid sequence. Nucleic Acids Res. 2005;33:1445–1453. doi: 10.1093/nar/gki282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwiller L, et al. Trawler: de novo regulatory motif discovery pipeline for chromatin immunoprecipitation. Nat. Methods. 2007;4:563–565. doi: 10.1038/nmeth1061. [DOI] [PubMed] [Google Scholar]
- Gupta S, et al. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauch R, et al. Crystal structure and DNA binding of the homeodomain of the stem cell transcription factor Nanog. J. Mol. Biol. 2008;376:758–770. doi: 10.1016/j.jmb.2007.11.091. [DOI] [PubMed] [Google Scholar]
- Kapushesky M, et al. Gene expression atlas at the European Bioinformatics Institute. Nucleic Acids Res. 2010;38:D690–D698. doi: 10.1093/nar/gkp936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassouf MT, et al. Genome-wide identification of TAL1's functional targets: Insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010;8:1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, et al. Ligand-induced transrepressive function of VDR requires a chromatin remodeling complex, WINAC. J. Steroid Biochem. Mol. Biol. 2007;103:372–380. doi: 10.1016/j.jsbmb.2006.12.038. [DOI] [PubMed] [Google Scholar]
- Lim LS, et al. The pluripotency regulator Zic3 is a direct activator of the Nanog promoter in embryonic stem cells. Stem Cells. 2010;28:1961–1969. doi: 10.1002/stem.527. [DOI] [PubMed] [Google Scholar]
- Linhart C, et al. Transcription factor and microRNA motif discovery: the Amadeus platform and a compendium of metazoan target sets. Genome Res. 2008;18:1180–1189. doi: 10.1101/gr.076117.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcellini S, et al. Evolution of the interaction between Runx2 and VDR, two transcription factors involved in osteoblastogenesis. BMC Evol. Biol. 2010;10:78. doi: 10.1186/1471-2148-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MJ, et al. Identification of context-dependent motifs by contrasting ChIP binding data. Bioinformatics. 2010;26:2826–2832. doi: 10.1093/bioinformatics/btq546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, et al. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan SV, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, et al. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Ko MSH. Exhaustive search for over-represented dna sequence motifs with CisFinder. DNA Res. 2009;16:261–273. doi: 10.1093/dnares/dsp014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Tompa M. YMF: a program for discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res. 2003;31:3586–3588. doi: 10.1093/nar/gkg618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- Tallack MR, et al. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 2010;20:1052–1063. doi: 10.1101/gr.106575.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolón RM, et al. Association with Ets-1 causes ligand- and AF2-independent activation of nuclear receptors. Mol. Cell Biol. 2000;20:8793–8802. doi: 10.1128/mcb.20.23.8793-8802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, et al. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Vidal M, et al. Stat1-vitamin D receptor interactions antagonize 1,25-dihydroxyvitamin D transcriptional activity and enhance stat1-mediated transcription. Mol. Cell Biol. 2002;22:2777–2787. doi: 10.1128/MCB.22.8.2777-2787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman IA, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbanks EG, Facciotti MT. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS One. 2010;5:e11471. doi: 10.1371/journal.pone.0011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T, et al. Runx1 is involved in primitive erythropoiesis in the mouse. Blood. 2008;111:4075–4080. doi: 10.1182/blood-2007-05-091637. [DOI] [PubMed] [Google Scholar]
- Zakrzewska A, et al. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood. 2010;116:e1–e11. doi: 10.1182/blood-2010-01-262873. [DOI] [PubMed] [Google Scholar]