Summary
Considerable details about microRNA (miRNA) biogenesis and regulation have been uncovered, however, little is known about the fate of the miRNA subsequent to target regulation. To gain insight into this process, we carried out kinetic analysis of a miRNA’s turnover following termination of its biogenesis, and during regulation of a target that is not subject to Ago2-mediated catalytic cleavage. By quantitating the number of molecules of the miRNA and its target in steady-state, and in the course of its decay, we found that each miRNA molecule was able to regulate at least 2 target transcripts, providing in vivo evidence that the miRNA is not irreversibly sequestered with its target, and that the non-slicing pathway of miRNA regulation is multiple-turnover. Using deep-sequencing, we further show that miRNA recycling is limited by target regulation, which promotes post-transcriptional modifications to the 3′ end of the miRNA, and accelerates the miRNA’s rate of decay. These studies provide new insight into the efficiency of miRNA regulation, which help to explain how a miRNA can regulate a vast number of transcripts, and identify one of the mechanisms that impart specificity to miRNA decay in mammalian cells.
Results
While considerable details about miRNA biogenesis have been uncovered, little is known about miRNA turnover and decay[1,2,3,4]. One outstanding question is whether a miRNA is recycled following regulation of its target. Early studies revealed that when a miRNA binds to a target with perfect complementarity, the transcript is cleaved between the nucleotides pairing with position 10 and 11 of the miRNA[5], and this process is multiple-turnover, enabling a single miRNA (or siRNA) to regulate multiple transcripts[6,7]. When a target site does not pair with the miRNA at nucleotides 10, 11, creating a bulge, catalytic cleavage is prevented, but translation of the transcript is still blocked[8]. The miRISC complex, it is believed, then transports the bound transcript to the p-body[9,10], and other cellular compartments, where the mRNA is deadenylated and rapidly decays[8]. The fate of the miRNA subsequent to this process still remains poorly understood.
Stability helps to determine the relative abundance of a miRNA compared to its host mRNA
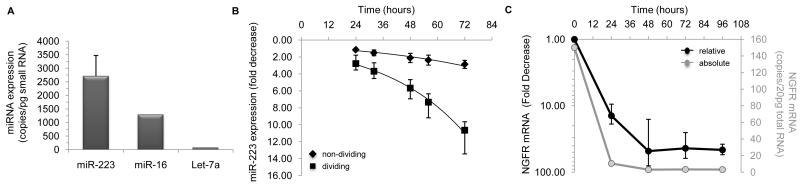
To establish a system to study miRNA turnover in vivo, we made use of a lentiviral vector (LV) encoding hsa-miR-223 under the control of a tetracycline-dependent promoter (Figure S1A). This vector expresses a pri-miR-223 transcript within an intron, which is upstream of a reporter encoding a truncated nerve growth factor receptor (NGFR). The vector constitutively expresses a transactivator (tTA-2S) that drives transcription of miR-223 and NGFR[11]. In the presence of Doxycycline (Dox), tTA-2S is blocked, and transcription is terminated. Human 293T cells, which do not express miR-223, were transduced with the vector, and the cells were FACS sorted to isolate NGFR-positive cells, and generate a stable population of conditional miR-223 expressing cells (c223 cells) with 1 vector copy/cell. The absolute steady-state level of miR-223 expression in the cells was determined by quantitative real-time PCR (qPCR) using a standard curve to extrapolate the values, and found to be an average of 2,700±750copies/pg of small RNA (approximately 1 cell)(Figure 1A). This is similar to miR-223 levels in U937 monocytic cells[12], where miR-223 is naturally expressed. Conditional gene expression was validated by monitoring the decrease in NGFR expression by FACS at 24, 48, 72, and 96 hours after the addition of Dox, and found to be uniform within the population of c223 cells (Figure S1B).
Figure 1. Monitoring the kinetics of miRNA decay in mammalian cells.
(A) Absolute level of miR-223 expression in c223 cells (n=40). To derive the absolute levels of expression, a standard curve was generated using synthetic miR-223 RNA oligonucleotides corresponding to the 22 and 23 nucleotide forms of miR-223, and CT values were extrapolated from the curve. Note that 1 pg of small RNA is approximately the amount of small RNA in a single cell. The level of miR-16 and Let-7a expression is derived from their expression relative to miR-223, as determined by qPCR. (B) Analysis of miR-223 decay in dividing (square) and non-dividing (diamond) c223 cells at the indicated time point after addition of Dox. Each value is the mean±SD (n=4). (C) The kinetics of NGFR RNA decay was monitored by treating c223 cells with Dox, and measuring the decrease in NGFR transcripts by qPCR (black, left axis). A standard curve for NGFR was generated, and the absolute levels of NGFR mRNA were extrapolated from the curve (gray, right axis). Values are the mean±SD (n=3). For additional related data see Figure S1.
To monitor miR-223 decay we extracted small RNA from the Dox-treated cells at the same times, and measured miR-223 expression by qPCR. For each time point we also had a matching sample in which Dox was not added. Under steady-state conditions (i.e. no Dox), miR-223 levels did not change over time, whereas addition of Dox led to a slow decrease in miR-223 expression. At 24 hours miR-223 levels were almost 2.8-fold lower compared to untreated cells, and at 48 hours miR-223 levels were on average a further 2.8-fold lower (5.67±1.26-fold compared to steady-state), and this trend continued over the 96 hours of study (Figure 1B). By extrapolating from the exponential curve, we determined the decay constant, λ, of miR-223 to be 0.028±0.003 hr−1, corresponding to a half-life of 25 hours (Table 1).
Table 1.
miR-223 Decay Rates in Target-Expressing Cells
| Target | Target Concentration | Cell Status | Decay Constant (hr−1) | Half-Life (hr) | Significance (p value) |
|---|---|---|---|---|---|
| − | N/A | dividing | 0.028 ± 0.003 | 25 | − |
| 19aPT | N/A | dividing | 0.024 ± 0.003 | 29 | 0.4706 |
| 223PT | moderate | dividing | 0.030 ± 0.005 | 23 | 0.2303 |
| 223PT | high | dividing | 0.047 ± 0.004 | 15 | 0.0001 |
| 223BT1 | high | dividing | 0.037 ± 0.003 | 19 | 0.0040 |
| 223-5’PT | high | dividing | 0.034 ± 0.002 | 20 | 0.0074 |
| − | N/A | nondividing | 0.015 ± 0.002 | 46 | − |
| 223PT | high | nondividing | 0.027 ± 0.001 | 26 | 0.0004 |
| 223BT1 | high | nondividing | 0.019 ± 0.001 | 37 | 0.0257 |
The decay constant (λ) was extrapolated from the exponential decay curves of each biological replicate (n = 4–5 per condition), and the mean±SD is shown. The half-life in hours (hr) was calculated by the formula ln2/λ. The significance was determined by performing an unpaired t test between the miR-223 decay constant in c223 cells and each of the other conditions.
To understand whether the rate of miR-223 decay was dependent on cell division we treated the c223 cells with mitomycin C (MMC) to arrest cell cycle, and repeated the experiment described above. In steady-state, the absolute expression of miR-223 did not significantly differ between untreated and MMC-treated c223 cells. When we treated the non-dividing c223 cells with Dox, and monitored miR-223 expression, the miRNA’s abundance decreased more slowly than in dividing cells (Figure 1B), and the miRNA’s half-life was 46 hours (Table 1). However, given that there was no appreciable difference in miR-223 levels in steady-state between dividing and non-dividing cells, and each c223 cell divides in two every 20 - 24 hours, the ~2-fold difference in half-life is likely due to dilution of the miRNA during cell division, and thus, at least for miR-223, the cell cycle itself does not change the miRNA’s decay rate.
In parallel, we measured the changes in NGFR mRNA, which is derived from the same precursor transcript as miR-223. Based on a standard curve, we determined there were on average 105 NGFR transcripts per 20pg total RNA (approximately 1 cell). Following addition of Dox, the concentration of NGFR mRNA decreased more than 10-fold within 24 hours, and reached its lowest level by 48 hours (Figure 1C). The shorter half-life of NGFR mRNA compared to miR-223 helps to explain how the mature miR-223 RNA is >20 times more abundant in c223 cells than the NGFR mRNA despite their co-expression. These results suggest that differences in stability, in addition to differences in processing efficiency[13], between an intron-embedded miRNA and its host mRNA, which are often transcribed from the same promoters[14], enable the miRNA to accumulate to a higher concentration within a cell. This is significant for the miRNA’s function, since a high concentration is a key determinant of effective target suppression[12,15].
miRNA regulation of a bulge target is multiple-turnover
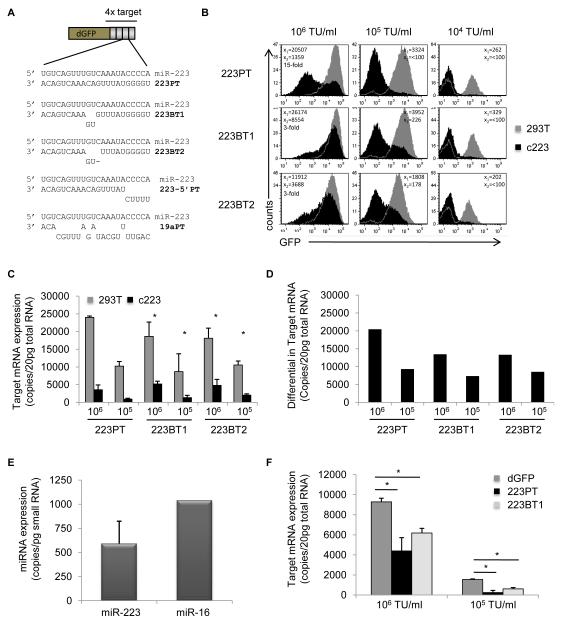
The long half-life of miR-223 suggests that target regulation will be prolonged after transcription and biogenesis are terminated, however this will also depend on the turnover capacity of the miRNA. To address this issue we sought to introduce a well defined target of the miRNA, and evaluate its impact on miRNA turnover. To do so, we generated lentiviral vectors encoding perfect (PT) or bulge (BT) targets for miR-223 downstream of destabilized GFP (GFP), and expressed from the strong CMV/β-actin promoter (CAG). Two different BT vectors were designed to prevent Ago2-mediated slicing. In one, the target sites mismatch with miR-223 at nucleotides 10 and 11 (223BT1), and in the other, the target sites mismatch at nucleotides 10 - 12 (223BT2) (Figure 2A). c223 and 293T cells were transduced with 104, 105, or 106 TU/ml of the 223PT, 223BT1, or 223BT2 vectors and passaged for at least 2 weeks to establish stable expression. Deep-sequencing confirmed that the perfect target site was subject to slicing, and the bulge target was not (Figure S2A and S2B).
Figure 2. Quantification of miR-223-mediated protein and RNA suppression in c223 cells.
(A) Schematic of the sequences of the targets used, and their respective pairing to mature miR-223. Each vector contained 4 copies of the indicated target site. (B) Comparison of miR-223 target expression between 293T (gray) and c223 (black) cells. GFP expression was analyzed by FACS. Representative histograms are shown from 3 independent transductions. The mean florescence intensity of GFP in 293T and c223 cells is indicated by x1 and x2, respectively. (C) Quantitation of target mRNA expression in 293T and c223 cells. GFP mRNA was quantitated by qPCR from the indicated cells, and the absolute value was extrapolated from a standard curve generated using in vitro transcribed GFP RNA. Values are the mean±SD (n=3). (D) Absolute differential in target mRNA transcripts between 293T and c223 cells. The average concentration of GFP mRNA in c223 cells was subtracted from the average concentration of GFP mRNA in 293T cells from (C). (E) miR-223 expression in human THP-1 monocytic cells. miR-223 was measured in THP-1 cells by qPCR, and compared against a standard curve. Values are the mean±SD (n=6). miR-16 expression is derived from its value relative to miR-223. (F) Quantitation of target mRNA expression in THP-1 cells stably expressing the indicated target. Values are the mean±SD (n=3). *P< 0.005 unpaired t-test. For additional related data see Figure S2.
To determine the level of suppression mediated by miR-223, we compared GFP expression between the cell lines by FACS. At 104 TU/ml, all three targets were fully suppressed by the miRNA (Figure 2B). As we increased vector concentration, and thus target concentration, the extent of target suppression decreased for all three targets. The effect was particularly acute for the bulge targets where suppression went from 10-fold to 3-fold at the highest vector concentration, suggesting that the miRNA was starting to be saturated.
To investigate further, we quantitated the number of target transcripts being regulated by miR-223 by comparing the absolute level of GFP mRNA between the stably transduced 293T and c223 cells. qPCR was used to measure gene expression, and a standard curve for GFP was used to extrapolate the absolute number of target transcripts. In 293T cells transduced with 105 TU/ml, we detected between 10,000 – 13,000 target transcripts/20pg RNA (approximately 1 cell). In c223 cells, transduced with the same vector concentration, there were an average of 1,300 223PT transcripts/cell, and 1,800 and 2,600 223BT1 and 223BT2 transcripts/cell, respectively (Figure 2C). This was a deficit of approximately 8,700 223PT transcripts and ~7,000 223BT1 and 223BT2 transcripts (Figure 2D).
At the highest vector concentration, we measured between 15,000 – 25,000 target transcripts per 293T cell being produced from the CAG-driven vectors (Figure 2C). In c223/223PT cells, we detected an average of 4,100 target transcripts/cell, which is a deficit of more than 15,000 transcripts compared to 293T/223PT cells. In c223 cells expressing the 223BT1 and 223BT2 bulge targets we detected an average of 5,700 transcripts/cell, which was a reduction of ~10,000 transcripts/cell compared to their expression in 293T cells (Figure 2D). This deficit is triple the number of miR-223 molecules being expressed in the cells (see above), and thus indicates that each miR-223 molecule is regulating multiple transcripts bearing the bulge target.
To understand if miR-223 would behave similarly in its natural context, we quantitated the level of miR-223 expression and target regulation in THP-1 human monocytic cells. qPCR analysis determined that THP-1 cells naturally express ~590±232 copies/cell of mature miR-223 (Figure 2E), and deep-sequencing revealed that the abundance and relative frequency of miR-223 isoforms was similar in THP-1 cells to c223 cells (Figure S2C). Next, we transduced the THP-1 cells with 105 or 106 TU/ml of the 223PT or 223BT2 vectors, or a control vector that expressed GFP from the CAG promoter, but does not contain miR-223 target sites in its 3′ UTR (dGFP). After more than 1 week in culture, we measured GFP expression, and found that both 223PT and 223BT2 were well suppressed by the endogenous miR-223, as indicated by FACS (Figure S2D). In THP-1 cells transduced with the higher concentration of the control vector, we detected 9,300 copies of dGFP mRNA per 20pg of total RNA. Whereas, there were on average 4,500 copies of the perfect target, and 6,100 copies of the bulge target (Figure 2F). Thus, up to 3000 bulge transcripts were subject to miR-223 regulation in steady-state, which is more than the number of miR-223 molecules in steady-state. This provides additional evidence that a single miRNA is capable of regulating multiple transcripts.
Target regulation accelerates the miRNA’s rate of decay
Our studies in steady-state indicate that the miRNA is recycled, but because the miRNA is continually produced in these conditions, we cannot distinguish between ‘naïve’ and ‘target-experienced’ miRNA. To determine the fate of the miRNA subsequent to target regulation, we treated c223 cells stably expressing the perfect or bulge target with Dox to terminate miR-223 biogenesis, and followed miRNA and target expression over time. As an additional control, we utilized c223 cells stably expressing target sites for miR-19a (c223/19aPT), a miRNA that is highly expressed in c223 cells. The 19aPT has little complementarity to miR-223 (Figure 2A), and is regulated by endogenous miR-19a in c223 cells.
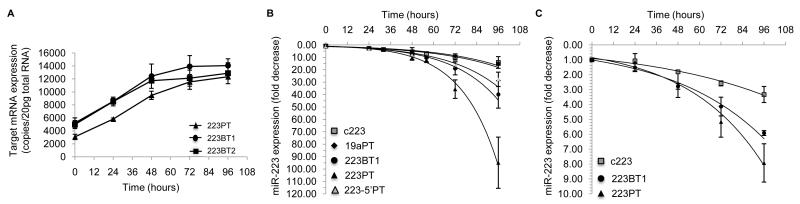
Twenty-four hours after adding Dox we observed a small (<2-fold) increase in GFP protein expression in the c223 cells expressing the 223PT, 223BT1, or 223BT2 target (Figure S3A), but no change in the miR-19a-regulated reporter (Figure S3B). GFP continued to slowly increase in the c223/223PT, c223/223BT1, and c223/223BT2 cells, and did not reach maximal expression until 96 hours after adding Dox. At the RNA level, we also observed a relatively slow rate of increase in target concentration. At 24 hours post-Dox, the number of GFP transcripts increased from an average of 3100 to 5800 copies/cell in c223/223PT cells, and from 5000 to 8500 copies/cell in both c223/223BT1 and c223/223BT2 cells (Figure 3A). The maximum number of target transcripts, which was 12,000 – 15,000 copies/cell, was not reached until between 48 and 72 hours after miR-223 biogenesis was terminated. This increase of 7,000 – 9,000 transcripts in the c223/223BT1 and c223/223BT2 cells confirms our findings in steady-state, which indicated that the ratio of regulated bulge target transcripts to miR-223 molecules was greater than 1.
Figure 3. Evaluating the dynamics of target regulation and miR-223 decay.
(A) Kinetics of target mRNA de-repression during miR-223 decay. The quantity of GFP mRNA was determined by qPCR at each time point after Dox. Values are the mean±SD (n=3). (B) Analysis of miR-223 decay in target expressing c223 cells. Cells stably expressing the indicated target were treated with Dox, and miR-223 expression was monitored at the indicated times. Each value is the mean±SD (n=≥4). (C) Analysis of miR-223 decay in non-dividing, target expressing c223 cells. One day after treatment with mitomycin C, cells were treated with Dox, and miR-223 expression was monitored. Each value is the mean±SD (n=3). For additional related data see Figure S3.
Next, we monitored the kinetics of miR-223’s decay. In c223/223PT cells expressing a moderate level of target (~2600 transcripts/cell, data not shown), the decay of miR-223 was similar to the rate observed in the c223 cells without the exogenous target, with a half-life of 23 hours (Table 1). This was similar to the level in control cells that did not overexpress a synthetic target, and in c223/19aPT cells, which overexpressed an irrelevant target. Strikingly, in c223 cells expressing a high concentration of the perfect or bulge target we observed a substantial increase in the rate of miR-223 decay (p<0.01, Table 1). In the case of the perfect target, at 48 hours post-Dox miR-223 levels declined an average of 10-fold, and by 96 hours the levels were >80-fold lower than steady-state levels (Figure 3B). In cells overexpressing the bulge target miR-223 levels declined by 40- fold by 96 hours. In these cells the miR-223 decay constant was 0.037 hr−1 or a half-life of approximately 19 hours, which is faster than in c223 cells that were not overexpressing target (p<0.01, Table 1). A similar reduction in the miRNA’s half-life was observed in non-dividing cells overexpressing the miRNA’s target. In MMC-treated c223/223PT, miR-223’s half-life was 26 hours, and in MMC-treated c223/223BT1 cells the miRNA’s half-life was 37 hours, which is almost 10 hours less than in non-dividing c223 cells that did not overexpress the miR-223 targets (Figure 3C and Table 1).
Although both the perfect and bulge target accelerated miR-223’s decay, the rate was faster in the presence of the perfect target. To understand if this was related to the slicing pathway, we engineered c223 cells to stably express a target site that is perfectly complementary with nucleotides 1 – 17 of miR-223, and mismatches with nucleotide 18 – 23 (223-5′PT)(Figure 2A). c223/223-5′PT cells were treated with Dox, and miR-223 levels were monitored over time. At 48 hours, miR-223 levels decreased by 7.9-fold, and by 96 hours the levels were 28-fold lower than steady-state levels (Figure 2B). Extrapolating from the decay curve, we determined that the half-life of miR-223 was 20 hours in the presence of the 223-5′PT site (Table 1), which, while still accelerated compared to the control, was less than the perfect target. Consistent with this result, there was a slower increase in 223-5′PT target expression compared to 223PT target expression after addition of Dox, as measured by FACS analysis (Figure S3C).
Thus, while our results indicate that miRNA regulation of a bulge target is multiple-turnover, target regulation can increase the miRNA’s rate of decay in a manner that is dependent on target concentration and complementarity.
Target regulation results in uridylation of the cognate miRNA
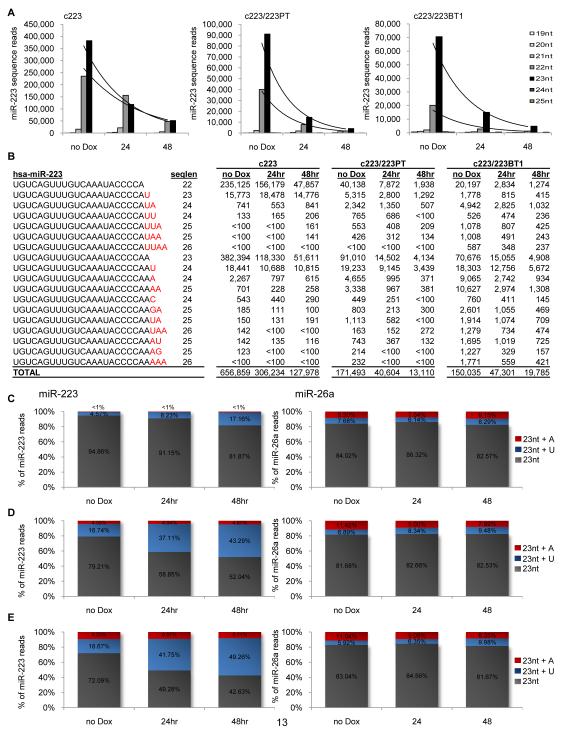
The ability to terminate miR-223 biogenesis provided a unique opportunity to visualize a miRNA during its decay. To do so, we deep-sequenced c223 cells in steady-state and 24 and 48 hours after the addition of Dox. Sequences were mapped and normalized as we previously described[16], and more than 15,000,000 reads corresponded to miRNA genes. In steady-state c223 cells, 635,654 reads mapped precisely to the miR-223 gene (2.95% of total), with the majority being the 23 nucleotide form (Figure 4A). In Dox-treated cells, the overall levels of miR-223 decreased over the 48 hours, consistent with the qPCR data. There was an indication of shortening from the 3′ end of miR-223, as shorter isoforms were enriched at later time points. We did not observe shortening from the 5′ end of the miRNA during decay, but it is possible that these products were degraded too rapidly to detect. There were no appreciable changes in the levels of expression of endogenously expressed miRNAs in the cells (Figure S4A and S4B).
Figure 4. Enumerating miR-223 isoforms during decay and following target regulation.
The small RNA fraction of c223 cells, and c223 cells stably expressing either the perfect target (c223/dGFP.223PT) or bulge target (c223/dGFP.223BT1) in steady-state (no Dox), and 24 and 48 hours after the addition of Dox were deep-sequenced to identify and quantitate miR-223 isoforms. (A) Comparison of miR-223 isoforms during decay in the indicated cells. Sequence reads mapping to the miR-223 gene with lengths varying from 19 – 25 nucleotides starting from the 5′ end of the miRNA were quantitated and graphed. An exponential curve is plotted in black. (B) Sequence and abundance of miR-223 isoforms in the indicated cells. Highlighted in red are non-templated nucleotides. (C-E) Relative contribution of mature miR-223 (5′- UGUCAGUUUGUCAAAUACCCCAA-3′) to adenylated (+A) and uridylated (+U) forms of miR-223 during decay in c223 cells (C) or c223 cells ovexpressing the perfect (D) or bulge target (E) to miR-223. The percent contribution of each isoform to the total is shown on the graph. As a comparison, a similar analysis was performed for miR-26a from the same samples (right). miR-26a was selected as a control because the summed reads were the most similar to miR-223, and unambiguous addition of a uridine or adenine on the most abundant isoform could be discerned. The total number of reads mapping to miRNAs were used to normalize all the data. For additional related data see Figure S4.
Next we examined the pool of sequence reads from each time point that did not map precisely to the genome. These reads represent either sequencing errors or post-transcriptional modifications to the miRNA. Interestingly, during decay there was a consistent increase in the proportion of miR-223 reads that contained a single, non-templated, 3′ uridine (Figure 4B and Figure S4C). In steady-state, 4.57% of the canonical UGUCAGUUUGUCAAAUACCCCAA miR-223 reads contained an additional uridine, whereas by 48 hours the frequency of miR-223 sequences with an extra uridine increased to 17% (Figure 4C). Other modifications were less frequent (<2%) and did not change over time. Moreover, an increase in non-templated uridines was not observed on miR-26a (Figure 4C) and other similarly expressed miRNAs over the 48 hours (Figure S4B). To gain a better understanding of the effect of target regulation on the miRNA, we deep-sequenced the c223 cells overexpressing the perfect (223PT) and bulge (223BT1) target in steady-state and 24 and 48 hours after terminating miR-223 biogenesis. In support of the qPCR data, the rate of miR-223 decay was faster in the cells overexpressing the miRNA’s target compared to the cells without target (Figure 4A). Within 24 hours of adding Dox, the number of mature miR-223 reads decreased from 131,148 to 22,374, or 5.8-fold, in cells overexpressing the perfect target. The bulge target had a similar effect, with the number of miR-223 reads decreasing by more than double the rate in c223 cells.
Most strikingly, the relative abundance of miR-223 molecules containing a non-templated uridine was higher in cells overexpressing the perfect and bulge target (Figure 4B) compared to c223 cells without the target, and this trend was even more exaggerated during decay. Indeed, by 48 hours, nearly 50% of canonical miR-223 reads contained a non-templated uridine in cells overexpressing the miRNA’s target (Figure 4D and 4E). In comparison, the addition of a guanine or cytosine was <1% in steady-state, and did not increase during the course of decay. A single or double non-templated, adenine on the 3′ end of miR-223 was more frequent in the target expressing cells, particularly in c223 cells expressing the bulge target, where 17.5% of the mature miR-223 had non-templated adenines. The frequency of these isoforms did not change during miR-223’s decay, suggesting that this modification occurs as a consequence of target regulation, but is not related to the miRNA’s decay. Instead, the increase in frequency of post-transcriptionally uridylated miRNA during decay suggests that the addition of a uridine may be a de-stabilizing modification.
Discussion
Here we followed the turnover of a single miRNA in mammalian cells, and examined the effect of target regulation on this process. The Zamore laboratory first showed that miRNA regulation of a perfect target is multiple-turnover[6], enabling a single miRNA molecule to catalyze cleavage of up to 50 target transcripts[17]. This is a salient feature of RNAi because it lowers the effective dose of a siRNA or miRNA. However, most natural targets are not perfectly complementary to a miRNA, and thus, are not subject to Ago2-mediated cleavage. To investigate whether regulation of a bulge target is also multiple turnover, we established an in vivo model where we could control and quantitate the concentration of a miRNA and its target within a cell. In steady-state, we found that the number of transcripts being regulated was greater than the number of miRNA molecules, implying that the miRNA was being recycled. This helps to explain the capacity of miRNA to regulate a vast number of transcripts.
Kinetic analysis provided additional support that the miRNA was recycled by showing that the miRNA was not destroyed in the process of regulating the bulge target, and that suppression continued during the miRNA’s decay. That is, when miRNA biogenesis was terminated, the number of miR-223 molecules within each cell was ~2700±750. Since the bulge target was at saturating levels at this time, and continually being expressed, this means virtually all the miRNA molecules would be bound to a target transcript. If the miRNA was not recycled after regulation, a more immediate reduction in the miRNA’s concentration and/or a rapid increase in target expression would have occurred. Instead, we observed a relatively slow rate of decay, even in the presence of overwhelming amounts of target, and target regulation continued to occur for over 48 hours after biogenesis was terminated.
Previous studies have shown that miRNA suppression of a bulge target is less efficient than suppression of a perfect target[18], and that expression of a bulge target can saturate the miRNA at a lower concentration than a perfect target[12,19]. However, because the stoichiometry between the miRNA and target were not specifically determined, and the studies were carried out in steady-state, it was not known if this was because the miRNA irreversibly lost activity after regulation of the bulge target, or because the catalytic rate of the miRNA is slower when regulating a bulge target. Our studies indicate the latter, and even though this process was less efficient compared to regulation of a perfect target, it does provide a mechanism to reduce the sensitivity of miRNA regulation to the overall abundance of mRNA target transcripts.
Although the miRNA persisted for several days after biogenesis was terminated, we found that target regulation accelerated the miRNA’s rate of decay. Thus, while the miRNA can accelerate the mRNA’s decay, it appears that the inverse is also true, and this may serve as a mechanism to control the recycling of the miRNA. Interestingly, Chatterjee and Grosshans found that target could stabilize a miRNA by protecting it from 5′ → 3′ exonuclease digestion[3]. We did not find evidence of this in our studies, but it may be an alternative pathway, which we were unable to detect using our system, or it could be a species-specific difference in how miRNA decay is regulated since their studies were done using C. elegans cell extracts. Recently, Ameres et al. showed that the steady-state levels of a miRNA are lower in Drosophila cells overexpressing a perfect target[20]. While they did not look at the decay kinetics of the miRNA in their model, we would speculate that the decreased miRNA abundance they detected, and the accelerated rate of decay we observed, is due to a similar mechanism. As they note, this may be caused by target interaction releasing the miRNA′s 3′ end from protection by Ago[21], and enabling nuclease digestion of the miRNA.
By quantitating the absolute level of expression of the miRNA and the target within the cells, our studies provide novel insight into the physiological relevance of the target’s effect on miRNA turnover. We found that only when the miRNA was saturated did the miRNA’s rate of decay become appreciably accelerated. Since the more active miRNAs are expressed at >100 copies per cell[12], and the majority of mRNAs are <100 copies per cell[22], a change in the expression level of any one endogenous target gene may be insufficient to have a significant impact on a miRNA’s steady-state level. However, since a miRNA may have 100 or more different target genes, and each of these targets may be expressed at 50 - 100 transcripts per cell (>5,000 molecules of substrate), if the miRNA is regulating at a 1:2 ratio, this would help to explain why the level of suppression mediated by a miRNA on an individual gene is often modest[23,24], and why miRNA activity is dependent on a high abundance of the miRNA[25].
Our studies, the first to examine the structure of a miRNA during its decay, provide insight into the impact of target regulation on the miRNA. We found that target regulation resulted in the post-transcriptional addition of a non-templated uridine to the cognate miRNA, which increased in proportion as the pool of miR-223 molecules was exhausted. Although our studies only provide a correlation between uridylation and miRNA decay, because the frequency of uridylated miR-223 was higher in cells overexpressing the miRNA’s target, and the rate of the miRNA’s decay was faster in these cells, this suggests a mechanism whereby target interaction can lead to uridylation of the miRNA and this accelerates its decay.
Other studies have observed that non-templated uridines are frequently found on the 3′ end of the miRNA, and studies in plants[26], and more recently Drosophila[20], suggested that this is a mark of instability. There has been discrepancy about the significance of this modification to miRNA stability[1], and in mammalian cells overexpression of the uridyltransferase Zcchc11 did not lead to decreased levels of miRNA[27]. Further studies will be needed to determine the actual function of uridylation in miRNA stability. It may be that the type of modifications made to the miRNA are related to the specific miRNA, the cellular context, or the target transcript under regulation, which could recruit particular factors that impact the miRNA differently[2,28].
Experimental Procedures
Supplementary Material
Acknowledgements
We would like to thank Luigi Naldini, Mario Amendola, Albert Ruzo, Emily Bernstein, Andrea Ventura, and Evelyn Yao for helpful discussions. BDB is supported by the National Institutes for Health (NIH) Pathfinder Award (DP2DK083052-01). This work was supported by 1R56AI081741-01A1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010;17:5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 4.Bail S, Swerdel M, Liu H, Jiao X, Goff LA, et al. Differential regulation of microRNA stability. Rna. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. Embo J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 7.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 8.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 11.Amendola M, Passerini L, Pucci F, Gentner B, Bacchetta R, et al. Regulated and multiple miRNA and siRNA delivery into primary cells by a lentiviral platform. Mol Ther. 2009;17:1039–1052. doi: 10.1038/mt.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 13.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, et al. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. Rna. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 17.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 18.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, et al. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 23.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 25.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, et al. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.