Abstract
Glaucoma is one of the most common neurodegenerative diseases. Despite this, the earliest stages of this complex disease are still unclear. This study was specifically designed to identify early stages of glaucoma in DBA/2J mice. To do this, we used genome-wide expression profiling of optic nerve head and retina and a series of computational methods. Eyes with no detectable glaucoma by conventional assays were grouped into molecularly defined stages of disease using unbiased hierarchical clustering. These stages represent a temporally ordered sequence of glaucoma states. We then determined networks and biological processes that were altered at these early stages. Early-stage expression changes included upregulation of both the complement cascade and the endothelin system, and so we tested the therapeutic value of separately inhibiting them. Mice with a mutation in complement component 1a (C1qa) were protected from glaucoma. Similarly, inhibition of the endothelin system with bosentan, an endothelin receptor antagonist, was strongly protective against glaucomatous damage. Since endothelin 2 is potently vasoconstrictive and was produced by microglia/macrophages, our data provide what we believe to be a novel link between these cell types and vascular dysfunction in glaucoma. Targeting early molecular events, such as complement and endothelin induction, may provide effective new treatments for human glaucoma.
Introduction
Recently developed genomic technologies promise a deeper understanding of human diseases, but challenges remain. Despite great advances in gene expression studies, the large volume of data make it difficult to identify specific molecular changes that are relevant to a disease of interest. It also has been difficult to distinguish primary versus secondary changes. The challenges are especially great for complex diseases such as glaucoma that involve multifactorial genetic and environmental contributions and asynchronous changes within affected tissues. Particularly little progress has been made in identifying the earliest processes in these complex diseases.
Glaucoma is a heterogeneous group of diseases involving retinal ganglion cell (RGC) loss and degeneration of the optic nerve. Glaucoma is one of the most common neurodegenerative diseases, affecting an estimated 70 million people (1). Elevated intraocular pressure (IOP) is a major risk factor for glaucoma, but disease outcome is affected by many other factors whose molecular effects are not well defined (reviewed in ref. 2). Current treatments for glaucoma aim to lower IOP but are not always effective and have various associated side effects (3–5). To improve patient care, more treatments are needed. A key step toward developing new treatments is determining the primary molecular mechanisms of glaucoma pathogenesis.
Models for studying glaucoma exist in different species and are instrumental in advancing understanding of disease processes. Studies of humans and non-human primates established that early RGC damage occurs in the axon segment in the optic nerve head (ONH) (6–14). The same is true in glaucomatous DBA/2J mice, where RGC axons are insulted in the optic nerve (15). While it is clear that direct axonal injury occurs in glaucoma, other insults to different compartments of the RGC may occur (including cell body, axon, dendrites, and synapses) (2). It is not clear whether early disease-specific changes in the retina are dependent or independent of the initial insult(s) in the ONH. For the optic nerve and retina, the temporal order of specific events and the potential roles of non-neuronal cell types including various types of glial cells, immune cells, and endothelial cells require further characterization.
Experimentally induced models of glaucoma (16–18) can provide a degree of control that should facilitate identification of early glaucoma changes in the ONH and retina. However, the method of inducing the glaucoma varies and may impact outcomes, with findings in a particular model not being reproduced in other models (19–21). Inherited models, such as DBA/2J mice, are valuable because they have features of the human disease including age-related, variable onset of IOP elevation, progressive optic nerve axon damage, and RGC loss. However, the asynchronous and variable nature of these models makes it particularly difficult to identify early molecular events and to temporally stage changes.
Gene expression profiling is a powerful tool for assessing molecular changes associated with disease. Microarray studies have previously evaluated various induced and inherited models of glaucoma and have highlighted molecules and pathways for further mechanistic evaluation (22–27). Although important, none of these studies was specifically designed to assess and temporally stage early molecular changes that precede detectable RGC and axon loss. Additionally, none assessed changes in both the retina and ONH of the same eye.
Clustering genes in microarray datasets according to their expression profiles is a powerful approach that has identified molecular signatures that predict cancer progression (28, 29). Clustering methods group the most similar samples based on the expression profiles of many relevant genes. Comparing clustered groups of samples enables identification of a greater number of expression differences and more sensitive interrogation of disease pathways than comparing nonclustered groups — a feature we exploit in our current study. As demonstrated here, clustering allowed identification of many small but important gene expression changes between disease states, and thus can provide valuable insight into complex disease processes. If only large expression differences are considered, these insights would be missed. Clustering of samples based on gene expression data has not been previously reported for glaucoma.
Here, we present the use of microarray-generated gene expression data, and a series of clustering methods to identify early molecular stages of glaucoma. We generated gene expression profiles for a set of eyes from the DBA/2J strain of mice, which is widely used as an inherited, later-onset, complex, and chronic model of glaucoma. Mutations in two genes (Gpnmb and Tyrp1) result in IOP elevation and subsequent glaucoma in this strain (30). To study the timing of and relationships between events in the retina and ONH, we assessed both tissues from each eye as separate samples. We analyzed the eyes in two separate ways. First, we identified differentially expressed (DE) genes by comparing eyes grouped based on the extent of optic nerve damage (referred to as morphologically defined groups). Comparison of samples grouped by optic nerve damage did not allow for sensitive detection of the early molecular events that precede detectable axon damage. To identify these early molecular changes, we compared eyes that were grouped based on the similarity of gene expression profiles for either the ONH or the retina. These eyes had been grouped using unbiased hierarchical clustering. The analyses identified early molecularly defined stages of glaucoma. Eyes in these stages were previously indistinguishable from control eyes based on optic nerve damage. Early events include activation of the complement cascade and upregulation of endothelin 2 (Edn2) in microglia/macrophages. Separately disrupting either of these two pathways was profoundly protective from glaucoma in DBA/2J mice.
Results
Genome-wide assessment of gene expression changes was performed in DBA/2J mice. The ONH and retina from 40 DBA/2J eyes at 10.5 months of age were separately profiled. These eyes were selected because they encompassed a range of glaucoma severity (based on a previously validated method for determining the degree of optic nerve damage; see Methods). Selected eyes had the following degrees of damage: no detectable axon damage compared with controls (no or early [NOE]); 10%–50% of axons lost (moderate [MOD]), and greater than 50% axons lost (severe [SEV]). As the main focus of this study was to identify early molecular events in glaucoma, we included 21 eyes with NOE glaucoma (no detectable damage), 9 eyes with MOD glaucoma, and 10 eyes with SEV glaucoma. To keep numbers of eyes in each group relatively balanced, the 21 NOE eyes were divided into two independent groups (NOE1 n = 10, and NOE2 n = 11). Two control groups were also included: 10 eyes from 10.5-month-old DBA/2J-Gpnmb+/Sj (D2-Gpnmb+) mice (age and strain matched, no-glaucoma control) and 10 eyes from 4.5-month-old DBA/2J mice (young, pre-glaucoma control).
Initially, pairwise comparisons were performed between the 4 morphologically defined groups (NOE1, NOE2, MOD, and SEV groups) and the D2-Gpnmb+ control group (dataset 1, Figure 1A). For the ONH, 223 and 456 probe sets were DE (q ≤ 0.05) in the NOE1 and NOE2 groups, respectively, compared with control. Importantly, the vast majority of probes DE by at least 2-fold were the same in these independent groups. The two most DE genes were lipocalin 2 (Lcn2; DE 8.2-fold NOE1 and 12.7-fold NOE2) and orosomucoid 1 (Orm1; DE 5.8-fold NOE1 and 6.1-fold NOE2), two immunomodulatory genes. For the retinas of these same eyes, only 7 (NOE1) and 24 (NOE2) probe sets were DE compared with controls, including glial fibrillary acidic protein (Gfap; 2.1-fold NOE2) a marker of reactive astrocytosis. Many more probe sets were DE in the MOD and SEV groups, including decreased detection of genes expressed in RGCs (e.g., Sncg, –1.5-fold MOD and –6.3-fold SEV; Nefl, –1.7-fold MOD and –11.0-fold SEV; and Thy1, –1.3-fold MOD and –3.0-fold SEV).
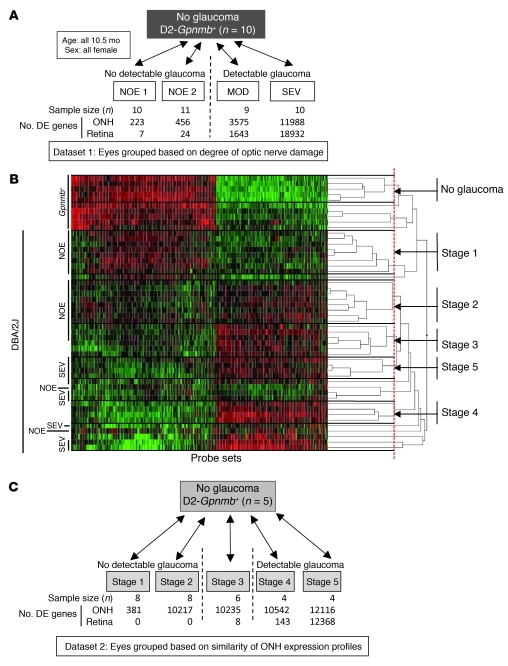
Figure 1. Early stages of glaucoma identified by hierarchical clustering.
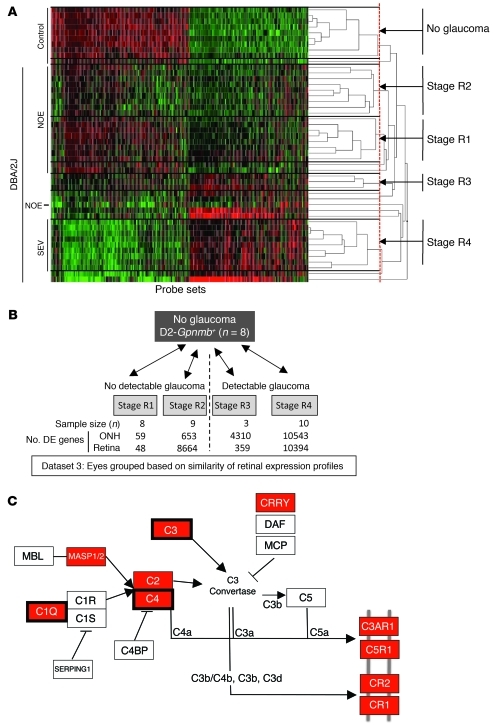
(A) Pairwise comparisons for eyes grouped by conventional morphological assessment of optic nerve damage (Dataset 1). The numbers of DE genes compared with controls are shown for both the ONH and retina. NOE eyes had no detectable glaucoma; MOD and SEV eyes had readily detectable damage and significant axon loss (Supplemental Figure 1). A total of 40 DBA/2J eyes were compared with the 10 controls. (B) A glaucoma-relevant probe set (see Methods and Supplemental Figure 1) was used to cluster the same eyes into molecular stages based on expression profiles of ONH samples. The expression data for all 50 eyes are shown. Five molecularly defined stages containing at least 4 eyes were identified (ONH stages 1–5). The optic nerve damage level is indicated on the left for control, NOE, and SEV eyes. All unlabeled eyes had MOD glaucoma. Black horizontal lines indicate borders between stages. The red line indicates the cutoff level for inclusion in a stage. Raw normalized intensity values are represented as green to black to red. For each probe set, green represents the lower normalized intensity values across all eyes and red the higher intensity values. All normalized intensity values for each probe set are available in Geo DataSets (GSE26299). (C) Pairwise comparisons for these molecular stages (Dataset 2). Stage 3 contains 3 eyes with no detectable glaucoma and 3 eyes with detectable glaucoma. The numbers of DE genes for the ONH and retina for each stage are shown.
Hierarchical clustering allows early disease stages to be identified.
Comparison of eyes grouped by conventional morphological criteria did not allow for sensitive detection of early gene expression changes. We reasoned that grouping the same set of eyes by molecular clustering of gene expression profiles and then comparing these groups would allow more sensitive detection of early changes in glaucoma. Since a critical insult and early damage are known to occur in the optic nerve, clustering was first used to group eyes based on the similarity of gene expression profiles in the ONH. Unbiased hierarchical clustering of the 40 DBA/2J ONH samples and 10 D2-Gpnmb+ control samples was performed using the gene expression values of 570 disease-relevant probe sets (Methods and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI44646DS1). Hierarchical clustering first linked the most similar eyes, and continued until all eyes were linked in a dendrogram (Figure 1B and Methods). As the least similar eyes were linked at the highest branch points, a cutoff (threshold of relatedness) was applied to separate groups of eyes into independent disease stages (Methods). We selected a cutoff that made biological sense based on conventional morphological criteria, e.g., Gpnmb+ controls were grouped into their own stage, and SEV eyes were separate from NOE eyes. We only considered groups containing at least 4 eyes as a stage. Using these criteria, 5 new, molecularly defined, stages of glaucoma were identified (dataset 2, stage 1 to stage 5; Figure 1, B and C). Not surprisingly, 10 of the 40 eyes did not fit into these 5 stages. This reflects the large number of possible molecular states for such a variable disease.
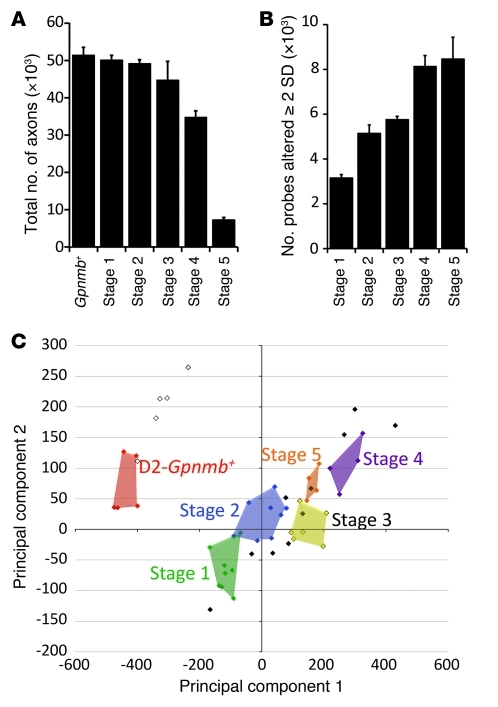
The 5 stages were ordered based on two criteria: (a) Number of DE genes compared with no-glaucoma control: stage 1 had the smallest number of DE genes when stages 1–5 were compared with the Gpnmb+ control group (381 DE genes, Figure 1C) and was considered to be the earliest molecular stage. (b) Previously determined morphological damage: although stages 2–4 had a similar number of DE genes (approximately 10,000), only stages 1 and 2 contained eyes with no detectable glaucoma (NOE). In comparison, stage 3 contained 3 eyes with NOE glaucoma and 3 eyes with MOD glaucoma. Stage 4 contained 4 eyes with MOD glaucoma. Therefore, stage 3 was determined to be between stage 2 and stage 4. Stage 5 contained only eyes with SEV glaucoma. As expected, axon number decreased with increasing glaucoma stage (Figure 2A). Also, the number of genes whose expression value was DE by at least 2 SDs from the average of Gpnmb+ controls increased from stages 1 to 5 (Spearman’s correlation 0.9, P < 10–100) (Figure 2B).
Figure 2. Differences between the molecularly defined ONH stages.
(A) Overall, axon number decreased, from stage 1 to 5. However, for stages 1–3, there was no significant axon loss with respect to D2-Gpnmb+ control eyes (P > 0.1). (B) The number of probe sets altered by at least 2 SDs (with respect to D2-Gpnmb+ control) increased with disease stage. (C) PCA showed that stages 1–5 occupy essentially non-overlapping territories. D2-Gpnmb+ control samples that did not cluster (open diamonds, upper left quadrant) were clearly distinct from DBA/2J glaucoma samples (filled colored diamonds). DBA/2J samples that did not cluster are shown as filled black diamonds.
Based on optic nerve damage, 3 of the 6 eyes in stage 3 were indistinguishable from no-glaucoma control eyes. The other 3 eyes had MOD glaucoma (Supplemental Figure 2). This indicates that the molecular changes determining the similarity of eyes in stage 3 must have occurred prior to detectable glaucomatous damage. In addition to RGCs, these early molecular changes may be occurring in a variety of other cell types in the ONH, such as astrocytes, microglia, and endothelial cells.
Principal component analysis (PCA) is a powerful method for reducing the dimensionality of data and is used to facilitate microarray experiments (31). Here, PCA was used to independently assess whether the 5 identified molecular stages are distinct. The reduced dimensional position of the 40 DBA/2J and 10 control eyes was plotted using the first two principal component vectors calculated for the 570 disease-relevant probe sets (see Methods). As expected, eyes in each of the 5 molecular stages occupied essentially distinct territories (P < 0.01, Figure 2C). Therefore, PCA supported the hierarchical clustering.
Molecularly defined ONH stages increase sensitivity to detect DE genes in the ONH.
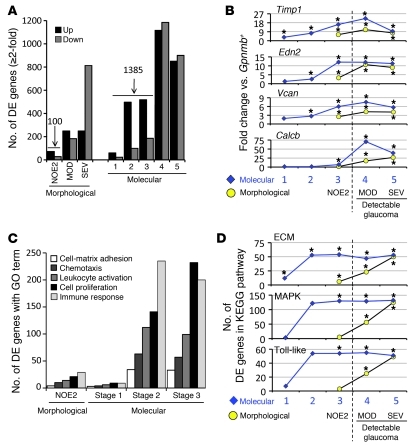
To identify DE genes, molecularly defined stages 1–5 were compared with the Gpnmb+ control group (Figure 3). DE genes were defined as being those with a q value (false discovery rate [FDR]) of 0.05 or less. Significantly more DE genes were detected in the ONH for the molecularly defined stages than for the morphological groups (Figure 3A). Considering groups with eyes that had no detectable optic nerve damage and only probe sets that were DE at least 2-fold, 1,385 probe sets were DE in molecularly defined stages 1–3 versus 100 in morphologically defined NOE2 eyes (Figure 3A) and 55 in NOE1 eyes (data not shown). Additionally, many specific genes, such as Timp1, Edn2, Vcan, and Calcb, had a greater fold change in the molecularly as compared with morphologically defined stages (Figure 3B).
Figure 3. Clustered stages allow sensitive detection of early changes in the ONH.
(A) The number of DE genes in morphologically defined dataset 1 was compared with the number of DE genes in molecularly defined dataset 2. 1,385 genes were DE at least 2-fold compared with controls in molecularly defined stages 1–3. This is in comparison to only 100 genes for the NOE2 group. Importantly, the expression patterns that define stages 1–3 represent changes that occur prior to detectable morphological damage. (B) The fold changes for individual genes were often 2 or 3 times greater for the molecularly compared with morphologically defined groups. For this figure, stage 3 is included to the left of the line indicating detectable glaucoma. This is because the gene expression differences that determine inclusion in this stage occur prior to detectable optic nerve damage. (C) The molecularly defined stages allow identification of more DE genes with a particular GO term than the morphologically defined data. The expression of genes with immune-modulating functions, including chemotaxis and leukocyte activation, change early in the ONH. (D) KEGG pathway analysis suggests that ECM-receptor interactions, MAPK signaling, and Toll-like receptor signaling pathways are among the earliest pathways to become activated. *q ≤ 0.05 compared with Gpnmb+ controls.
ECM and immune changes in the ONH.
We used a variety of software-based annotation tools to provide functional insight into the ONH expression differences detected in this study (see Methods). As it is not possible to show all analyses in this article, we highlight a few enriched gene ontology terms, pathways, and networks that are likely to be important during early stages of glaucoma.
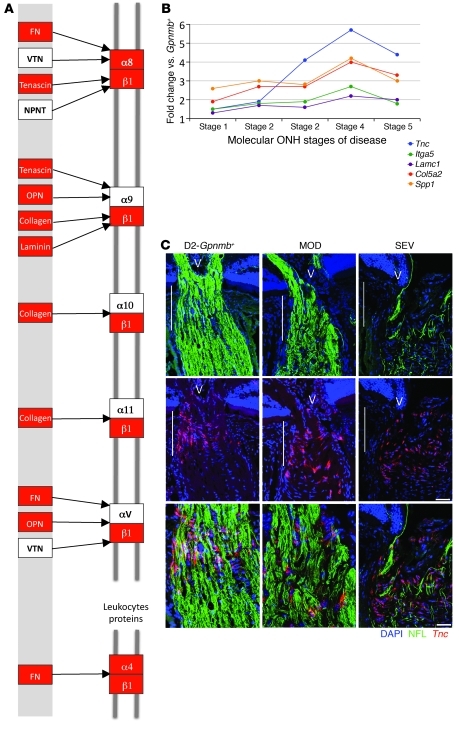
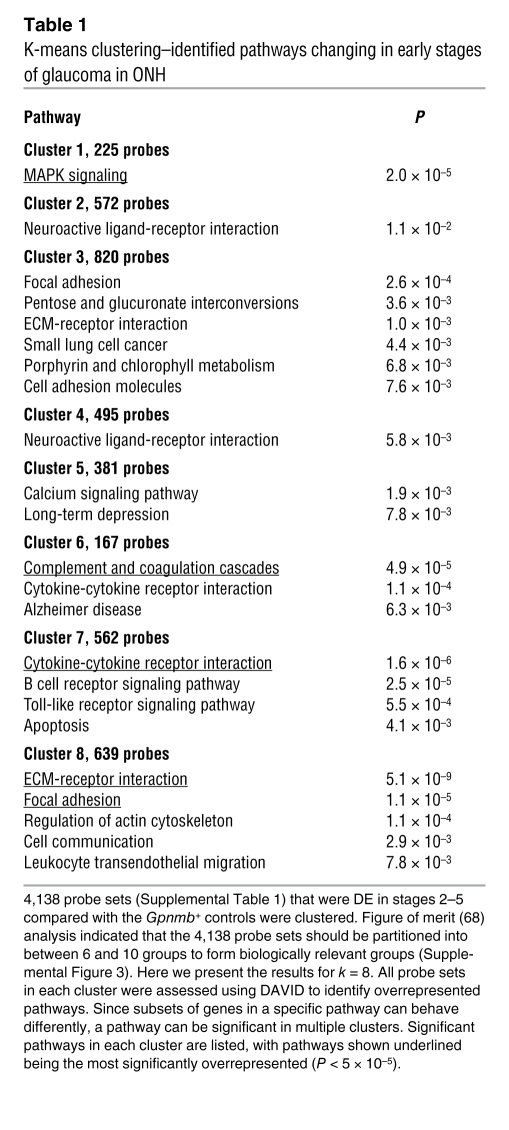
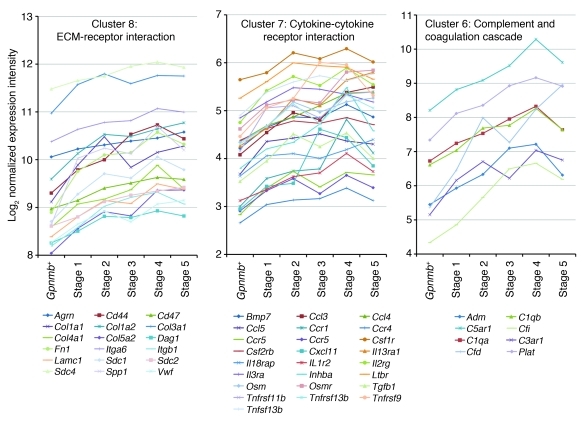
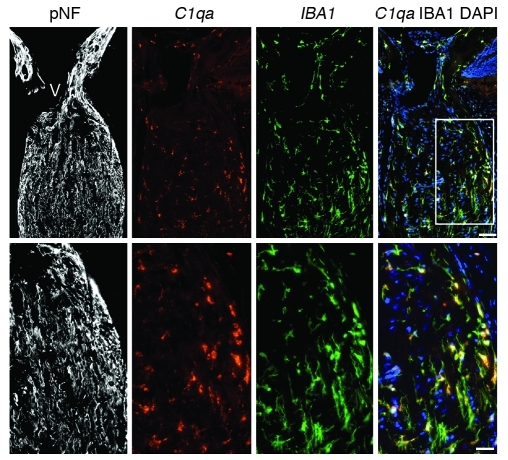
Gene ontology (GO) analysis (performed by DAVID; see Methods) revealed that a significant number of genes with the GO terms “immune response” (Gene Ontology Database 0008283), “leukocyte activation” (GO 0045321), and the related term “chemotaxis” (GO 0006935) were DE in the early molecularly defined stages of disease (Figure 3C). The majority of these changes were not detected in the morphologically based NOE groups. Genes belonging to pathways that are known to affect these same processes were also identified by independent Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Figure 3D). Two related KEGG pathways that were significantly overrepresented are “ECM-receptor interactions” (KEGG mmu04512; Figure 4, A and B) and “focal adhesions” (KEGG mmu04510). Neither of these pathways was found to be important in either of the NOE groups or the MOD group (dataset 1). ECM-receptor interactions are mediated by transmembrane molecules such as integrins and proteoglycans and lead to direct or indirect control of cellular activities that include adhesion, migration, and proliferation. These pathway changes may represent early changes to the glial-vascular unit or altered interactions between glia and neurons in the ONH. One member of this pathway, tenascin C (Tnc) is 1.9-fold elevated in stage 2, 4.1-fold elevated in stage 3, and 5.7-fold elevated in stage 4 (Figure 4B) and is localized to the glial lamina region of the ONH (Figure 4C). Showing the human relevance of our findings, expression of TNC was increased in glaucomatous compared with normal human ONHs (32). The importance of ECM-receptor interactions is also supported by computationally distinct k-means clustering, which groups genes that behave similarly across disease stages (Table 1 and Figure 5). K-means clustering also suggested that the complement cascade is an important pathway activated early in the ONH. C1qa is a component of the C1q complex, an early effector in the complement cascade. C1QA is expressed in microglia/macrophages in the ONH during early stages of glaucoma (Figure 6).
Figure 4. Genes in the ECM-receptor interaction pathway are upregulated in the ONH.
(A) DE genes in the ECM-receptor interaction pathway that are upregulated in stage 2 are shown in red. These genes remain upregulated through stage 5. Reproduced with permission from KEGG ( http://www.genome.jp/kegg/kegg1.html) (B) Fold changes for 5 genes within the ECM pathway. Lamc1, Col5a2, and Spp1 are DE (q ≤ 0.05) across all stages. Itga5 and Tnc are DE across stages 2–5. (C) RNA in situ hybridization for Tnc, the most highly expressed gene in the ECM-receptor interaction pathway. Tnc is largely expressed in the glial lamina (indicated by vertical white lines) of the ONH and is likely present in astrocytes. V, vessel; NFL, neurofilament (detected with an antibody; see Methods). Scale bars: top 2 rows, 50 μm; bottom row, 20 μm.
Table 1 .
K-means clustering–identified pathways changing in early stages of glaucoma in ONH
Figure 5. K-means clustering identified pathways changing in early stages of glaucoma.
K-means clustering was used to partition genes that change in the ONH into clusters (Table 1). Genes within a cluster behaved in a similar fashion as glaucoma progressed. Gene expression patterns in 3 overrepresented pathways are shown. Profiles are based on normalized raw expression values across D2-Gpnmb+ control and glaucoma stages 1–5.
Figure 6. C1qa expression in microglia in the ONH.
C1qa expression (red, riboprobe) occurs in microglia (IBA1 marker, green, antibody) in the ONH during DBA/2J glaucoma. All panels show the same eye, which had no detectable optic nerve damage (NOE). The boxed area is enlarged in the lower panels. Phosphorylated neurofilament (pNF) labels healthy axons throughout the ONH. DAPI labels nuclei. Scale bars: top panels, 50 μm; bottom panels, 20 μm. Supplemental Figure 3 shows other complement genes that are DE in the ONH.
Hubs and transcription factors.
Ingenuity pathway analysis (IPA) identifies molecular networks and biological pathways that are overrepresented in a given set of DE genes (see Methods). IPA allows the identification of key genes (or hubs) that lie at the center of any network or subnetwork. Therapeutic modulation of any hub has strong potential to impact a variety of network participants, making hubs attractive targets for future neuroprotective efforts. We separately analyzed the DE genes for stages 1–5 (dataset 2) by IPA. One of the most statistically significant networks included various members of the ECM-receptor interaction pathway and proinflammatory cytokines (Supplemental Figure 3A). Hubs within this network include IL-1β (encoded by Il1b, shown as IL1 in IPA network) and integrin β3 (Itgb3), molecules that are reported to affect ECM composition and metabolism (33–35). The network also includes various other integrins, fibronectin 1 (Fn1), matrix metalloproteinase 19 (Mmp19), tissue inhibitors of matrix metalloproteinases (Timp1 and Timp2), and a number of collagens. Of interest, caspase-1 (Casp1) was upregulated in the earliest glaucoma stage and continued to be increased at later stages (Supplemental Figure 3B). CASP1 has interleukin-converting enzyme activity, and its increase preceded the upregulation of Il1b and Itgb3 genes (Supplemental Figure 3B). Thus, proinflammatory responses are among the earliest changes in the ONH in glaucoma.
Clustering identifies early stages of glaucoma in the retina.
Clustering eyes based on morphological criteria (dataset 1) or the similarity of ONH gene expression profiles (dataset 2) was not sensitive for identifying early changes in the retina. No DE genes were identified in the retina in molecular stages 1 and 2, and only 8 DE genes were identified in stage 3 (Figure 1C). This may be because early changes in the ONH occur prior to changes in the retina, and/or because our sensitivity to detect retinal changes was lower as whole retinas were used (<5% of total retinal cells are RGCs). Alternatively, it is possible that grouping the eyes based on ONH expression patterns is not a sensitive way to determine early glaucoma changes in the retina. It is possible that early expression changes in the retina are completely or partially independent of events in the optic nerve. Thus, we separately clustered the same set of 40 DBA/2J eyes based on the similarity of their retinal gene expression profiles (dataset 3). We used the same methods as were used for the ONH-based clustering and similar criteria for determining stages (see Methods). Retinal clustering divided the eyes into a Gpnmb+ control cluster and 4 glaucoma stages (R1–R4) that made biological sense (dataset 3, Figure 7, A and B). Stages R1 and R2 contained only eyes that were previously indistinguishable from each other and controls by conventional morphological criteria. Retinal clustering increased the sensitivity of detecting DE genes at early stages of disease (Figure 7B). Compared with the D2-Gpnmb+ control stage, 48 probe sets were DE in stage R1 and 8,664 in stage R2. Many fewer DE genes were detected in the ONH of the eyes in these retinal clusters (59 DE genes, dataset 3, stage R1, compared with 381 DE genes, dataset 2, stage 1; Figure 1C). Since clustering using either the ONH or retina tissues is not sensitive at detecting early DE genes in the other tissue, our analyses suggest that events in the retina and ONH occur asynchronously and are not completely interdependent.
Figure 7. Molecular clustering identifies early stages of glaucoma in the retina.
(A) 600 glaucoma-relevant probe sets that were DE in retina were used to cluster samples into molecularly defined stages (retinal stages R1–R4). 48 of the 50 eyes in the study were clustered (quality control failed for 2 retinal microarrays; see Methods). Expression data for all 48 eyes are shown (normalized intensity values have been submitted to GEO). Although eyes are grouped based on the similarity of gene expression, the damage level based on morphological analysis is indicated on the left for control, NOE, and SEV eyes. Unlabeled eyes had MOD glaucoma. Black horizontal lines indicate borders between stages. The red line indicates the cutoff level for inclusion in a stage. (B) Pairwise comparisons for the new molecularly defined retinal stages. The number of DE genes for ONH and retina for each stage are shown. Importantly, stages R1 and R2 were previously indistinguishable, with no detectable glaucomatous optic nerve damage. Compared with the other stages, stage R3 lacks power, as it contains 3 eyes. (C) Pathway analysis (using DAVID, see Methods) of the DE genes in stage R1 and R2 (compared with control) identified the complement cascade as the most significantly overenriched pathway (KEGG mmu04610). Genes shown in red have greater expression in molecularly defined retinal stage R2 than Gpnmb+ controls. Three of these genes were DE in stage R1 (surrounded by thick black boxes).
Analyses of the 48 DE probe sets in stage R1 identified the complement cascade as the only significantly overrepresented pathway (P = 5 × 10–4). The DE genes in stage R1 included C1qa, C1qb, and C1qc. In stage R2, 12 members of the complement cascade were DE (Figure 7C). We have found that C1qa is expressed in RGCs and localizes to synapses in the retina during early stages of glaucoma (36). The complement cascade was also implicated in the ONH by k-means clustering (Table 1 and Figure 5, cluster 6), where it was expressed in microglia (Figure 6).
Early DE genes provide molecular markers to assess glaucoma status.
Genes shown to be DE early in glaucoma may allow the development of marker sets to group eyes with no detectable optic nerve damage into those undergoing early glaucoma and those with no glaucoma. Such markers may also be used to determine the degree of glaucoma or to assess the effectiveness of tested treatments. Since many molecular changes occur in glaucoma and specific changes will vary from eye to eye, a reasonable approach is to assess genes from different pathways and to track the number of DE genes for each of these pathways as a measure of glaucoma status.
To assess the feasibility of this approach, we assessed genes from pathways that were activated early in the ONH or retina in the microarray study (Supplemental Tables 2 and 3). These pathways included ECM-receptor interactions, MAPK signaling, and Toll-like receptor signaling for the ONH tissue and the complement cascade for the retina tissue. We assessed completely new sets of 10-month-old DBA/2J and D2-Gpnmb+ control eyes. As we were most interested in eyes with no conventionally detectable glaucoma, we separately assessed the ONH and retina for 21 NOE and 3 MOD DBA/2J eyes, as well as 11 D2-Gpnmb+ control eyes. To further refine the location of retinal changes compared with the microarray study, the retinal tissue was enriched for the RGC layer and lacked the inner nuclear layer and photoreceptor layers (see Methods). For each eye, a gene was classified as DE only if the normalized expression level was at least 2 SDs from the average of the 11 control eyes. This approach ensured that only robust expression changes were regarded as DE. For each eye, we determined the total number of DE genes as well as the number of DE genes in each pathway (Supplemental Figure 5). A pathway was considered “activated” in eyes that had at least 2 times the number of DE genes for that pathway relative to any of the controls.
In both ONH and retina, this analysis clearly distinguished the vast majority of eyes with NOE glaucoma from controls (Supplemental Figure 5A). Individual eyes had differing numbers of DE genes, with eyes classified with MOD glaucoma having the greatest total number of DE genes. For the ONH, ECM-receptor interactions, MAPK signaling, and Toll-like receptor signaling pathways were activated in 18 of the 21 eyes with NOE glaucoma and all 3 eyes with MOD glaucoma. For the retina, the complement cascade was activated in 13 of 21 NOE eyes and 3 of 3 eyes with MOD glaucoma. In the NOE eyes, there was no clear relationship between the number of DE genes in the ONH and retina. Most NOE eyes had high numbers of DE genes in the ONH, with lower numbers activated in the retina (16 of 21 having ≥60% the number of genes activated in moderate for ONH compared with only 3 of 21 for retina). However, the reverse was true in some eyes. Most strikingly, eye number 3 had a large number of DE genes in the retina (similar to eyes with MOD glaucoma), but a low number in the ONH (Supplemental Figure 5). This further suggests that early changes in the retina and ONH are not completely dependent on each other and can occur asynchronously. These marker genes (Supplemental Table 2), along with approaches for assessing glaucoma status in individual eyes, are likely to be valuable for future studies.
C1QA-deficient mice are protected from glaucoma.
Changes in the complement cascade have been reported in animal models and human glaucoma. However, it has not been determined whether these changes occur very early and prior to detectable axon loss, or if they are later and possibly secondary (22, 24, 36, 37). Our current study demonstrates that complement cascade changes occur very early and prior to detectable glaucoma by conventional assays, with complement genes being among the first genes to change in the retina (Figure 7C). Given the very early occurrence of these changes and the prior association of complement cascade changes with human glaucoma, we tested the importance of this cascade in DBA/2J mice.
To test the functional importance of the complement cascade in this inherited glaucoma, we generated and analyzed mice with a mutation in the C1qa gene (Figure 8). Clinical examinations of mice from 6.0 to 12.0 months of age assessed the severity of the pigment-dispersing iris disease, which precedes the glaucoma (30, 38). Both C1QA-deficient and wild-type DBA/2J mice developed prominent pigment dispersion and iris atrophy with a similar age of onset, but progression to the most severe iris stage was slightly delayed in C1QA-deficient mice. Subsequent to the iris disease and despite a modest delay in some C1QA-deficient DBA/2J eyes, IOP became elevated in mice of each genotype, and their IOP distributions overlapped extensively (Figure 8A). We previously detected an alteration of IOP similar to that in the C1qa mutants in BAX-deficient DBA/2J mice (39). In these Bax mutants, this IOP change did not profoundly alter the degree of optic nerve damage.
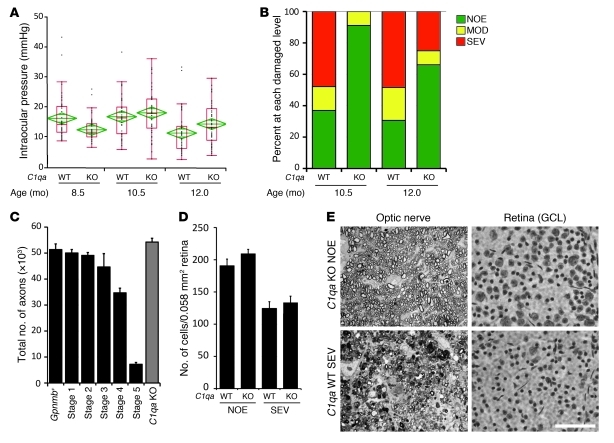
Figure 8. C1QA deficiency protects against DBA/2J glaucoma.
(A) Despite a delay in IOP elevation in some eyes, IOPs of C1qa WT and homozygous mutant (KO) mice overlap extensively (WT versus KO: 8.5 months, P = 0.026; 10.5 months, P = 0.34). Number of assessed eyes was at least 43 for all groups (B) The vast majority of optic nerves from C1qa mutant mice are indistinguishable from control nerves with no detectable glaucoma (NOE, green bars) at 10.5 months of age, and retain a lower glaucoma incidence than WT mice at 12 months of age. Sample sizes: 46 WT, 45 KO at 10.5 months; 62 WT, 56 KO at 12 months. P < 1 × 10–8 at each age comparing KO with WT distributions. (C) C1qa KO eyes with NOE nerve damage (n = 10) have axon number similar to that of no-glaucoma Gpnmb+ controls (n = 11, P = 0.26). For ease of comparison, axon counts for stages 1–5 are repeated from Figure 1. (D) In the ganglion cell layer (GCL), RGC somata are also spared in C1qa mutants with NOE nerve damage, but the number of RGC soma lost in KO eyes with severe optic nerve damage was similar to that in WT counterparts. Amacrine cells make up the majority of remaining GCL cells in severely affected eyes. (E) The most common optic nerve and GCL phenotypes for 12-month-old mice of each genotype are shown. Optic nerve cross sections were stained with PPD and flat mounted retinas stained with cresyl violet. Scale bar: 50 μm.
C1qa mutant mice were protected from glaucomatous damage to the optic nerve and retina at both 10.5 and 12.0 months of age (Figure 8B), two key ages in this strain (40). Importantly this protection was very profound at 10.5 months of age, with only 9% of mutant eyes having detectable glaucoma compared with 63% of wild-type eyes (P = 5 × 10–100, Figure 8B). None of the mutant eyes had SEV glaucoma, while 48% of wild-type eyes were severely affected. At 12.0 months of age, only 25% of mutant eyes had SEV glaucoma compared with 48% of wild-type eyes. The percentage of eyes with no detectable glaucoma was more than doubled in C1qa mutants at 12.0 months (66% versus 30%, P = 2.2 × 10–9). Protection in C1qa mutant mice was confirmed by axon and soma counts (Figure 8, C–E). Thus, although future experiments are required to completely define the roles of C1QA in ocular drainage and neural tissues during DBA/2J glaucoma, it is likely to have an important role in glaucomatous neurodegeneration. Irrespective of the relative role in different tissues, our findings indicate that complement pathway inhibition has potential as a potent treatment to protect against glaucoma.
Inhibiting the endothelin system protects from glaucoma in DBA/2J mice.
Our current study demonstrates that the Edn2 gene is upregulated in both the retina and the ONH very early in glaucoma (Figure 3B). The endothelin system was previously found to be upregulated in human and animal models of glaucoma (26, 41–43), but our study indicates that the upregulation of Edn2 is a very early event. EDN2 localized to microglia/macrophage-like cells in the nerve fiber layer of DBA/2J retinas (Figure 9, A–D).
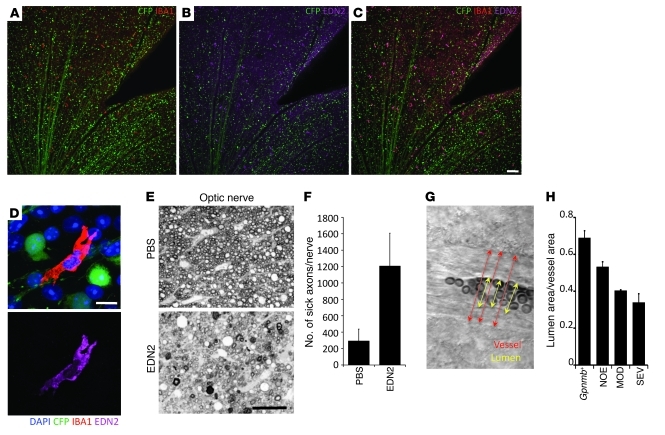
Figure 9. EDN2 is expressed in retinal microglia and may mediate early vascular damage in glaucoma.
(A–C) Images of a retina from a glaucomatous DBA/2J eye with fluorescently marked RGCs (D2.Thy1-CFP mice; RGCs and their axons are green). EDN2 (magenta) is present in IBA1-positive cells (red) in the RGC and nerve fiber layers. IBA1 is a marker of microglia and macrophages. (D) High-resolution images of an EDN2-positive microglia (IBA1, upper panel; EDN2, lower panel). (E–F) Intravitreal injection of EDN2 peptide causes RGC axon damage. EDN2 peptide (500 μm) or vehicle (PBS) was injected into the vitreous and RGC axon damage assessed 4 weeks later. There was a significant increase in the level of RGC axon damage in EDN2-injected eyes compared with PBS-treated control eyes (n = 5 control, n = 8 EDN2 eyes, P = 0.0074). (G) To assess vascular changes, retinas were visualized by DIC, and the ratio of lumen area (calculated from the lumen diameter, yellow arrows; see Methods) to total vessel area (calculated from vessel diameter, red arrows) was measured. (H) There is a significant decrease in the lumen/vessel ratio in eyes with increasing levels of glaucoma (P < 0.001). Scale bars: A–C, 100 μm, D, 10 μm, E, 50 μm.
Administration of EDN1, which shares receptors with EDN2, is known to kill RGCs. In contrast, EDN2 was suggested to protect photoreceptors from stress (44). To initially assess whether EDN2 damages RGCs, we injected EDN2 peptide into young DBA/2J eyes and found that it damaged both RGCs and the optic nerve (Figure 9, E and F). EDN1 is reported to be harmful to neurons by constricting blood vessels, triggering reactive astrocytosis, and/or inhibiting axon transport, but effects on astrocytes and axon transport may be secondary to altered blood flow (45–47). Since EDN2 is a potent vasoactive peptide, we hypothesized that EDN2 induces vasoconstriction in the ONH and retina during glaucoma. Even if subtle, this vasoconstriction may metabolically stress RGCs and contribute to their demise in glaucoma. In support of this vasoconstriction hypothesis, we identified a decrease in the ratio of lumen area to total blood vessel area in retinas of DBA/2J eyes at early and later stages of glaucoma (see Methods, Figure 9, G and H, and Supplemental Figure 6). This decrease was not uniform in any of the eyes investigated, suggesting local differences that may affect some but not all RGCs. The decreased lumen area is likely a result of vasoconstriction, but an EDN2-induced thickening of the vascular smooth muscle (48) cannot be ruled out. Either way, decreased lumen area is likely to negatively impact perfusion and RGC survival.
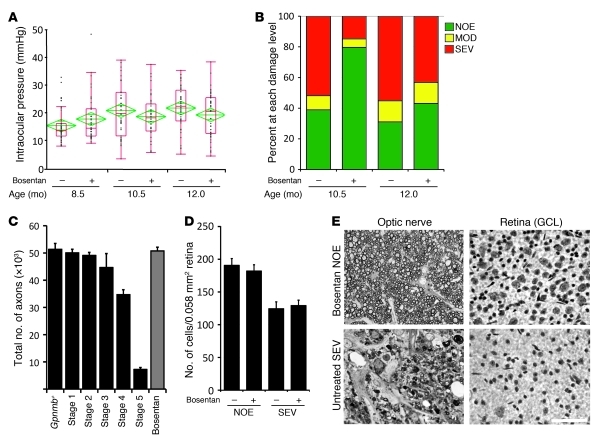
To functionally assess the role of the endothelin system in glaucoma, we administered bosentan, an endothelin receptor antagonist, to DBA/2J mice starting at 6.0 months of age (Figure 10). We selected bosentan because it antagonizes both types of endothelin receptors and does not alter blood pressure (see Methods). Also, bosentan has been shown to increase ocular blood flow in human glaucoma patients (49). Administration of bosentan did not alter the onset or progression of either the iris disease or IOP elevation (Figure 10A). However, bosentan significantly reduced glaucoma at both 10.5 and 12.0 months of age (Figure 10, B–E). The protection was especially strong at 10.5 months of age, when 80% of treated eyes had no detectable glaucoma, compared with only 39% of untreated eyes (P = 1.7 × 10–26). At 12.0 months, 43% of treated eyes had no detectable glaucoma compared with only 31% of untreated eyes (P = 0.01). These experiments implicate early activation of the endothelin system as pathogenic in this inherited model of glaucoma. Endothelin receptor antagonists offer promise as new treatments to alleviate glaucoma.
Figure 10. Bosentan robustly protects from glaucoma in DBA/2J glaucoma.
(A) Administration of bosentan (an endothelin receptor antagonist) does not alter IOP elevation in DBA/2J mice. Sample size: 39–47 per group. (B) Bosentan administration protects significant numbers of eyes from glaucoma (10.5 months, P = 1 × 10–26; 12 months, P = 0.01; sample sizes: 54 untreated, 54 treated at 10.5 months and 58 untreated, 58 treated at 12 months). (C) Randomly selected mice that were treated with bosentan and had the NOE nerve damage level have an axon number similar to that in no-glaucoma Gpnmb+ controls (11 treated vs. 13 Gpnmb+, P = 0.5) and in DBA/2J mice prior to glaucomatous axon loss. For ease of comparison, axon counts for stages 1–5 are repeated from Figure 1. (D) There is no significant soma loss in bosentan-protected eyes (NOE). Not unexpectedly, Bosentan did not save RGC soma in eyes with severe axon loss (SEV). (E) Representative optic nerve and retinal phenotypes for mice of each treatment group are shown. Optic nerve cross sections were stained with PPD; retinas with cresyl violet. Scale bar: 50 μm.
Discussion
We used cluster analyses to identify early stages of the later-onset glaucoma that occurs in DBA/2J mice. Importantly, these early stages precede morphologically detectable glaucomatous damage to the optic nerve and retina. Previously indistinguishable eyes with no detectable glaucoma were subdivided into molecularly defined stages that represent progression from control to more advanced disease states. Our cluster-based analyses proved more sensitive at detecting expression differences than more conventional analyses of the same samples. We separately analyzed gene expression in the ONH and retina of each eye, and our data suggest that events in these tissues are not completely dependent upon each other. For this study, the entire retina was profiled, but additional information may be achieved by assessing specific retinal cell types or regions. All our raw and processed data have been submitted to GEO (GSE26299). In addition, we hope that our experiments will be useful to guide future experiments, and thus we are developing an online environment that allows in-depth visualization and interrogation of these data (Glaucoma Discovery Platform, http://www.simonjohnlab.org).
A number of experimental design features allowed us to molecularly characterize early glaucoma. To estimate normal variation and provide a robust estimate of disease-specific changes, we separately assessed samples from two different no-glaucoma control groups. The first control set was age matched to all of the glaucomatous DBA/2J mice but lacked the glaucoma causing Gpnmb mutation (D2-Gpnmb+ mice). We produced and characterized this control strain for this as well as other studies (50). Although these mice genetically differ from the glaucomatous mice only at the Gpnmb locus, mice of this control strain rarely (if ever) develop glaucoma in our colony (50). The second control group were 4.5-month-old DBA/2J mice that did not differ in their Gpnmb genotype. Mice of this age have never demonstrated high IOP or glaucoma in our colony (40). To allow identification and staging of gene expression data associated with early glaucoma, we studied more than 20 eyes that had no detectable glaucoma by conventional assays. To minimize non-disease-related differences, all mice were sex- and strain-matched, including the two sets of control mice.
To identify disease-relevant genes, we first compared all glaucoma test eyes with both sets of controls and generated a list of genes with disease-specific expression changes. The disease-relevant genes were then used to cluster all samples into groups representing disease stages in an unbiased, unsupervised manner. The clustering outcomes made biological sense when compared with groups of eyes with morphologically recognizable stages of glaucoma. Clustering analyses have proven useful for identifying gene expression differences that distinguish various known types and stages of cancer (28, 29). To our knowledge, clustering analysis has not been previously used to distinguish and order early states of a complex multifactorial disease (or any other disease) in samples that are morphologically similar to unaffected controls.
A significant issue in analyzing complex diseases such as glaucoma is the asynchrony of pathogenic events and large phenotypic and molecular variability from eye to eye. Simply analyzing groups of eyes can miss key changes that are important in disease manifestation but are not uniform within assessed groups. Our results show that microarray-based cluster analyses can help by grouping eyes with the most similar patterns of gene expression. However, microarray analysis is not practical for determining relative disease status in large numbers of individual eyes in subsequent experiments. To address this, we used quantitative PCR to assess the expression levels of specific sets of genes identified in the microarray study in an independent set of individual eyes. Since many molecular changes occur in glaucoma and specific changes vary from eye to eye, we assessed a variety of genes from multiple pathways as a measure of glaucoma status. We defined altered gene expression in individual DBA/2J eyes as changes that were greater than or equal to 2 SDs from the average of D2-Gpnmb+ control eyes. This promising approach successfully distinguished the majority of DBA/2J from control eyes. Although further refinement is needed, our methodology may also be relevant to other complex diseases such as Alzheimer disease and age-related macular degeneration.
Publicly available tools were used to identify pathways that are overrepresented among the DE genes (see Methods). The earliest changes in the ONH include upregulation of ECM-receptor interactions and focal adhesion pathways. Genes in these pathways include collagens and laminins, important components of the basal lamina surrounding ONH axons and microcapillaries. Changes in the expression of these genes may indicate early damaging changes to the glial-axon or glial-vascular unit (51). Additionally or alternatively, ECM changes may reflect protective mechanisms induced by the stress of raised IOP. Focal adhesion molecules such as integrins are mechanosensors known to sense changes in pressure-induced deformation and trigger increases in ECM components including tenascin C (33, 35). The expression of many immune/inflammatory genes also changed in early glaucoma. These inflammatory processes may be either damaging or protective (52, 53), and future experiments are needed to determine the roles of specific molecules. It is difficult to compare our data with previous gene expression profiling studies of human glaucoma and animal models of glaucoma, as the complete datasets are not always available, and criteria for determining DE genes can vary (22, 24–26, 54). Nevertheless some similarities are evident, including changes in immune/inflammatory and ECM genes. Current and future experiments that profile gene expression changes in defined cell types from glaucomatous eyes (55–58) will enhance sensitivity and contribute to the assignment of different events to specific cell types. Important cell types to profile include astrocytes, endothelial cells, microglia, Müller cells, and RGCs.
A reasonable approach toward developing new treatments to intervene in glaucoma pathogenesis is to develop treatments that target pathways that change early in glaucoma, prioritizing pathways that are known to change in multiple animal models of glaucoma and/or human glaucoma. In this study, we demonstrate that both the endothelin system and complement cascade change early in DBA/2J glaucoma. The complement cascade mediates innate immunity, contributes to elimination of synapses during development (36), and is suggested to participate in Alzheimer disease (reviewed in ref. 59). Endothelins induce vasoconstriction, inhibit axonal transport, activate glial cells, and can kill RGCs (refs. 45–47 and our present results). Changes in both the complement and endothelin systems were previously reported in animal models of glaucoma and human glaucoma (22–24, 26, 36, 37, 41–43). Thus, we tested the effects of separately manipulating these two pathways. Glaucoma was ameliorated in C1QA-deficient mice, with a very substantial lowering of glaucoma incidence at 10.5 months. Inhibition of the endothelin system using bosentan also significantly ameliorated glaucoma in DBA/2J mice, again with the greatest effect at 10.5 months. These results suggest that inhibition of these pathways may provide valuable new treatments against human glaucoma. A full investigation of the glaucoma-relevant mechanisms influenced by these pathways, including genetic manipulation of different pathway components in different cell types during different stages of disease, is warranted.
In conclusion, we have used clustering analyses of ONH and retina gene expression to identify early stages of glaucoma. Our successful implementation of clustering methodologies to an inherited, later-onset, complex glaucoma suggests that similar approaches will prove valuable for uncovering early processes in other diseases and for identifying novel therapeutic targets.
Methods
Mouse strains, breeding, and husbandry.
All experiments were conducted in accordance with the Association for Research in Vision and Ophthalmology statement on the use of animals in ophthalmic research. All mice were housed in a 14-hour light/10-hour dark cycle under previously described conditions (60). The Jackson Laboratory Animal Care and Use Committee approved all of the experiments in this study. All mice in the gene expression profiling study were female and housed under the same conditions in same animal facility, minimizing environmental differences. Our DBA/2J (D2) mouse colony is routinely crossed with DBA/2J mice from The Jackson Laboratory production facility to prevent genetic drift. The D2-Gpnmb+ strain (DBA/2J-Gpnmb+/Sj), which rarely if ever develops glaucoma in our colony, was used as a control strain for DBA/2J glaucoma studies (50). For the gene expression analyses, more than 500 mice were aged and analyzed to produce sufficient eyes with each glaucoma stage. To generate D2.C1qa+/– mice, the C1qa+/– allele (61) was backcrossed to D2 mice for 8 generations. The C1qa mutation was selected due to its established role in retinogeniculate refinement (36). D2.C1qa+/– mice were assessed with 102 genetic markers evenly distributed across the mouse genome, and only one marker closely linked to the C1qa gene was positive for C57BL/6J (this represents flanking sequence closely linked to the C1qa mutation). The remaining 101 markers were positive for DBA/2J. This indicates that greater than 99% of the genetic background of D2.C1qa+/– mice is derived from DBA/2J. To generate DBA/2J mice deficient in C1QA and littermate controls, we intercrossed D2.C1qa+/– mice.
Analysis of glaucomatous damage.
Intracranial portions of optic nerves were processed and analyzed as previously described (38–40). Briefly, optic nerves were fixed in situ for 48 hours, dissected free, processed, and embedded in plastic. One-micrometer-thick cross sections of optic nerve from behind the orbit were cut and stained with paraphenylenediamine (PPD). PPD darkly stains the myelin sheaths and axoplasm of sick or dying axons, but not healthy axons (62). PPD staining allows sensitive determination of disease stage for large numbers of nerves. Three observers blinded to the experimental protocol determined the degree of nerve damage as previously described (38–40, 63). The 3 clearly distinguishable stages of damage were as follows: (a) NOE — less than 5% axons damaged. This amount of damage is seen in age-matched mice of various strains that do not develop glaucoma. It is called “no or early” because some of these eyes are undergoing early molecular stages of disease, but they are not distinguishable from eyes with no glaucoma by conventional morphologic analyses of axon number and nerve damage. (b) MOD — many damaged axons throughout the nerve, with an average of 30% axon loss. (c) SEV — substantial axon loss (>50% lost) and damage. To ensure very distinct stages for this study, we selected severely affected eyes with very few remaining axons (>90% loss). For C1qa and bosentan experiments, differences in optic nerve damage distributions between groups were compared using χ2 tests.
Tissue harvesting, RNA isolation, and processing for microarrays.
Following anesthesia, the left eye was dissected free, and the cornea and lens were removed. The optic nerve was cut close to the back of the eye. The ONH was excised using a sharpened 1-mm glass capillary centered over the optic nerve. The punched-out nerve head was immediately placed in RNAlater (Ambion). A small portion of central retina as well as a small portion of choroid and sclera were included in this tissue. This tissue is substantially enriched for the glial lamina. The retina (containing no ONH) was then dissected free from the back of the eye and placed in a separate vial containing RNAlater and stored at 4°C until required. The process was repeated for the right eye. Dissection of each eye was completed in 5 minutes or less (10 minutes per mouse).
For RNA preparation, tissue was homogenized in RLT Buffer (QIAGEN). Total RNA was isolated using the RNeasy Micro Kit (QIAGEN) according to the manufacturer’s protocols, and quality was assessed using a 2100 Bioanalyzer instrument and RNA 6000 Nano LabChip assay (Agilent Technologies). Tissue harvesting and RNA preparation were completed for all mice aged for the gene expression studies. Samples with only the highest-quality RNA were used.
RNA labeling and microarray processing.
Biotin-labeled cDNA was synthesized separately from 20 ng of each total RNA sample per the manufacturer’s protocols (Ovation Biotin System, NuGEN). 2.5 μg of each biotin-labeled and fragmented cDNA sample was then hybridized onto Mouse Genome 430 version 2.0 GeneChip arrays (Affymetrix). Post-hybridization staining and washing were performed according to the manufacturer’s protocols using a Fluidics Station 450 instrument (Affymetrix). Finally, the arrays were scanned with a GeneChip Scanner 3000 laser confocal slide scanner. The images were quantified using GeneChip Operating Software (GCOS) version 1.2. Probe level data were imported into the R statistical software environment ( http://www.r-project.org), and expression values for each probe set on the array were summarized using the robust multichip average (RMA) method with quantile normalization in the R/affy package (64, 65).
A total of 110 arrays were used, assessing 60 ONHs and 50 retinas. The ONHs included 10 D2-Gpnmb+ control eyes at 10.5 months, 40 DBA/2J eyes at 10.5 months, and 10 DBA2J eyes at 4.5 months. The 50 retinas included 10 D2-Gpnmb+ control eyes at 10.5 months and 40 DBA/2J eyes at 10.5 months. Only 2 arrays failed the quality control; both were 10.5-month DBA/2J MOD retinas. All 110 arrays were processed at the same time. To allow future comparisons, quantile normalization of all of the arrays was performed. All individual gene expression profiles are available in GEO datasets (NCBI, accession number GSE26299).
Statistics for differential gene expression.
Differential gene expression between any two morphologically defined groups or molecularly defined stages of glaucoma was assessed by gene-specific fixed-effect ANOVA methods using the R/maanova package (64). The generalized 1-way ANOVA model can be written as Yi = μ + GROUP + εi, where Yi represents the log-transformed expression measure of probe set i, μ is the mean for each array, GROUP represents the fixed effect under comparison, and εi captures random error absorbing both array and biological components. Given the ANOVA model, all statistical tests were performed using Fs, a modified F statistic incorporating shrinkage estimates of variance components (66). All P values were calculated by permuting model residuals 1,000 times. To address the problem of multiple testing, the permutation P values were adjusted using the FDR method of Storey (67) implemented in R/q value. The resulting q values estimated the proportion of false positives in the list of DE genes and were used to select suitable candidate gene lists for subsequent analysis. Unless otherwise noted, statistical significance was determined at q ≤ 0.05 (FDR <5%). After statistical assessment, probe sets were annotated using Netaffx software (Affymetrix). A wide selection of the DE genes were confirmed to be DE by real-time PCR, including genes with a fold change greater than 2.
Defining early stages of disease using hierarchical clustering.
To generate a list of disease-specific probe sets to cluster the ONH samples, we first made a pairwise comparison between 10 non-glaucomatous D2-Gpnmb+ and 40 glaucomatous DBA/2J mice (which have the GpnmbR150X mutation) that were 10.5 months of age (Supplemental Figure 1A). As all of these mice were age-, sex-, and strain-matched, genes on this first list were DE due to effects of glaucoma or Gpnmb genotype (glaucoma and genotype list). To avoid DE genes due to Gpnmb genotype, we next generated a genotype-related list of probes that were DE between the 10.5-month-old D2-Gpnmb+ and the ten 4.5-month-old DBA/2J ONH samples. Importantly, since neither 4.5-month-old DBA/2J nor D2-Gpnmb+ mice have glaucoma in our colony, this second genotype list will not contain genes that are DE due to glaucoma. However, it may contain some genes that differ due to age. The final disease-specific gene list was then generated by subtracting the probe sets on the genotype list from the glaucoma and genotype list (570 DE probe sets, q < 0.001). The 570 probe sets were used to perform hierarchical clustering of ONH samples (using JMP Statistical Discovery version 7.0 software; SAS). Using the average distance metric, hierarchical clustering started with each eye as its own cluster. At each step, the clustering algorithm calculated the distance between each cluster and combined the two clusters that were closest together. This combining continued until all the points were in one final cluster or dendrogram. A threshold or cutoff, based on known differences between control, MOD, and SEV groups, was applied to identify 5 molecular stages that made biological sense. Since the filters that we used to identify disease-specific probe sets were not perfect, all genes other than Gpnmb were included in subsequent group comparisons. PCA was performed using MultiExperiment Viewer (TM4 version 4.5.1; www.http://www.tm4.org/mev) on the 10 D2-Gpnmb+ and 40 DBA/2J eyes using the 570 probe sets used for hierarchical clustering.
For the retina, 600 probe sets were DE (q < 0.01) when comparing the 10 D2-Gpnmb+ control samples with the 38 DBA/2J retina samples (2 of the 40 retina samples failed quality control). No subtraction was performed on the retina samples, as only Gpnmb was DE when D2-Gpnmb+ eyes were compared with either of the NOE groups at 10.5 months. Since Gpnmb clearly differed due to genotype, this gene was not counted as DE due to glaucoma.
A second clustering experiment was performed using probe sets DE between control and MOD eyes to cluster eyes with NOE glaucoma. Providing confirmation of our first clustering experiment, hierarchical clustering placed the NOE eyes in the same stages (data not shown).
Functional analyses and gene set enrichment of DE genes.
The Database for Annotation, Visualization and Integrated Discovery ( DAVID, http://david.abcc.ncifcrf.gov/; ref. 68) was used to add functional annotation to gene lists and provide statistical assessment of the annotations. We focused on the KEGG pathways (69) and GO terms (70). For each list of DE genes, DAVID determines the number of genes in each KEGG pathway and uses a Fisher exact test with Benjamini correction to determine the probability that the number of genes in each pathway would have occurred by chance (68). A corrected P value of 0.05 or less was used to identify significant pathways. Pathways with gene expression changes represented by colors (red: upregulated, green: downregulated) were generated using the KEGG mapper tool ( http://www.genome.jp/kegg/tool/color_pathway.html) (69).
We performed k-means clustering in R/maanova to identify clusters of probe sets whose expression changed in a similar pattern when considering stages 2–5 from the molecularly determined ONH groups. The earliest stage (stage 1) was not included, as there were too few DE probe sets in this stage. The normalized expression values (generated by RMA) of 4,138 probe sets (DE in each of the stages 2, 3, 4, and 5) were clustered into 8 groups (see Supplemental Table 2). Figure of merit analysis was used to determine the most meaningful number of clusters (see Supplemental Figure 4 and ref. 71). We performed k-means clustering using k = 4, 6, 8, 10, and 12. Based on pathway enrichment analysis, 4, 10, and 12 were not appropriate, and there was little difference between the results for 6 and 8. Therefore, we show the results for k = 8.
To assess gene networks and hubs, we used IPA (Ingenuity Systems).
Quantitative real-time PCR.
A completely independent set of eyes from 10-month-old mice were obtained. To avoid RNA degradation, two investigators harvested the eyes of each mouse in parallel. All dissections were performed in an RNase-free environment. Eyes were dissected free (including a significant portion of the myelinated optic nerve) and immediately placed in sterile 1× PBS. The retina with ONH attached was removed from the eye. To separate ONH from retina, an incision was made in the retina as close to the ONH as possible. Choroidal and scleral tissues, as well as the distal portion of the optic nerve, were removed from the ONH (approximately 100 μm of the myelinated nerve was left attached to the ONH). The ONH tissue was then placed immediately into RNA isolation buffer (Ambion) and stored at –20°C. The retina was then placed flat (ganglion cell layer up) onto the sterile membrane in a culture plate insert (Millipore, PR01715) ensuring no bubbles between the retina and membrane. The membrane was cut from the insert and placed flat (retina side up) onto a previously prepared flat surface of frozen OCT (Tissue-Tek). Freezing was instantaneous. The uppermost 80 μm of retina, containing the vast majority of the ganglion cell layer and some inner plexiform layer, was isolated by cryosectioning (4 × 20–μm sections) and immediately placed in RNA isolation buffer and stored at –20°C. For each mouse, the total dissection time never exceeded 15 minutes. RNA was extracted using an RNAeasy Micro Kit (QIAGEN), and RNA quality was assessed using a bioanalyzer (Agilent). Only samples with more than 5 ng of material with no sign of degradation were subjected to further analysis. cDNA synthesis was performed using the WT-Ovation Pico RNA Amplification System (NuGEN). In total, 12 D2-Gpnmb+ control and 23 DBA/2J eyes passed quality control.
Expression levels for 91 genes (69 ONH and 22 RGC layer of retina) and 6 normalizers were assessed. The normalizer genes were selected because they had the most consistent expression level, irrespective of damage level, among a previously tested panel of 194 genes (tested in DBA/2J retina and ONH; data not shown). The normalizers were 5730453I16Rik, NM_080456.1, and Slc11a2 for RGC layer and Etfb, Odc1, and Rpn2 for ONH. All primers were designed using Primer3 (version 0.4.0; http://frodo.wi.mit.edu/primer3/) with product sizes of 100–120 bp and optimum melting temperature of 60°C. All sequences are available as supplemental material (Supplemental Table 3). Real-time PCR reactions were prepared using Quant-iT RiboGreen RNA assay kit (Invitrogen) and run on the 7900HT Fast Real-Time PCR System (Applied Biosystems).
For each sample, each gene was interrogated in triplicate. Any technical replicate that was greater than 1.5 cycle threshold (CT) values away from the other 2 replicates was removed. For each gene, the CTs were calculated as the average of the successful replicates. For normalization, ΔCT values were calculated as CT (gene of interest) – geometric mean of CTs for the normalizers. The average and standard deviation of the D2-Gpnmb+ control eyes were calculated. The ΔΔCT was defined as the ΔCT (gene of interest) – ΔCT (control average). The fold change was calculated as 2–ΔΔCT (upregulated) or –2ΔΔCT (downregulated). A gene was considered DE if the fold change was greater than 2 SDs away from the average fold change for the control eyes. One control eye was removed from the study because it clearly was a far outlier.
RNA in situ hybridization and immunofluorescence.
DBA/2J.Thy1-CFP mice were used to visualize RGCs (15). Immunofluorescence to visualize EDN2 and IBA1 was performed as follows. Eyes from DBA/2J.Thy1-CFP mice were fixed overnight at 4°C in 4% PFA. Retinas were dissected free and incubated in primary antibody diluted in antibody buffer (5% BSA, 1% Triton X-100) for 5 days, with shaking at 4°C. Primary antibodies used were anti-EDN2 (Santa Cruz Biotechnology Inc., 1:100) and anti-IBA1 (Wako, 1:200). Retinas were washed in antibody buffer for at least 6 hours, with shaking at 4°C (changing wash solution at least 5 times). Retinas were incubated in secondary antibodies (Invitrogen, diluted in antibody buffer, 1:1,000) for a further 5 days, with shaking at 4°C. Retinas were rinsed in antibody buffer, washed in 1× PBS, mounted onto a slide in mounting media, and coverslipped. Fluorescence was visualized using a SP5 confocal microscope (Leica).
RNA in situ hybridization was performed as previously described, using 4% PFA perfusion-fixed, 12-μm-thick optic nerve cryostat sections (72). Digoxigenin-labeled (DIG-labeled) riboprobes for C1qa and Tnc were transcribed from cDNA clones (Open Biosystems clone ID: 3592169 and 40099589, respectively). The plasmids were digested with EcoRI (C1qa) and with SpeI (Tnc), and in vitro transcription was performed with T7 polymerase. T3 polymerase was used to generate the control sense probes. For both C1qa and Tnc, the sense probes produced no signal. The detection of hybridized mRNA in sections was performed using the Cy-3 Tyramide Signal Amplification System (PerkinElmer). After in situ hybridization, the sections were incubated in the primary antibodies: mouse anti-phosphorylated neurofilament (2F11; 1:1,000; Dako) and rabbit anti-IBA1 (1:500, Wako). Primary antibodies were diluted in a solution of 10% normal goat serum, 0.5% Triton X-100, and 0.5% BSA in 0.1 M PBS. For the secondary antibodies, goat anti-mouse Alexa Fluor 647 and goat anti-rabbit Alexa Fluor 488 were used at a 1:200 dilution (Invitrogen). The sections were then incubated with DAPI (Invitrogen) and mounted in Fluoromount (Sigma-Aldrich). Fluorescence was visualized using a SP5 confocal microscope.
For all protein and RNA localization, at least 3 sections (ONH) or whole retinas from 10 DBA/2J eyes and 5 D2-Gpnmb+ control eyes were assessed.
Vessel measurements and intravitreal injections of EDN2.
Retinas were fixed in 4% PFA overnight and placed flat on a slide and coverslipped. Major arterioles were measured from 24 imaged fields that were evenly distributed around the retina and 1 mm out from the ONH. Images were taken using differential interference contrast (DIC) on an SP5 confocal microscope. For each vessel, the diameter of the both the lumen and the outer vessel wall (taken from widest part of the vessel present in the field) was calculated as the average of 3 measurements. The ratio of the areas of the lumen to the total vessel area was calculated (πr2).
For intravitreal injections of EDN2, 500 μM of EDN2 peptide (Invitrogen, dissolved in 2 μl of sterile 1× PBS vehicle) was injected into the vitreous chamber using a Hamilton syringe with a 35 gauge needle. Vehicle alone was injected into other eyes as a control. After 4 weeks, optic nerves were assessed for optic nerve damage (see Analysis of glaucomatous damage).
Bosentan administration.
Bosentan, an endothelin receptor antagonist, was provided by Actelion Pharmaceuticals. Bosentan was incorporated into standard mouse chow (100 mg/kg, Test Diet). DBA/2J mice were administered bosentan from 6 months of age. Control animals were aged in the same environment on the same mouse chow (minus bosentan). Although bosentan is not reported to affect blood pressure, as it is an inhibitor of both the EDNRA and EDNRB endothelin receptors, some endothelin system inhibitors do affect blood pressure. To ensure that bosentan would not affect blood pressure in DBA/2J mice, a separate cohort of 8 mice was administered bosentan for 2 weeks, and then blood pressures was measured using previously described procedures (73). Blood pressures were not affected by bosentan administration (BP, mmHg ± SEM: 113.5 ± 0.67 treated; 112.7 ± 1.2 control, P = 0.8).
Clinical examination, IOP measurements, and RGC soma assessment.
DBA/2J mice develop a disease of the iris that leads to intraocular pressure elevation and glaucoma. Clinical examinations (30) and IOP measurements (74, 75) were performed as previously described. For clinical examinations, at least 20 eyes from each genotype or treatment group were examined at 6, 8, 10, and 12 months of age. RGC layer soma counts were preformed as described previously (15). Two-tailed Student t tests were performed to determine statistical significance. P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
The authors thank Amy Bell for intraocular pressure measurements, Adrienne Mehalow for tissue harvesting and technical support, and K. Saidas Nair, Krish Kizhatil, and Mimi de Vries for critical comments on the manuscript. We also acknowledge Scientific Services at The Jackson Laboratory, particularly Gene Expression, Histology, and the Computational Sciences. This work was funded in part by National Eye Institute (NEI) grant EY011721 (to S.W.M. John), The Barbara and Joseph Cohen Foundation, donors of ADR — a program of the American Health Assistance Foundation (to G.R. Howell and S.W.M. John), NEI EY018606 (to R.T. Libby), NIH T32 NS51112-05 (to I. Soto), and a Research to Prevent Blindness Career Development Award (to R.T. Libby). S.W.M. John is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(4):1429–1444. doi:10.1172/JCI44646.
Benjamin L. King’s present address is: Mount Desert Island Biological Laboratory, Salisbury Cove, Maine, USA.
References
- 1.Quigley HA. Number of people with glaucoma worldwide. Br J Ophthalmol. 1996;80(5):389–393. doi: 10.1136/bjo.80.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitmore AV, Libby RT, John SW. Glaucoma: thinking in new ways-a role for autonomous axonal self-destruction and other compartmentalised processes? Prog Retin Eye Res. 2005;24(6):639–662. doi: 10.1016/j.preteyeres.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Hodge WG, et al. The efficacy and harm of prostaglandin analogues for IOP reduction in glaucoma patients compared to dorzolamide and brimonidine: a systematic review. Br J Ophthalmol. 2008;92(1):7–12. doi: 10.1136/bjo.2007.123737. [DOI] [PubMed] [Google Scholar]
- 4.Katz LJ, Simmons ST, Craven ER. Efficacy and safety of brimonidine and dorzolamide for intraocular pressure lowering in glaucoma and ocular hypertension. Curr Med Res Opin. 2007;23(12):2971–2983. doi: 10.1185/030079907X242476. [DOI] [PubMed] [Google Scholar]
- 5.Rolim de Moura C, Paranhos A, Jr, Wormald R. Laser trabeculoplasty for open angle glaucoma. Cochrane Database Syst Rev. 2007;2007(4):CD003919. doi: 10.1002/14651858.CD003919.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson DR, Hendrickson A. Effect of intraocular pressure on rapid axoplasmic transport in monkey optic nerve. Invest Ophthalmol. 1974;13(10):771–783. [PubMed] [Google Scholar]
- 7. Anderson DR. Is Ischemia the villain in glaucomatous cupping and atrophy? In: Brockhurst RJ, et al.Controversy in Ophthalmology . Philadelphia, Pennsylvania, USA: WB Saunders Company; 1977:312–319. [Google Scholar]
- 8.Quigley HA, Guy J, Anderson DR. Blockade of rapid axonal transport. Effect of intraocular pressure elevation in primate optic nerve. Arch Ophthalmol. 1979;97(3):525–531. doi: 10.1001/archopht.1979.01020010269018. [DOI] [PubMed] [Google Scholar]
- 9.Quigley H, Anderson DR. Cupping of the optic disc in ischemic optic neuropathy. Trans Sect Ophthalmol Am Acad Ophthalmol Otolaryngol. 1977;83(5):755–762. [PubMed] [Google Scholar]
- 10.Quigley H, Anderson DR. The dynamics and location of axonal transport blockade by acute intraocular pressure elevation in primate optic nerve. Invest Ophthalmol. 1976;15(8):606–616. [PubMed] [Google Scholar]
- 11.Quigley HA, Addicks EM. Chronic experimental glaucoma in primates. II. Effect of extended intraocular pressure elevation on optic nerve head and axonal transport. Invest Ophthalmol Vis Sci. 1980;19(2):137–152. [PubMed] [Google Scholar]
- 12.Quigley HA, Flower RW, Addicks EM, McLeod DS. The mechanism of optic nerve damage in experimental acute intraocular pressure elevation. Invest Ophthalmol Vis Sci. 1980;19(5):505–517. [PubMed] [Google Scholar]
- 13.Quigley HA, Addicks EM. Regional differences in the structure of the lamina cribrosa and their relation to glaucomatous optic nerve damage. Arch Ophthalmol. 1981;99(1):137–143. doi: 10.1001/archopht.1981.03930010139020. [DOI] [PubMed] [Google Scholar]
- 14.Quigley HA, Hohman RM, Addicks EM, Massof RW, Green WR. Morphologic changes in the lamina cribrosa correlated with neural loss in open-angle glaucoma. Am J Ophthalmol. 1983;95(5):673–691. doi: 10.1016/0002-9394(83)90389-6. [DOI] [PubMed] [Google Scholar]
- 15.Howell GR, et al. Axons of retinal ganglion cells are insulted in the optic nerve early in DBA/2J glaucoma. J Cell Biol. 2007;179(7):1523–1537. doi: 10.1083/jcb.200706181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKinnon SJ, Schlamp CL, Nickells RW. Mouse models of retinal ganglion cell death and glaucoma. Exp Eye Res. 2009;88(4):816–824. doi: 10.1016/j.exer.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang IH, Clark AF. Rodent models for glaucoma retinopathy and optic neuropathy. J Glaucoma. 2007;16(5):483–505. doi: 10.1097/IJG.0b013e3181405d4f. [DOI] [PubMed] [Google Scholar]
- 18.Sappington RM, Carlson BJ, Crish SD, Calkins DJ. The microbead occlusion model: a paradigm for induced ocular hypertension in rats and mice. Invest Ophthalmol Vis Sci. 2010;51(1):207–216. doi: 10.1167/iovs.09-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libby RT, et al. Inducible nitric oxide synthase, Nos2, does not mediate optic neuropathy and retinopathy in the DBA/2J glaucoma model. BMC Neurosci. 2007;8:108. doi: 10.1186/1471-2202-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neufeld AH. 2004. Pharmacologic neuroprotection with an inhibitor of nitric oxide synthase for the treatment of glaucoma. Brain Res Bull. 2004;62(6):455–459. doi: 10.1016/j.brainresbull.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Pang IH, et al. Evaluation of inducible nitric oxide synthase in glaucomatous optic neuropathy and pressure-induced optic nerve damage. Invest Ophthalmol Vis Sci. 2005;46(4):1313–1321. doi: 10.1167/iovs.04-0829. [DOI] [PubMed] [Google Scholar]
- 22.Johnson EC, Jia L, Cepurna WO, Doser TA, Morrison JC. Global changes in optic nerve head gene expression after exposure to elevated intraocular pressure in a rat glaucoma model. Invest Ophthalmol Vis Sci. 2007;48(7):3161–3177. doi: 10.1167/iovs.06-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed F, Brown KM, Stephan DA, Morrison JC, Johnson EC, Tomarev SI. Microarray analysis of changes in mRNA levels in the rat retina after experimental elevation of intraocular pressure. Invest Ophthalmol Vis Sci. 2004;45(4):1247–1258. doi: 10.1167/iovs.03-1123. [DOI] [PubMed] [Google Scholar]
- 24.Steele MR, Inman DM, Calkins DJ, Horner PJ, Vetter ML. Microarray analysis of retinal gene expression in the DBA/2J model of glaucoma. Invest Ophthalmol Vis Sci. 2006;47(3):977–985. doi: 10.1167/iovs.05-0865. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, et al. Changes in gene expression in experimental glaucoma and optic nerve transection: the equilibrium between protective and detrimental mechanisms. Invest Ophthalmol Vis Sci. 2007;48(12):5539–5548. doi: 10.1167/iovs.07-0542. [DOI] [PubMed] [Google Scholar]
- 26.Panagis L, Zhao X, Ge Y, Ren L, Mittag TW, Danias J. Gene expression changes in areas of focal loss of retinal ganglion cells (RGC) in the retina of DBA/2J mice. Invest Ophthalmol Vis Sci. 2010;51(4):2024–2034. doi: 10.1167/iovs.09-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson EC, et al. Cell proliferation and interleukin-6 type cytokine signaling are implicated by gene expression responses in early optic nerve head injury in rat glaucoma. Invest Ophthalmol Vis Sci. 2011;52(1):504–518. doi: 10.1167/iovs.10-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knosel T, et al. Immunoprofiles of 11 biomarkers using tissue microarrays identify prognostic subgroups in colorectal cancer. Neoplasia. 2005;7(8):741–747. doi: 10.1593/neo.05178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fevre-Montange M, et al. Microarray gene expression profiling in meningiomas: differential expression according to grade or histopathological subtype. Int J Oncol. 2009;35(6):1395–1407. doi: 10.3892/ijo_00000457. [DOI] [PubMed] [Google Scholar]
- 30.Anderson MG, et al. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet. 2002;30(1):81–85. doi: 10.1038/ng794. [DOI] [PubMed] [Google Scholar]
- 31.Ringner M. What is principal component analysis? Nat Biotechnol. 2008;26(3):303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 32.Pena JD, Varela HJ, Ricard CS, Hernandez MR. Enhanced tenascin expression associated with reactive astrocytes in human optic nerve heads with primary open angle glaucoma. Exp Eye Res. 1999;68(1):29–40. doi: 10.1006/exer.1998.0577. [DOI] [PubMed] [Google Scholar]
- 33.Chiquet M. Regulation of extracellular matrix gene expression by mechanical stress. Matrix Biol. 1999;18(5):417–426. doi: 10.1016/s0945-053x(99)00039-6. [DOI] [PubMed] [Google Scholar]
- 34.Lockwood CJ, et al. Matrix metalloproteinase 9 (MMP9) expression in preeclamptic decidua and MMP9 induction by tumor necrosis factor alpha and interleukin 1 beta in human first trimester decidual cells. Biol Reprod. 2008;78(6):1064–1072. doi: 10.1095/biolreprod.107.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarasa-Renedo A, Chiquet M. Mechanical signals regulating extracellular matrix gene expression in fibroblasts. Scand J Med Sci Sports. 2005;15(4):223–230. doi: 10.1111/j.1600-0838.2005.00461.x. [DOI] [PubMed] [Google Scholar]
- 36.Stevens B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131(6):1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 37.Stasi K, et al. Complement component 1Q (C1Q) upregulation in retina of murine, primate, and human glaucomatous eyes. Invest Ophthalmol Vis Sci. 2006;47(3):1024–1029. doi: 10.1167/iovs.05-0830. [DOI] [PubMed] [Google Scholar]
- 38.Anderson MG, et al. Genetic context determines susceptibility to intraocular pressure elevation in a mouse pigmentary glaucoma. BMC Biol. 2006;4:20. doi: 10.1186/1741-7007-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Libby RT, et al. Susceptibility to neurodegeneration in a glaucoma is modified by Bax gene dosage. PLoS Genet. 2005;1(1):17–26. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Libby RT, et al. Inherited glaucoma in DBA/2J mice: pertinent disease features for studying the neurodegeneration. Vis Neurosci. 2005;22(5):637–648. doi: 10.1017/S0952523805225130. [DOI] [PubMed] [Google Scholar]
- 41.Krishnamoorthy RR, Rao VR, Dauphin R, Prasanna G, Johnson C, Yorio T. Role of the ETB receptor in retinal ganglion cell death in glaucoma. Can J Physiol Pharmacol. 2008;86(6):380–393. doi: 10.1139/Y08-040. [DOI] [PubMed] [Google Scholar]
- 42.Noske W, Hensen J, Wiederholt M. Endothelin-like immunoreactivity in aqueous humor of patients with primary open-angle glaucoma and cataract. Graefes Arch Clin Exp Ophthalmol. 1997;235(9):551–552. doi: 10.1007/BF00947082. [DOI] [PubMed] [Google Scholar]
- 43.Tezel G, Kass MA, Kolker AE, Becker B, Wax MB. Plasma and aqueous humor endothelin levels in primary open-angle glaucoma. J Glaucoma. 1997;6(2):83–89. [PubMed] [Google Scholar]
- 44.Rattner A, Nathans J. The genomic response to retinal disease and injury: evidence for endothelin signaling from photoreceptors to glia. J Neurosci. 2005;25(18):4540–4549. doi: 10.1523/JNEUROSCI.0492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sasaoka M, Taniguchi T, Shimazawa M, Ishida N, Shimazaki A, Hara H. Intravitreal injection of endothelin-1 caused optic nerve damage following to ocular hypoperfusion in rabbits. Exp Eye Res. 2006;83(3):629–637. doi: 10.1016/j.exer.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 46.Stokely ME, Yorio T, King MA. Endothelin-1 modulates anterograde fast axonal transport in the central nervous system. J Neurosci Res. 2005;79(5):598–607. doi: 10.1002/jnr.20383. [DOI] [PubMed] [Google Scholar]
- 47.Taniguchi T, Shimazawa M, Sasaoka M, Shimazaki A, Hara H. Endothelin-1 impairs retrograde axonal transport and leads to axonal injury in rat optic nerve. Curr Neurovasc Res. 2006;3(2):81–88. doi: 10.2174/156720206776875867. [DOI] [PubMed] [Google Scholar]
- 48.Bouallegue A, Daou GB, Srivastava AK. Endothelin-1-induced signaling pathways in vascular smooth muscle cells. Curr Vasc Pharmacol. 2007;5(1):45–52. doi: 10.2174/157016107779317161. [DOI] [PubMed] [Google Scholar]
- 49.Resch H, et al. Effect of dual endothelin receptor blockade on ocular blood flow in patients with glaucoma and healthy subjects. Invest Ophthalmol Vis Sci. 2009;50(1):358–363. doi: 10.1167/iovs.08-2460. [DOI] [PubMed] [Google Scholar]
- 50.Howell GR, et al. Absence of glaucoma in DBA/2J mice homozygous for wild-type versions of Gpnmb and Tyrp1. . BMC Genet. 2007;8:45. doi: 10.1186/1471-2156-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Del Zoppo GJ, Milner R, Mabuchi T, Hung S, Wang X, Koziol JA. Vascular matrix adhesion and the blood-brain barrier. Biochem Soc Trans. 2006;34(pt 6):1261–1266. doi: 10.1042/BST0341261. [DOI] [PubMed] [Google Scholar]
- 52.Perry VH, Nicoll JA, Holmes C. Microglia in neurodegenerative disease. Nat Rev Neurol. 2010;6(4):193–201. doi: 10.1038/nrneurol.2010.17. [DOI] [PubMed] [Google Scholar]
- 53.Block ML, Hong JS. Microglia and inflammation-mediated neurodegeneration: multiple triggers with a common mechanism. Prog Neurobiol. 2005;76(2):77–98. doi: 10.1016/j.pneurobio.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Nikolskaya T, et al. Network analysis of human glaucomatous optic nerve head astrocytes. BMC Med Genomics. 2009;2:24. doi: 10.1186/1755-8794-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo Y, Cepurna WO, Dyck JA, Doser TA, Johnson EC, Morrison JC. Retinal cell responses to elevated intraocular pressure: a gene array comparison between the whole retina and retinal ganglion cell layer. Invest Ophthalmol Vis Sci. 2010;51(6):3003–3018. doi: 10.1167/iovs.09-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roesch K, et al. The transcriptome of retinal Muller glial cells. J Comp Neurol. 2008;509(2):225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang DY, et al. Global gene expression changes in rat retinal ganglion cells in experimental glaucoma. Invest Ophthalmol Vis Sci. 2010;51(8):4084–4095. doi: 10.1167/iovs.09-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexander JJ, Anderson AJ, Barnum SR, Stevens B, Tenner AJ. The complement cascade: yin-yang in neuroinflammation — neuro-protection and -degeneration. J Neurochem. 2008;107(5):1169–1187. doi: 10.1111/j.1471-4159.2008.05668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith RS, et al. Haploinsufficiency of the transcription factors FOXC1 and FOXC2 results in aberrant ocular development. Hum Mol Genet. 2000;9(7):1021–1032. doi: 10.1093/hmg/9.7.1021. [DOI] [PubMed] [Google Scholar]
- 61.Botto M, et al. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat Genet. 1998;19(1):56–59. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- 62.Sadun AA, Smith LE, Kenyon KR. Paraphenylenediamine: a new method for tracing human visual pathways. J Neuropathol Exp Neurol. 1983;42(2):200–206. doi: 10.1097/00005072-198303000-00009. [DOI] [PubMed] [Google Scholar]
- 63.Anderson MG, Libby RT, Gould DB, Smith RS, John SW. High-dose radiation with bone marrow transfer prevents neurodegeneration in an inherited glaucoma. Proc Natl Acad Sci U S A. 2005;102(12):4566–4571. doi: 10.1073/pnas.0407357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Churchill GA. Using ANOVA to analyze microarray data. Biotechniques. 2004;37(2):173–175. doi: 10.2144/04372TE01. [DOI] [PubMed] [Google Scholar]
- 65.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy — analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 66.Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6(1):59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- 67.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sherman BT, et al. DAVID Knowledgebase: a gene–centered database integrating heterogeneous gene annotation resources to facilitate high–throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34(Database issue):D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeung KY, Haynor DR, Ruzzo WL. Validating clustering for gene expression data. Bioinformatics. 2001;17(4):309–318. doi: 10.1093/bioinformatics/17.4.309. [DOI] [PubMed] [Google Scholar]
- 72.Soto I, et al. Retinal ganglion cells downregulate gene expression and lose their axons within the optic nerve head in a mouse glaucoma model. J Neurosci. 2008;28(2):548–561. doi: 10.1523/JNEUROSCI.3714-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krege JH, Hodgin JB, Hagaman JR, Smithies O. A noninvasive computerized tail-cuff system for measuring blood pressure in mice. Hypertension. 1995;25(5):1111–1115. doi: 10.1161/01.hyp.25.5.1111. [DOI] [PubMed] [Google Scholar]
- 74.John SW, Hagaman JR, MacTaggart TE, Peng L, Smithes O. Intraocular pressure in inbred mouse strains. Invest Ophthalmol Vis Sci. 1997;38(1):249–253. [PubMed] [Google Scholar]
- 75.Savinova OV, et al. Intraocular pressure in genetically distinct mice: an update and strain survey. BMC Genet. 2001;2:12. doi: 10.1186/1471-2156-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.