Pah1p promotes lipid droplet assembly independent of its role in triacylglycerol synthesis.
Abstract
Lipins are phosphatidate phosphatases that generate diacylglycerol (DAG). In this study, we report that yeast lipin, Pah1p, controls the formation of cytosolic lipid droplets. Disruption of PAH1 resulted in a 63% decrease in droplet number, although total neutral lipid levels did not change. This was accompanied by an accumulation of neutral lipids in the endoplasmic reticulum (ER). The droplet biogenesis defect was not a result of alterations in neutral lipid ratios. No droplets were visible in the absence of both PAH1 and steryl acyltransferases when grown in glucose medium, even though the strain produces as much triacylglycerol as wild type. The requirement of PAH1 for normal droplet formation can be bypassed by a knockout of DGK1. Nem1p, the activator of Pah1p, localizes to a single punctum per cell on the ER that is usually next to a droplet, suggesting that it is a site of droplet assembly. Overall, this study provides strong evidence that DAG generated by Pah1p is important for droplet biogenesis.
Introduction
Cytoplasmic lipid droplets are found in virtually all eukaryotic cells, where they provide a dense source of energy. In higher metazoans, many droplets reside in specialized tissues and provide energy for the entire organism (Murphy, 2001; Guo et al., 2009). The neutral lipids, mainly triacylglycerols (TAGs) and steryl esters (StEs), are surrounded by a single phospholipid leaflet (Tauchi-Sato et al., 2002) in which a unique constellation of proteins resides (Murphy, 2001; Beller et al., 2006; Binns et al., 2006; Bartz et al., 2007). Although many of these proteins catalyze the synthesis or breakdown of the core lipids (Goodman, 2009), others are involved in organelle trafficking and interorganellar communication (Goodman, 2008; Granneman and Moore, 2008; Murphy et al., 2009; Zehmer et al., 2009). Still, other proteins use lipid droplets as depots for storage (Cermelli et al., 2006).
Enlargement of lipid droplets and proliferation of adipose tissue are central to the obesity epidemic (Faust et al., 1978). On the other side of the spectrum is lipodystrophy, in which adipose tissue fails to develop normally as a result of defects in adipogenesis (Agarwal and Garg, 2006). Several transcription factors in the adipogenic pathway have been identified and ordered temporally (Rosen and MacDougald, 2006).
Compared with our knowledge of the transcriptional control of adipogenesis, we know considerably less about the pathway of lipid droplet assembly. There is general (but not unanimous; Robenek et al., 2006) consensus that droplets originate as depositions of neutral lipids between the leaflets of the ER (Martin and Parton, 2006). Somehow, they grow out toward the cytoplasm as buds and perhaps separate from the ER. However, the details of these steps are not clear. Roles for exchangeable perilipin-ADRP-Tip47 family proteins in droplet budding have been proposed (Wolins et al., 2006) with support from recent data (Skinner et al., 2009), and phospholipase D1 and extracellular signal–regulated kinase 2 are required in a cell-free droplet budding assay (Andersson et al., 2006). Genome-wide screens for aberrant droplet morphologies in knockdown and deletion libraries of flies and yeast have implicated the retrograde pathway, and COPI function in particular, in droplet budding, although it is not clear whether the effect is direct or indirect (Szymanski et al., 2007; Beller et al., 2008; Guo et al., 2008; Soni et al., 2009). In yeast droplet morphology screens, lipodystrophy proteins have also been implicated in droplet assembly or maintenance, particularly seipin and lipin (Szymanski et al., 2007; Fei et al., 2008).
The first lipin gene, Lpin1, was originally identified by positional cloning from a spontaneously mutated mouse that showed an 80% decrease in adipose tissue mass, transient fatty liver, and peripheral neuropathy (Klingenspor et al., 1999; Reue et al., 2000; Péterfy et al., 2001). The lipin family in mammals is now known to consist of three genes and five proteins, lipin-1a, -1b, and -1c (alternatively spliced products) and lipin-2 and lipin-3, with different tissue distributions (Péterfy et al., 2001; Donkor et al., 2007). Saccharomyces cerevisiae has one homologue originally named SMP2 (Irie et al., 1993; Péterfy et al., 2001) and then renamed PAH1 when lipins were found to encode phosphatidic acid phosphohydrolases that generate DAG (Fig. 1 A; Han et al., 2006). Pah1p localization is regulated by phosphorylation; it is dephosphorylated by a microsomal Nem1p–Spo7p phosphatase complex and phosphorylated by Cdc28p (Cdk1) kinase (Santos-Rosa et al., 2005; O’Hara et al., 2006). The dephosphorylation by Nem1p results in binding to the membrane bilayer through an amphipathic helix (Karanasios et al., 2010; Choi et al., 2011). Lipins are largely cytosolic but also colocalize, at least in animal cells, to the nucleus. In that compartment, they regulate adipogenesis and phospholipid biosynthesis through transcriptional modulation; this has been directly demonstrated for lipin-1 (Reue and Zhang, 2008). Thus, the lack of adipose tissue in the lipin-1–deficient mouse is thought to be caused by a combination of transcriptional deficiency and a lack of triglycerides as a result of a low concentration of the DAG precursor.
Figure 1.
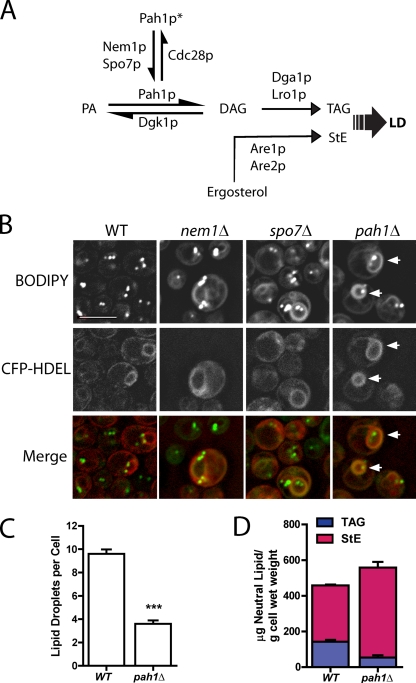
pah1Δ has fewer lipid droplets but a similar amount of neutral lipid. (A) Abbreviated diagram showing the final steps in neutral lipid synthesis. Pah1p*, phosphorylated cytosolic Pah1p; LD, lipid droplet; TAG, triacylglycerol; StE, steryl ester. (B) BODIPY-stained ER rings (arrows). Fluorescence images of cells of the indicated strains containing the ER marker CFP-HDEL, which were cultured in minimal dextrose medium (SD) and stained with BODIPY. Note the fewer droplets in pah1Δ. Bar, 5 µm. (C) Comparison of droplet number in wild-type (WT) and pah1Δ cells. ***, P < 0.0001; droplets in cells (examining z-section series) from four independent experiments, at least 20 cells per field; SEM shown. (D) Total neutral lipid content from mass spectrometry. Means and ranges are shown from two independent experiments.
However, DAG itself may play a more direct role in droplet biogenesis. A recent study (Skinner et al., 2009) showed that stabilizing DAG in membranes recruited perilipin-3, which associated with newly emerging droplets at the ER.
Here, we report that Pah1p, probably working through DAG, is important for droplet assembly independent of the generation of neutral lipids. In the absence of Pah1p, neutral lipids, particularly StEs, accumulate in the ER. We further show that Nem1p, which targets Pah1p to membranes, is usually adjacent to a droplet, consistent with a role in providing nascent droplets with DAG. Finally, we demonstrate that droplets are not synthesized in cells lacking both Pah1p and the steryl acyltransferases Are1p and Are2p when cultured in glucose medium, although the cells are replete in TAG.
Results
Decrease in droplets but not neutral lipids in pah1Δ cells
We previously identified 59 yeast gene disruption strains with altered lipid droplet morphology based on staining with the neutral lipid dye BODIPY 493/503 (Szymanski et al., 2007). In two of these strains, nem1Δ and spo7Δ, BODIPY-stained rings were apparent in a minor fraction (∼30%) of these cells (Fig. 1 B). These rings resembled the ER, and indeed, they colocalized with an ER marker, CFP-HDEL (pseudocolored red in the merge images for better visualization; Fig. 1 B).
The corresponding proteins Nem1p and Spo7p form a complex in the ER (Siniossoglou et al., 1998) and recruit yeast lipin, Pah1p (Santos-Rosa et al., 2005), to convert phosphatidate (PA) to DAG. To determine whether the ring phenotype was related to activation of Pah1p, we generated the deletion strain, pah1Δ, which was missing in the library we originally screened. In this strain, BODIPY-staining rings were apparent in every cell and again colocalized with the ER marker. Strikingly, there were only 37% of lipid droplets in this strain compared with wild type (Fig. 1, B and C). Droplets were more heterogeneous in size in pah1Δ than in wild type.
Cell TAG is known to decrease in pah1Δ because PAH1 encodes the main PA phosphatase that normally provides its precursor, DAG (Han et al., 2006). To determine whether the decrease in lipid droplets in pah1Δ can be attributed to an overall decrease in neutral lipids, we measured both TAG and StE in log-phase cells, where the decrease in TAG is not as severe as in stationary phase (Han et al., 2006). (All fluorescence experiments were also performed in log phase.) As expected, there was a decrease in TAG in pah1Δ cells of comparable magnitude to the decrease of lipid droplets, but there was a compensatory increase in StE so that the sum of these two neutral lipid classes, which normally are packaged into droplets, was at least as high in the mutant (Fig. 1 D). Therefore, the decrease in lipid droplets cannot be attributed to a decrease in overall neutral lipid.
Neutral lipid accumulation in the ER in pah1Δ
The decrease in lipid droplet number without a concomitant decrease in total neutral lipid suggests that there is a defect in droplet biogenesis in pah1Δ. If this were the case, neutral lipids should accumulate in the ER or other membranes in this strain. We first considered that the BODIPY-stained ER rings in the strain reflected neutral lipid accumulation. However, the ER proliferates in pah1Δ as a consequence of derepression of phospholipid biosynthesis, resulting in nuclear envelope deformation and the appearance of ER stacks (Santos-Rosa et al., 2005). To determine whether the BODIPY-stained rings in pah1Δ might be a result of altered ER morphology rather than neutral lipid accumulation, we stained cells that were disrupted in both PAH1 and the four acyltransferases that generate TAG and StE (Oelkers et al., 2002; Petschnigg et al., 2009). BODIPY weakly stained the cytosol in the acyltransferase-deficient “quad” mutant (Fig. 2 A, 4KO). In contrast, membrane rings were indeed observed when pah1Δ was additionally knocked out, generating a “quint” mutant (Fig. 2 A, 5KO). (The intensities of staining among strains can be directly compared because the absolute intensity range for all images of an experiment was fixed in microscope software.) We conclude that BODIPY 493/503 can stain membrane stacks even devoid of neutral lipids as well as it stains lipid droplets in wild type; thus, it cannot be used to detect membrane neutral lipids in pah1Δ.
Figure 2.
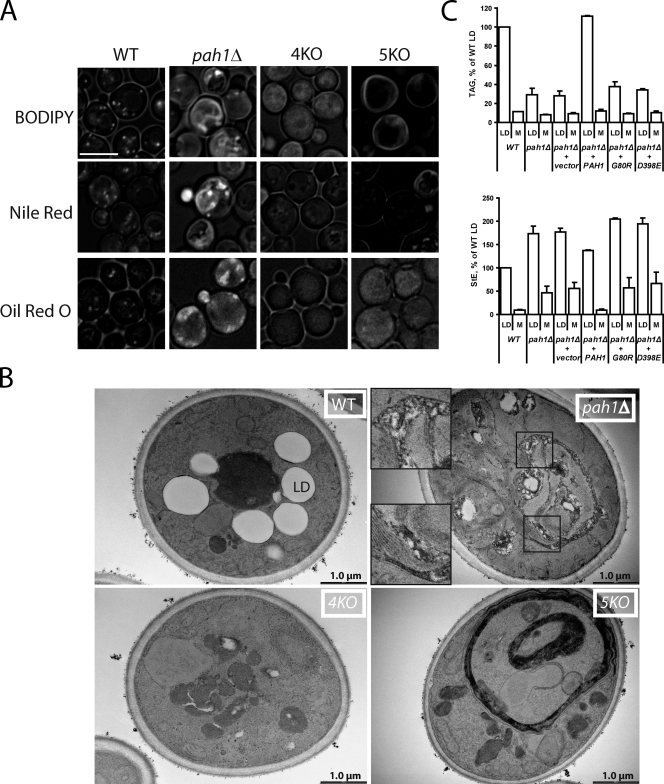
Neutral lipids accumulate in the ER in the absence of PAH1. (A) BODIPY, Nile red, and Oil red O staining of wild type (WT), pah1Δ, 4KO (dga1Δlro1Δare1Δare2Δ), and 5KO (pah1Δdga1Δlro1Δare1Δare2Δ) strains grown in SD medium. Images incorporate both brightfield and fluorescent channel for the indicated lipophilic dyes. Bar, 5 µm. (B) Inclusions in pah1Δ membranes seen with transmission electron microscopy. The indicated strains were cultured overnight in oleic acid medium before fixation. pah1Δ insets are higher magnifications of boxed areas and are shown to illustrate membrane inclusions. Note the membrane proliferation but no inclusions in the 5KO stain. LD, lipid droplet. (C) Droplet (LD) and membrane (M) levels of TAG and StE in the indicated strains. Postnuclear extracts from spheroplasts (grown in oleate) were fractionated by centrifugation. An identical percentage of the floating droplets and membrane pellets, normalized to total protein in the extracts, was subjected to TLC. Error bars indicate the range of values from two independent experiments.
However, both Nile red and Oil red O better differentiated neutral lipids from membrane stacks. Nile red intensely stained both membranes and droplets in pah1Δ; however, the dye also stained membranes in some cells of the quint mutant, regardless of the emission wavelength used (Fig. 2 A and not depicted; Wolinski and Kohlwein, 2008). In contrast, Oil red O, which is traditionally used to label neutral lipid in plants and animals (Ramírez-Zacarías et al., 1992), yielded strong patchy staining in pah1Δ that was quite different from the punctate appearance of droplets in wild type. In contrast, Oil red O did not stain membranes of the quint mutant at all. Although this result suggested neutral lipid staining in membranes in the pah1Δ strain, we could not visualize ER labeled with CFP-HDEL in Oil red O–stained cells to assess colocalization because isopropanol, which destroyed ER morphology, was required for Oil red O staining.
To gain better evidence for ER accumulation of neutral lipids in pah1Δ, we cultured the strains in medium with oleic acid to stimulate neutral lipid synthesis and subjected them to electron microscopy. As expected, wild-type cells contained large lipid droplets, which were absent in the quad mutant (Fig. 2 B). The proliferated ER was clearly seen in pah1Δ. Significantly, the membrane stacks contained many electron-transparent inclusions consistent with neutral lipid in this compartment. Sometimes, larger inclusions were seen within the proliferated ER, as if tiny lipid droplets had coalesced (Fig. S1). The bilayer structure could not be clearly visualized adjacent to the inclusions, and it was impossible to verify that the inclusions were between leaflets of the bilayer. Nevertheless, these membrane inclusions were entirely absent in the proliferated ER of the quint strain.
To biochemically verify membrane accumulation of neutral lipids in pah1Δ, we separated membranes and lipid droplets by ultracentrifugation from postnuclear supernatants of wild-type and pah1Δ cells cultured in oleic acid. In wild-type cells, 10.2% of TAG was found in the membrane fraction (Fig. 2 C). In contrast, 22.0% of TAG was found in membranes in pah1Δ cells, although the absolute amount of TAG in membranes was actually decreased 28% in the mutant. Thus, the membrane inclusions in pah1Δ were likely not predominantly TAG. In contrast, there was 5.2 times more membrane-associated StE in pah1Δ compared with wild type. Based on the large decrease in lipid droplet number with little change in total neutral lipids, membrane staining with Nile red and Oil red O, transparent inclusions seen by electron microscopy, and accumulation of StEs in pah1Δ membranes, we conclude that Pah1p is somehow important for assembly of droplets.
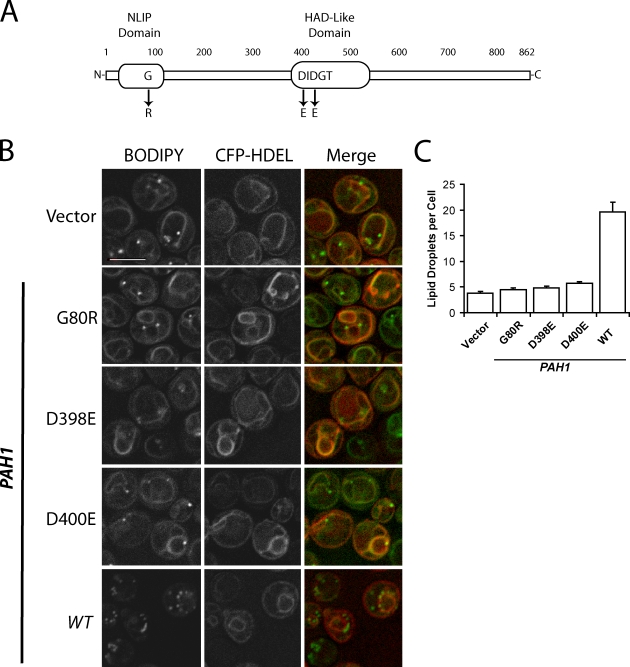
It has previously been shown that PA phosphatase activity of Pah1p depends on the intact N-terminal lipin homology domain and haloacid dehalogenase-like domain of the protein (Fig. 3 A; Han et al., 2006, 2007). To determine whether the low droplet phenotype of pah1Δ is caused by the absence of catalytic activity by itself, three point mutants were generated that are known to disrupt activity, and the pah1Δ strain was transformed with plasmids encoding intact or mutated Pah1p. Although the wild-type copy of Pah1p was able to revert both the ring (Fig. 3 B) and droplet phenotype (Fig. 3 C) to wild type, none of the three point mutants were able to complement the strain. Similarly, only the wild-type allele was able to reverse the accumulation of StE in isolated membrane fractions (Fig. 2 C). We conclude that the PA phosphatase activity of Pah1p is essential for its effect on droplet biogenesis.
Figure 3.
PA phosphatase activity of Pah1p is required for efficient formation of lipid droplets. (A) Diagram of Pah1p primary structure (modified from Han et al., 2007). NLIP, N-terminal lipin homology domain; HAD, haloacid dehalogenase-like domain. (B) The pah1Δ strain expressing the indicated PAH1 alleles on a pRS313 vector. The ER marker CFP-HDEL was expressed on a plasmid and driven by the PGK1 promoter. Lipid droplets were stained with BODIPY. Cells were observed in SD medium. Bar, 5 µm. (C) Droplet number per cell in strains shown in B. Error bars represent SEM based on three fields (∼50 cells per field) from three independent experiments. WT, wild type.
PAH1 and StEs in combination are essential for droplet formation
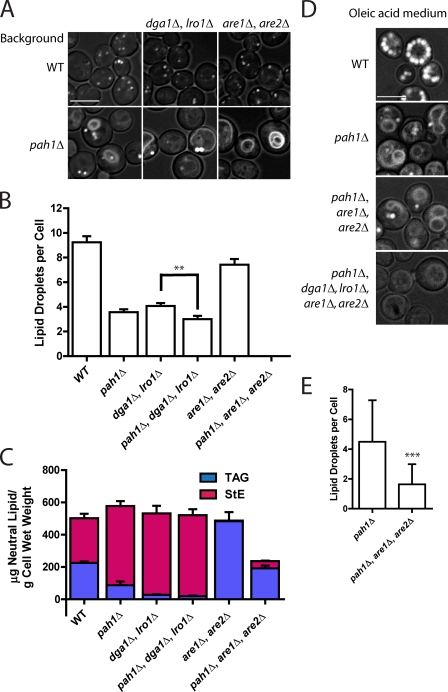
Although total neutral lipid is not decreased in pah1Δ, we next asked whether Pah1p was particularly important for packaging TAG versus StE. To address this, we knocked out PAH1 in cells that either lacked the genes for the two DAG acyltransferases (DGA1 and LRO1) or for the two steryl acyltransferases (ARE1 and ARE2; Yang et al., 1996; Kohlwein, 2010). As reported previously (Garbarino et al., 2009), droplets are formed in the absence of either TAG or StE (Fig. 4 A), although numbers are reduced, indicating that neither TAG nor StE formation is essential for droplet formation. There was a small (26%) but highly reproducible decrease in droplet number when PAH1 was disrupted in dga1Δlro1Δ cells, even though there was no decrease in total StE in this strain (Fig. 4, B and C; also see Fig. S2 and Tables S2 and S3 for distribution of acyl chains). If expressed in terms of cell volume (disruption of PAH1 results in a calculated 96% increase in volume; compare cell sizes in Fig. 4 A), the decrease in droplet number is 51%. The effect of disrupting PAH1 in the are1Δare2Δ background is much more extreme: no droplets at all were detected in pah1Δare1Δare2Δ, even though the cells contained about as much TAG as wild type. In cells cultured in medium containing oleate, the large fatty acid load resulted in some droplet formation in the pah1Δare1Δare2Δ strain, but droplets were far fewer in number even compared with pah1Δ (Fig. 4, D and E). Perhaps the presence of droplets in this strain growing in oleic acid medium is caused by reaching saturation for storing neutral lipids in the ER bilayer. We conclude that in the absence of StE, Pah1p is required to form droplets containing TAG in the absence of exogenous fatty acids, although it also facilitates the packaging of StE in the absence of TAG.
Figure 4.
Knockout of PAH1 and steryl acyltransferases abolishes lipid droplets in cells grown in glucose medium. (A) BODIPY staining of the indicated strains in SD. DGA1 and LRO1 encode DAG acyltransferases, and ARE1 and ARE2 encode steryl acyltransferases. Images incorporate both brightfield and FITC channel (for lipid droplets). Bar, 5 µm. (B) Lipid droplet number in strains shown in A. Droplets in ∼30 cells were counted in z-section series from three independent experiments (∼90 cells total per strain). Mean ± SEM is shown. Knockout of pah1Δ always resulted in significantly lower droplets, although the significance in only the middle pair is shown for clarity; **, P < 0.001. (C) Quantification of TAG and StE from mass spectrometry analysis for the indicated strains grown in SD. Means and ranges from two independent experiments are shown. The values for wild type (WT) and pah1Δ are also shown in Fig. 1 D. (D) BODIPY staining of the indicated strains grown in medium containing oleic acid. Bar, 5 µm. (E) A comparison of droplet number in pah1Δ and pah1Δare1Δare2Δ grown in oleic acid. Error bars represent standard deviation; ***, P < 0.0001; based on two fields (∼50 cells per field) from three independent experiments.
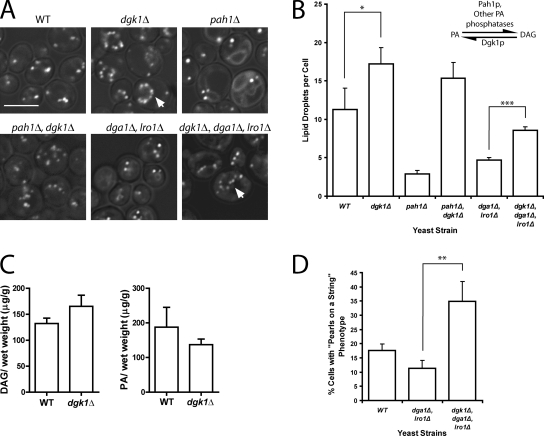
Evidence that DAG is important for droplet formation
Thus far, our data suggest that the DAG generated by Pah1p is important for droplet formation independent of its role as a precursor of TAG. This is consistent with a recent study showing that manipulation of DAG levels in mammalian cells can drastically affect droplet formation (Skinner et al., 2009). If DAG is required for the formation of droplets, overproduction of DAG might be able to bypass the role of Pah1p. To test this idea, we forced more DAG formation by eliminating the backward reaction to PA in a DAG kinase knockout strain, dgk1Δ. DGK1 has recently been shown to be important for providing PA for phospholipid synthesis during the lag phase (Fakas et al., 2011). We confirmed an increase in DAG and decrease in PA in this strain (Fig. 5 C), as has been previously reported (Han et al., 2008; Fakas et al., 2011). As predicted, the dgk1Δ strain alone had significantly more lipid droplets than wild type (Fig. 5, A and B), which were often in a “pearls on a string” (defined as four or more lipid droplets in a row spaced ≤0.5 µm apart) on the perinuclear ER (Fig. 5 A, arrows). The increase in droplets completely bypassed the role of PAH1 as shown in the pah1Δdgk1Δ strain. (In the absence of PAH1, DAG can be generated by another Mg2+-dependent PA phosphatase activity whose identity has not yet been established [Han et al., 2006].) Even if TAG synthesis was completely blocked, the knockout of DGK1 resulted in more droplets (Fig. 5 B, compare droplet number between dga1Δlro1Δ and dgk1Δdga1Δlro1Δ) and an enhancement of the pearls-on-a-string phenotype (Fig. 5 D). These data again demonstrate that the production of DAG itself is probably important for droplet formation in yeast.
Figure 5.
Knockout of DAG kinase leads to more droplets and a bypass in Pah1p function. (A) Knockout of DGK1 increases droplet number independent of TAG. The indicated strains were grown in SD, and lipid droplets were stained with BODIPY. Images incorporate brightfield channel. Arrows illustrate pearls-on-a-string phenotype. Bar, 5 µm. (B) Droplet number increases in dgk1Δ even in the absence of TAG. The inset illustrates the enzymes that catalyze PA-DAG interconversions; the source for the other PA phosphatase activity is unknown. Droplets were counted in at least three random fields, ∼20 cells per field; mean ± SEM is shown. *, P < 0.05; ***, P < 0.0001. (C) Comparison of DAG and PA levels of wild type (WT) and dgk1Δ, showing an increase in DAG and decrease in PA in the mutant. Error bars are SEM based on measurements from three independent experiments. (D) Histogram showing the percentage of cells with pearls-on-a-string lipid droplet phenotype; mean ± SEM (**, P < 0.001) based on three random fields (at least 20 cells per field) from three independent experiments.
The Pah1p phosphatase Nem1p is close to droplets
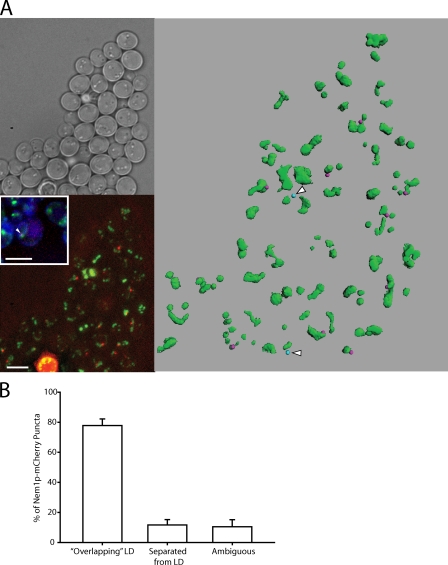
DAG should be produced at the site of droplet formation for it to be active because otherwise it would rapidly diffuse laterally in the membrane. Pah1p is dephosphorylated on the ER by the membranous Nem1p–Spo7p complex; Nem1p is the catalytic subunit of the activating phosphatase (Santos-Rosa et al., 2005), which results in membrane localization of Pah1p, where it is active (Karanasios et al., 2010; Choi et al., 2011). Both Spo7p-GFP and GFP-Nem1p localize to the ER when exogenously expressed (Siniossoglou et al., 1998), whereas GFP knocked in behind the NEM1 reading frame revealed puncta of uncertain origin (Huh et al., 2003). To more closely examine the localization of Nem1p, we tagged the chromosomal copy of NEM1, keeping the endogenous promoter intact, with mCherry. We confirmed that Nem1p forms puncta, and they colocalized with the ER marker CFP-HDEL (Fig. 6 A, bottom left inset). (As expected, GFP-tagged Spo7p colocalized in puncta with dtTomato-tagged Nem1p [Fig. S3].) Interestingly, only one Nem1p-mCherry punctum was usually found per cell (viewing through z sections); only 3% of cells had two puncta, and none had more. Most of the puncta were in close proximity to BODIPY-stained droplets (Fig. 6 A). By scoring the proximity of Nem1p-mCherry to droplets in three dimensions, 77% of them appeared to be in contact with droplets, whereas only 13% could clearly be resolved from droplets (Fig. 6 B). The proximity to droplets was clearly visualized in a computer-aided projection of a field of cells (Fig. 6 A, right; and Video 1). 10 out of 12 Nem1p-mCherry puncta within the field were within 0.65 µm of a droplet, the limit of resolution in this method.
Figure 6.
Nem1p localizes next to lipid droplets on the ER. (A) Nem1p-mCherry was chromosomally expressed at the NEM1 locus. Lipid droplets were stained with BODIPY. (top left) Brightfield image. (bottom left) Projection image of BODIPY-stained cells showing Nem1p-mCherry localization (bottom left inset). The arrowhead is an example of colocalization of CFP-HDEL (ER), BODIPY (lipid droplet), and Nem1p-mCherry. Bars, 5 µm. (right) Three-dimensional reconstruction of the same field of cells. Green, droplets; pink (10 dots), Nem1p within 0.65 µm of a droplet; blue (two dots with arrowheads), Nem1p-mCherry beyond 0.65 µm of a droplet. (B) Juxtaposition of Nem1p-mCherry puncta with lipid droplets (LD). Nem1p-mCherry puncta were counted from seven separate groups of cells and scored for their observed association with lipid droplets. Mean ± SEM was calculated across all seven groups.
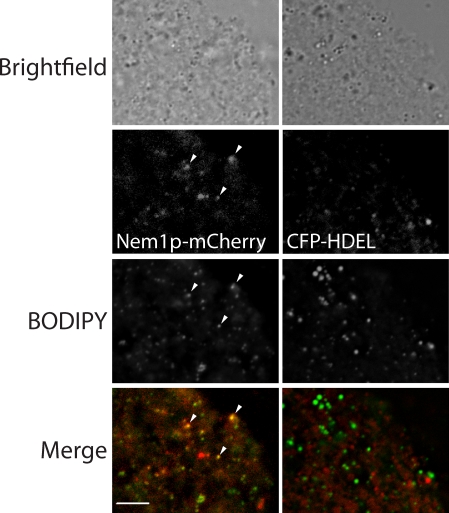
To observe the colocalization of Nem1p with droplets in another way, we subjected membranes derived from a postnuclear supernatant of cells expressing Nem1p-mCherry in the genome to fluorescence microscopy. When grown on glucose medium, droplets were small, and some remained attached to the ER even after centrifugation; these can be visualized as puncta with BODIPY in this preparation (Fig. 7). Most of these spots also coincided with mCherry staining. In contrast, when membranes containing the CFP-HDEL marker were analyzed, there was no colocalization with lipid droplets. The proximity of Nem1p to droplets suggests that Pah1p docks onto the Nem1p–Spo7p complex, where it provides DAG (and later TAG) during droplet production. In yeast, we have observed rapid lateral movement of droplets on the surface of the ER, but never dissociation (Video 2). Perhaps the ∼20% of Nem1p puncta that are clearly separated from droplets are cycling to a new droplet after one is fully formed. In summary, our data strongly suggest that DAG is produced by Pah1p at the site of droplet formation, where it plays an important role in biogenesis of cytoplasmic lipid droplets.
Figure 7.
Colocalization of Nem1p-mCherry and droplets in isolated membranes. Membranes derived from postnuclear supernatant of yeast strains containing chromosomally expressed Nem1p-mCherry (left) or plasmid-expressed 3-phosphoglycerate kinase (PGK) CFP-HDEL (right) were stained with BODIPY. The arrowheads point to three examples of colocalization of Nem1p-mCherry and BODIPY puncta on membranes. Bar, 5 µm.
Discussion
The fatty liver dystrophy mouse, the result of a spontaneous mutation in lipin-1, displays a serious defect in adipogenesis (Péterfy et al., 2001). The defect is consistent with the known roles that lipin-1 plays as a PA phosphatase and a transcriptional regulator of lipid metabolism, both of which lead to low cellular TAG levels (Reue and Zhang, 2008). In this study, we add another likely function for at least the yeast lipin homologue, as a facilitator in lipid droplet assembly through its generation of DAG, independent of TAG formation.
We have presented several lines of evidence that support this novel activity of Pah1p. (a) The absence of PAH1 results in low droplet number, whereas total neutral lipids are not decreased. This results in accumulation of neutral lipids in membranes. (b) PA phosphatase catalytic activity is required for this function. (c) There is a decrease in droplet assembly in pah1Δ regardless of whether cells can synthesize only TAG or StE, although the effect is most profound in the absence of StE. (d) The importance of DAG in droplet formation is suggested by the increase in droplets if DAG concentration is not regulated in dgk1Δ cells, even if no TAG is synthesized. (e) The activator of Pah1p, the protein phosphatase Nem1p, is localized next to droplets 80–85% of the time.
The neutral lipid identity of the electron-transparent inclusions observed for pah1Δ is supported by an increase in membrane StEs (Fig. 2 C). The biochemical analysis of the membrane fraction may underestimate the membrane neutral lipids in the strain. Because droplets remain attached to the ER, it requires high centrifugal force (∼200,000 g) to separate most neutral lipids from the membranes of wild-type cells (unpublished data). This high force may also remove neutral lipids stored in the membrane of the pah1Δ mutant.
We report that both tagged Nem1p and Spo7p yield a punctate appearance in cells, in contradiction to a previous finding (Siniossoglou et al., 1998) in which the ER and other membranes were stained. Expression levels and background strains may contribute to this difference because Siniossoglou et al. (1998) could not detect Nem1p driven by its endogenous promoter. Although that group detected ER-stained Spo7p expressed from its own promoter, they used a different background strain.
The demonstration of one punctum of Nem1p-mCherry per cell that is next to droplets was unexpected. The localization of its mammalian homologue (Dullard) is punctate, but in multiple regions around and inside the nucleus (Kim et al., 2007). The unique localization of Nem1p in yeast may provide an opportunity for understanding the biogenesis of lipid droplets. Its single punctum is in contrast to seipin, another lipodystrophy protein, which is found on the ER next to most, if not all, droplets (Szymanski et al., 2007; Fei et al., 2008). Whether seipin and Spo7p–Nem1p physically interact is unknown; we have failed thus far to detect binding (unpublished data). The single Nem1p punctum may mark a developing lipid droplet, whereas seipin may be required by all droplets and serve a maintenance role. Our ability to visualize the Nem1p puncta by normal epifluorescence microscopy indicates that several copies of the protein must localize there, possibly resulting in several associated Pah1p molecules and a high local concentration of DAG. The 15–20% of the Nem1p-mCherry puncta that is well resolved from lipid droplets may indicate inactive protein, association with nascent droplets not yet visualized with BODIPY, or possibly lateral movement of droplets away from the Nem1p puncta; such rapid movement of droplets can be observed in Video 2. When DAG concentration is uncontrolled in dgk1Δ, spontaneous droplet formation may occur, leading to the pearls-on-a-string droplet phenotype (Fig. 5).
Although it is attractive to consider Nem1p (in combination with Spo7p) as a dock for Pah1p, it is unknown whether the latter actually remains attached once binding initially occurs. Recent experiments show that dephosphorylation of Pah1p results in exposure of an N-terminal amphipathic helix, which can bind to membranes independent of Nem1p (Karanasios et al., 2010). If Pah1p diffused away from Nem1p after activation, it is hard to imagine that DAG would be as effective in droplet formation, assuming that the Nem1p punctum concentrates Pah1p at that site. However, under conditions that favor Nem1p-independent binding of Pah1p to membranes, it still colocalizes with tagged Spo7p, suggesting a more permanent complex (Karanasios et al., 2010).
The single Nem1p punctum per cell also provides an avenue for studying the dynamics of lipid droplet formation. For example, if Nem1p-mCherry marks only nascent droplets, it should colocalize with the first droplet produced in a system in which droplet formation can be regulated. This hypothesis also predicts that multiple droplets do not develop simultaneously in yeast.
The inability of cells to form lipid droplets in the absence of StE and Pah1p-derived DAG in glucose-grown cells was surprising. One explanation might be that there is an accumulation of unesterified sterol that stiffens the membrane (Bacia et al., 2005), preventing droplet formation. This appears unlikely because droplets form in the are1Δare2Δ mutant, although in this case, DAG from Pah1p may overcome this effect. Another possibility is that there is a threshold of total cell neutral lipids before a droplet can form and that the sum of TAG and StE in are1Δare2Δpah1Δ, which was about half of that in the other strains in this experiment, is below that threshold.
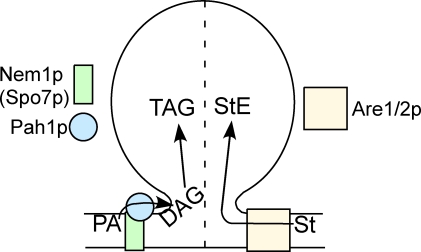
We favor a third explanation, shown in the simple working model of Fig. 8. There are two pathways to droplet formation in this model, one requiring DAG from Pah1p (unless it is overproduced in dgk1Δ) and the other requiring Are1p and/or Are2p. Some droplets will form if either Pah1p or Are1/2p is active, but none will form in the absence of both unless exogenous fatty acids are provided. For the first arm, our data indicate that DAG rather than TAG facilitates droplet formation, as discussed in Results. The effect of DAG in perturbing phospholipid bilayers, promoting high curvature of leaflets, and conversion to hexagonal phase structures is well known (Goñi and Alonso, 1999). DAG clearly promotes budding and fission of vesicles from membranes, such as the Golgi complex (Roth, 1999; Asp et al., 2009). Although DAG can bend a membrane even more than PA because of its smaller head group, it is unstable in a bilayer and can rapidly flip-flop (Pagano and Longmuir, 1985), leading to membrane instability that may promote bud formation. Our knowledge of the roles of DAG in diverse membrane-localized cell physiology continues to expand rapidly (Carrasco and Mérida, 2007). It is possible that the role of DAG is indirect, for example, in recruiting other molecules important for trafficking, such as ARF GTPase-activating protein (Antonny et al., 1997) or, in mammalian cells, perilipin-3 (Skinner et al., 2009). In our in vivo approach, we also cannot rule out potential effects of other lipids, the concentration of which are altered in the pah1Δ strain.
Figure 8.
Working model. Droplets are controlled by Pah1p-generated DAG and Are1p/Are2p-generated StEs in normal glucose medium. The dotted line indicates the possibility that these two inputs control two populations of droplets. St, sterol.
Regarding the second arm of the model, how can Are1/2p promote droplet formation? Perhaps the combination of a local decrease in sterol as a result of the action of Are1/2p and an accumulation of StE between the monolayers of the membrane, forcing them apart, is sufficient to nucleate a droplet.
Our model shows the input of DAG and Are1/2p in the formation of a single droplet. A local high concentration of both DAG and StE may function synergistically in droplet assembly. The dashed line that bisects the droplet in Fig. 8 is meant to indicate the possibility that these two inputs control the assembly of different droplets, one predominantly composed of TAG and the other composed of StE. Although there is evidence for a droplet containing both TAG and StE (Czabany et al., 2008), there also is ample evidence for droplet heterogeneity (Ducharme and Bickel, 2008; Horn et al., 2011).
The current model for droplet formation envisions a lens of neutral lipid developing between the ER leaflets that enlarges to produce a droplet bud that evaginates from the plane of the ER (Martin and Parton, 2006; Wolins et al., 2006). A direct role of DAG, produced by Pah1p at the ER membrane, in promoting the formation of droplets is consistent with this model. Now that several other factors for droplet assembly have been implicated through genetic screens, proteomics, and other approaches, future efforts will focus on how these factors coordinate droplet assembly.
Materials and methods
Reagents
Restriction enzymes, DNA ligases, and other reagents for DNA manipulation were purchased from New England Biolabs, Inc., Roche, and Bio-Rad Laboratories. Neutral lipid standards were obtained from Nu-Chek Prep, Inc. Zymolyase 100T was purchased from US Biologicals, and hygromycin was bought from A.G. Scientific. All other chemical reagents were obtained from Sigma-Aldrich or Thermo Fisher Scientific.
Yeast strains and growth conditions
All yeast strains and plasmids used in this study are listed in Table S1. Strains SCY1703, SCY1998, and SCY328 were gifts from S. Sturley (Columbia University, New York, NY). To generate strain BY4742-KS001, we used the cassette within pGH317 provided by G.S. Han and G. Carman (Rutgers University, New Brunswick, NJ), and we followed their procedure to generate a PAH1 knockout from the cassette (Han et al., 2006) in BY4742. In brief, the cassette consisted of the URA3 gene flanked by ∼400 bp of upstream and downstream PAH1 sequence.
Cells were cultured at 30°C in 2% glucose or in oleic acid (Fig. 2, B and C; and Fig. 4, D and E) as previously described (Hettema et al., 2000). All cultures were harvested during log-phase growth.
DNA manipulations
All plasmids used in this study are listed in Table S1. Standard recombinant DNA techniques were used. Bacterial strain DH10β (Invitrogen) was used for all bacterial transformations. Site-directed mutations to plasmids were performed using the QuikChange site-directed mutagenesis kit (Agilent Technologies). PAH1 knockout in OAS001, OAS002, and OAS003 strains was performed by homologous recombination using a DNA cassette containing ∼500 bp of PAH1 sequence (from the 5′ and 3′ untranslated region flanking the PAH1 ORF) ligated to opposite ends of a TRP1 sequence. The BY4742-SL001 strain was generated by homologous recombination with the use of a DNA cassette containing mCherry followed by the PGK1 3′ terminator and URA3 to the 3′ end of the NEM1 ORF. BY4741-Spo7p-GFP-Nem1p-tdTomato was generated similarly from BY4741-Spo7p-GFP except that the cassette contained tdTomato instead of mCherry and the hygromycin-resistance marker (a gift from B. Tu, University of Texas Southwestern, Dallas, TX) instead of URA3; selection of transformants was performed on rich plates containing 300 µg/ml hygromycin. Yeast strains were transformed by the lithium acetate method (Ito et al., 1983). DNA manipulations resulting in changes to coding sequence as well as changes to genome were verified by sequence performed by the McDermott Center for Human Growth and Development.
Fluorescence and electron microscopy
All microscopy was performed at room temperature. For the acquisition of static images, strains were stained with BODIPY 493/503 (Invitrogen; Szymanski et al., 2007) to visualize lipid droplets and transformed (Figs. 1 B, 3 B, and 7) with pRS315-PGK-CFP-HDEL to visualize the ER. Cells in log phase were removed from growth medium, concentrated by centrifugation (3,000 g for 5 min at room temperature), and suspended on microscope slides in growth medium containing 1% agar (Kron, 2002). All images were acquired in Slidebook (version 4.1.0.3; Intelligent Imaging Innovations) using a microscope (Axiovert 200M; Carl Zeiss, Inc.) with a 100× 1.3 NA oil immersion objective lens (plan-Neofluar) equipped with a digital camera (Sensican; Cooke). BODIPY images were acquired using the fluorescein isothiocyanate filter set, CFP with the CFP filter set, and mCherry with the CY3 filter set (Chroma Technology Corp.). Z stacks were obtained at 0.3-µm spacing, and Slidebook commands of deconvolution (nearest neighbor option) were used to build images through cells. For Fig. 2 A and Fig. 4 (A and D), the range of pixel intensities used for the control strain to clearly visualize droplets was fixed and used for the other strains in the series so the images could be directly compared.
To observe movement of lipid droplets on the ER membrane, BY4742-DB001 expressing the ER marker GFP-HDEL was grown in glucose to log phase and subjected to spinning disk confocal microscopy using a spinning disk confocal system (Ultraview ERS; PerkinElmer) set up on a microscope (Axiovert 200M) with a camera (Orca; Hamamatsu Photonics) in the University of Texas Southwestern Live Imaging Core. Lipid droplets and ER were followed for 4.5 min by saving images over 32 planes (0.2 µm between planes), cycling every 5 s. To generate the QuickTime videos, data were collected over five continuous planes (0.2 µm between them) every 4–5 s and projected into one image per time point.
To visualize Nile red–stained cells, samples were processed and observed as previously published (Wolinski and Kohlwein, 2008). In brief, cells were fixed in 2% vol/vol formaldehyde, incubated for 1 min at room temperature, and washed twice in 50 mM Tris buffer, pH 7.5, before observation. Oil red O–stained cells were processed and observed as previously described (Binns et al., 2006). In brief, cells were washed with H2O and resuspended in freshly filtered Oil red O solution (three parts of 1% stock in isopropanol and two parts H2O). After 10 min of stain, cells were washed twice and observed. Three-dimensional reconstruction (Fig. 6 A, right) was performed using Imaris software (Bitplane AG).
For electron microscopy, yeast cells were grown in 0.1% oleate medium and processed in the University of Texas Southwestern Molecular and Cellular Imaging Facility as previously described (Wright, 2000; Binns et al., 2006). In brief, cells were fixed in potassium permangenate, dehydrated, and postfixed in uranylacetate before sectioning.
Lipid analysis
Lipid extraction and one-dimensional TLC analysis were performed as previously described (Binns et al., 2006) with few modifications. Yeast cell pellets were treated with Zymolyase 100T at 4 mg/1,000 OD600 for 40 min at 30°C. Spheroplasts were weighed and subjected to hot isopropanol and chloroform extraction. Thin layer plates were charred with 3% cupric acetate and 8% phosphoric acid solution, and lipid bands were digitally scanned and analyzed using ImageJ (v.1.41; National Institutes of Health).
Samples subjected to mass spectrometric analysis were spiked with an internal standard mixture composed of 2 µg tri-TAG (15:0), 4 µg tri-TAG (21:0), 2 µg cholesteryl ester (13:0), and 4 µg cholesteryl ester (19:0). Samples were suspended in 0.4 ml chloroform and fractionated by silica gel column chromatography (6 ml Supelco Discovery DSC-Si and 500 mg of solid-phase extraction cartridges). Columns were preconditioned with hexane, and lipids were separated by sequential elution with solvents (hexane/diethyl ether [4:1, vol/vol], hexane/diethyl ether [1:1, vol/vol], methanol, and finally chloroform; Laffargue et al., 2007). Solvents were evaporated off with N2 before suspending neutral lipid in 0.1 ml chloroform. Neutral lipid fractions were diluted into chloroform-methanol [1:1, vol/vol] plus 10 mM aqueous ammonium acetate. Samples were introduced by direct infusion at a flow rate of 750 µl/h into the electrospray source of liquid chromatography tandem mass spectrometry (Waters Micromass Quattro Ultima Triple Quadrupole). TAGs and StEs were quantified in full-scan mode. Neutral loss scans for TAGs and StE were used separately to verify acyl chain compositions and head group. The collision energies, with argon in the collision cell, were 20 V for TAGs and StE. Full and neutral loss/precursor scans were acquired in continuum mode and averaged over 60 and 30 s, respectively. Conditions for acquisition were all under a positive capillary voltage of 3.50 kV, cone gas of 80 ml/h, desolvation gas of 200 ml/h, 80°C source temperature, and 200°C desolvation temperature. The background of each spectrum was subtracted using a solvent blank, the peak areas integrated, isotopic overlap and response correction factors applied, and peaks quantified against their respective internal standards using in-house Visual Basic macros developed in Microsoft Excel.
Cell lysis and organelle fractionation
Yeast microsomes were prepared as described previously (Brodsky et al., 1993) with few modifications. The postnuclear supernatant in yeast lysis buffer (20 mM Hepes, pH 7.4, 200 mM sorbitol, 50 mM KOAc, 2 mM EDTA, and protease inhibitor cocktail: 0.2 mM 4-[2-aminoethyl]benzenesulfonyl fluoride hydrochloride, 1.2 µg/ml pepstatin A, 1.2 µg/ml leupeptin, 12 µg/ml p-toluenesulfonyl-l-arginine methyl ester, 12 µg/ml tosyl-lysyl-chloromethylketone, 12 µg/ml p-nitroenzoyl-l-arginine methyl ester, and 12 µg/ml soybean trypsin inhibitor) was loaded into a tube (SW60; Beckman Coulter), layered with a 1-ml Hepes cushion (20 mM Hepes, pH 7.4, 100 mM KCl, and 2 mM MgCl2), and centrifuged at 38,000 rpm for 1 h at 4°C in a rotor (SW60). Floating lipid droplet and membrane pellets were collected and taken through the lipid analysis procedure as described in the previous section. For Fig. 7, membrane pellets of the required yeast strain were resuspended gently in 0.5 ml lysis buffer and stained for 10 min with BODIPY 493/503 before observation by fluorescence microscopy.
Online supplemental material
The online supplemental material contains a table of strains and plasmids used in this study (Table S1), mass spectroscopic analysis of TAG chain length and StE sterol subtypes in various strains (Tables S2 and S3 and Fig. S2), electron micrographs showing neutral lipid inclusions in pah1Δ (Fig. S1), localization of tagged Pah1p and colocalization of tagged Nem1p and Spo7p (Fig. S3), animation of the proximity of Nem1p with lipid droplets (Video 1), and movement of droplets on ER membranes (Video 2). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201010111/DC1.
Acknowledgments
Thanks to Gil-Soo Han and George Carman for providing a pah1Δ strain used in preliminary experiments and materials for a PAH1 knockout used throughout and to Steve Sturley for the acyltransferase knockout strains. Thanks also to Chris Hilton in the lab for help with lipid separation and analysis, the Goodman lab members for useful discussions, and George Carman for a critical review of the manuscript before submission.
This work was supported by the National Institutes of Health grant GM084210 (to J.M. Goodman), U.S. Department of Energy contract DE-SC0000797 (to K.D. Chapman), a doctoral fellowship from the University of North Texas (to P.J. Horn), and the University of Texas Southwestern Science Teacher Access Resources at Southwestern program (to A. Chandrahas).
Footnotes
Abbreviations used in this paper:
- PA
- phosphatidate
- PGK
- 3-phosphoglycerate kinase
- StE
- steryl ester
- TAG
- triacylglycerol
References
- Agarwal A.K., Garg A. 2006. Genetic basis of lipodystrophies and management of metabolic complications. Annu. Rev. Med. 57:297–311 10.1146/annurev.med.57.022605.114424 [DOI] [PubMed] [Google Scholar]
- Andersson L., Boström P., Ericson J., Rutberg M., Magnusson B., Marchesan D., Ruiz M., Asp L., Huang P., Frohman M.A., et al. 2006. PLD1 and ERK2 regulate cytosolic lipid droplet formation. J. Cell Sci. 119:2246–2257 10.1242/jcs.02941 [DOI] [PubMed] [Google Scholar]
- Antonny B., Huber I., Paris S., Chabre M., Cassel D. 1997. Activation of ADP-ribosylation factor 1 GTPase-activating protein by phosphatidylcholine-derived diacylglycerols. J. Biol. Chem. 272:30848–30851 10.1074/jbc.272.49.30848 [DOI] [PubMed] [Google Scholar]
- Asp L., Kartberg F., Fernandez-Rodriguez J., Smedh M., Elsner M., Laporte F., Bárcena M., Jansen K.A., Valentijn J.A., Koster A.J., et al. 2009. Early stages of Golgi vesicle and tubule formation require diacylglycerol. Mol. Biol. Cell. 20:780–790 10.1091/mbc.E08-03-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacia K., Schwille P., Kurzchalia T. 2005. Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc. Natl. Acad. Sci. USA. 102:3272–3277 10.1073/pnas.0408215102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz R., Zehmer J.K., Zhu M., Chen Y., Serrero G., Zhao Y., Liu P. 2007. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6:3256–3265 10.1021/pr070158j [DOI] [PubMed] [Google Scholar]
- Beller M., Riedel D., Jänsch L., Dieterich G., Wehland J., Jäckle H., Kühnlein R.P. 2006. Characterization of the Drosophila lipid droplet subproteome. Mol. Cell. Proteomics. 5:1082–1094 10.1074/mcp.M600011-MCP200 [DOI] [PubMed] [Google Scholar]
- Beller M., Sztalryd C., Southall N., Bell M., Jäckle H., Auld D.S., Oliver B. 2008. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 6:e292 10.1371/journal.pbio.0060292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D., Januszewski T., Chen Y., Hill J., Markin V.S., Zhao Y., Gilpin C., Chapman K.D., Anderson R.G., Goodman J.M. 2006. An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173:719–731 10.1083/jcb.200511125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J.L., Hamamoto S., Feldheim D., Schekman R. 1993. Reconstitution of protein translocation from solubilized yeast membranes reveals topologically distinct roles for BiP and cytosolic Hsc70. J. Cell Biol. 120:95–102 10.1083/jcb.120.1.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco S., Mérida I. 2007. Diacylglycerol, when simplicity becomes complex. Trends Biochem. Sci. 32:27–36 10.1016/j.tibs.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Cermelli S., Guo Y., Gross S.P., Welte M.A. 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16:1783–1795 10.1016/j.cub.2006.07.062 [DOI] [PubMed] [Google Scholar]
- Choi H.S., Su W.M., Morgan J.M., Han G.S., Xu Z., Karanasios E., Siniossoglou S., Carman G.M. 2011. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER(602), THR(723), and SER(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286:1486–1498 10.1074/jbc.M110.155598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabany T., Wagner A., Zweytick D., Lohner K., Leitner E., Ingolic E., Daum G. 2008. Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 283:17065–17074 10.1074/jbc.M800401200 [DOI] [PubMed] [Google Scholar]
- Donkor J., Sariahmetoglu M., Dewald J., Brindley D.N., Reue K. 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282:3450–3457 10.1074/jbc.M610745200 [DOI] [PubMed] [Google Scholar]
- Ducharme N.A., Bickel P.E. 2008. Lipid droplets in lipogenesis and lipolysis. Endocrinology. 149:942–949 10.1210/en.2007-1713 [DOI] [PubMed] [Google Scholar]
- Fakas S., Konstantinou C., Carman G.M. 2011. DGK1-encoded diacylglycerol kinase activity is required for phospholipid synthesis during growth resumption from stationary phase in Saccharomyces cerevisiae. J. Biol. Chem. 286:1464–1474 10.1074/jbc.M110.194308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust I.M., Johnson P.R., Stern J.S., Hirsch J. 1978. Diet-induced adipocyte number increase in adult rats: a new model of obesity. Am. J. Physiol. 235:E279–E286 [DOI] [PubMed] [Google Scholar]
- Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., Yang H. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180:473–482 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino J., Padamsee M., Wilcox L., Oelkers P.M., D’Ambrosio D., Ruggles K.V., Ramsey N., Jabado O., Turkish A., Sturley S.L. 2009. Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J. Biol. Chem. 284:30994–31005 10.1074/jbc.M109.050443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goñi F.M., Alonso A. 1999. Structure and functional properties of diacylglycerols in membranes. Prog. Lipid Res. 38:1–48 10.1016/S0163-7827(98)00021-6 [DOI] [PubMed] [Google Scholar]
- Goodman J.M. 2008. The gregarious lipid droplet. J. Biol. Chem. 283:28005–28009 10.1074/jbc.R800042200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman J.M. 2009. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J. Lipid Res. 50:2148–2156 10.1194/jlr.R001446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman J.G., Moore H.P. 2008. Location, location: protein trafficking and lipolysis in adipocytes. Trends Endocrinol. Metab. 19:3–9 10.1016/j.tem.2007.10.006 [DOI] [PubMed] [Google Scholar]
- Guo Y., Walther T.C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J.S., Vale R.D., Walter P., Farese R.V. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 453:657–661 10.1038/nature06928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Cordes K.R., Farese R.V., Jr, Walther T.C. 2009. Lipid droplets at a glance. J. Cell Sci. 122:749–752 10.1242/jcs.037630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.S., Wu W.I., Carman G.M. 2006. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281:9210–9218 10.1074/jbc.M600425200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.S., Siniossoglou S., Carman G.M. 2007. The cellular functions of the yeast lipin homolog PAH1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282:37026–37035 10.1074/jbc.M705777200 [DOI] [PubMed] [Google Scholar]
- Han G.S., O’Hara L., Siniossoglou S., Carman G.M. 2008. Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase. J. Biol. Chem. 283:20443–20453 10.1074/jbc.M802866200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Girzalsky W., van Den Berg M., Erdmann R., Distel B. 2000. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19:223–233 10.1093/emboj/19.2.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn P.J., Ledbetter N.R., James C.N., Hoffman W.D., Case C.R., Verbeck G.F., Chapman K.D. 2011. Visualization of lipid droplet composition by direct organelle mass spectrometry. J. Biol. Chem. 286:3298–3306 10.1074/jbc.M110.186353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Irie K., Takase M., Araki H., Oshima Y. 1993. A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol. Gen. Genet. 236:283–288 10.1007/BF00277124 [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E., Han G.S., Xu Z., Carman G.M., Siniossoglou S. 2010. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. USA. 107:17539–17544 10.1073/pnas.1007974107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Gentry M.S., Harris T.E., Wiley S.E., Lawrence J.C., Jr, Dixon J.E. 2007. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc. Natl. Acad. Sci. USA. 104:6596–6601 10.1073/pnas.0702099104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenspor M., Xu P., Cohen R.D., Welch C., Reue K. 1999. Altered gene expression pattern in the fatty liver dystrophy mouse reveals impaired insulin-mediated cytoskeleton dynamics. J. Biol. Chem. 274:23078–23084 10.1074/jbc.274.33.23078 [DOI] [PubMed] [Google Scholar]
- Kohlwein S.D. 2010. Triacylglycerol homeostasis: insights from yeast. J. Biol. Chem. 285:15663–15667 10.1074/jbc.R110.118356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron S.J. 2002. Digital time-lapse microscopy of yeast cell growth. Methods Enzymol. 351:3–15 10.1016/S0076-6879(02)51838-3 [DOI] [PubMed] [Google Scholar]
- Laffargue A., de Kochko A., Dussert S. 2007. Development of solid-phase extraction and methylation procedures to analyse free fatty acids in lipid-rich seeds. Plant Physiol. Biochem. 45:250–257 10.1016/j.plaphy.2007.01.012 [DOI] [PubMed] [Google Scholar]
- Martin S., Parton R.G. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7:373–378 10.1038/nrm1912 [DOI] [PubMed] [Google Scholar]
- Murphy D.J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40:325–438 10.1016/S0163-7827(01)00013-3 [DOI] [PubMed] [Google Scholar]
- Murphy S., Martin S., Parton R.G. 2009. Lipid droplet-organelle interactions; sharing the fats. Biochim. Biophys. Acta. 1791:441–447 [DOI] [PubMed] [Google Scholar]
- Oelkers P., Cromley D., Padamsee M., Billheimer J.T., Sturley S.L. 2002. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277:8877–8881 10.1074/jbc.M111646200 [DOI] [PubMed] [Google Scholar]
- O’Hara L., Han G.S., Peak-Chew S., Grimsey N., Carman G.M., Siniossoglou S. 2006. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281:34537–34548 10.1074/jbc.M606654200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano R.E., Longmuir K.J. 1985. Phosphorylation, transbilayer movement, and facilitated intracellular transport of diacylglycerol are involved in the uptake of a fluorescent analog of phosphatidic acid by cultured fibroblasts. J. Biol. Chem. 260:1909–1916 [PubMed] [Google Scholar]
- Péterfy M., Phan J., Xu P., Reue K. 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27:121–124 10.1038/83685 [DOI] [PubMed] [Google Scholar]
- Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C.F., Natter K., Kohlwein S.D. 2009. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 284:30981–30993 10.1074/jbc.M109.024752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Zacarías J.L., Castro-Muñozledo F., Kuri-Harcuch W. 1992. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry. 97:493–497 10.1007/BF00316069 [DOI] [PubMed] [Google Scholar]
- Reue K., Zhang P. 2008. The lipin protein family: dual roles in lipid biosynthesis and gene expression. FEBS Lett. 582:90–96 10.1016/j.febslet.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reue K., Xu P., Wang X.P., Slavin B.G. 2000. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J. Lipid Res. 41:1067–1076 [PubMed] [Google Scholar]
- Robenek H., Hofnagel O., Buers I., Robenek M.J., Troyer D., Severs N.J. 2006. Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J. Cell Sci. 119:4215–4224 10.1242/jcs.03191 [DOI] [PubMed] [Google Scholar]
- Rosen E.D., MacDougald O.A. 2006. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7:885–896 10.1038/nrm2066 [DOI] [PubMed] [Google Scholar]
- Roth M.G. 1999. Lipid regulators of membrane traffic through the Golgi complex. Trends Cell Biol. 9:174–179 10.1016/S0962-8924(99)01535-4 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24:1931–1941 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., Hurt E. 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17:6449–6464 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J.R., Shew T.M., Schwartz D.M., Tzekov A., Lepus C.M., Abumrad N.A., Wolins N.E. 2009. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J. Biol. Chem. 284:30941–30948 10.1074/jbc.M109.013995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni K.G., Mardones G.A., Sougrat R., Smirnova E., Jackson C.L., Bonifacino J.S. 2009. Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 122:1834–1841 10.1242/jcs.045849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., Goodman J.M. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277:44507–44512 10.1074/jbc.M207712200 [DOI] [PubMed] [Google Scholar]
- Valachovic M., Bareither B.M., Shah Alam Bhuiyan M., Eckstein J., Barbuch R., Balderes D., Wilcox L., Sturley S.L., Dickson R.C., Bard M. 2006. Cumulative mutations affecting sterol biosynthesis in the yeast Saccharomyces cerevisiae result in synthetic lethality that is suppressed by alterations in sphingolipid profiles. Genetics. 173:1893–1908 10.1534/genetics.105.053025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins N.E., Brasaemle D.L., Bickel P.E. 2006. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 580:5484–5491 10.1016/j.febslet.2006.08.040 [DOI] [PubMed] [Google Scholar]
- Wolinski H., Kohlwein S.D. 2008. Microscopic analysis of lipid droplet metabolism and dynamics in yeast. Methods Mol. Biol. 457:151–163 [DOI] [PubMed] [Google Scholar]
- Wright R. 2000. Transmission electron microscopy of yeast. Microsc. Res. Tech. 51:496–510 [DOI] [PubMed] [Google Scholar]
- Yang H., Bard M., Bruner D.A., Gleeson A., Deckelbaum R.J., Aljinovic G., Pohl T.M., Rothstein R., Sturley S.L. 1996. Sterol esterification in yeast: a two-gene process. Science. 272:1353–1356 10.1126/science.272.5266.1353 [DOI] [PubMed] [Google Scholar]
- Zehmer J.K., Huang Y., Peng G., Pu J., Anderson R.G., Liu P. 2009. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 9:914–921 10.1002/pmic.200800584 [DOI] [PMC free article] [PubMed] [Google Scholar]