Abstract
Electron cryo-microscopy (cryo-EM) has played an increasingly important role in elucidating the structure and function of macromolecular assemblies in near native solution conditions. Typically, however, only non-atomic resolution reconstructions have been obtained for these large complexes, necessitating computational tools for integrating and extracting structural details. With recent advances in cryo-EM, maps at near-atomic resolutions have been achieved for several macromolecular assemblies from which models have been manually constructed. In this work, we describe a new interactive modeling toolkit called Gorgon targeted at intermediate to near-atomic resolution density maps (10-3.5 Å), particularly from cryo-EM. Gorgon's de novo modeling procedure couples sequence-based secondary structure prediction with feature detection and geometric modeling techniques to generate initial protein backbone models. Beyond model building, Gorgon is an extensible interactive visualization platform with a variety of computational tools for annotating a wide variety of 3D volumes. Examples from cryo-EM maps of Rotavirus and Rice Dwarf Virus are used to demonstrate its applicability to modeling protein structure.
Keywords: cryo-EM, Gorgon, modeling, protein structure, near-atomic resolution
INTRODUCTION
From cell motility to signal transduction, large macromolecular assemblies participate in a highly coordinated fashion within the cell. Accordingly, one of the outstanding challenges in biology is the quantitative description of these assemblies and their role in cellular processes (Sali et al., 2003; Sali, 2003; Sali and Kuriyan, 1999). Electron cryomicroscopy (cryo-EM) is capable of imaging large assemblies in discrete physiological states at near-atomic resolutions (Baker et al., 2010; Zhou, 2008) though visualization and analysis tools, from feature detection to domain localization, are required to interpret the complex density maps (Baker et al., 2010).
Fitting of known atomic models within a cryo-EM density map is a relatively common approach for building models of entire assemblies (Rossmann et al., 2005). Computational fitting tools range from simple rigid-body localization of protein structures, such as Situs (Wriggers et al., 1999), Foldhunter (Jiang et al., 2001) and Mod-EM (Topf et al., 2005), to complex and dynamic flexible fitting algorithms that morph known structures to a density map like NMFF (Tama et al., 2004), Flex-EM (Topf et al., 2008), MDFF (Trabuco et al., 2009) and DireX (Schröder et al., 2007; Zhang et al., 2010a). As the structure of individual protein structures are often solved alone or in small complexes, significant differences in the atomic models may be present when placed in the context of the entire macromolecular assembly.
When an atomic model is not known, cryo-EM density maps can be used in building and/or evaluation of structural models from a gallery of potential models constructed computationally (Baker et al., 2006; DiMaio et al., 2009; Topf et al., 2005; Topf et al., 2006; Zhu et al., 2010). The caveat here is that either a related template structure must be known for constrained comparative modeling or, for constrained ab initio modeling, the fold to be modeled must be relatively small.
Significant structural and functional information can also be mined directly from the density map itself (Chiu et al., 2005). At 5-10 Å resolutions, some secondary structure elements (SSEs) are visible in cryo-EM density maps; α-helices appear as cylinders, while β-sheets appear as thin, curved plates (Baker et al., 2002; Böttcher et al., 1997; Conway et al., 1997; Jiang et al., 2003; Zhou et al., 2000; Zhou et al., 2001). These SSEs can be reliably identified and quantified using feature recognition tools to describe protein structure or infer function of individual proteins (Baker et al., 2007; Jiang et al., 2001; Kong and Ma, 2003; Kong et al., 2004).
Until recently, cryo-EM was unable to achieve the resolution needed to build structural models directly from the density map without an initial template. Recently, several cryo-EM structures have reported near-atomic resolutions (3-5 Å), at which point the pitch of α-helices, separation of β-strands, as well as the densities that connect them, can be visualized unambiguously (Chen et al., 2009; Cheng et al., 2010; Cong et al., 2010; Jiang et al., 2008; Liu et al., 2010; Ludtke et al., 2008; Wolf et al., 2010; Yu et al., 2008; Zhang et al., 2010a; Zhang et al., 2008; Zhang et al., 2010b). It is worth noting that while these near-atomic resolution density maps report different resolutions, partially due to different resolution criteria, similar structural features are observed.
While density for bulky sidechains can often be seen at near-atomic resolutions, cryo-EM density maps often lack the necessary resolution to use standard X-ray crystallography model building tools. As with the early X-ray crystallographic structures, models built de novo, i.e. without reference to a known or homologous structural template, for near-atomic resolution density maps have relied almost entirely on visual interpretation of the density and manual structural assignment (Jiang et al., 2008; Liu et al., 2010; Ludtke et al., 2008; Yu et al., 2008).
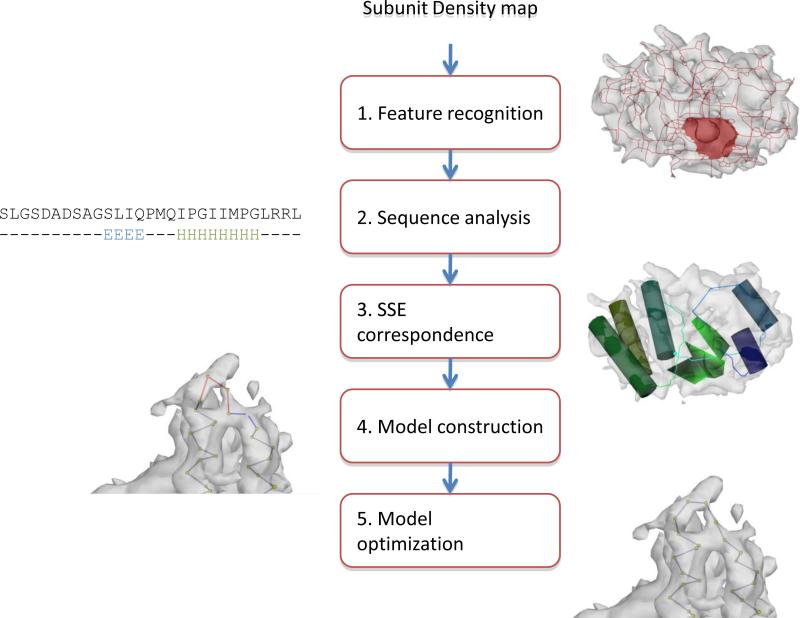
De novo model building in cryo-EM can be divided into five general steps: feature recognition, sequence analysis, SSE correspondence, Cα placement and model optimization (Figure 1). However, individual projects may utilize slightly modified procedures depending on the resolvability of features and available structural information. Secondary structure identification programs like SSEHunter provide a semi-automated mechanism for detecting and displaying these visually observable SSEs in a density map (Baker et al., 2007). Generally, SSEs are depicted as simple geometric objects (i.e. cylinders and surfaces) and contain no information about sequence, direction or connectivity though some topological information can be inferred with a density skeleton (Ju et al., 2007). Similarly, sequence-based secondary structure predictions offer an assignment of SSEs to the sequence but have no spatial localization within a cryo-EM density. Registration of SSEs in the sequence and structure, combined with geometric and biophysical information, can be used to anchor the protein backbone in the density map (Abeysinghe et al., 2008a; Jiang et al., 2008; Ludtke et al., 2008). This sequence-to-structure correspondence relates the observed SSEs in the density to those predicted in the sequence. No longer are SSEs simply geometrical objects; rather they relate the positions of amino acids within the density. Once a correspondence has been determined, Cα atoms can then be assigned to the density beginning with α-helices and followed by β-strands and loops. In the final steps, Cα positions are interactively adjusted such that they fit the density optimally while maintaining reasonable geometries and eliminating clashes within the model. This model can be further optimized using computational modeling such as Rosetta (DiMaio et al., 2009).
Figure 1.
Modeling pathway. An overall outline of the de novo modeling pathway as implemented in Gorgon is shown. Images of the data at each are shown.
In the development of the first de novo models (Jiang et al., 2008; Ludtke et al., 2008) and supporting model building utilities, no single software toolkit was available. Rather a collection of software was used: EMAN for density map segmentation and manipulation, SSEHunter (Baker et al., 2007) to detect secondary structure elements, visualization in UCSF's Chimera (Pettersen et al., 2004) and atom manipulation in Coot (Emsley et al., 2010). With each of these tools, data exchange and interoperability was problematic.
Due to the complexity of de novo modeling at non-atomic resolutions and lack of a unified software toolkit, we have created Gorgon, an interactive molecular modeling toolkit targeted towards near-atomic resolution density maps from cryo-EM and X-ray crystallography (http://gorgon.wustl.edu). Gorgon is built around the aforementioned de novo modeling protocol, utilizing pattern matching and geometry processing algorithms to quickly and accurately model protein structure (Baker et al., 2010; Baker et al., 2010). Gorgon also incorporates several unique utilities that leverage the information at near-atomic resolutions. In this work, we describe the architecture of Gorgon and its utilities for model construction. Furthermore, we have included examples using the publically available density maps at different resolutions for Rotavirus VP6 and Rice Dwarf Virus P8 to demonstrate the accuracy and efficiency for model building.
METHOD
As described, construction of a de novo model from a near-atomic resolution cryo-EM density map requires a set of diverse computational tools not previously found in any single molecular modeling toolkit. The following sections describe the overall design of Gorgon and its set of unique model building tools to accomplish de novo modeling at near-atomic resolutions.
GORGON DESIGN
Gorgon utilizes an increasingly common architecture that divides the core-processing routines from the user-interaction layer. The result is a sophisticated low-level core C++ layer for the infrastructure and computationally intensive algorithms. While this allows for high-performance computations, it does not offer an easy-to-use model for building a user-interface. The user interface layer is written in Python (http://python.org) and coupled with PyQT (http://www.riverbankcomputing.co.uk). The use of Python also allows for scripting support which lets users write their own tools as well as interface with external software.
Gorgon also utilizes a number of publicly available libraries such as OpenGL (http://www.opengl.org/) and PyOpenGL (http://pyopengl.sourceforge.net/) for visualization, Alglib (http://www.alglib.net/) and FFTW (http://www.fftw.org/) for math routines and Boost-Python (http://www.boost.org/) to interface between the C++ and Python layers. For each element in the modeling pipeline (volume, skeleton, SSEs, Cα atoms), we constructed a visualizer that is responsible for the user-interface elements and a renderer that is responsible for the OpenGL rendering routines.
Stable and nightly builds for Windows (32/64-bit XP/Vista/7), Mac Os X (Leopard and Snow Leopard) and Linux (32/64-bit CentOS) are freely available to download at http://gorgon.wustl.edu.
A short snipet of code from the main viewer window and update checker using the API are shown in Supplemental Figure 1.
Input/Output
Gorgon utilizes a number of standard file formats for reading in and exporting data (found under the “File” menu option). Density maps can be loaded in the MRC, CCP4 and RAW formats while skeleton files can be loaded into Gorgon in the MRC and OFF formats. Gorgon also writes density and skeleton maps using those same formats. SSEs identified in the density are uploaded to Gorgon as VRML or DeJa Vu (Kleywegt and Jones, 1997) SSE files. Sequence and secondary structure files are read in as a single text file in either a Gorgon-specific format or a PDB-style header. All atomic coordinates are imported and exported as standard PDB files. A summary of the supported file formats can be found in Table 1.
Table 1.
Supported file formats. A list of all currently supported file formats in Gorgon are shown in Table 1.
| Type | Description | Input | Output |
|---|---|---|---|
| Density maps | |||
| MRC | Cryo-EM density map | ✓ | ✓ |
| CCP4 | X-ray crystallography/ cryo-EM density maps | ✓ | ✓ |
| MAP | X-ray crystallography density map | ✓ | ✓ |
| RAW | Raw, 3D float arrays | ✓ | ✓ |
| BMP | Bitmap image set | ✓ | |
| OFF | Iso-surface mesh | ✓ | |
| Geometric Skeletons | |||
| MRC | Binary density volume | ✓ | ✓ |
| OFF | Curves and surfaces | ✓ | ✓ |
| SSE Annotations | |||
| VRML | VRML cylinders and surfaces | ✓ | ✓ |
| SSE | DeJa Vu secondary structure annotations | ✓ | ✓ |
| Sequence | |||
| SEQ | Annotated sequence information | ✓ | ✓ |
| PDB | Protein Data Bank coordinates | ✓ | ✓ |
| Atom Locations | |||
| PDB | Protein Data Bank coordinates | ✓ | ✓ |
Sessions
In addition to the various supported input and output file formats, Gorgon also features a utility for saving a session. Sessions are text files that log the progress of the model building procedure and save the current state of Gorgon. They can also be used as a means to build models collaboratively with multiple users.
Gorgon User Interface
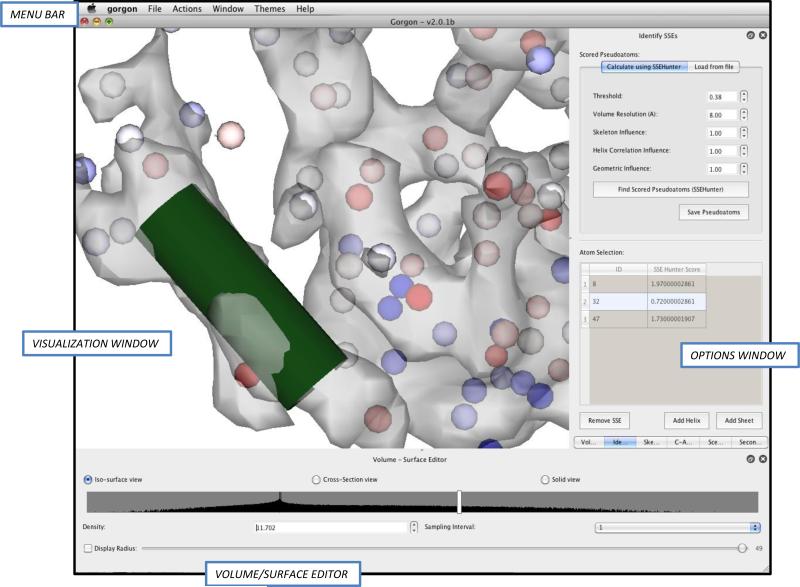
Upon loading a density map into Gorgon, the user will observe four primary environments. Figures 2-4 show the user interfaces for several steps in the modeling procedure. The main environment is an interactive visualization window. All 3-D data such as density maps, SSE annotations, and atoms appear in this window. Directly below the visualization window is the main volume/surface editor window. Here, the user can select the rendering type (surface, cross-section or solid) and set the display parameters. Above the visualization window, a top-level menu bar provides access to all of Gorgon's utilities, including file I/O, modeling tools and help functions. When a tool is selected from the menu bar, it will appear in the options window immediately to the right of the visualization window. As new tools are opened, a sub-menu will appear at the bottom of the options window to allow users to select open tools. A list of open and active tools can also be found under the “window” tab in the top-level menu bar.
Figure 2.
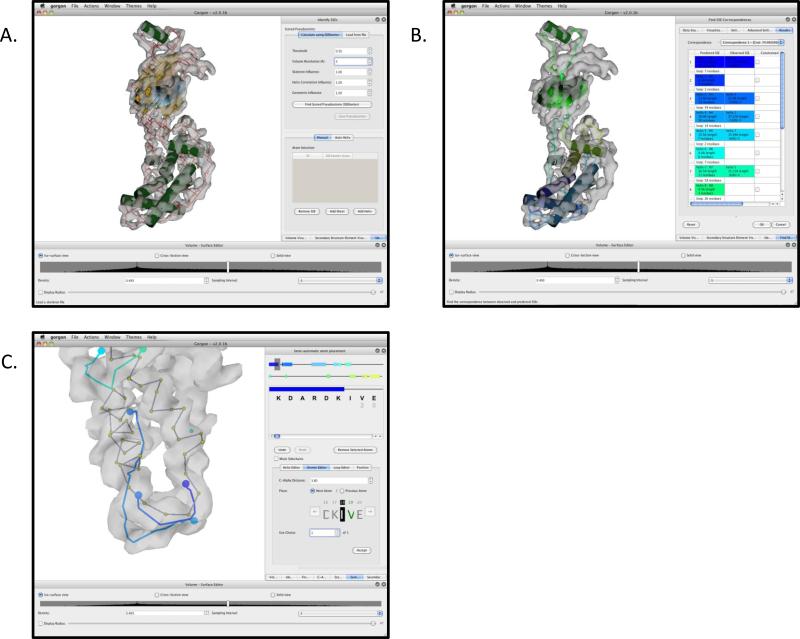
Building SSEs with Gorgon. The basic Gorgon interface is divided into four key sections: the menu bar at top, the visualization window in the middle, the volume/surface editor at the bottom and the option panel on the right. The SSEHunter result (red and blue pseudoatoms) for the apical domain from the 4.2Å resolution cryo-EM structure of GroEL in Gorgon is shown. A single helix has been annotated (green).
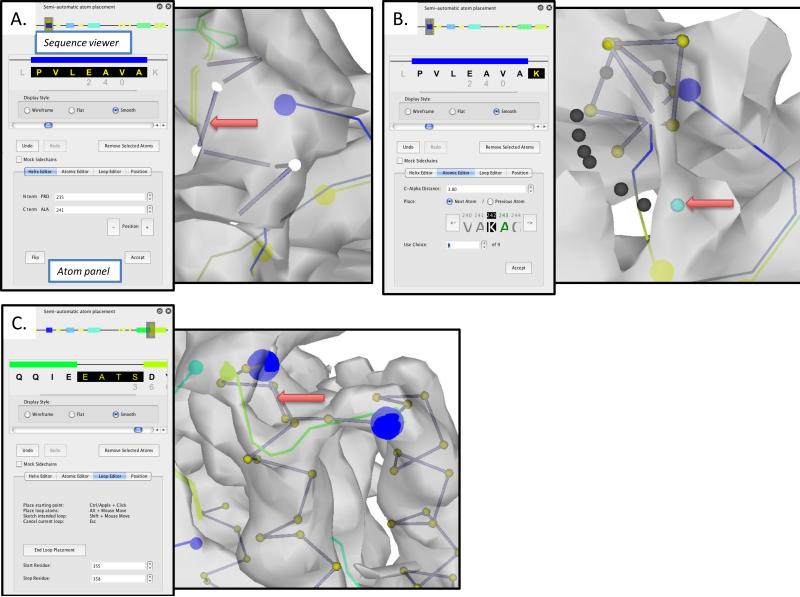
Figure 4.
Atom placement with Gorgon. The “Semi-Automatic atom placement” tool is divided into two panes: a sequence viewer, which displays both global and local sequence views and the atom panel, which contains four separate tools for placing and adjusting atom positions in the density map. In (A), the Helix Editor is being used to assign the highlighted sequence in the sequence viewer to the selected VRML helix in the visualization window. In (B), the “Atomic Editor” is being used to walk along the density skeleton of the GroEL apical domain density map. The currently selected residue is highlighted in the sequence viewer; the atom to be placed is shown in green in the “Atomic editor”. In the visualization window, the currently selected residue is highlighted; potential positions for the next residue are shown as a dark grey or cyan sphere. In (C), the “Loop Editor” is being used to sketch a path through the density map and connect two adjoining helices. The start/stop end points of the loop are shown as large blue spheres.
GORGON UTILITIES
Gorgon contains a set of unique utilities designed specifically for intermediate and near-atomic resolution density maps found under the “Actions” menu. In addition to these utilities, Gorgon contains an extensible framework with an API that allows others to develop plug-in modules for Gorgon. Currently, this framework is used to check for new updates of the software. As part of our future work, the framework will be utilized to build user-specific functionality as well as to integrate with other molecular modeling systems such as Rosetta (Bradley et al., 2005) and Modeller (Sali et al., 1995).
Density Skeletonization
SSE identification and correspondence both utilize a density skeleton, which is a compact geometrical representation of the density maps using curves and surfaces. This type of skeleton has been designed specifically for intermediate and near-atomic resolution density maps, in which the surfaces represent β sheets and curves represent helices and loops (Baker et al., 2007; Ju et al., 2007). Gorgon currently supports three different mechanisms for calculating a density skeleton (“Actions>Volume>Skeletonization”), two of which are unique to Gorgon. Any of these three can be used as input in SSEHunter, defining secondary structure correspondences and in the model building procedures described later.
The binary skeletonization routine is the skeletonization routine implemented in the original EMAN version of SSEHunter (Ju et al., 2007). This skeletonization requires the user to select an iso-surface value based on the density map. Typically, the proper setting is the highest iso-surface value of the density map that still maintains connectivity of all of the elements in the density map. At this point, SSEs should be readily apparent in the density map.
The grayscale skeletonization routine, unique to Gorgon, expands on the binary skeletonization routine and eliminates the need for the user to provide a threshold for skeleton generation (Abeysinghe et al., 2008b). Rather, the user provides a starting threshold or a previously built skeleton as an initial reference. This skeletonization routine is not very sensitive to the user-input threshold; this threshold can be any iso-surface value at which the density map can be seen without noise. Generally, grayscale skeletons are more robust to noise and variation in density within a density map.
The final skeletonization method found in Gorgon is an interactive sketching tool, which is also unique to the graphical interface of Gorgon (Abeysinghe and Ju, 2009). The user can select a starting point anywhere on a density map and then “sketch” a path using the mouse. Branch points can be quickly created and multiple paths explored. Unlike the previous two routines, the interactive routine produces a density skeleton consisting of only curves (which can be useful for backbone modeling in later steps).
Identifying SSEs
SSEHunter, a feature recognition program, has been successfully used on numerous subnanometer resolution cryo-EM density maps to reliably identify α-helices longer than two turns and β-sheets with three or more strands (Baker et al., 2007). SSEHunter has been incorporated into Gorgon (“Actions>Secondary Structure Element>Identify SSE”) (Figure 2). Unlike most of the other Gorgon processes, this step is not interactive and requires significant time to compute. Typically, map sizes between 483 and 1603 require less than 15 minutes to run on a modern desktop computer.
In addition to providing a visual interface for SSEHunter, several new features have been added to its performance in Gorgon. Improvements in the scoring routines and the availability to interactively set weights to the various SSEHunter sub-routines have been added. A re-factoring of the code to include the Gorgon computational routines has also resulted in a speed up as compared to the previous EMAN versions. As previously mentioned, the user can select any one of the three skeleton types or provide their own density skeleton as input for the Gorgon version of SSEHunter, an option not found in the EMAN version.
Building SSEs
SSEHunter returns a set of Cα atoms (pseudoatoms in PDB format) with values assigned between -3 and 3. These values represent the likelihood of a density region to be either α-helix (0 - 3) or β-sheet (-3 - 0). The intensity of the color reflects the score and thus the confidence of the prediction (Figure 2). To annotate these pseudoatoms and display stylized VRML SSEs within the density map, the user must select related, neighboring pseudoatoms and group them into SSEs (“Actions>Secondary Structure Element>Identify SSE”).
In Gorgon, a new interactive utility has been created to add/delete and modify these assignments. Additionally, a computational routine for grouping similarly scored helical pseudoatoms has been incorporated for automated SSE assignment (Auto Helix). Also unique to Gorgon is a new utility that allows the user to optimally fit VRML helices to the density (“Actions>Secondary Structure Element>Fit Selected Helices”). This option attempts to optimally align a VRML helix created in the previous step to a local region of the density map.
Secondary Structure Prediction
In Gorgon, model building is accomplished by generating a sequence-to-structure correspondence using SSEs. Gorgon contains a utility for loading the primary sequence, launching web-based secondary structure prediction queries and saving the results in a Gorgon-specific text file (“Actions>Secondary Structure Elements>Predict SSE from Sequence”).
SSE Correspondence
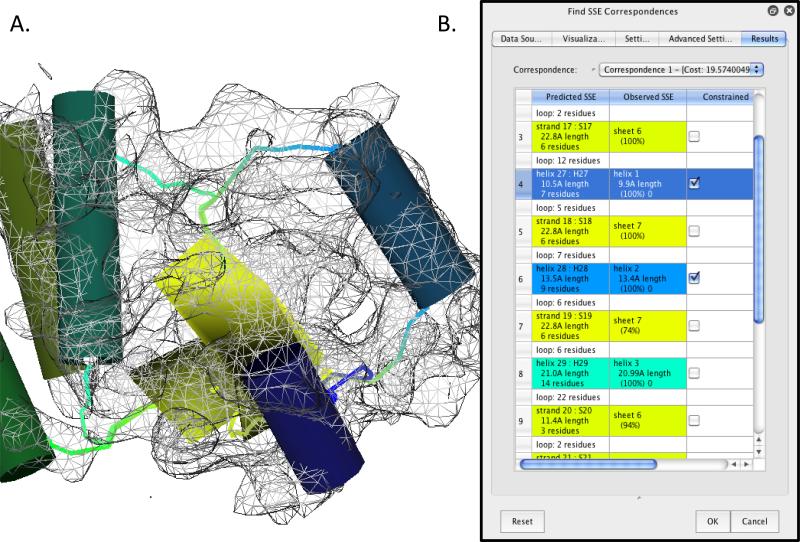
In Gorgon, the SSE correspondence process (“Actions>Secondary Structure Elements>Find SSE Correspondence) is dynamic and interactive, allowing users to rapidly setup, build, evaluate and modify model topologies. This utility is unique to Gorgon, the SSE correspondence tool and uses an efficient graph matching approach to match SSEs detected by SSEHunter to SSEs found by secondary structure prediction. Building on our original SSE correspondence search algorithm (Abeysinghe et al., 2008a), Gorgon's latest correspondence routine utilizes both helices and strands to improve the mapping of sequence elements to secondary structure elements seen in the density map, resulting in more accurate topological models. This topology generated by the SSE correspondence search is then used as the starting point for placing Cα atoms and tracing a protein backbone in the density map.
To calculate a correspondence in Gorgon, the user needs to provide four files: helix and sheet locations (in VRML format), a cryo-EM density skeleton and the sequence prediction from the previous step. These files can either be calculated directly from within Gorgon or loaded individually. From these inputs, correspondences are then calculated and shown under the “Results” tab. The best scoring correspondences are listed first in the drop-down results list. Below this list, the actual correspondences are displayed in a table (Figure 3), showing which SSE elements are matched (second and third columns in the table), as well as the confidence of the matching (the percentile in the cells in the third column). Individual SSE correspondences can be constrained by selecting the SSE in either the correspondence table (fourth column) or the SSE in the main viewer panel. As the correspondence calculation is relatively quick (<15sec for most cases), constraints can be added or removed repeatedly to new correspondence calculations until the user is satisfied with the results.
Figure 3.
Correspondence search with Gorgon. A sequence-to-structure correspondence search for the apical domain of GroEL is shown. Helices in the visualization window (A), shown as cylinders, are colored coded to the corresponding sequence element in the “Find SSE correspondence” tool in the option panel (B). Sheets are shown as yellow planes. Connectivity between the secondary structure elements are represented in the visualization window by curve segments using color gradients based on the two connecting SSEs.
In building de novo models, the SSE correspondence is crucial and, in practice, may require the most user time and input. Even with the latest improvements to the algorithm, the top correspondence may not be the correct correspondence, though typically the true SSE correspondence is ranked in the top ten correspondences. Therefore, the user must carefully examine all of the potential correspondences before choosing one and proceeding to the model building stage. In selecting a SSE correspondence, the best correspondence is typically judged by three criteria: 1) matching of SSE lengths (expressed in residues and Å in the results window), 2) the “rate” at which an SSE appears in the same position in the ensemble of correspondences (expressed as a percentage in the correspondence search results) and 3) the connectivity of SSEs along the topological path, shown as connecting lines in Gorgon's main display window. If a single correspondence does not meet these criteria, the user can select a few SSEs that meet these criteria, lock them into place and re-run the search. This procedure can be iteratively re-run to refine a correspondence, particularly when a large number of SSEs are present.
Beyond the basic functions, the SSE correspondence tool contains several advanced options that are further detailed on the Gorgon website. Some of the more useful options allow the user to hide or show different features. The “Settings” tab controls the weights associated with the various SSEs when calculating the correspondence. Of note is the “Include Sheets” option. When unselected, this allows the user to run a helix only correspondence. This is often necessary when calculating a correspondence with a large number of elements, due to the higher computational cost associated with strand matching. The “Advance Settings” tab contains additional options for expanding or restricting the search, controlling the number of missing SSE elements and options for strand matching.
Atom placement
A correspondence produces a topological mapping of SSEs but does not place any atoms in the density map. Gorgon features several semi-automated atom placement options beginning with a correspondence or with any previously placed atoms (“Actions>C-Αlpha Atoms>Semi-automatic Atom Placement”). Like the SSE correspondence tool, the atom placement tools are uniquely designed to take advantage of the features at near-atomic resolution and are not found in any other software toolkit.
The semi-automated atom placement module is divided into two primary sections: the sequence viewer panel (top) and the atom panel (bottom) (Figure 4A). The sequence viewer panel is further divided into two views of the sequence and predicted secondary structure. A global view of the predicted secondary structure is represented in the topmost portion of the sequence viewer. The grey box in the global view scrolls through the local sequence view just below the global view. In the local view, clicking on one of the α-helices will highlight the corresponding sequence; clicking on a helix, loop or atoms in the main viewer will select the corresponding sequence element.
The atom panel contains four separate utilities for placing atoms in the density map. The “Helix Editor” builds atoms into helices where the SSE correspondence search identified a matching sequence/helix pair; unmatched helices are colored gray in the sequence viewer panel (Figure 4A). Individual helices are selected from the local sequence viewer and constructed by pressing “Accept”. Cα atoms are then placed on the position of the cylinder corresponding to the helix. Note, the sequence in the local sequence viewer turns black to indicate atoms have been placed in the density map for these residues. The user may also want to adjust helix length before assigning the helix atoms by adjusting the starting and stopping residues of the helix. It may also be necessary to flip the direction of the helix using the “Flip” option.
Extending from the assigned residues in the α-helices, assignment of the remaining residues is accomplished with the “Atomic Editor” and “Loop Editor” modules in the atom panel. With the “Atomic Editor”, unassigned residues are added serially, beginning at a previously assigned atom position (Figure 4B). Walking down the density skeleton, individual Cα atoms are sequentially added. The next unassigned residue is shown in green in the atom panel and a list of possible positions along the density skeleton that satisfy a Cα-Cα distance is shown as a set of dark gray spheres in the main viewer window. The current selected choice is highlighted in cyan; the user can either select the atom in the main viewer window or toggle through the choices with the “Use choice” selector in the “Atomic Editor” panel. Once the desired position is found, clicking “Accept” will accept the placement and increment the atom selector. Note, atoms are placed along the skeleton that runs through the local extrema of the density; the Cα distance may need to be adjusted to ~3.5 Å initially to complete a trace.
While the “Atomic Editor” places residues one at a time, the “Loop Editor” allows the interactive assignment of an entire loop (Figure 4C). By selecting a set of unassigned residues in the local sequence viewer, the “Loop Editor” will construct a loop that can be interactively adjusted in the density map. In both the “Loop Editor” and the “Atomic Editor”, Gorgon's density skeleton is used to help place atoms making these processes optimal and unique to Gorgon.
The fourth module of the semi-automated atom placement utility is the “Position Editor”, which allows the user to select one or more assigned residues and adjust their positions within the density map. This is accomplished using three rotation and three translation options.
In each of these modules, Cα-Cα distances are indicated by color in Gorgon. Red bond distances are too long, blue bonds are too short and gray bonds are approximately the right length (3.8Å ± 0.5Å). Additionally, the size and properties of individual amino acid sidechains may be displayed in the main viewer window by selecting the “Mock Sidechains” option (Figure 4A).
Fitting Atomic Models of Structural Homolog
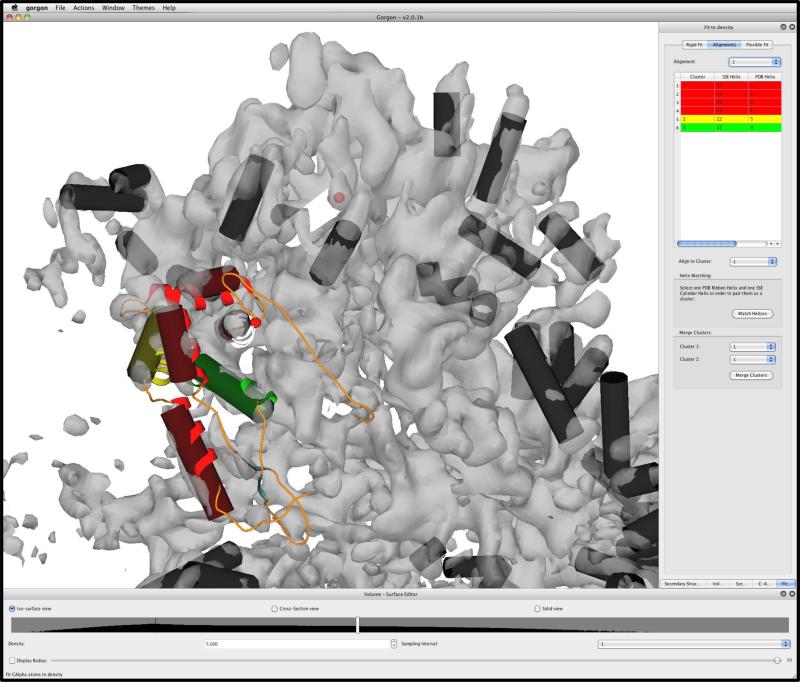
In addition to building Cα models, Gorgon features a rapid method for fitting atomic models to a cryo-EM density map (“Actions>C-Alpha Atoms>Fit to density”). This novel method utilizes the position of the density-map derived SSEs to guide the placement of an atomic model within a density map by determining the best local alignment of SSEs using a fast-clique finding algorithm (Abeysinghe et al., 2010). As such, fits based on Gorgon or user-derived sets of SSEs can be rapidly evaluated. In the case of the 30S subunit from T. thermophilus 70S ribosome at 6.4 Å resolution (PDBID: 3FIC, 3FIN; EMDBID: 5030) (Schuette et al., 2009) with 65 helices, Gorgon was used to compute the optimal solution in less than 1 second on a modern desktop computer (Figure 5, Supplemental Movie 1).
Figure 5.
Model fitting with Gorgon. 65 helices were identified in the 30S subunit structure of the T. thermophilus 70S ribosome (6.4Å resolution) using SSEHunter in Gorgon. Chain C of the 30S crystal structure (PDB ID: 3FIC) was fit to the density map based on the positions of the helices. The clique matching method for this fitting routine identified three groups of matching helices, annotated in red, yellow and green in the upper right hand corner and in the main viewer window.
RESULTS
Various pieces of Gorgon have been used during its development with several near-atomic resolution density maps, including the 4.2 Å resolution structure of Mm-cpn (Zhang et al., 2010a) and the structure of bacteriophage P22 (4.0 and 3.8Å resolution) (Chen et al., 2011). In the following sections, model building and assessment are described. Though the structures of these proteins were either known or previously modeled, they were not used in the construction of their respective models presented here.
Modeling at near-atomic resolution
As part of a recent modeling in cryo-EM workshop challenge (http://ncmi.bcm.edu/challenge), we built models from several near atomic resolution density maps (Figure 6) using Gorgon's de novo modeling process, including Rotavirus VP6 at 3.8Å resolution (EMDB ID: 1461) (Zhang et al., 2008), Mm-cpn at 4.3Å resolution (EMDB ID: 5137) (Zhang et al., 2010a), GroEL at 4.2Å resolution (EMDB ID: 5001) (Ludtke et al., 2008), Aquaporin at 3.8Å resolution (PDB ID: 1FQY) (Murata et al., 2000) and bacteriophage ε15 gp7 at 4.5Å resolution (EMDB ID: 5003) (Jiang et al., 2008). The complete model building and assessment process for Rotavirus VP6 at 3.8Å resolution (EMDBID: 1461) (Zhang et al., 2008) is detailed below (Figures 7, 8). Results for all of these models, including density maps, SSEs, models and fit X-ray structures, are available through the challenge workshop website.
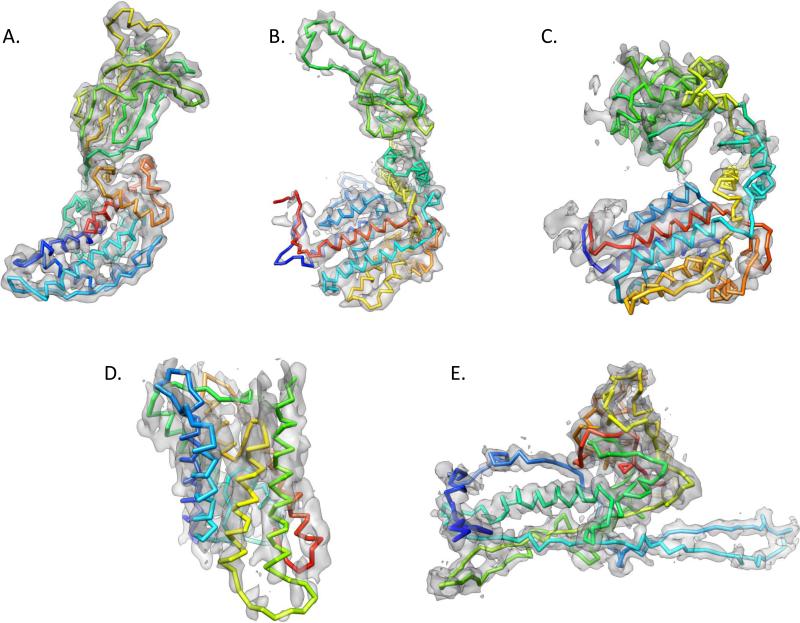
Figure 6.
Modeling with Gorgon. A gallery of models constructed with Gorgon is shown. The models include (A) Rotavirus VP6 at 3.8 Å resolution (EMDB ID: 1461), (B) Mm-cpn at 4.3 Å resolution (EMDB ID: 5137), (C) GroEL at 4.2 Å resolution (EMDB ID: 5001), Aquaporin at 3.8 Å resolution (PDB ID: 1FQY) and (E) bacteriophage ε15 gp7 at 4.5 Å resolution (EMDB ID: 5003). The backbone traces are shown superimposed on the corresponding density colored from N- (blue) to C- (red) terminus.
Figure 7.
Modeling Rotavirus VP6 with Gorgon. (A) The result of Gorgon's “Identify SSE” tools on the 3.8Å map of Rotavirus VP6 is shown. SSEs are shown as green cylinders (helices) and cyan planes (sheets). The density skeleton, colored in red (for curves) and yellow (for surfaces), is shown in the density and connects the visible SSEs. (B) The result of a helix-only SSE correspondence is shown. In the right panel, the sequence-to-structure assignment can be seen. Helices and sheets in this panel are colored correspondingly in the main Gorgon viewer. (C) Model construction is shown using the “Semi-Automatic atom placement” tool. In this example, the first two helices and their connecting loop have been built using the Helix and Atom Editor functions, respectively.
Figure 8.
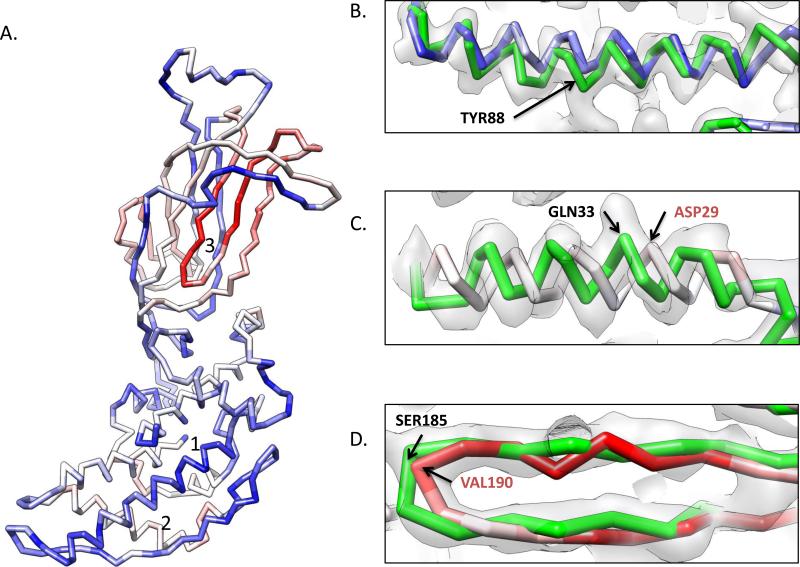
Errors in modeling VP6. The final VP6 model is shown in (A). Positions with a low-RMSD when compared to the crystal structure are shown in blue, while high RMSD regions are shown in red. Three regions are labeled and shown in zoomed in views in (B), (C) and (D) along with the X-ray structure (green model), the sequence (top line), secondary structure from the crystal structure (second line), JPRED secondary structure prediction (third line) and the final model's secondary structure. In (B), helix 1 fits well to the density and agrees well with the X-ray structure. In (C), an error in the starting residues of the sequence prediction of helix 2 resulted in a sequence shift during modeling. While the helix fits the density well, the model is off by approximately one turn, illustrated by GLN33 of the X-ray structure and ASP29 of the model occupying the same position in the density map. In (D), long loops between anchor points resulted in modeling errors in the large β-sheet region, labeled as region “3” in (A). This region is shifted by ~5 amino acids, resulting in the highest RMS deviations in the model.
For modeling VP6, model generation began with the identification of SSEs and generation of a density skeleton in the density map (Supplemental Movie 2). Using Gorgon's “Identify SSEs” tool, nine α-helices and two β-sheets were identified (Figure 7A, Supplemental Movie 3). Calculation of the scored pseudoatoms took ~5 minutes for the segmented map of VP6 with 96×96×96 pixels. Manual selection of the SSEs took ~5 minutes as well. A consensus secondary structure prediction (from SSPro (Pollastri et al., 2002), JPred (Cole et al., 2008) and PsiPred (McGuffin et al., 2000) was then obtained from the primary sequence of VP6 (Uniprot: P04509). Using this prediction and the identified SSEs within the density map, a SSE correspondence was calculated based on helices alone with the “Find SSE Correspondence” tool (Figure 7B, Supplemental Movie 4). This step required less than two seconds to compute. The top correspondence appeared to be the correct solution as all SSEs appeared to have similar sizes and reasonable connectivity. From this correspondence, the “Semi-automatic Atom Placement” was used to assign Cα positions (Supplemental Movie 4). First, the “Helix editor” module was used to assign the primary sequence to the helices. Once helices were assigned, the “Atomic editor” module was used to place and connect the remaining Cα positions (Figure 7C). Assignment of these positions started at the termini of the helices and continued until either another assigned atom was reached or an ambiguous path was found. Once all obvious paths were assigned, model building proceeded at the ambiguous regions. For an experienced user, it took approximately 2 hours to place all 397 amino acids in VP6. Finally, to fix bad bond distances and geometry it required an additional ~30 minutes. Complete model construction and optimization for VP6 was accomplished in an afternoon though model building times will vary greatly with experience and the quality of the density map.
Visual inspection of the Gorgon model versus the crystal structure (PDBID: 1QHD) (Mathieu et al., 2001) revealed that the models essentially had the same fold or topology. Overall the Gorgon model had an RMS deviation of 3.34 Å when compared to the crystal structure. In Figure 8A, the Gorgon model is colored based on individual Cα RMS deviation. The majority of the differences in the model appear to be localized to the upper domain β-sheets, where there are few anchor points for tracing the protein's backbone. In the lower helical domain of VP6, RMS deviation is considerably better where the abundances of helices helps to register the backbone trace.
Examining the Gorgon model more closely revealed that nearly 50% of all Cαs were within 3 Å of their counterpart in the X-ray structure. In fact 72.5% of all Cαs were within 5Å and 88.9% were within 8 Å. Furthermore, the average difference, calculated ignoring the sequence assignment for each Cα in the Gorgon backbone trace and the Cα positions of the X-ray structure, was determined to be 1.7 Å. This suggests that the overall topology or fold of the VP6 model was correct, though amino acid assignment and registration were off in certain places.
To better understand the differences in the models, a detailed analysis of the sequence, structure and density were carried out for three discrete regions of secondary structure (Figure 8B-D). In Figure 8B, a helix containing residues 83-95 is shown. The Gorgon model (blue) and the X-ray structure (green) appear to be very similar and fit the density. In terms of the sequence (top line), the secondary structure prediction from JPred3 (third line) also agrees well with the secondary structure from the crystal structure (second line), though the predicted helical length was longer. The Gorgon model was initially constructed with this longer helix. Clearly visible in the density map, several large aromatic sidechains, particularly TYR188, were used to register the helix position and orientation in the density map. To better fit the density the last turn was eventually made into a loop, resulting in a final secondary structure assignment (fourth line) identical to that of the crystal structure.
In a second helix (residues 25-41), a more pronounced difference was seen. The residue in this helix had a larger RMS difference (~3.5-4.5 Å; grey to red color) when compared to the X-ray structure. In examining the sequence, secondary structure prediction shows this helix should start at SER25, but the crystal structure shows the helix starting at residue SER28. Without clearly visible sidechain densities to anchor this region of the trace, the addition of the one helical turn causes a shift in the sequence assignment.
The upper domain of P8 is primarily composed of β-sheets and loops. The anchor points for model building are located in the helices in the lower domain. As such, any errors, like those found in Figure 8C, are propagated and magnified due to distance between anchor points during the trace of this domain. In one portion of the upper domain β-sheet, the crystal structure and model differ by up to 14 Å due to such errors. Shown in Figure 8D, the backbone trace for P8 is off by five amino acids. The errors can be further seen by examining the sequence, in which the secondary structure prediction, model and X-ray structure all differ by several amino acids.
Similar results were obtained with other aforementioned near atomic resolution density maps. In the models, the RMS deviation varied from 4-9 Å, while the overall topological difference between the models and known structures were ~1.4-2.0 Å. Again, analysis of the sequence, structure, model and map revealed that small differences in secondary structure assignment were the major cause of the relatively high RMS deviations and not necessarily the placement of individual Cα atoms.
Based on our earlier models such as GroEL (Ludtke et al., 2008), this type and level of error is typical for the models obtained by de novo modeling techniques. The important point here is that de novo modeling in Gorgon can properly build initial backbone models with the correct fold/topology. Further model optimization with cryo-EM density constraints, as implemented in Rosetta, can correct for errors, restore proper sequence-structure registration and generate refined all-atom models (DiMaio et al., 2009).
Modeling at subnanometer resolutions
Gorgon is primarily targeted at near-atomic resolution density maps though many of its tools, such as fitting and feature detection, are applicable to density maps between 10 and 4 Å resolution. However, limited resolution does not preclude the use of Gorgon's modeling tools. In addition to the aforementioned near atomic resolution density maps, a partial model for the virus capsid protein P8 from the ~7Å resolution cryo-EM density map of Rice Dwarf Virus (Zhou et al., 2001) was also constructed with Gorgon (Figure 9).
Figure 9.
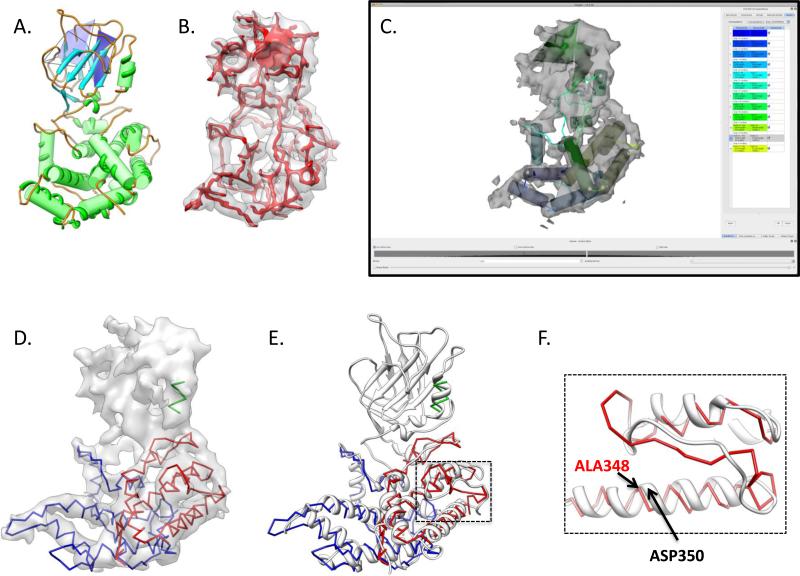
Modeling Rice Dwarf Virus P8 at ~7Å resolution. A model for Rice Dwarf Virus VP6 was constructed using the de novo modeling tools in Gorgon despite its low resolution. In (A), Gorgon's secondary structure detection routines were able to identify all twelve helices and both large β-sheets. However, the density skeleton (in red in B), contained a large number of breaks and branch points, particularly in the upper domain. Gorgon's SSE correspondence routine was able to correctly identify a correspondence that matched all twelve helices with those identified by secondary structure prediction. A final model for the N- (blue) and C- (red) terminal portions of the lower domain of P8 was constructed (D). A single helix in the upper domain (green) was also modeled but not connected to the two termini. The model for the lower domain agreed well with the X-ray structure of P8 (grey ribbon, PDB ID: 1UF2) (E). In examining the model, many of the loops connecting helices varied somewhat from the X-ray structure (F). Additionally, the inability to align the helices to the density resulted in modeling errors, shown by ALA348 (model) and ASP350 (X-ray structure) in (F).
From the ~7 Å Rice Dwarf Virus density map (EMDB ID: 1060), a single P8 monomer was extracted using UCSF's Chimera. Using the same protocol as described for VP6, secondary structure elements were detected using Gorgon. Twelve helices and two sheets were identified, which agreed well with the known X-ray structure (PDB ID:1UF2) (Figure 9A) (Nakagawa et al., 2003). Unlike the VP6 density map, individual β-strands were not visible; the two upper domain β-sheets were visible as two thin planes (Figure 9B). A correspondence was then generated using a secondary structure prediction derived from the JPred3 web service. Gorgon's correspondence search top result matched ten of the twelve helices were assigned correctly; during this initial run, only α-helices were considered in the search. Two helices in the lower domain, helix 1 and helix 3 in the correspondence were swapped. After examining the density map and constraining five helices (helices 7, 9, 10, 11 and 12), the correspondence search was re-run. These five helices were selected based on the match of lengths and the number of times they were identified in alternate correspondences (100%). During this run, sheets were also considered and the global search parameters were relaxed (border margin threshold was set to 10 and Maximum Euclidean distance was set to 30) to allow for more possible SSE correspondences. This correspondence resulted in the correct matching of all α-helices (Figure 9C).
From this correspondence, a model was constructed for P8. Beginning at the N- and C- termini, atoms were added consecutively to complete a backbone trace for P8 from residues 1-141 and 304-421 (Figure 9D). Residues 142-303 contained the two β-sheets, corresponding to the upper domain portion of the P8 protein. This region of the density map did not have enough resolution or anchor points to properly trace the backbone. The lower domain of P8, even at ~7Å resolution, did however have enough anchor points to produce a reasonably good topological trace of the protein containing primarily α-helices. When compared to the X-ray structure, this portion of the model had a 4.71 Å RMS deviation. Like the Rotavirus VP6 example, differences in secondary structure prediction resulted in mis-assignments and register shifts (Figure 9E). Additionally, the low resolution made modeling loops particularly difficult.
DISCUSSION
Gorgon occupies a unique area in interactive modeling tools. It is the only software toolkit that has been developed exclusively for modeling protein structure directly from non-atomic resolution density maps. Like many other interactive modeling tools, such as Chimera (Pettersen et al., 2004), UROX (Siebert and Navaza, 2009), COOT (Emsley et al., 2010) and Sculptor (Birmanns et al., 2010), Gorgon has a modular design that utilizes a set of menu-driven processes to manipulate density maps and/or models. Not only does Gorgon offer tools for fitting known structures but also has a set of unique utilities, including feature recognition and correspondence searches, for skeleton-based model construction at near atomic resolution density maps. Currently, no other modeling toolkit provides such a complete interactive environment for modeling protein structure directly from a density map at non-atomic resolutions.
It should also be noted that Gorgon and the de novo modeling process results in a Cα-only model without considerations for mainchain and sidechain constraints. These types of model are less detailed than those generally obtained from Coot or other modeling tools targeted at higher resolution crystallographic data that include additional density and biophysical constraints. The utilities in Gorgon allow a user to rapidly and robustly build “first-approach” models that generally have the correct topology or protein fold but non-optimized atom positioning and/or assignment. Again, utilities commonly found in atomic modeling software, such as Ramachandran plots and rotamer libraries, are not used for Cα backbone construction and thus not part of Gorgon's current toolkit. In the event a density map does have sufficient resolution to visualize the majority of sidechains, Gorgon's initial model can be further optimized using conventional modeling and refinement software designed for X-ray crystallography.
Although Gorgon is designed around building de novo models directly from the cryo-EM density map, components of Gorgon can be used independently. As an example, secondary structure identification can be performed on nearly all subnanometer resolution density maps without using any of Gorgon's additional features. However, it is important to emphasize that all of Gorgon tools are focused on subnanometer resolution density maps. As such, Gorgon's “Fit to Density” tool is extremely quick and accurate for fitting atomic models to a density map, but requires the presence of SSEs and thus, would not work on low-resolution density maps.
The extensible framework of Gorgon provides users with a convenient and feature-rich environment for structure annotation at intermediate resolutions. This flexible framework allows for the integration of additional software packages, such as molecular dynamics and modeling programs, as well as facilitating the rapid development of new algorithms and tools for use within Gorgon's user interface. We envision that this design will allow for the incorporation of new tools, such as flexible fitting and segmentation routines, as the number of subnanometer resolution structures continues to grow.
Pitfalls in de novo modeling
While Gorgon was designed to provide a complete modeling environment for subnanometer to near-atomic resolution density maps, there are certain caveats. Ultimately, Gorgon's utilities are limited by the resolvability of map features and not by the stated map resolution. With the utilities in Gorgon, a density map is processed to produce an annotation, such as a skeleton, SSEs, a correspondence or a model. Therefore, even if the map has sufficient resolution but is of low quality, accurate results cannot be obtained from Gorgon, or that matter, any other modeling software.
In Gorgon, the de novo modeling protocol is based on establishing a sequence to structure correspondence using SSEs. This correspondence is based on the ability to accurately predict SSEs in sequence and identify them in the density map. Secondary structure predictions typically have a success rate of ~80%. Often, consensus sequence predictions are built from many different predictions to reduce potential errors. However, even small errors in secondary structure prediction can result in small register shifts, mis-identification of lengths or even missing SSEs. During model building, these errors are propagated throughout the model resulting in improper sequence to structure assignments. Whether it is from a consensus prediction or better prediction methods, improvements in the prediction will translate directly to more accurate models.
Another potential hazard is the lack of anchor points in model building. A correspondence search provides the user with a set of SSE visualized in the density that can be assigned to sequence. These SSEs serve as anchor points from which a model is “grown” along a skeleton path connecting these elements. To assist in growing and anchoring the backbone trace, sidechain density can also be used, though it is often only sporadically visible in the best cryo-EM density maps at the present time. As such, regions with no or few anchor points, such as long extended loops, can result in less accurate Cα placement.
Illustrating the aforementioned sources of error, the average error for the backbone trace of Rotavirus VP6 was ~1.7 Å, indicating a high level of accuracy in describing the fold of the protein. While, nearly 50% of all Cα atoms were within 3 Å of their corresponding position in the X-ray structure, ~11% of the Cα atoms had deviations greater than 8 Å. These deviations were often associated with either long loops between anchor points or in regions where the secondary structure prediction was inaccurate. At higher resolutions, sidechain density can aid in the placing of Cα atoms and help eliminate some of these outliers.
As already mentioned, a backbone trace is built along a path defined by the density skeleton, which is a simplistic geometrical representation of the medial axis of the density. This means that atoms are placed linearly in all regions except for helices. β strands do not contain the zig-zag like appearance found in atomic resolution models, but rather are just a linear array of Cαs.
Finally, no explicit refinement tools are used in generating models in Gorgon. Simple distance warnings in Gorgon and visual inspection of density occupancy and bond geometry are essentially the only mechanisms of assessing model quality. As no computational refinements are done from directly inside Gorgon, no force fields or atomistic prosperities are considered during model building.
Despite their drawbacks, “first-approach” de novo models are topologically equivalent to a fully refined protein structure. Combined with constraints derived from their density map, these de novo models can be computationally refined with additional modeling software, such as Rosetta, to produce more accurate and stereochemically correct all-atom models.
Modeling and resolution
As all maps vary in composition, quality and resolution, it is difficult to assign an exact resolution cut-off for building models. This is in part due to the various resolution definitions and variability in resolvability of maps at ostensibly the same resolution. Clearly though, model building is easier and more reliable at higher resolution, though still possible even at lower resolutions (Böttcher et al., 1997; Conway et al., 1997; Zhou et al., 2001). Regardless of resolution, for building de novo backbone models, the density map must contain SSEs that can be clearly identified.
As demonstrated with the Rotavirus VP6 and Rice Dwarf Virus P8 capsid proteins examples presented here, models could be constructed from their respective density maps despite the difference in resolution. Even where loops were somewhat ambiguous in the case of P8, the presence of well-defined helices allowed for short loops to be built between the helices in the lower domain. However, in the larger β-sheet rich upper domain of P8, no clear path could be found due to the lack of resolution and identifiable anchor points. In VP6, which has ostensibly the same protein fold as P8 but nearly 3 Å better in resolution, the β-strands were clearly visible making construction of a complete model possible. It should also be noted that a potential complication in higher resolution (>4Å) structures is feature detection with SSEHunter, as the β-sheets begin to look like a series of parallel densities rather than a thin flat plate. As such, β-sheets at this resolution can simply be treated as a series of loops during modeling.
Conclusion
Model building requires a significant time investment and an understanding of density features at near-atomic resolutions. Size, complexity and quality of the density map all affect the model building process. Even the most experienced users may not be able to build a reliable model in poorly resolved regions of density maps. Therefore, the time and ease of building a de novo model is related to the map quality, motif of the target protein and experience of the user. Gorgon attempts to integrate and streamline the model building process in a user-friendly environment. Anecdotally, the GroEL model required approximately three months to construct by hand (Ludtke et al., 2008). With Gorgon, the construction of a similarly size Rotavirus VP6 took a single afternoon by an experienced Gorgon user.
Since Gorgon's initial release, it has grown to include numerous utilities dedicated to annotating subnanometer resolution density maps from cryo-EM and other structural methodologies. In addition to these utilities, walkthroughs, video tutorials, bug-tracking and biological/computational references have been added to the Gorgon website (http://gorgon.wustl.edu), which details the tools and approaches found in Gorgon. As the number of subnanometer resolution structures continues to grow, we anticipate that the Gorgon user-base will continue to grow rapidly.
Supplementary Material
SUPPLEMENTAL FIGURES
Figure S1. A short snipet of code from the main window viewer (A) and update checker using the Gorgon API (B) are shown.
SUPPLEMENTAL MOVIES
Supplemental Movie 1. Model fitting in Gorgon with the T. thermophilus 70S ribosome at 6.4Å resolution structure. 65 helices were identified in the 30S subunit using SSEHunter in Gorgon. Chain C was extracted from the crystal structure (3FIC, chain G) and fit to the density map using helix positions.
Supplemental Movie 2. SSEHunter in Gorgon with Rotavirus VP6 density map.
Supplemental Movie 3. SSE building in Gorgon with Rotavirus VP6 density map.
Supplemental Movie 4. SSE correspondence and model building in Gorgon with Rotavirus VP6 density map.
ACKNOWLEDGEMENTS
We would like to thank Dr. K. Murata for providing the density map for Aquaporin 1, (PDB ID: 1FQY). Additionally we would like to thank Brian Chen and Paul Heider for their contributions to the Gorgon source code. This research is supported by grants from NIH through the National Center for Research Resources (P41RR002250), National Institute of General Medical Science (R01GM079429) and National Science Foundation (IIS-0705644, IIS-0705474, IIS-0705538). MM was supported by a training fellowship from the Keck Center Biomedical Discovery Training Program of the Gulf Coast Consortia (NIH Grant No. 1 T90 DA022885-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abeysinghe S, Baker ML, Chiu W, Ju T. Semi-isometric registration of line features for flexible fitting of protein structures. Computer Graphics Forum (Proceedings of Pacific Graphics 2010) 2010. [DOI] [PMC free article] [PubMed]
- Abeysinghe S, Ju T, Baker ML, Chiu W. Shape modeling and matching in identifying 3D protein structures. COMPUT AIDED DESIGN. 2008a;40:708–720. [Google Scholar]
- Abeysinghe SS, Baker ML, Chiu W, Ju T. Segmentation-free skeletonization of grayscale volumes for shape understanding. IEEE International Conference on Shape Modeling and Applications. 2008b:63–71. [Google Scholar]
- Abeysinghe SS, Ju T. Interactive skeletonization of intensity volumes. The Visual Compter. 2009;25:627–635. [Google Scholar]
- Baker ML, Baker MR, Hryc CF, Dimaio F. Analyses of subnanometer resolution cryo-EM density maps. Methods Enzymol. 2010;483:1–29. doi: 10.1016/S0076-6879(10)83001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Wedemeyer WJ, Rixon FJ, Baker D, Chiu W. Ab initio modeling of the herpesvirus VP26 core domain assessed by CryoEM density. PLoS Comput Biol. 2006;2:e146. doi: 10.1371/journal.pcbi.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Ju T, Chiu W. Identification of secondary structure elements in intermediate-resolution density maps. Structure. 2007;15:7–19. doi: 10.1016/j.str.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Serysheva II, Sencer S, Wu Y, Ludtke SJ, Jiang W, Hamilton SL, Chiu W. The skeletal muscle Ca2+ release channel has an oxidoreductase-like domain. Proc Natl Acad Sci U S A. 2002;99:12155–12160. doi: 10.1073/pnas.182058899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Zhang J, Ludtke SJ, Chiu W. Cryo-EM of macromolecular assemblies at near-atomic resolution. Nat Protoc. 2010;5:1697–1708. doi: 10.1038/nprot.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmanns S, Rusu M, Wriggers W. Using Sculptor and Situs for simultaneous assembly of atomic components into low-resolution shapes. J Struct Biol. 2010 doi: 10.1016/j.jsb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- Bradley P, Malmström L, Qian B, Schonbrun J, Chivian D, Kim DE, Meiler J, Misura KMS, Baker D. Free modeling with Rosetta in CASP6. Proteins 61 Suppl. 2005;7:128–134. doi: 10.1002/prot.20729. [DOI] [PubMed] [Google Scholar]
- Chen DH, Baker ML, Hryc CF, Dimaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JZ, Settembre EC, Aoki ST, Zhang X, Bellamy AR, Dormitzer PR, Harrison SC, Grigorieff N. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci U S A. 2009;106:10644–10648. doi: 10.1073/pnas.0904024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhu J, Hui WH, Zhang X, Honig B, Fang Q, Zhou ZH. Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics. J Mol Biol. 2010;397:852–863. doi: 10.1016/j.jmb.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Baker ML, Jiang W, Dougherty M, Schmid MF. Electron cryomicroscopy of biological machines at subnanometer resolution. Structure. 2005;13:363–372. doi: 10.1016/j.str.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, et al. 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Natl Acad Sci U S A. 2010;107:4967–4972. doi: 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Ludtke SJ, Chiu W. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J Mol Biol. 2001;308:1033–1044. doi: 10.1006/jmbi.2001.4633. [DOI] [PubMed] [Google Scholar]
- Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- Jiang W, Li Z, Zhang Z, Booth CR, Baker ML, Chiu W. Semi-automated icosahedral particle reconstruction at sub-nanometer resolution. J Struct Biol. 2001;136:214–225. doi: 10.1006/jsbi.2002.4439. [DOI] [PubMed] [Google Scholar]
- Ju T, Baker ML, Chiu W. Computing a family of skeletons of volumetric models for shape description. Comput Aided Des. 2007;39:352–360. doi: 10.1016/j.cad.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 1997;277:525–545. doi: 10.1016/s0076-6879(97)77029-0. [DOI] [PubMed] [Google Scholar]
- Kong Y, Ma J. A structural-informatics approach for mining beta-sheets: locating sheets in intermediate-resolution density maps. J Mol Biol. 2003;332:399–413. doi: 10.1016/s0022-2836(03)00859-3. [DOI] [PubMed] [Google Scholar]
- Kong Y, Zhang X, Baker TS, Ma J. A Structural-informatics approach for tracing beta-sheets: building pseudo-C(alpha) traces for beta-strands in intermediate-resolution density maps. J Mol Biol. 2004;339:117–130. doi: 10.1016/j.jmb.2004.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baker ML, Chen D-HH, Song J-LL, Chuang DT, Chiu W. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Mathieu M, Petitpas I, Navaza J, Lepault J, Kohli E, Pothier P, Prasad BV, Cohen J, Rey FA. Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion. EMBO J. 2001;20:1485–1497. doi: 10.1093/emboj/20.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Miyazaki N, Taka J, Naitow H, Ogawa A, Fujimoto Z, Mizuno H, Higashi T, Watanabe Y, et al. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure. 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pollastri G, Przybylski D, Rost B, Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Morais MC, Leiman PG, Zhang W. Combining X-ray crystallography and electron microscopy. Structure. 2005;13:355–362. doi: 10.1016/j.str.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Glaeser R, Earnest T, Baumeister W. From words to literature in structural proteomics. Nature. 2003;422:216–225. doi: 10.1038/nature01513. [DOI] [PubMed] [Google Scholar]
- Sali A, Kuriyan J. Challenges at the frontiers of structural biology. Trends Cell Biol. 1999;9:M20–M24. [PubMed] [Google Scholar]
- Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- Sali A. NIH workshop on structural proteomics of biological complexes. United States: 2003. Structure. [UNKNOWN REFERENCE TYPE] [DOI] [PubMed] [Google Scholar]
- Schröder GF, Brunger AT, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette JC, Murphy FV, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert X, Navaza J. UROX 2.0: an interactive tool for fitting atomic models into electron-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2009;65:651–658. doi: 10.1107/S0907444909008671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tama F, Miyashita O, Brooks CL. Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. J Struct Biol. 2004;147:315–326. doi: 10.1016/j.jsb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Topf M, Baker ML, John B, Chiu W, Sali A. Structural characterization of components of protein assemblies by comparative modeling and electron cryo-microscopy. J Struct Biol. 2005;149:191–203. doi: 10.1016/j.jsb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Topf M, Baker ML, Marti-Renom MA, Chiu W, Sali A. Refinement of protein structures by iterative comparative modeling and CryoEM density fitting. J Mol Biol. 2006;357:1655–1668. doi: 10.1016/j.jmb.2006.01.062. [DOI] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular Dynamics Flexible Fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- Yu X, Jin L, Zhou ZH. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Baker ML, Schröder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, et al. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010a;463:379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010b;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr Opin Struct Biol. 2008;18:218–228. doi: 10.1016/j.sbi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Baker ML, Jiang W, Dougherty M, Jakana J, Dong G, Lu G, Chiu W. Electron cryomicroscopy and bioinformatics suggest protein fold models for rice dwarf virus. Nat Struct Biol. 2001;8:868–873. doi: 10.1038/nsb1001-868. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 A. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- Zhu J, Cheng L, Fang Q, Zhou ZH, Honig B. Building and Refining Protein Models within Cryo-electron Microscopy Density Maps Based on Homology Modeling and Multiscale Structure Refinement. J Mol Biol. 2010;397:835–851. doi: 10.1016/j.jmb.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL FIGURES
Figure S1. A short snipet of code from the main window viewer (A) and update checker using the Gorgon API (B) are shown.
SUPPLEMENTAL MOVIES
Supplemental Movie 1. Model fitting in Gorgon with the T. thermophilus 70S ribosome at 6.4Å resolution structure. 65 helices were identified in the 30S subunit using SSEHunter in Gorgon. Chain C was extracted from the crystal structure (3FIC, chain G) and fit to the density map using helix positions.
Supplemental Movie 2. SSEHunter in Gorgon with Rotavirus VP6 density map.
Supplemental Movie 3. SSE building in Gorgon with Rotavirus VP6 density map.
Supplemental Movie 4. SSE correspondence and model building in Gorgon with Rotavirus VP6 density map.