Abstract
Suppose that under the conventional randomized clinical trial setting, a new therapy is compared with a standard treatment. In this article, we propose a systematic, 2-stage estimation procedure for the subject-level treatment differences for future patient's disease management and treatment selections. To construct this procedure, we first utilize a parametric or semiparametric method to estimate individual-level treatment differences, and use these estimates to create an index scoring system for grouping patients. We then consistently estimate the average treatment difference for each subgroup of subjects via a nonparametric function estimation method. Furthermore, pointwise and simultaneous interval estimates are constructed to make inferences about such subgroup-specific treatment differences. The new proposal is illustrated with the data from a clinical trial for evaluating the efficacy and toxicity of a 3-drug combination versus a standard 2-drug combination for treating HIV-1–infected patients.
Keywords: Cross-validation, HIV infection, Non-parametric function estimation, Personalized medicine, Subgroup analysis
1. INTRODUCTION
One of the major components for modern evidence-based medicine is to utilize the patient's “baseline” information for personalized disease management and treatment selection. For instance, in a recent study, it was demonstrated that the benefit of giving toxic chemotherapy prior to hormone therapy with tamoxifen for postmenopausal women with lymph node–negative breast cancer varies depending on the estrogen receptor (ER) status of the tumor. Those with ER-negative tumors benefited substantially from chemotherapy, while those with ER-positive tumors did not benefit as compared to receiving tamoxifen alone (Goldhirsch and others, 2002). In another example with an observational study, Sabine (2005) suggested that the efficacy and toxicity profiles for the highly active antiretroviral therapy vary markedly across different subgroups of HIV-infected patients and recommended certain subject-specific treatment strategies. These individualized decision rules can be extremely useful in practice. However, there are numerous examples that the results from the so-called subgroup analyses, which were not properly planned or executed subgroup analyses, could not be validated (Rothwell, 2005, Pfeffer and Jarcho, 2006, Wang and others, 2007).
In this paper, we consider the case that a new therapy was compared with a control under the standard randomized comparative clinical trial setting. Generally, the main goal of a randomized clinical trial is to make inferences about an “overall” treatment difference with respect to efficacy and toxicity. On the other hand, a “positive” trial does not imply that all future patients would benefit from the new treatment. Moreover, a “negative” study does not mean that all patients should be treated by the standard therapy. In fact, based on the extensive collection of the study patient's baseline information from a clinical trial, it would be valuable to utilize such information to make inferences about the individual-level treatment efficacy.
In practice, a commonly employed first step to perform subgroup analyses is to examine whether there are statistically significant interactions between the treatment assignment indicator and various baseline covariates via, for example, parametric regression models (Byar, 1985). This ad hoc “fishing expedition” approach may not accurately or efficiently identify proper subgroups of patients who would benefit from the new treatment. Recently, Song and Pepe (2004) proposed a novel procedure to identify a critical value for a “single” covariate, which may guide us to split the future population into 2 groups. Patients in one group would be treated by the new therapy and those in the other group would take the control. Although such a approach is interesting with respect to an overall utility for the entire population, it does not provide a treatment choice scheme at a subject-specific level. Moreover, if the treatment difference is not monotonically associated with this single covariate, such a procedure may not be appropriate. To examine the treatment difference at a subject-level with a single covariate (Bonetti and Gelber, 2000, Bonetti and Gelber, 2005) grouped the patients with their covariate values and utilized a moving average procedure to make inferences about the profile of the treatment differences over the covariate. Recently, in her unpublished thesis, Park (2004) proposed a procedure to identify patients who may or may not benefit from the new treatment based on generalized linear models and the Cox proportional hazards models. The validity of her procedure heavily relies on the model assumptions.
In this paper, we consider the case that each patient has “multiple” baseline covariates and propose a systematic, 2-stage method to identify patients who would benefit from the new treatment. Specifically, for the first stage, we use a parametric or semiparametric model to estimate the subject-specific mean response for each treatment group. We then utilize the resulting difference of these 2 estimates as an index scoring system to group patients. That is, subjects in each subgroup would have the same parametric index score. For the second stage, we calibrate the estimated treatment differences with a consistent, nonparametric function estimation procedure and provide valid “pointwise” and “simultaneous” inferences about the true average treatment difference for each group of patients by controlling the desirable confidence level locally and globally. The new proposal is illustrated with a data set from a clinical trial for evaluating a 3-drug combination versus a standard 2-drug combination for treating HIV-infected patients. For cost-benefit decision makings, we also provide the underlying mean treatment response for each treatment group over the index score and the estimated relative frequency for the parametric score.
2. POINT AND INTERVAL ESTIMATION FOR TREATMENT DIFFERENCES
Suppose that study subjects in a population of interest are randomly assigned to either a standard treatment denoted by G = 0 or an experimental treatment denoted by G = 1. Let U denote a vector of baseline covariates, Y denote the response variable, and Y(k) denote the potential outcome of an individual if he/she were assigned to treatment group G = k. Without loss of generality, we assume that a larger value of Y is beneficial. Now, suppose that for subjects with U = u, we are interested in estimating the treatment difference
Our data consist of n random vectors {(Yi,Ui,Gi),i = 1,…,n}, where we assume that {(Yi,Ui):Gi = k} are nk = ∑i = 1nI(Gi = k) independent and identically distributed random vectors, for k = 0,1. Furthermore, the ratio n1/n goes to a constant in the open interval (0,1) as n→∞. To estimate 𝒮(u), one may consider a nonparametric function procedure. In practice, however, when u is not univariate, generally it is difficult, if not impossible, to estimate 𝒮(U) well nonparametrically.
To reduce the complexity of the high-dimensional problem, conventionally one utilizes a parametric or semiparametric procedure to estimate 𝒮(u). Here, we consider the following generalized linear “working” model to approximate the mean function of Y(k):
| (2.1) |
where Z, a p×1 vector, is a function of U with first column being 1, gk is a known, strictly increasing smooth link function, and βk is an unknown vector of regression coefficients. Note that in (2.1), we only model the mean function of Y. To estimate the parameter vector βk in (2.1) without distribution assumptions about the response, one may use a solution 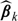 to the following estimating equation:
to the following estimating equation:
| (2.2) |
Using the arguments given in Tian and others (2007), one can show that 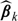 converges to a deterministic vector
converges to a deterministic vector 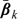 even when Model (2.1) is incorrectly specified. This stability property is crucial for developing our new procedure. It follows that for a given u, a parametric estimator for 𝒮(u) is
even when Model (2.1) is incorrectly specified. This stability property is crucial for developing our new procedure. It follows that for a given u, a parametric estimator for 𝒮(u) is
Note that when Model (2.1) is correctly specified, 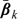 is the true parameter for Model (2.1) and
is the true parameter for Model (2.1) and 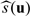 is a consistent estimator of 𝒮(u).
is a consistent estimator of 𝒮(u).
Let U0 be the baseline covariate vector for a future subject from the study population. If this subject is treated by treatment k, the response is Y(k)0,k = 0,1. For U0 = u0, we may use 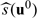 to decide which treatment this specific subject should be treated with. The adequacy of such a decision heavily depends on the appropriateness of Model (2.1). On the other hand,
to decide which treatment this specific subject should be treated with. The adequacy of such a decision heavily depends on the appropriateness of Model (2.1). On the other hand, 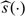 may be used as an index scoring system for grouping future subjects with potentially similar treatment differences. That is, we divide the future population into many strata based on the score
may be used as an index scoring system for grouping future subjects with potentially similar treatment differences. That is, we divide the future population into many strata based on the score 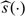 such that patients in the same subset
such that patients in the same subset 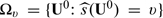 have the same parametric score value v. To employ such scoring system in clinical practice, one could create strata by segmenting all possible values of
have the same parametric score value v. To employ such scoring system in clinical practice, one could create strata by segmenting all possible values of 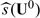 into small intervals.
into small intervals.
Now, for any given v, consider subjects in the subset Ωv. We propose to consistently estimate the average treatment difference for this subgroup,
based on a local likelihood approach (Tibshirani and Hastie, 1987, Fan and Gijbels, 1996), where μk(v) = E(Y(k)0|U0∈Ωv),k = 0,1, and the expectation is taken with respect to the data and (Y(k)0,U0). Specifically, we obtain the root 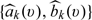 to the local weighted estimating equation,
to the local weighted estimating equation, 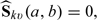 where
where
| (2.3) |
where h is the smoothing parameter chosen such that h = Op(n − ν) with 1/5 < ν < 1/2, Kh(x) = K(x/h)/h, K(x) is a symmetric kernel function with a finite support, 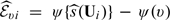 and ψ(·) is a suitably chosen, nondecreasing function. Here, g(x) = x if the response Y is continuous and g(x) = exp(x)/{1 + exp(x)} if Y is binary. Transforming the score via an appropriately chosen function, ψ(·) could potentially improve the performance of the kernel estimation (Wand and others, 1991, Park and others, 1997). The μk(v) can then be estimated by
and ψ(·) is a suitably chosen, nondecreasing function. Here, g(x) = x if the response Y is continuous and g(x) = exp(x)/{1 + exp(x)} if Y is binary. Transforming the score via an appropriately chosen function, ψ(·) could potentially improve the performance of the kernel estimation (Wand and others, 1991, Park and others, 1997). The μk(v) can then be estimated by 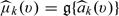 This estimator corresponds to the local linear least square estimator for continuous Y and local linear logistic likelihood estimator for binary Y. Subsequently, we estimate
This estimator corresponds to the local linear least square estimator for continuous Y and local linear logistic likelihood estimator for binary Y. Subsequently, we estimate 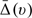 as
as
In Section A of the Supplementary Material available at Biostatistics online, we show that 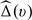 is uniformly consistent for
is uniformly consistent for 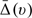 , for v∈𝒥 = [ρl,ρr], where 𝒥 is an interval properly contained in the support of
, for v∈𝒥 = [ρl,ρr], where 𝒥 is an interval properly contained in the support of 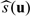 .
.
To construct confidence interval for 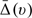 , the average treatment difference among the stratum Ωv, we show in Section B of the Supplementary Material available at Biostatistics online that for large n, the distribution of
, the average treatment difference among the stratum Ωv, we show in Section B of the Supplementary Material available at Biostatistics online that for large n, the distribution of 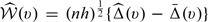 can be approximated by the conditional distribution of a mean-zero normal variable
can be approximated by the conditional distribution of a mean-zero normal variable
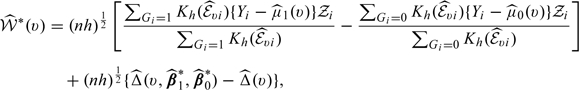 |
(2.4) |
given the data, where 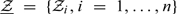 is a random sample from the standard normal variable and is independent of the data,
is a random sample from the standard normal variable and is independent of the data, 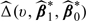 is obtained by replacing
is obtained by replacing 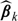 in
in 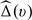 with
with 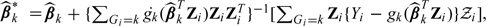 , for k = 0,1, where for any function g, g(·) denotes its derivative. Note that
, for k = 0,1, where for any function g, g(·) denotes its derivative. Note that 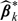 is a solution to the counterpart of estimating equation (2.2), which is perturbed with the same set of
is a solution to the counterpart of estimating equation (2.2), which is perturbed with the same set of 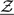 in (2.4). Also, note that
in (2.4). Also, note that 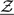 are the only random quantities in (2.4), whose distribution can be approximated easily by simulating
are the only random quantities in (2.4), whose distribution can be approximated easily by simulating 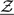 repeatedly. A (1 − α) pointwise confidence interval estimates for
repeatedly. A (1 − α) pointwise confidence interval estimates for 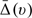 can be constructed via this large-sample approximation, which is
can be constructed via this large-sample approximation, which is 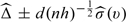 . Here,
. Here, 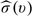 is the standard error estimate of
is the standard error estimate of 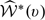 and d is the upper (1 − α/2) percentile of the standard normal.
and d is the upper (1 − α/2) percentile of the standard normal.
To select subgroups that may benefit from the new treatment, one may aim to identify regions of v such that 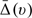 is above or below a certain threshold value. However, as for any subgroup analysis, it is crucial to control for the global error rate. For the present case, we may control for the overall error rate by constructing a simultaneous confidence band for
is above or below a certain threshold value. However, as for any subgroup analysis, it is crucial to control for the global error rate. For the present case, we may control for the overall error rate by constructing a simultaneous confidence band for 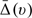 . Specifically, to make inference about the treatment differences over a range of v, one may construct a simultaneous confidence band for
. Specifically, to make inference about the treatment differences over a range of v, one may construct a simultaneous confidence band for 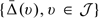 A conventional way to obtain such a confidence band is based on a sup-type statistic
A conventional way to obtain such a confidence band is based on a sup-type statistic
| (2.5) |
Although 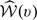 does not converge weakly as a process in v, we show in Section C of the Supplementary Material available at Biostatistics online that a standardized version of
does not converge weakly as a process in v, we show in Section C of the Supplementary Material available at Biostatistics online that a standardized version of 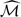 converges in distribution to a proper random variable. In practice, for large n, one can approximate the distribution of
converges in distribution to a proper random variable. In practice, for large n, one can approximate the distribution of 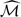 by
by 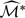 , the sup of the absolute value of
, the sup of the absolute value of 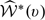 divided by
divided by 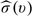 , with (2.4) perturbed by the same set of perturbation variables
, with (2.4) perturbed by the same set of perturbation variables 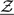 for all v∈𝒥. It follows that the 100(1 − α)% simultaneous confidence interval for
for all v∈𝒥. It follows that the 100(1 − α)% simultaneous confidence interval for 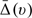 is
is
| (2.6) |
where the cutoff point γ is chosen such that pr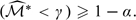
The simultaneous interval estimators for 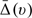 can be used to suggest treatment options for patient subgroups. For example, if there is little concern regarding the cost of the new treatment relative to the standard treatment, then one may suggest the new treatment to subgroups in
can be used to suggest treatment options for patient subgroups. For example, if there is little concern regarding the cost of the new treatment relative to the standard treatment, then one may suggest the new treatment to subgroups in 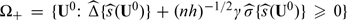 On the other hand, if the new treatment is expensive or has serious adverse risk, only patients with
On the other hand, if the new treatment is expensive or has serious adverse risk, only patients with 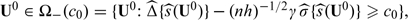 where c0 is a preselected cutoff value representing adequate benefit. Making decisions based on the simultaneous confidence intervals ensures that there is no inflated type I error associated with testing whether
where c0 is a preselected cutoff value representing adequate benefit. Making decisions based on the simultaneous confidence intervals ensures that there is no inflated type I error associated with testing whether 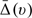 exceeds or is above a certain threshold value for a range of v. This is particularly important if
exceeds or is above a certain threshold value for a range of v. This is particularly important if 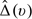 is a nonmonotone function crossing zero more than once.
is a nonmonotone function crossing zero more than once.
As for any nonparametric functional estimation problem, the choice of the smoothing parameter h for 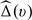 is crucial for making inferences about
is crucial for making inferences about 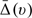 . We may choose a single smooth parameter for 2 nonparametric function estimates,
. We may choose a single smooth parameter for 2 nonparametric function estimates, 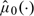 and
and 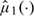 . Since each study subject is assigned to a single-treatment group, therefore, we cannot use the standard cross-validation method with the integrated mean square error criterion to choose h for
. Since each study subject is assigned to a single-treatment group, therefore, we cannot use the standard cross-validation method with the integrated mean square error criterion to choose h for 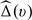 . A reasonable, feasible alternative to choose an “optimal” smooth parameter is minimizing an average squared distance between the observed “cumulative” treatment difference, and their predicted counterparts in the validation samples under the 𝒦-fold cross-validation setting. Noting that
. A reasonable, feasible alternative to choose an “optimal” smooth parameter is minimizing an average squared distance between the observed “cumulative” treatment difference, and their predicted counterparts in the validation samples under the 𝒦-fold cross-validation setting. Noting that
| (2.7) |
we propose to use empirical estimates of E(Yi|Gi = 1,Ui ≤ u) − E(Yi|Gi = 0,Ui ≤ u) as the observed cumulative treatment difference and the empirical counterpart of 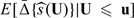 as the predicted cumulative treatment difference based on the proposed procedure. Here and in the sequel, for any 2 vectors U = (U1,…,Uq)T and u = (u1,…,uq)T, U ≤ u denotes {Uj ≤ uj,j = 1,…,q}. Specifically, we randomly split the data into 𝒦 disjoint subsets of about equal sizes, denoted by {ℐι,ι = 1,…,𝒦}. For each ι, we use all observations not in ℐι to obtain estimators for the parametric index score and
as the predicted cumulative treatment difference based on the proposed procedure. Here and in the sequel, for any 2 vectors U = (U1,…,Uq)T and u = (u1,…,uq)T, U ≤ u denotes {Uj ≤ uj,j = 1,…,q}. Specifically, we randomly split the data into 𝒦 disjoint subsets of about equal sizes, denoted by {ℐι,ι = 1,…,𝒦}. For each ι, we use all observations not in ℐι to obtain estimators for the parametric index score and 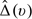 with a given h. Let the resulting estimators be denoted by
with a given h. Let the resulting estimators be denoted by 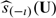 and
and 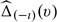 , respectively. We then use the observations from ℐι to calculate the empirical integrated cumulative square error
, respectively. We then use the observations from ℐι to calculate the empirical integrated cumulative square error
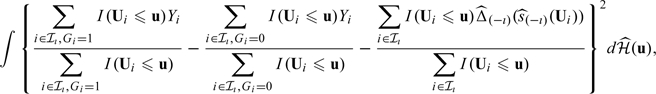 |
(2.8) |
where I(·) is the indicator function and 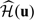 is an empirical weight function such as the empirical distribution function. Last, we sum (2.8) over ι = 1,…,𝒦, and then choose h by minimizing this sum. It is important to note that since the bandwidth is selected by minimizing a quantity which is composed of cumulative predicted errors, the order of such optimal bandwidth is expected to be n − 1/3 (Bowman and others, 1998). Thus, the selected bandwidth satisfies the condition required for the resulting functional estimator
is an empirical weight function such as the empirical distribution function. Last, we sum (2.8) over ι = 1,…,𝒦, and then choose h by minimizing this sum. It is important to note that since the bandwidth is selected by minimizing a quantity which is composed of cumulative predicted errors, the order of such optimal bandwidth is expected to be n − 1/3 (Bowman and others, 1998). Thus, the selected bandwidth satisfies the condition required for the resulting functional estimator 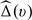 with the data-dependent smooth parameter to have the above desirable large-sample properties.
with the data-dependent smooth parameter to have the above desirable large-sample properties.
3. NUMERICAL STUDIES
3.1. Estimating treatment differences for HIV-infected patients
We use a data set from a clinical trial conducted by the AIDS Clinical Trials Group (ACTG) to illustrate the new proposal. Here, we present 2 cases, the first one is for a continuous response and the second case is for a binary end point. This study, ACTG 320, is one of early studies for examining the clinical added value from a protease inhibitor with 2 nucleoside analogues for treating human immunodeficiency virus type 1 infection (Hammer and others, 1997). A total of 1156 patients were randomized to 2 treatment groups. Here, treatment 0 is the 2-drug combination, zidovudine and lamivudine, and treatment 1 is the 3-drug combination consisting of the above 2 and indinavir. The study was terminated at the second formal interim analysis due to substantial overall improvements from the 3-drug combination over the standard 2-drug combination with respect to various study end points. However, even with this potent new treatment, some patients may not respond to the therapy but instead suffer from nontrivial toxicity. Therefore, for future patients' disease management, it is important to predict patient's treatment responses based on certain “baseline” markers.
For the first example of the illustration, we let the response Y be the patient's change of CD4 count at week 24 from the baseline level, an important end point for evaluating HIV treatments. In our analysis, we only considered subjects who had baseline covariate and CD424 values (n0 = 429, n1 = 427), where CD4k denote the CD4 count at week k. Here, the vector u of baseline covariates consists of CD40, log10RNA0, and age. To accommodate the potential nonlinear relationship between CD40 and the response, we let z in Model (2.1) be the vector (1,CD40,logCD40,log10RNA0,Age)T. Furthermore, we let g0(·) and g1(·) be the identity function. The estimated regression coefficients for fitted models via the estimating equations (2.2) are summarized in Table 1(a). It appears that the 2 resulting fitted regression functions 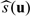 are rather different, indicating that there are potential interactions between the baseline covariates and the treatment response. It is interesting to note that for the 2-drug combination group, only age is marginally significant. On the other hand, the baseline CD4 and RNA are highly associated with the response for the 3-drug combination group. Moreover, note that younger patients with higher log10RNA0 tend to have high scores. For example, among the 44 patients with age below 40, log10RNA0 more than 5, and CD40 between 100 and 150, the average score is 95. On the other hand, among the 46 patients with age above 40, CD40 above 150, and log10RNA0 below 5, the average score is only 25.
are rather different, indicating that there are potential interactions between the baseline covariates and the treatment response. It is interesting to note that for the 2-drug combination group, only age is marginally significant. On the other hand, the baseline CD4 and RNA are highly associated with the response for the 3-drug combination group. Moreover, note that younger patients with higher log10RNA0 tend to have high scores. For example, among the 44 patients with age below 40, log10RNA0 more than 5, and CD40 between 100 and 150, the average score is 95. On the other hand, among the 46 patients with age above 40, CD40 above 150, and log10RNA0 below 5, the average score is only 25.
Table 1.
Estimated regression coefficients and their standard error estimates for the 2 treatment groups with the data from ACTG 320
| (a) Continuous outcome: change in CD4 from baseline to week 24 | ||||||
| Intercept | log 10RNA0 | CD40 | log CD40 | Age | ||
| Two drug | Estimate | − 18.86 | 3.47 | 0.04 | − 0.07 | 0.44 |
| Standard error | 24.04 | 3.40 | 0.07 | 3.91 | 0.26 | |
| Three drug | Estimate | − 118.09 | 28.65 | − 0.24 | 24.14 | − 0.22 |
| Standard error | 45.30 | 6.65 | 0.14 | 8.05 | 0.49 | |
| (b) Binary outcome: RNA at week 24 ≤ 500 copies/ml | |||||
| Intercept | log 10RNA0 | log CD40 | Age | ||
| Two drug | Estimate | 3.82 | − 1.14 | − 0.31 | 0.00 |
| Standard error | 1.73 | 0.22 | 0.16 | 0.02 | |
| Three drug | Estimate | − 0.20 | − 0.60 | 0.32 | 0.06 |
| Standard error | 1.13 | 0.18 | 0.10 | 0.01 | |
For the second step, we used this continuous score index 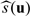 to group future patients and predict the true treatment difference
to group future patients and predict the true treatment difference 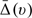 for patients with
for patients with 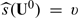 with
with 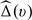 Here, since the estimated score has a left skewed distribution, we let ψ(v) = log( − log[Φ{(v − 70)/25}]) be the transformation for choosing the smooth parameter h. The kernel function K(·) is the Epanechnikov kernel, and the smoothing parameter h was obtained through a 10-fold cross-validation as a minimizer for the criterion (2.8). Furthermore, in (2.8), the weight function
Here, since the estimated score has a left skewed distribution, we let ψ(v) = log( − log[Φ{(v − 70)/25}]) be the transformation for choosing the smooth parameter h. The kernel function K(·) is the Epanechnikov kernel, and the smoothing parameter h was obtained through a 10-fold cross-validation as a minimizer for the criterion (2.8). Furthermore, in (2.8), the weight function 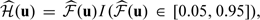 , where
, where 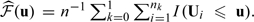 . The resulting bandwidth h = 0.93 for the transformed score. Here, the confidence intervals for
. The resulting bandwidth h = 0.93 for the transformed score. Here, the confidence intervals for 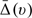 were constructed for the transformed score ψ(v)∈[ − 1.73,1.12]. This corresponds to v∈[30,94], which is
were constructed for the transformed score ψ(v)∈[ − 1.73,1.12]. This corresponds to v∈[30,94], which is 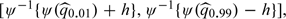 where
where 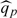 is the pth percentile of the observed
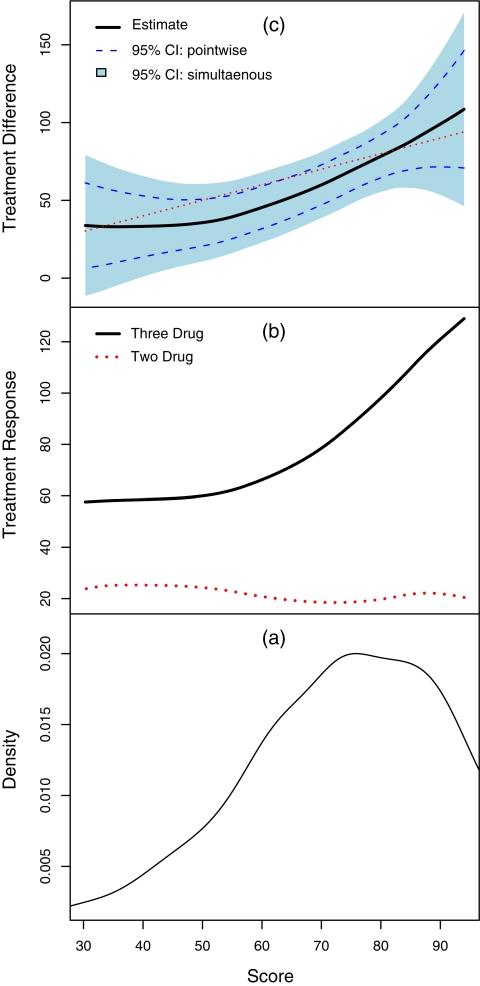
is the pth percentile of the observed 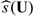 . In Figure 1(c), we present the estimate
. In Figure 1(c), we present the estimate 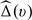 , the solid curve. This curve, in some region, seems markedly different from the 45-degree line. For example, subjects with score 60 have an estimated average treatment difference of 45, indicating that the parametric risk estimates need to be calibrated.
, the solid curve. This curve, in some region, seems markedly different from the 45-degree line. For example, subjects with score 60 have an estimated average treatment difference of 45, indicating that the parametric risk estimates need to be calibrated.
Fig. 1.
(a): The estimated density function of the parametric score with respect to week 24 CD4 changes; (b): Estimated group averages of week 24 CD4 changes over the score for 2- and 3-drug combination groups; (c): Estimated treatment differences (thick curve), 3-drug combo minus 2-drug combo, with respect to week 24 CD4 changes over the score and the corresponding 95% pointwise (dashed curve) and simultaneous (shaded region) confidence intervals.1
To make further inferences about 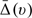 we approximated the distribution of the estimator
we approximated the distribution of the estimator 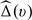 via the aforementioned perturbation-resampling method (2.4) with 500 independent realized normal samples of
via the aforementioned perturbation-resampling method (2.4) with 500 independent realized normal samples of 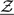 . In Figure 1(c), we present the resulting 0.95 pointwise intervals (bounded by the dotted curves) and its simultaneous band (gray area). For risk-cost-benefit decision makings, we also present the estimates
. In Figure 1(c), we present the resulting 0.95 pointwise intervals (bounded by the dotted curves) and its simultaneous band (gray area). For risk-cost-benefit decision makings, we also present the estimates 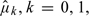 the underlying mean changes at week 24 for both treatment groups in Figure 1(b). We present the relative frequency of patients in the study population based on the index score
the underlying mean changes at week 24 for both treatment groups in Figure 1(b). We present the relative frequency of patients in the study population based on the index score 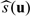 in Figure 1(a). Since most patients have estimated scores between 50 and 90, the estimation of the true treatment differences tends to be more precise in this region.
in Figure 1(a). Since most patients have estimated scores between 50 and 90, the estimation of the true treatment differences tends to be more precise in this region.
The average treatment response from the 3-drug combination group is uniformly higher than its counterpart from the 2-drug combination. However, the treatment differences do not change much when index score values are low but for score values higher than 50, the treatment differences appear to increase significantly. The confidence interval estimates displayed in Figure 1(c) play important roles for treatment selections, especially with additional information regarding the toxicity profiles over the index score.
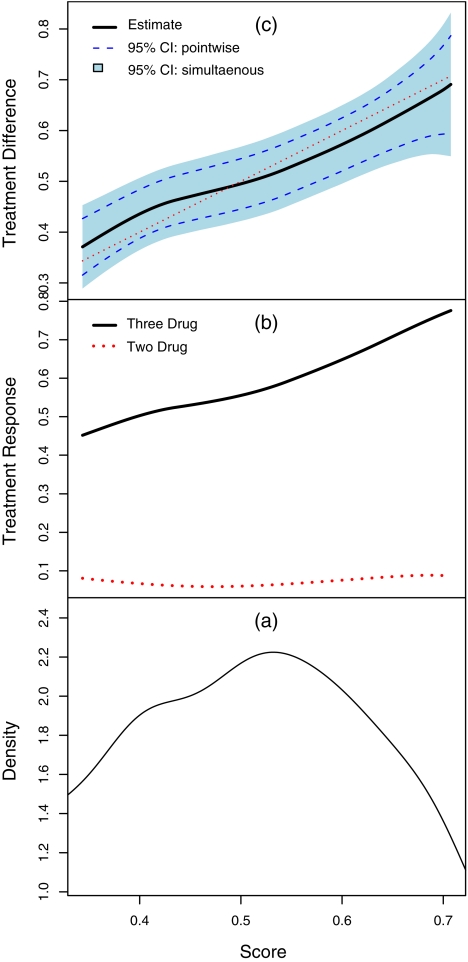
For the second example, we considered the patient's response being a binary variable, which is one if the week 24 RNA is below 500 copies per milliliter (the criterion for the RNA response used for ACTG 320). Since the linear effects of CD40 are almost 0 for both groups, we only included logCD40 for this analysis. It follows that z = (1,logCD40,log10 R N A 0, A g e )′ and g0(x) = g1(x) = exp(x)/{1 + exp(x)} for Model (2.1). Here, n0 = 408 and n1 = 416. In Table 1(b), we present the estimates for the regression parameters and their estimated standard errors. It appears that older patients with higher baseline CD4 tend to have higher score. The score appears to increase initially with log10RNA0 and drop after log10RNA0 exceeds 4.0. For the present case, the estimated score is between − 1 and 1, we applied a transformation ψ(x) = log{(x + 1)/(1 − x)} for choosing smooth parameter. The optimal bandwidth corresponding to the transformed score is 0.49 using the 10-fold cross-validation procedure as the one for the continuous response case. The confidence intervals for 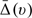 were constructed over ψ(v)∈[0.72,1.76] which corresponds to v∈[0.34,0.71]. In Figure 2(c), we present the estimated treatment differences (solid curve) over the index score. For this end point, the 3-drug combination is substantially and uniformly better than the 2-drug combination as shown in Figure 2(b). For instance, for patients with a score of 0.35, the probability of having RNA suppressed below the limit of quantification is 46% if treated with the 3-drug therapy and only 8% if treated with the 2-drug alternatives. For this subgroup, the true treatment difference was estimated as 0.38 with 95% pointwise confidence interval [0.32,0.43] and 95% simultaneous confidence interval [0.30,0.46].
were constructed over ψ(v)∈[0.72,1.76] which corresponds to v∈[0.34,0.71]. In Figure 2(c), we present the estimated treatment differences (solid curve) over the index score. For this end point, the 3-drug combination is substantially and uniformly better than the 2-drug combination as shown in Figure 2(b). For instance, for patients with a score of 0.35, the probability of having RNA suppressed below the limit of quantification is 46% if treated with the 3-drug therapy and only 8% if treated with the 2-drug alternatives. For this subgroup, the true treatment difference was estimated as 0.38 with 95% pointwise confidence interval [0.32,0.43] and 95% simultaneous confidence interval [0.30,0.46].
Fig. 2.
(a): The estimated density function of the parametric score with respect to week 24 HIV-RNA; (b): Estimated averages of week 24 RNA over the score for 2- and 3-drug combination groups; (c): Estimated treatment differences (thick curve), 3-drug combo minus 2-drug combo, with respect to week 24 RNA over the score and the corresponding 95% pointwise (dashed curve) and simultaneous (shaded region) confidence intervals.
3.2. Simulation studies
To assess the performance of the proposed procedure under practical settings, we conducted numerical studies to examine the finite-sample performance of our proposed point and interval estimators. Based on the results of the studies, we find that the new proposed estimation procedure has negligible bias and the coverage levels are very close to the nominal counterparts. For example, in one of our simulation studies, we simulated data with continuous CD4 response by mimicking the HIV study. Specifically, we generated CD424 from a log-normal model with a 3×1 covariate vector, U = (log10RNA0,Age,logCD40)T. First, for each subject, we generated U from a multivariate normal with mean and covariance matrix obtained from the observed HIV data. Then, for each given value of U, we generated CD424 from
where α1 = (0.13,0.0086, − 0.45)T, α0 = ( − 0.041,0.0033, − 0.24)T, σ1 = 0.69, and σ0 = 0.73. These parameters are also obtained from fitting the above model to the observed data.
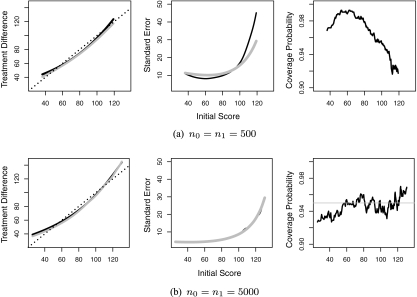
We considered a moderate sample size of n0 = n1 = 500 and relatively large-sample size of n0 = n1 = 5000. Since the estimated score has a left skewed distribution, we let ψ(v) = log( − log[Φ{(v − 76)/33}]) be the transformation for choosing the smooth parameter h. For computational ease, the bandwidth for constructing the nonparametric estimate was fixed at h = 0.95 for nk = 500 and 0.39 for nk = 5000. Here, these h's were chosen as the averages of the bandwidths selected based on (2.8) from 10 simulated data sets. For both settings, the nonparametric estimator has negligible bias, the estimated standard errors are close to their empirical counterparts, and the empirical coverage levels of the 95% confidence intervals are close to the nominal level. In Figure 3, we summarize the results of our findings performance for the point and interval estimation procedures for 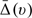 when nk = 500 in (a) and 5000 in (b). The proposed procedures have negligible bias and the estimated standard errors are close to the empirical standard errors. When the sample size is relatively small, the estimated standard errors are slightly lower than the empirical standard errors and the coverage levels are slightly lower than the nominal level when the score is near the upper boundary points. However, both the standard error estimates and the coverage levels improve when we increase the sample sizes to nk = 5000. For example, when v = 80, the coverage level for
when nk = 500 in (a) and 5000 in (b). The proposed procedures have negligible bias and the estimated standard errors are close to the empirical standard errors. When the sample size is relatively small, the estimated standard errors are slightly lower than the empirical standard errors and the coverage levels are slightly lower than the nominal level when the score is near the upper boundary points. However, both the standard error estimates and the coverage levels improve when we increase the sample sizes to nk = 5000. For example, when v = 80, the coverage level for 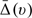 is 97.7% when nk = 500 and 95.4% when nk = 5000. When v = 115, the coverage level for
is 97.7% when nk = 500 and 95.4% when nk = 5000. When v = 115, the coverage level for 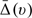 is 92.3% when nk = 500 increase to 94.5% when nk = 5000. The simultaneous confidence intervals have a coverage level of 95.9% when nk = 500 and 97.1% when nk = 5000.
is 92.3% when nk = 500 increase to 94.5% when nk = 5000. The simultaneous confidence intervals have a coverage level of 95.9% when nk = 500 and 97.1% when nk = 5000.
Fig. 3.
Summary of the simulation results for (a) n0 = n1 = 500 and (b) n0 = n1 = 5000. Left panel: the empirical average of the estimated treatment difference (solid black curve) versus the true treatment difference (solid gray curve). Shown also are the 45o reference line. Middle panel: the estimated (gray line) and empirical (black line) standard error. Right panel: empirical coverage level.
4. REMARKS
Under the parametric models given in the first stage, the estimated score 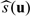 consistently estimates the true conditional treatment difference 𝒮(u) = E(Y(1) − Y(0)|U = u). Consequently, the calibrated estimator
consistently estimates the true conditional treatment difference 𝒮(u) = E(Y(1) − Y(0)|U = u). Consequently, the calibrated estimator 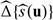 is also a consistent estimator of 𝒮(u). The nonparametric estimator may be less efficient than its parametric counterpart. When the parametric models fail to hold, inferences about the treatment differences based on
is also a consistent estimator of 𝒮(u). The nonparametric estimator may be less efficient than its parametric counterpart. When the parametric models fail to hold, inferences about the treatment differences based on 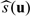 alone may be invalid. On the other hand,
alone may be invalid. On the other hand, 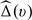 is always a consistent estimator of the average treatment effect for the subgroup with
is always a consistent estimator of the average treatment effect for the subgroup with 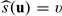 regardless the adequacy of the first-stage working models in (2.1). Nevertheless, how well
regardless the adequacy of the first-stage working models in (2.1). Nevertheless, how well 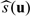 approximates 𝒮(U) does affect the effectiveness of the proposed procedure in selecting treatment options. When
approximates 𝒮(U) does affect the effectiveness of the proposed procedure in selecting treatment options. When 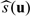 is a poor approximation to 𝒮(u), grouping patients based on
is a poor approximation to 𝒮(u), grouping patients based on 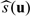 may not be optimal in terms of treatment selection and including additional covariates for constructing
may not be optimal in terms of treatment selection and including additional covariates for constructing 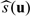 may yield a more informative scoring system. More flexible parametric models those considered in Royston and Sauerbrei (2004) can be used to improve the fitting in the first stage. Another important issue is the selection of predictive covariates for constructing
may yield a more informative scoring system. More flexible parametric models those considered in Royston and Sauerbrei (2004) can be used to improve the fitting in the first stage. Another important issue is the selection of predictive covariates for constructing 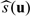 . This initial step could be carried out with careful exploratory analyses. When there are large number of candidate predictors, appropriate subset selection procedures such as the LASSO (Tibshirani, 1996) may be used. On the other hand, it is important to note that under correct specification of (2.1) with {g1,g0} being nonidendity functions, covariates predictive of the treatment difference 𝒮(U) are not only those significantly “interact” with the treatment assignment but also those relate to the outcome in either treatment groups. When the models are misspecified, interactions may not be well defined and ensuring the validity of the inferences about treatment difference under such settings becomes even more crucial.
. This initial step could be carried out with careful exploratory analyses. When there are large number of candidate predictors, appropriate subset selection procedures such as the LASSO (Tibshirani, 1996) may be used. On the other hand, it is important to note that under correct specification of (2.1) with {g1,g0} being nonidendity functions, covariates predictive of the treatment difference 𝒮(U) are not only those significantly “interact” with the treatment assignment but also those relate to the outcome in either treatment groups. When the models are misspecified, interactions may not be well defined and ensuring the validity of the inferences about treatment difference under such settings becomes even more crucial.
In practice, it is often not feasible to completely recover information on 𝒮(u) when the dimension of U is not small and the effects of U on Y are complex. An interesting and important question is how to evaluate the parametric index scoring system, which can “efficiently” group patients for the first stage of our proposal. Unlike the standard risk prediction problem, there is no obvious metric such as the receiver operating characteristic curve to evaluate the performance of the index system globally or locally. On the other hand, one may assess the effectiveness of 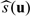 in grouping patients by examining how well
in grouping patients by examining how well 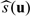 approximates 𝒮(u). To examine whether
approximates 𝒮(u). To examine whether 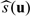 is an adequate approximation to 𝒮(u), one may construct cumulative residual processes based on (2.7) similar to those used in (2.8). One may expect due to the same reason, it is also important to compare different scoring systems. Intuitively, a scoring system
is an adequate approximation to 𝒮(u), one may construct cumulative residual processes based on (2.7) similar to those used in (2.8). One may expect due to the same reason, it is also important to compare different scoring systems. Intuitively, a scoring system 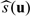 is good if the corresponding
is good if the corresponding 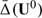 has big variation for U0 in the target population and thus can better differentiate different patients subgroups.
has big variation for U0 in the target population and thus can better differentiate different patients subgroups.
In practice, one may also be concerned about the efficiency in estimating 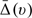 . It is possible to improve efficiency by augmenting the proposed estimators similar to those proposed in Zhang and others (2008). Specifically, using the fact that the treatment assignment G is independent of U, one may augment the proposed estimator
. It is possible to improve efficiency by augmenting the proposed estimators similar to those proposed in Zhang and others (2008). Specifically, using the fact that the treatment assignment G is independent of U, one may augment the proposed estimator 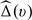 with estimating functions of the form
with estimating functions of the form
where p1 = P(G = 1) and ζ is a prespecified multivariate function of U. Let 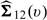 denote the covariance of
denote the covariance of 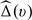 and
and 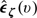 and
and 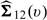 denote the variance–covariance matrix of
denote the variance–covariance matrix of 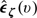 . Then the augmented estimator
. Then the augmented estimator 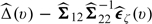 could be more efficient than
could be more efficient than 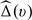 , especially if
, especially if 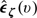 is significantly correlated with
is significantly correlated with 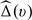 .
.
One may use the same approach presented in this article to estimate individual-specific treatment differences from an observational study. Such estimates, however, may not have the causal interpretation for the treatment intervention. The treatment difference may also be quantified using other measures for the contrast between 2 treatment groups, for example, the relative risk or odds ratio for binary response variable. Furthermore, using the same idea presented in this article, one may develop subject-level inference procedures for the treatment differences with a censored event time end point. For example, one may fit a proportional hazards working model for the 2 treatment groups and obtain the conditional hazard ratio as the scoring index. Subsequently, we may obtain risk differences within each subgroup based on conditional Kaplan–Meier estimates. When the variability or sample size of one treatment group is substantially larger than the other, one may allow the bandwidth h to depend on the treatment group. For such cases, optimal {h0,h1} can be chosen to minimize the integrated cumulative squared error in (2.8) with respect to both bandwidths. We provide theoretical justifications for the proposed procedures under this more general case in the Supplementary Material available at Biostatistics online.
SUPPLEMENTARY MATERIAL
Supplementary material is available at http://biostatistics.oxfordjournals.org.
FUNDING
U.S. National Institutes of Health (R01 GM079330, R01 HL089778, R01 AI052817, U54 LM008748-06).
Acknowledgments
The authors are grateful to the editors and the reviewers for their insightful comments on the article. Conflict of Interest: None declared.
References
- Bonetti M, Gelber RD. A graphical method to assess treatment-covariate interactions using the cox model on subsets of the data. Statistics in Medicine. 2000;19:2595–2609. doi: 10.1002/1097-0258(20001015)19:19<2595::aid-sim562>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Bonetti M, Gelber RD. Patterns of treatment effects in subsets of patients in clinical trials. Biostatistics. 2005;5:465–481. doi: 10.1093/biostatistics/5.3.465. [DOI] [PubMed] [Google Scholar]
- Bowman A, Hall P, Prvan T. Bandwidth selection for the smoothing of distribution functions. Biometrika. 1998;85:799–808. [Google Scholar]
- Byar DP. Assessing apparent treatment: covariate interactions in randomized clinical trials. Statistics in Medicine. 1985;4:255–263. doi: 10.1002/sim.4780040304. [DOI] [PubMed] [Google Scholar]
- Fan J, Gijbels I. Monographs on Statistics and Applied Probability. Volume 66. London: Chapman Hall; 1996. Local polynomial modelling and its applications. [Google Scholar]
- Goldhirsch ASC, Colleoni M, Nasi ML, Bernhard J, Zahrieh D, Bonetti M, Gelber RD, Italiana S, Bellinzona S, Coates AS and others. Endocrine responsiveness and tailoring adjuvant therapy for postmenopausal lymph node-negative breast cancer: a randomized trial. Journal of the National Cancer Institute. 2002;94:1054–1065. doi: 10.1093/jnci/94.14.1054. [DOI] [PubMed] [Google Scholar]
- Hammer S, Squires K, Hughes M, Grimes J, Demeter L, Currier J, Eron J, Feinberg J, Balfour H, Deyton L and others. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and cd4 cell counts of 200 per cubic millimeter or less. The New England Journal of Medicine. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- Park BU, Kim WC, Ruppert D, Jones MC, Signorini DF, Kohn R. Simple transformation techniques for improved non-parametric regression. Scandinavian Journal of Statistics. 1997;24:145–163. [Google Scholar]
- Park Y. Boston, MA: Harvard University; 2004. Semiparametric statistical inference in survival analysis, [PhD. Thesis] [Google Scholar]
- Pfeffer MA, Jarcho JA. The charisma of subgroups and the subgroups of CHARISMA. The New England Journal of Medicine. 2006;354:1744. doi: 10.1056/NEJMe068061. [DOI] [PubMed] [Google Scholar]
- Rothwell PM. Treating individuals 1 external validity of randomised controlled trials: “To whom do the results of this trial apply?”. Lancet. 2005;365:82–93. doi: 10.1016/S0140-6736(04)17670-8. [DOI] [PubMed] [Google Scholar]
- Royston P, Sauerbrei W. A new approach to modelling interactions between treatment and continuous covariates in clinical trials by using fractional polynomials. Statistics in Medicine. 2004;23:2509–2525. doi: 10.1002/sim.1815. [DOI] [PubMed] [Google Scholar]
- Sabine C. AIDS events among individuals initiating HAART: do some patients experience a greater benefit from HAART than others? AIDS. 2005;19:1995–2000. doi: 10.1097/01.aids.0000189858.59559.d2. [DOI] [PubMed] [Google Scholar]
- Song X, Pepe MS. Evaluating markers for selecting a patient's treatment. Biometrics. 2004;60:874–883. doi: 10.1111/j.0006-341X.2004.00242.x. [DOI] [PubMed] [Google Scholar]
- Tian L, Cai T, Goetghebeur E, Wei LJ. Model evaluation based on the sampling distribution of estimated absolute prediction error. Biometrika. 2007;94:297. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society, Series B (Methodological) 1996;58:267–288. [Google Scholar]
- Tibshirani RJ, Hastie TJ. Local likelihood estimation. Journal of the American Statistical Association. 1987;82:559–567. [Google Scholar]
- Wand MP, Marron JS, Ruppert D. Transformation in density estimation (with comments) Journal of the American Statistical Association. 1991;86:343–361. [Google Scholar]
- Wang R, Lagakos SW, Ware JH, Hunter DJ, Drazen JM. Statistics in medicine–reporting of subgroup analyses in clinical trials. The New England Journal of Medicine. 2007;357:2189. doi: 10.1056/NEJMsr077003. [DOI] [PubMed] [Google Scholar]
- Zhang M, Tsiatis AA, Davidian M. Improving efficiency of inferences in randomized clinical trials using auxiliary covariates. Biometrics. 2008;64:707–715. doi: 10.1111/j.1541-0420.2007.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.