SUMMARY
The activity of RING finger ubiquitin ligases (E3) is dependent on their ability to facilitate transfer of ubiquitin from ubiquitin-conjugating enzymes (E2) to substrates. The G2BR domain within the E3 gp78 binds selectively and with high affinity to the E2 Ube2g2. Through structural and functional analyses, we determine that this occurs on a region of Ube2g2 distinct from binding sites for ubiquitin-activating enzyme (E1) and RING fingers. Binding to the G2BR results in conformational changes in Ube2g2 that affect ubiquitin loading. The Ube2g2:G2BR interaction also causes an ~ 50-fold increase in affinity between the E2 and RING finger. This results in markedly increased ubiquitylation by Ube2g2 and the gp78 RING finger. The significance of this G2BR effect is underscored by enhanced ubiquitylation observed when Ube2g2 is paired with other RING finger E3s. These findings uncover a mechanism whereby allosteric effects on an E2 enhance E2-RING finger interactions and consequently ubiquitylation.
INTRODUCTION
Ubiquitylation occurs as the result of a hierarchical multi-enzyme process. Ubiquitin-activating enzyme (E1) activates ubiquitin, forming a thiolester linkage between the active site Cys of E1 and the C-terminus of ubiquitin. Ubiquitin is transferred to the conserved active site Cys of ubiquitin-conjugating enzymes (E2), of which there are over 30 in mammals. E2s bind to specific ubiquitin-protein ligases (E3s), which mediate the transfer of ubiquitin to primary amines on substrates or to growing chains of ubiquitin (polyubiquitin or multiubiquitin). In many cases the E3s also undergo auto- or self-ubiquitylation.
There are more than 500 E3s in mammals that can be divided into two major classes. The HECT E3s include ~30 E3s and are characterized by a conserved 350 amino acid catalytic domain. HECT E3s are catalytic intermediates in substrate ubiquitylation as a consequence of the transthiolation of ubiquitin from bound E2 to their conserved catalytic Cys (Fang and Weissman, 2004).
RING finger and RING finger-like E3s collectively represent the large majority of E3s. The RING finger is a compact Zn-binding domain of 40 to 100 amino acids. These domains generally bind E2s with low affinity and do not form catalytic intermediates with ubiquitin. It is generally believed that RING fingers either position E2~Ub to facilitate transfer to substrates or function as allosteric activators of E2~Ub (Lorick et al., 2006; Ozkan et al., 2005). Binding sites on E2s for RING fingers, HECT domains and E1 all overlap. Therefore, E2s must dissociate from ligase domains to reload with ubiquitin (Huang et al., 2005; Eletr et al., 2005). Interestingly, there are a few examples where other regions, either within a multi-subunit ubiquitin ligase complex or a single subunit E3, bind specific E2s through generally uncharacterized interactions (Madura et al., 1993; Hatakeyama et al., 1997; Wu et al., 2002; Biederer et al., 1997; Chen et al., 2006). This could increase the availability of E2~Ub and theoretically allow for reloading of E2 with ubiquitin without dissociation from the E3.
Ubiquitylation and proteasomal degradation perform critical functions in degradation of misfolded, unassembled and highly regulated proteins from the endoplasmic reticulum (ER). ER-associated degradation (ERAD) is a multi-step, highly coordinated process (Nakatsukasa and Brodsky, 2008). In mammals there are at least five known ER membrane-spanning ERAD E3s. Among these is gp78, also known as the human tumor autocrine motility factor receptor (AMFR). gp78 is implicated in degradation of T cell antigen receptor subunits, regulatory proteins in lipid metabolism (Kostova et al., 2007), CFTRΔ508 (Morito et al., 2008) and the metastasis suppressor KAI1 (CD82) (Tsai et al., 2007).
gp78 has a complex domain structure for a single subunit E3. In addition to its RING finger, it has at least three more C-terminal domains in its extended cytoplasmic tail (Figure 1A). Each of these is implicated in ubiquitylation and degradation of ERAD substrates. They include a ubiquitin-binding CUE (coupling of ubiquitin conjugation to ERAD) domain and a C-terminal binding site for p97. Unique to gp78 is a high affinity binding site for its cognate E2, Ube2g2. This Ube2g2 binding region (G2BR) is required for the function of gp78 in cells (Chen et al., 2006).
Figure 1.
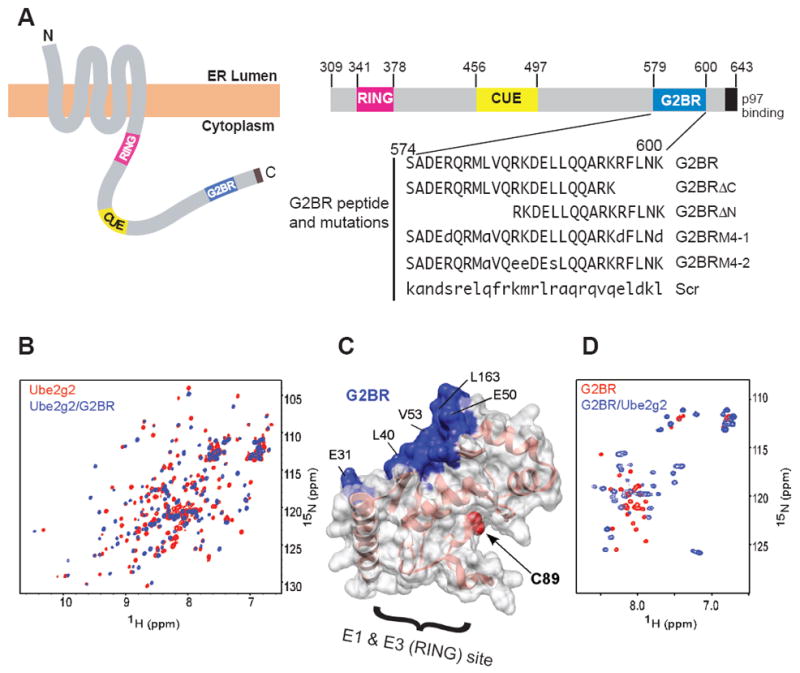
The G2BR and NMR-Determined Interactions with Ube2g2. (A) Schematic representation of gp78 in the ER membrane (left). To the right is a linear representation of gp78 cytoplasmic tail with amino acids (corresponding to the entire human gp78) indicated. Peptides used in this study corresponding to the G2BR, specific mutations/truncations and a scrambled (Scr) peptide control are shown below. Mutations are indicated in lower case. (B) Overlay of 15N-HSQC NMR spectra of Ube2g2 in free (red) and G2BR-bound (blue) form. (C) The contact residues observed in an intermolecular NOESY spectrum are painted blue on the free Ube2g2 structure (PDB entry 2CYX). Region of E1 and E3 binding based on other E2-E3 pairs is indicated by the bracket. The active site Cys (C89) is in red. (D) 15N-HSQC of isotopically-labeled G2BR in free form (red) and bound to Ube2g2 (blue).
We now report that the G2BR binds Ube2g2 through an extended interface distinct from sites of RING finger and E1 binding. This results in subtle changes in the Ube2g2 core that are manifested in functional alterations in loading with ubiquitin and a marked increase in affinity for the gp78 RING finger, which is reflected in enhanced ubiquitylation.
RESULTS
The G2BR is a High Affinity Binding Site for Ube2g2
To evaluate the interaction between the gp78 G2BR (Figure 1A) and Ube2g2, isothermal titration calorimetry (ITC) was employed (Supplemental Figure 1). This confirmed the direct, high affinity interaction of Ube2g2 with a 27 amino acid G2BR peptide. The 1:1 complex of E2 and G2BR has a dissociation constant (Kd) of 21(± 4) nM (Table 1). The reaction is exothermic and the significant free energy change (ΔG = -43.72 kJ mol-1) implies a very stable complex. The decrease in entropy reveals that part of the complex is losing some degree of freedom and likely folding into a regular structure.
Table 1.
Dissociation constants of E2-E3 interactions
| Complex | Kd | ΔH (kJ mol-1) | ΔS (J K-1 mol-1T-1) | Method | NMR Exchange timescale |
|---|---|---|---|---|---|
| Ube2g2:G2BR | 21(±4) nM | -56.3(±0.01) | -41.8(±0.04) | ITC | slow |
| Ube2g2:G2BRΔN | 740(±110) nM | -9.80(±0.40) | 83.6(±1.43) | ITC | slow |
| Ube2g2:G2BRΔC | 192(±22) μM | n.d.* | n.d. | NMR | fast |
| Ube2g2:G2BRM4–1 | 55 (±18) μM | n.d. | n.d. | NMR | fast |
| Ube2g2:G2BRM4-2 | 9.5 (±4) μM | n.d. | n.d. | NMR | fast |
| Ube2g2:gp78-RING | 144(±10) μM | n.d. | n.d. | NMR | fast |
| (Ube2g2:G2BRΔN):gp78-RING | 29(±5) μM | n.d. | n.d. | NMR | fast |
| (Ube2g2:G2BR):gp78-RING | 3(±1) μM | n.d. | n.d. | NMR | fast |
n.d. = not determined
‘Backside Binding’ to Ube2g2 Induces G2BR Folding
To identify the binding surface between Ube2g2 and G2BR, the interaction was examined by monitoring the NMR spectra of each molecule independently. First, the Ube2g2 preparation was validated by confirming the resonance assignments for Ube2g2 compared to a previous study (Briggman et al., 2005). A secondary structure analysis based on chemical shifts (Wishart and Sykes, 1994) combined with distance information from a 15N-edited NOESY-HSQC spectrum confirmed that our Ube2g2 preparation matched the reported crystal structure (Arai et al., 2006). Titration of G2BR into isotopically-labeled Ube2g2 (Figure 1B) yielded slow exchange NMR spectra and indicated a Kd «1μM, consistent with the ITC data. An unexpectedly large number of the Ube2g2 HN-N peaks shifted in the presence of G2BR, requiring reassignment. The 13C-edited HSQC spectra of the methyl resonances of Ile, Leu, and Val residues (data not shown) indicated a localized binding site, which correlates with the largest shifts observed in Figure 1B. The secondary structure based on chemical shifts of Ube2g2:G2BR was identical to free Ube2g2 and NOESY spectra of Ube2g2:G2BR exhibited equivalent patterns of connectivities between HN- HN, HN-methyl, and methyl-methyl protons compared to free Ube2g2, implying that the overall structure of Ube2g2 did not change significantly.
The binding surface was determined by chemical shift mapping on Ube2g2 and by intermolecular NOESY experiments. Chemical shift mapping for Ube2g2 indicates the primary binding site is on beta-strands β1-β3 (Supplemental Figure 2). In addition, the C-terminal regions of helices α1 and α4 are perturbed by G2BR binding. Intermolecular NOESY crosspeaks were observed between G2BR and the methyl and amide protons of Ube2g2 (Supplemental Figure 3) that correspond to the surface indicated in blue on unbound Ube2g2 in Figure 1C. These NOEs refined the contact surface to residues V25, A26, E31, E38, L40-M42, E45, E50, F51-V53, V159, L163 and L165. This surface includes the hydrophobic patch formed by V25, A26, L40-M42 and F51-V53 that is mostly conserved in other E2s (Hamilton et al., 2001; Moraes et al., 2001; Brzovic et al., 2006). The G2BR binding site also includes negatively charged residues flanking the hydrophobic core. From the perspective of the G2BR, 15N-HSQC spectra (Figure 1D) of a biosynthetically-prepared G2BR exhibited poor chemical shift dispersion, consistent with a random-coil conformation. The backbone chemical shifts were assigned in the free G2BR peptide and, except for indications of a nascent four-residue helical turn at V583-K586, the peptide lacks any regular secondary structure. The spectrum of G2BR in the presence of Ube2g2 exhibits a dramatic increase in spectral dispersion and resonance linewidths consistent with a 1:1 complex of Ube2g2:G2BR (Figure 1D). This suggests that the G2BR must fold into a compact regular secondary structure utilizing all 27 residues to fit on the defined E2 binding site. Based on this, we redefine the G2BR as amino acids 574-600 of gp78 instead of 579-600 as originally defined (Chen et al., 2006).
The Ube2g2:G2BR Crystal Structure Reveals Distant Changes that Impact E2 Loading of Ubiquitin
The Ube2g2:G2BR complex was crystallized, forming rod-shaped crystals (0.1 mm × 0.3 mm × 0.1 mm). These diffracted to 1.8 Å resolution (Table 2). The structure shows that the Ube2g2 backbone is largely unchanged in the presence of G2BR (Figure 2A), as predicted by NMR measurements. The conserved secondary structural elements include α1 = T4-L18, β1 = G23-P28, β2 = E36-M42, β3 = V53-S59, β4 = K70-F73, 310 helix = S91-L93, α2 = V116-A128, α3 = V138–D146, and α4 = R148-L163. Strands β1-β4 form an anti-parallel β-sheet (Figure 2A and 2B). A dynamic region in the β4α2 loop is present between residues H94 and W110. This corresponds to an acidic extension (aa 96-108) that, in mammals, is limited to Ube2g2 and two other E2s [Ube2g1 and Ube2r1 (Cdc34 in yeast)]. The backbone root-mean-square deviation (rmsd) between the free and bound Ube2g2 is 1.8 Å over all residues and decreases to 0.9 Å if residues 96-108 in the β4α2 loop are excluded.
Table 2.
Data collection and refinement statistics
| Data Collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 48.92, 60.15, 61.64 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 50-1.8 (1.86-1.8)a |
| No. of observations | 105668 (5851) |
| No. of unique reflections | 16657 (1272) |
| bRmerge | 7.8 (43.7) |
| I/σ | 22.4 (2.7) |
| Completeness (%) | 95.4 (74.2) |
| Redundancy | 6.4 (4.6) |
| Refinement | |
| Resolution (Å) | 43-1.8 (1.86-1.8) |
| cRworking (%) | 19.3 (23.1) |
| dRfree (%) | 24.1 (27.4) |
| No. of Atoms / B factors (Å2) | |
| Protein | 1318 / 33.1 |
| Peptide | 260 / 34.4 |
| Water | 164 / 40.7 |
| r.m.s.d | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.92 |
| Ramachandran plot | |
| Most favored region (%) | 87.7 |
| Additional allowed region (%) | 12.3 |
| Generously allowed region (%) | 0 |
| Disallowed region (%) | 0 |
Values in parentheses are for the highest resolution shell.
Rmerge = Σ|(I − <I>)|/σ(I), where I is the observed intensity.
R-factor = Σhlk| |Fo| - |Fc| | / Σhkl |Fo|, calculated from working dataset.
Rfree is calculated from 5% of data randomly chosen and not included in refinement.
Figure 2.
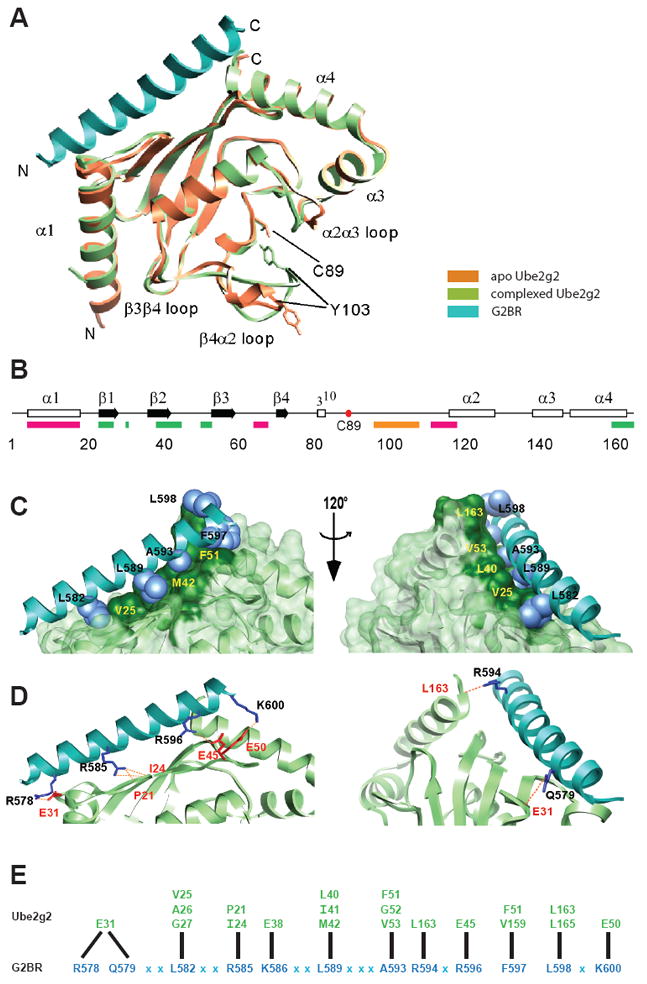
Crystal Structure of the Ube2g2:G2BR Complex. (A) Ribbon representation of the superimposed Ube2g2:G2BR complex with free Ube2g2 (PDB entry 2CYX). G2BR is cyan and Ube2g2 is light green in the complex. Free Ube2g2 is in orange. (B) A linear representation of the primary sequence, secondary structure, and binding regions of Ube2g2. α-helices are represented by open rectangles, β-strands by filled arrows, and the active site Cys by a red dot. Binding regions are indicated below the sequence line as RING finger (magenta) and G2BR (green). The position of Ube2g2’s extended dynamic loop is indicated in orange. (C) Hydrophobic sidechains of G2BR (light blue, residues in black) lock into the hydrophobic surface of Ube2g2 (dark green, residues in yellow). (D) Inter-molecular hydrogen bonds and salt bridges between G2BR and Ube2g2 are shown. Sidechains of G2BR are in blue and residues are in black, the Ube2g2 contact sidechains and residues are red. The P21 and I24 side-chains of Ubeg2g2 were not displayed, since the interaction is through backbone hydrogen bonds. (E) Contacts between Ube2g2 (green) and G2BR (blue) indicating residues involved in hydrogen bonds, salt-bridges and hydrophobic interactions. Black lines link each residue to its reciprocal contact and X denotes G2BR residues that do not have direct contacts in the interface.
Key structural changes were observed in Ube2g2 near the active site upon binding the G2BR. Access to the active site Cys (C89) is via a channel flanked on either side by β4α2 loop and the α2α3 loop (Figure 2A). The β4α2 loop was observed to exhibit dynamic behavior for aa 96-108 in the crystal structure of free Ube2g2 (Arai et al., 2006). In fact, there were three molecules in the asymmetric unit, and the major difference between the molecules was the conformation of the β4α2 and α2α3 loops (Figure 3A). These conformations are ordered; however, they exhibit higher B-factors than the rest of the structure (Supplemental Figure 2). The dynamic region of the β4α2 loop contains a short 310 helix and generally extends away from the protein in the three conformations in the free form; however, in the Ube2g2:G2BR complex presented here, the 310 helix is gone and a region of the loop (P100-Y103) orients back towards C89. The multiple conformations of these loops, which appear to shift to a new average state in the Ube2g2:G2BR complex, and the high B-factors in the crystal structures are consistent with dynamic averaging as suggested by chemical shift effects observed in solution (Supplemental Figure 2). The present structure may represent a low energy population of the average, in which the proximity of Y103 and C89 changes from 20.8 Å in unbound Ube2g2 to 3.8 Å in the complex. In addition, the α2α3 loop (comprising N131-G135) approaches C89 in the complex. This inward conformation is stabilized in the 1.8 Å crystal structure by a new network of water-mediated hydrogen bonding, including Y103, C89, and Y83 as well as a network including E133 and S91. The structural modifications of these two loops decrease accessibility around the active site (compare Figure 1C to 4B and Figure 3B to 3C). These changes are accompanied by an altered orientation of the C89 sidechain from pointing towards the β4α2 loop in the unbound form to pointing away from the loop (~80% probability) with bound G2BR (Figure 2A and compare Figure 3B to 3C).
Figure 3.
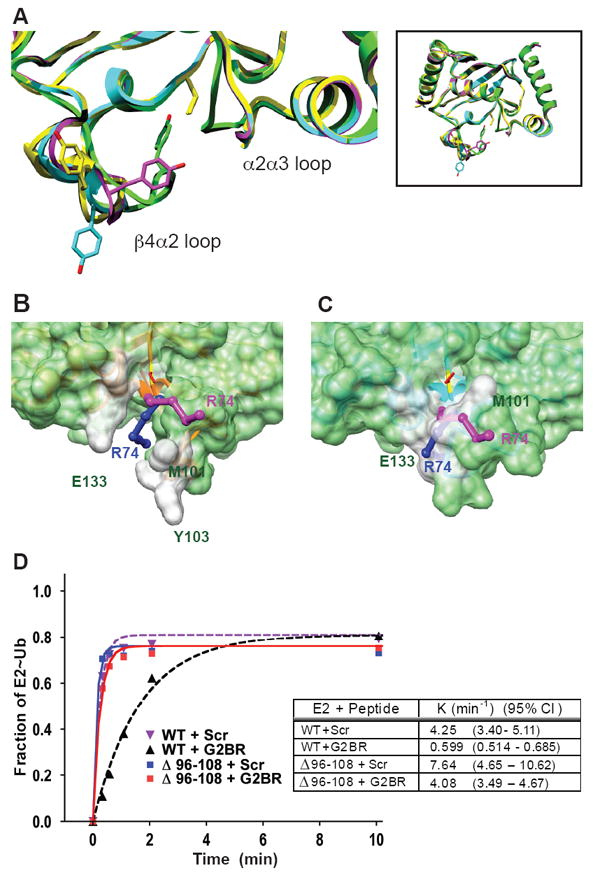
G2BR-Induced Structural Changes in Ube2g2 Occlude the Area Around the Active Site and Correlate with Ubiquitin Loading. (A) Superposition of active site region (full structures shown in small inset) for four Ube2g2 molecules (yellow, cyan, and magenta from the ligand-free Ube2g2 in PDB entry 2CYX and green in Ube2g2:G2BR) showing the conformational flexibility of the β4α2 loop. (B and C) Images showing the surface rendering of Ube2g2 and Ube2g2:G2BR combined with the potential orientation of ubiquitin chains, based on known E2-Ub structures. Ube2g2 was superimposed onto the E2 coordinates of two published E2-Ub structures; Ubc1-Ub (PDB entry 1FTX, rmsd=1.8Å) and Ubc13-MMs2-Ub (PDB entry 2GMI, rmsd=1.4Å). We display only the surface of Ube2g2 and the C-terminal end of the ubiquitin molecule (in ball and stick form) for Ubc1-Ub (blue) and Ubc13-MMs2-Ub (magenta). In (B), the free Ube2g2 structure is rotated relative to the orientation of Figure 1C by 180° about the vertical axis and zoomed in on the active site. The active site Cys (C89) backbone is yellow and its sidechain is red. Some residues of Ube2g2 that play a role in the allosteric change are labeled in green. R74 of both ubiquitin tails are indicated, the C-terminal di-glycine approaches C89. In (C), the G2BR bound form of Ube2g2 in an identical orientation as in (B). (D) 35S-labelled Ube2g2 or Ube2g2Δ96-108 generated by in vitro translation in E. coli lysate was incubated with 100 nM E1 and ubiquitin lacking lysines (Ub K0), so as to avoid formation of polyubiquitin chains. The formation of thiolester-linked Ube2g2 (E2~Ub) was assessed at 30°C with saturating (4 μM) G2BR peptide or a control ‘scrambled’ peptide (Scr). Shown is the average of two experiments for each condition. Rate constant, K, and 95% confidence index are shown for each condition.
Figure 4.
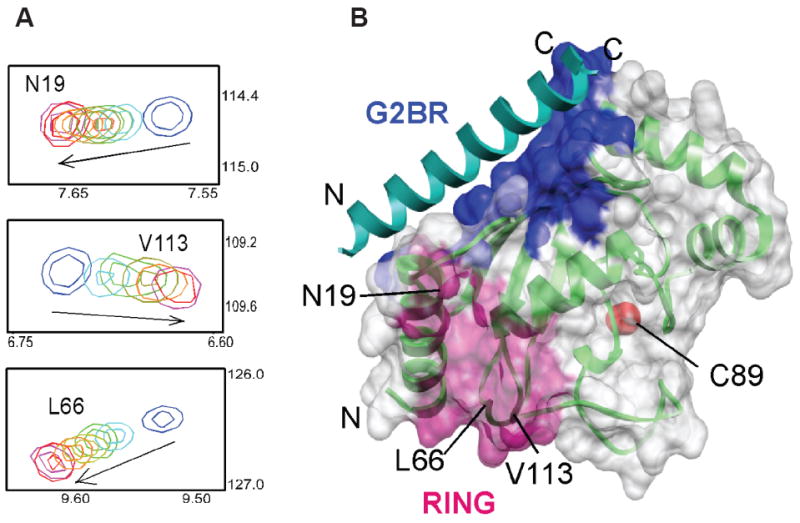
Interactions Between gp78 RING finger and Ube2g2:G2BR Complex. (A) Ube2g2:G2BR complex was titrated with gp78 RING finger followed by acquisition of 15N-HSQC spectra of isotopically-labeled Ubeg2g2 at each titration point. 15N chemical shifts in parts per million (ppm) are indicated on the y-axis, and the 1H chemical shifts in ppm are on the x-axis. Three of the affected residues of Ube2g2 (N19, L66 and V113) are shown. The peaks shift from the free (blue) form towards the bound form (magenta) with addition of gp78 RING finger. The dissociation constant was determined by fitting the peak positions against ligand:protein concentrations. (B) The surface representation of Ube2g2:G2BR shows the G2BR interface (blue), RING interface (magenta) and the active site cysteine (red).
The multiple states surrounding the active site seen in the crystal structures and the shift in conformational averaging in the Ube2g2:G2BR complex in solution suggest that these changes may impact interactions with E1 or ubiquitin bound to C89. To further evaluate this, we compared the structures of Ube2g2 and Ube2g2:G2BR with two reported E2-Ub structures (Hamilton et al., 2001; Eddins et al., 2006). Superposition of free Ube2g2 with the E2 component of Ubc1-Ub, which forms K48 ubiquitin chains (Chen and Pickart, 1990) and Ubc13-MMs2-Ub, which forms K63 chains (Deng et al., 2000), (Figure 3B) reveals that the C-termini of either of the two bound ubiquitins can readily be accommodated in the channel surrounding C89. However, superposition of Ube2g2:G2BR with the same two structures (Figure 3C) indicates that the area around the active site becomes occluded due to rearrangements of residues M101-Y103 and E133-G135 (within the β4α2 loop and α2α3 loop, respectively), resulting in steric clashes with the C-termini of the two ubiquitins. In Figure 3C, Y103 is buried and is not visible. These structure-based models predict direct consequences for loading of G2BR-bound Ube2g2 with ubiquitin from E1. This was tested by comparing ubiquitin loading of Ube2g2 in the presence of either a control scrambled peptide (Scr) (Figure 1A) or the G2BR. The G2BR resulted in a marked decrease in the rate of E2 loading at 30°C (Figure 3D) and almost no detectable loading at 12°C (Supplemental Figure 4). Deletion of the extended acidic portion of the β4α2 loop (aa 96-108; Ube2g2Δ96-108), which is the major contributor to steric crowding around the active site, decreased the G2BR effect by almost 75%, while G2BR binding was retained (Figure 3D; Supplemental Figure 5).
Ube2g2-Bound G2BR Forms an α-Helix with Extensive Contacts Across the Backside of the E2
The crystal structure of Ube2g2:G2BR demonstrates that the G2BR assumes a completely α-helical fold in the 1:1 complex and binds to a surface comprising the beta sheet (β1-β3) and the C-termini of α1 and α4 (Figure 2A), which is consistent with the solution NMR data (Figure 1C). The buried surface at the interface of Ube2g2 and G2BR is ~1950 Å2. The entire peptide is well ordered, and there is an extensive network of contacts between the G2BR and Ube2g2, comprising both hydrophobic and ionic or hydrogen bonding contacts (Figure 2C, 2D and 2E and Supplemental Table 1). The contact network is consistent with the thermodynamic data for formation of this complex and suggests that the recognition is highly specific. In the G2BR α-helix, there is evidence for 24 hydrogen bonds between all i and i+4 pairs beginning with the Ser 574 carbonyl oxygen to Arg 578 NH and running through Arg 596 carbonyl oxygen to Lys 600 NH. As amino acids 574-600 extends completely across the backside of Ube2g2, this suggests that extensions beyond this region do not interact with Ube2g2. This is in agreement with previous data (Chen et al., 2006) and NMR studies of aa 574-643 of gp78 (our unpublished results).
The presence of extensive contacts between Ube2g2 and G2BR suggest that single point mutations will not disrupt binding or activity. The initial characterization of the G2BR (Chen et al., 2006) indicated that the N- and C-terminal ends of the G2BR were each critical both for binding Ube2g2 and for the cellular function of gp78. To examine this observation, binding of G2BRΔN and G2BRΔC (Figure 1A) to Ube2g2 was assessed. G2BRΔC binds Ube2g2 with relatively weak affinity [Kd = 192 (± 22) μM] (Table 1). The affinity between G2BRΔN and Ube2g2 is significantly stronger [Kd = 740 (±110) nM], but nonetheless exhibits a ~35-fold decrease in affinity compared to wild-type G2BR. This confirms that the N and C terminal regions of G2BR both contribute directly to the high affinity binding observed with the intact domain. Binding of either G2BRΔN or G2BRΔC to Ube2g2 causes a subset of the observed chemical shift changes seen with the G2BR, consistent with their predicted contacts (data not shown).
On the basis of the Ube2g2:G2BR structure, we designed two peptides with four mutations each to assess the most important interactions, G2BRM4-1 and G2BRM4-2 (Figure 1A). These peptides were 15N-labeled and examined by NMR (Supplemental Figure 6). The 15N-HSQC spectra of both peptides indicated random-coil conformations characteristic of wild-type G2BR. Titration of G2BRM4-1 and G2BRM4-2 with unlabeled Ube2g2 yielded Kds of 55 (±18) μM and 9.5 (±4) μM respectively (Table 1). These findings, combined with the data for G2BRΔN and G2BRΔC, underscore the highly distributed nature of the contacts in Ube2g2:G2BR binding.
The G2BR Enhances the Affinity of Ube2g2 for the gp78 RING Finger
The positioning of the Ube2g2:G2BR interface on the ‘backside’ of Ube2g2, suggests that this interaction should not present a direct steric impediment to the interaction of Ube2g2 with the gp78 RING finger binding site (Zheng et al., 2000; Brzovic et al., 2003; Dominguez et al., 2004) (Figure 2A). To evaluate possible allosteric effects of G2BR on RING finger binding, the interaction of isotopically-labeled Ube2g2 with gp78 RING finger was monitored by NMR (Figure 4). The gp78 RING finger binding interface on Ube2g2 consists of the N-terminal end of α1, the β3β4 loop (F62-P69) and part of the β4α2 loop (W110-S115; see Figure 2A and 2B for secondary structure elements). The solution structure of the gp78 RING finger is similar to other RING finger structures (Das & Byrd, unpublished data) and a reverse labeling experiment, using isotopically-labeled gp78 RING finger, confirmed that the gp78 RING finger side of the binding surface is similar to other RING finger:E2 interactions. The interaction exhibited fast exchange (Figure 4A) and a Kd of 144 (±10) μM (Table 1, Supplemental Figure 7). This is consistent with the low affinity of many RING finger:E2 interactions (Lorick et al., 2006; Christensen et al., 2007). The affinity of the gp78 RING finger for the Ube2g2:G2BR complex was also determined. While the binding interface was unchanged and remained in fast exchange, strikingly, the Kd decreased by ~48-fold to 3 (±1) μM (Table 1, Supplemental Figure 7). Separate binding experiments confirmed that the G2BR and RING finger do not directly interact in the gp78 RING finger:Ube2g2:G2BR complex (data not shown). The allosteric relationship of Ube2g2:gp78 RING finger affinity to Ube2g2:G2BR association was tested by measuring the affinity of gp78 RING finger for Ube2g2 in the presence of G2BRΔN, which exhibits an affinity for Ube2g2 intermediate between the full G2BR and G2BRΔC. The measured Kd was 29 (±5) μM (Table 1), which indicates that even a smaller fragment of G2BR has effects that translate through Ube2g2 and increase its affinity for the gp78 RING finger.
G2BR Enhances RING Finger-Dependent Ubiquitylation by Ube2g2
gp78 is a substrate for its own ubiquitylation in cells (Fang et al., 2001; Chen et al., 2006). To begin to assess the effect of the G2BR on ubiquitylation, a GST fusion of the cytoplasmic domain of gp78 (GST-gp78C) was employed in an autoubiquitylation assay (Lorick et al., 1999). Ube2g2 was provided in ~5-fold molar excess relative to glutathione Sepharose-immobilized GST-gp78C and ubiquitylation of bead-bound material was assessed. Ubiquitylation was totally dependent on an intact gp78 RING finger (Figure 5A), and mutations of two key residues in the G2BR (L582S/L589S; gp78CL582,589S) significantly decreased ubiquitylation. A truncated form of GST-gp78C lacking the G2BR (GST-gp78CΔ577-643) also demonstrated markedly diminished ubiquitylation (Figure 5B), consistent with G2BR-dependent increased affinity of Ube2g2 for the gp78 RING finger. However, when G2BR peptide was provided ‘in trans’ there was a marked increase in ubiquitylation of truncated gp78 (Figure 5B). This rescue of activity was not observed with the G2BRΔN, G2BRΔC, G2BRM4-1, or G2BRM4-2 peptides (Figure 5C and 5D). To determine whether the G2BR peptide also enhances ubiquitylation of heterologous proteins, we took advantage of gp78’s function as an E4 in ubiquitylating proteins already modified with a single ubiquitin (Morito et al., 2008). A fusion protein of ubiquitin and GFP with a C terminal His6 tag (Ub-GFP-His6) was ubiquitylated by GST-gp78CΔ577-643 as assessed using Flag-tagged ubiquitin (Figure 5E). As with gp78 autoubiquitylation, ubiquitylation of Ub-GFP-His6 by GST-gp78CΔ577-643 was substantially increased in the presence of G2BR. Thus, the effect of the G2BR applies to ubiquitylation of a heterologous substrate as well as to autoubiquitylation.
Figure 5.
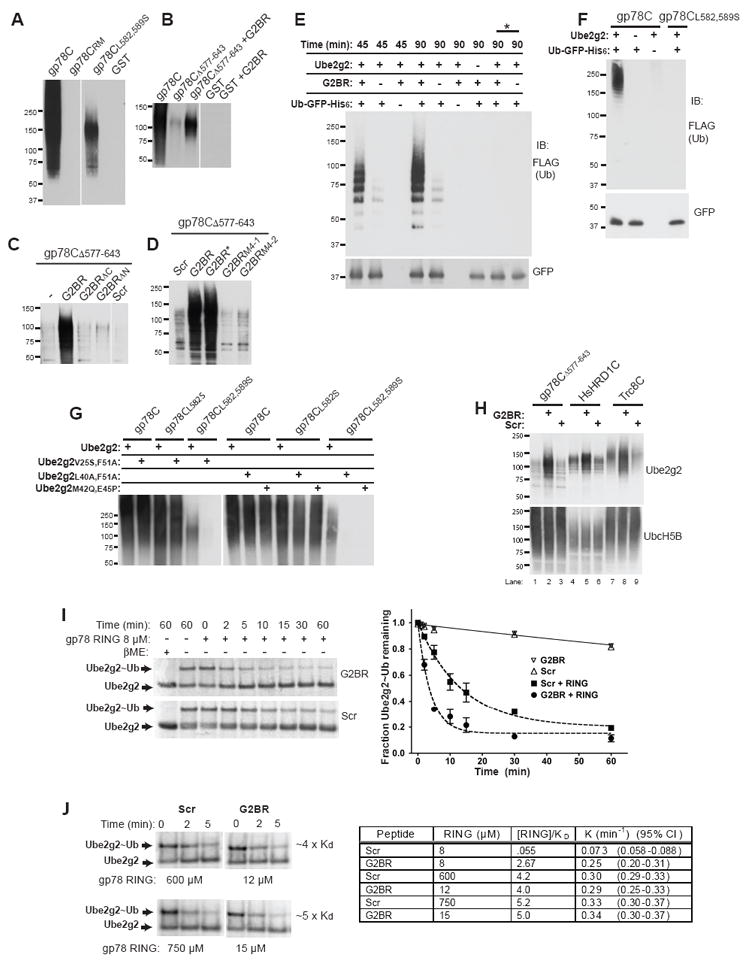
G2BR-mediated Increase in Ube2g2:gp78 RING finger Affinity Results in Enhanced Ubiquitylation. (A) Glutathione Sepharose-bound GST fusions of the entire gp78 cytoplasmic tail (aa 309-643; GST-gp78C), an inactivating mutation in the RING finger (GST-gp78CRM) or in the G2BR (GST-gp78CL582,589S) or GST alone were incubated for 90 min in the presence of Ube2g2 and E1. After washing, ubiquitylated bead-bound material was assessed by SDS-PAGE and immunoblotting with anti-ubiquitin. See Supplemental Figure 10 for Coomassie Blue stain of fusion proteins. (B) Glutathione Sepharose-bound GST-gp78C, a truncation at amino acid 577 at the beginning of the G2BR (GST-gp78CΔ577-643) or GST alone was incubated with or without G2BR peptide as indicated and assessed in (A). (C) and (D) were carried out as in (A) using GST-gp78CΔ577-643 and the indicated synthetic or recombinant peptides. Recombinant wild-type G2BR peptide is indicated by (*) to distinguish from the synthetic wild type peptide. (E) Ubiquitylation of Ub-GFP-His6 was carried out under the indicated conditions. The (*) denotes two control samples in which Ub-GFP-His6 was added after the reaction was first terminated by the addition of 10% SDS for 10 min followed by 8 M urea. Following addition of urea, Ub-GFP-His6 was purified on Ni+ Sepharose beads and samples were resolved by SDS-PAGE. (F) Ubiquitylation of Ub-GFP-His6 was carried out as in (E) for 60 min. (G) Ubiquitylation reactions were carried out with GST-gp78C or the indicated Leu to Ser mutations together with wild-type Ube2g2 or the indicated double mutations of Ube2g2. (H) GST fusions of the cytoplasmic tails of HsHRD1, Trc8 or truncated gp78 were incubated as in (A) with the indicated peptides and E2s. (I) 35S-labelled Ube2g2 generated as in Figure 3D was loaded with wild-type ubiquitin for 10 min followed by inactivation of E1 by NEM (5 mM). Discharge of ubiquitin from Ube2g2 +/- G2BR and +/- RING finger was monitored by measuring the fraction of Ube2g2~Ub remaining at each time point. Shown on the left is a representative experiment in which 8 μM gp78 RING finger was utilized with addition of either G2BR or Scr at a final concentration of 4 μM and Ube2g2 at <100 nM. To the right is the average and standard deviation of three independent experiments with or without RING finger. (J) Discharge experiments as in (I) were carried out for 2 or 5 min at the indicated concentrations of gp78 RING finger. Shown on the left are images demonstrating loss of ubiquitin from Ube2g2. The insert on the right summarizes data from discharge experiments from both (I) and (J). Discharge rate constants, K, are shown for each experiment.
To evaluate the effect of G2BR on substrate ubiquitylation using the full length gp78 cytoplasmic tail, gp78C and gp78CL582,589S were compared. gp78C resulted in easily detectable high molecular weight ubiquitylated Ub-GFP-His6, which was not evident with gp78CL582,589S (Figure 5F). As gp78CL582,589S showed some persistent autoubiquitylation in Figure 5A, these reactions were carried out by combining G2BR mutants with double mutations of Ube2g2 in key contact points (Figure 5G; see Figure 2 and Supplemental Table 1 for Ube2g2:G2BR contacts). While none of the double mutants of Ube2g2 substantially decreased ubiquitylation with either gp78C or a single mutation (gp78CL582S), combining any of these E2 mutants with gp78CL582,589S resulted in a complete loss of ubiquitylation. Collectively these findings demonstrate the functional importance of structurally-defined key contact residues in the Ube2g2-G2BR interface.
G2BR Enhances Ubiquitylation by Ube2g2 with other RING Fingers
Ube2g2 also functions with other RING finger ERAD E3s (our unpublished observations). Two of these, HsHRD1 and Trc8, are implicated in human disease (Kostova et al., 2007). These E3s also function, at least in vitro, with members of the UbcH5 (Ube2d1-3) E2 family (Lorick et al., 1999; Nadav et al., 2003; Kikkert et al., 2004). HsHRD1 and Trc8 lack identifiable regions that are analogous to the G2BR. We therefore asked whether the enhanced ubiquitylation observed with the G2BR was unique to the gp78 RING finger. Addition of the G2BR to ubiquitylation reactions utilizing GST-fusions of the C-terminal RING finger-containing regions of either HsHRD1 or Trc8 resulted in a marked increase in RING finger-dependent autoubiquitylation (Figure 5H upper panel lanes 5 and 8 and Supplemental Figure 8). Ubiquitylation mediated by these E3s as well as by GST-gp78CΔ577-643 was not increased by the G2BR when UbcH5B was used as the E2 (Figure 5H, lower panel). Similarly, critical mutations in the G2BR do not decrease ubiquitylation by UbcH5B (Supplemental Figure 9). Thus, changes in the ERAD E2, Ube2g2, induced by binding of the G2BR at a site distant from where this E2 interacts with RING fingers, enhances ubiquitylation with at least three different mammalian E3s. This indicates both the specificity of the G2BR for Ube2g2 and its apparent effect on increasing productive interactions between Ube2g2 and RING fingers.
Increased Affinity of Ube2g2 for the gp78 RING Finger Accounts for the Enhanced Ubiquitylation
To further evaluate the effect of the G2BR, the rate of discharge of ubiquitin from Ube2g2 was assessed, based on a previously used approach (Petroski and Deshaies, 2005), in the presence of saturating amounts of G2BR or an equal concentration of Scr. In these reactions acceptors for ubiquitin are other proteins in the reaction mix, including bacterial proteins and excess added ubiquitin. In the absence of the RING finger, discharge of ubiquitin from Ube2g2~Ub was slow and unaffected by G2BR peptide (Figure 5I). In the presence of 8 μM gp78 RING finger, loss of Ub-bound Ube2g2 was markedly accelerated. This was significantly increased by saturating amounts of G2BR (Figure 5I). We wished to determine if this apparent increased rate of discharge can be accounted for by an increased population of Ube2g2:RING finger due to the presence of G2BR (< 5% Ube2g2 bound with Scr; > 50% bound with G2BR), or as yet unknown allosteric effects on the Ube2g2:gp78 RING finger complex mediated by the G2BR. Therefore, experiments were carried out with concentrations of gp78 RING finger at which approximately equal amounts of RING finger are bound to Ube2g2. Under these conditions no significant increase in discharge rate was observed in the presence of G2BR (Figure 5J). Thus, the G2BR-mediated increase in affinity of Ube2g2 for the RING finger provides a molecular basis for the accelerated discharge of ubiquitin from Ube2g2~Ub and, by extension, for the observed enhancement of ubiquitylation.
DISCUSSION
Structural Basis for the High Affinity G2BR Binding to Ube2g2
The NMR and X-ray crystallographic determination of the G2BR binding site on Ube2g2 provides an explanation for its high affinity interaction with gp78. The G2BR binds Ube2g2 through an extended region of the core UBC domain of Ube2g2 that includes the C-terminal ends of both the α1 and α4 helices and extensive contacts with the intervening β sheets (β1-3). This backside binding substantially overlaps the non-covalent binding site for ubiquitin on UbcH5C (Brzovic et al., 2006). While ubiquitin binds UbcH5C with a Kd of ~300 μM, which is similar to the analogous binding of ubiquitin to Ube2g2 (Das and Byrd, unpublished observations), the binding of the G2BR to Ube2g2 is of higher affinity by over 10,000-fold. The basis for this difference is explained by the interacting interfaces. Ubiquitin, characterized by a stable, ordered structure, interacts with E2s through its hydrophobic face centered on I44. The G2BR, on the other hand, folds from an unstructured state into a well-ordered α-helix upon interacting with Ube2g2. At their interface, hydrophobic sidechains along the spine of the G2BR α-helix interlock with Ube2g2 in a fashion similar to the UbcH5C:Ub complex. Additionally, charged and polar residues distributed along the G2BR α-helix, on either side of the hydrophobic spine (Figure 2C and 2D) form salt-bridges and hydrogen bonds with Ube2g2, resulting in a high-affinity complex with a Kd of ~21 nM.
Implications of Ube2g2:G2BR for Ubiquitylation and ERAD
Until now, the existence of the G2BR as a high affinity binding site for Ube2g2 had been assumed to primarily provide a means to increase the level of Ube2g2~Ub in proximity to gp78 and to enhance ubiquitylation (Chen et al., 2006). We have now uncovered additional effects of the G2BR. This highly-specific 27 aa binding site for Ube2g2 has the striking effect of increasing the affinity of Ube2g2 for the gp78 RING finger as a consequence of conformational/allosteric changes in Ube2g2. The basis for the dramatic change in Kd, from 144 μM to 3 μM, is not yet fully understood and awaits further structural studies. A striking consequence of this increased affinity is the increased association of Ube2g2 with the gp78 RING finger, thereby facilitating the discharge of ubiquitin from Ube2g2~Ub through as yet undefined RING finger-mediated effects (Ozkan et al., 2005; Petroski and Deshaies, 2005). This is reflected in enhanced ubiquitylation even when the G2BR peptide is provided ‘in trans’ to Ube2g2 together with the cytoplasmic domain of gp78 lacking the G2BR sequence. Significantly, at least in vitro, the G2BR also increases ubiquitylation by heterologous ERAD RING finger E3s. In the cell, this potential for cross-talk between different ERAD ubiquitin ligases has the potential to contribute to associations between ERAD RING finger E3s and more importantly to promote synergistic interactions among these proteins (Ye et al., 2005; Morito et al., 2008). Similarly, if the reported gp78-oligomerization-dependent formation of K48 ubiquitin chains on C89 of Ube2g2 occurs in cells (Li et al., 2009), this will be facilitated by the enhanced affinity of Ube2g2 for the gp78 RING finger with G2BRs. This domain can serve both as platforms for Ube2g2 bearing nascent ubiquitin chains and as a means to enhance transfer of ubiquitin from heterologous Ube2g2s.
In addition to the increase in RING finger binding, the Ube2g2:G2BR structure, compared to apo-Ube2g2, reveals a conformation in which there is narrowing of the channel projecting from the active site Cys89. This is largely due to repositioning of the extended mobile acidic portion of the β4α2 loop including a dramatic alteration in orientation of sidechains of M101 and Y103. All of the available structural data indicates that this region is dynamically disordered (Arai et al., 2006; Li et al., 2009; this study). One consequence of this repositioning is a substantial delay in formation of Ube2g2~Ub in the presence of G2BR. Depending on the rate limiting step(s) in vivo, slowing of E2 loading with ubiquitin in the presence of the G2BR could allow for ‘proofreading’ by deubiquitylating enzymes in either substrate selection or chain linkage. The region analogous to the β4α2 loop is important in interactions between other E2-E3 pairs (Lorick et al., 2006) as well as for Ube2g2:gp78 RING finger (this study). The acidic extension in this loop is also found in Cdc34. Mutations in acidic residues in this region in both Ube2g2 and Cdc34 translate into decreased RING finger-dependent formation of K48-linked polyubiquitin chains (Li et al., 2007; Petroski and Deshaies, 2005). Moreover, Cdc34-Ub recently has been shown to manifest ~2 fold lower Kd for the core SCF E3 than Cdc34 (Saha and Deshaies, 2008). As the net effect of the G2BR, either in cells or in vitro, is to enhance ubiquitylation, potential effects of this dynamic loop on modulating transfer of ubiquitin from Ube2g2~Ub, formation of K48 ubiquitin chains, or in differential RING finger binding of Ube2g2~Ub versus Ube2g2 may be of equal or greater significance than the pronounced inhibition of loading with ubiquitin that we observe. Addressing these possibilities and the unresolved question of whether ubiquitin can be transferred from E1 to Ube2g2 bound either to the full cytoplasmic tail of gp78 or the isolated G2BR are important questions in ERAD and in E2 function that will require additional in vitro and cellular approaches.
Existence and Implications of E2 Binding Sites Distinct from Ligase Domains
E2s interact with E1 as well as with HECT, RING and RING finger-like domains on E3s. The question now arises as to the prevalence of other functionally significant E2 interactions. A well-characterized example is the E2-like molecule Mms2 (UEV1a), which binds Ubc13 through a region distinct from the G2BR-Ube2g2 interface and facilitates formation of K63-linked ubiquitin chains (Eddins et al., 2006). Another example, mentioned above, is ubiquitin, which plays a role in the processivity of BRCA1-mediated ubiquitin chain formation, presumably through alignment of multiple UbcH5C~Ub complexes (Brzovic et al., 2006; Christensen et al., 2007). The high affinity interaction of Ube2g2:G2BR precludes backside ubiquitin binding in the context of Ube2g2 bound to gp78. However, it does not exclude that alignment of multiple E2~Ub complexes (not necessarily Ube2g2) in vivo could occur in the context of a G2BR-bound Ube2g2~Ub and play a role in chain formation.
There are several examples where regions of E3s, which are not included in their canonical ligase domains, bind E2s, e.g. Nedd4 and Ubr1p. For these, neither the sites of interaction on their respective E2s nor the functional effects of this binding are known (Madura et al., 1993; Hatakeyama et al., 1997). There is evidence that SCF E3s recruit Cdc34 through part of the C-terminal extension of this E2 (Wu et al., 2002). Yeast Ubc7p, which is the ortholog of Ube2g2, is recruited by Cue1p to the ER membrane to function with the ERAD E3s Hrd1p and Doa10p (Kostova et al., 2007). Recent findings suggest that the 180 amino acid cytoplasmic domain of Cue1p activates Ubc7p in vitro in a RING finger-independent manner through unknown mechanisms (Bazirgan and Hampton, 2008; Kostova et al., 2009). Moreover, a ~50 aa domain in Cue1p, with some sequence homology to G2BR, directly binds Ubc7p. This domain is sufficient to activate ERAD by Hrd1p in vivo, and stimulates ubiquitylation in vitro, analogous to the G2BR (Kostova et al., 2009) and thus may represent a yeast equivalent of the G2BR.
A possible parallel to gp78 and Ube2g2 comes from the SUMO E3 Nup358/RanBP2. This E3 binds the SUMO E2 (Ubc9) through regions on Ubc9 analogous to interactions between ubiquitin E3s and E2s. However, this E3 also contacts Ubc9 through a second region with similarity to the Ube2g2:G2BR interface (Reverter and Lima, 2005). As pointed out by Reverter and Lima, mutations in this interface impact Ubc9 binding and Nup358/RanBP2 function (Pichler et al., 2004; Tatham et al., 2005). It would be of interest as to whether this second site of E2:E3 interaction has analogous effects to those observed in this study.
It seems reasonable to postulate that E2 binding sites within E3 complexes, but distinct from canonical ligase domains, may be a property of a substantial fraction of E3s. To what extent these result in functionally significant allosteric effects on E2s, as demonstrated herein, now becomes an exciting area for future research.
EXPERIMENTAL PROCEDURES
NMR Spectroscopy
NMR samples were prepared in 50 mM Tris, 2 mM TCEP, pH 7.5 buffer and experiments were performed at 25°C. NMR spectra were acquired on 600 and 800MHz Varian INOVA spectrometers equipped with triple resonance gradient cryoprobes. Details of resonance assignments, binding titrations and intermolecular NOESY experiments are provided in the Supplemental Experimental Methods.
X-ray Diffraction
Crystallization screens were carried out with a Phoenix robot (Art Robbins Instruments). The crystals were flash-frozen in liquid N2 after a short soak in a cryo-protection solution. A native dataset was collected at beamline 22-ID of the Advanced Photon Source, Chicago. The structure was solved by molecular replacement. The initial Fo–Fc map revealed the electron density for the G2BR. Details of crystallization, data acquisition, structure solution, model building and structure refinement are provided in Supplemental Experimental Methods.
Isothermal Titration Calorimetry
ITC was carried out using a VP-ITC microcalorimeter (MicroCal LLC, Northampton, MA) at 25°C. The typical experiment included injection of 25-27 aliquots (10 μL each) of 0.1-0.5 mM peptide solution into a 0.01-0.10 mM protein solution in the ITC cell (volume ~1.4 ml) stirring at 300 rpm. Additional details are provided in Supplemental Experimental Methods.
In Vitro Ubiquitylation, E2 loading and Discharge
Autoubiquitylation was carried out as described (Kostova et al., 2009; Lorick et al., 1999). Ubiquitylation of bacterially expressed purified Ub-GFP-His6 (~500 nM) was carried out using Glutathione Sepharose 4B bound GST-gp78CΔ577-643. Reactions were terminated by 2% SDS (final). After 10 min samples were diluted with 8 M urea in 50 mM Tris pH 7.4 to 0.1% SDS. Ub-GFP-His6 was isolated on Ni2+ beads and eluted with SDS-PAGE sample buffer after washing. 35S-labeled Ube2g2 was translated in the S30 T7 bacterial TnT system (Promega) and used at <100 nM. In loading experiments Ub K0 was used. For discharge of ubiquitin from E2, E2 was loaded with wild-type ubiquitin for 10 min. Reactions were quenched with 5 mM NEM and the buffer exchanged to 50 mM Tris (pH 7.4) with Ub (80 μM), peptide (4 μM) with or without purified gp78 RING finger (aa 313-380); discharge was monitored at 25°C. Additional details are provided in Supplemental Experimental Methods.
Supplementary Material
Acknowledgments
We thank Stanley Lipkowitz and Philip Ryan for critical reading of this manuscript, Mei Yang and Prasenjit Bhawmik for technical assistance, Kevin Lorick for assistance in initial studies and Robert Gemmill, Kazuhiro Iwai, Amy Lam and Emmanuel Wiertz for reagents. X-ray diffraction data were collected at the 22-ID beamline of SER-CAT, Advanced Photon Source, Argonne National Laboratory. Characterization of all proteins (CD and mass spectroscopy) and calorimetry were performed in the Biophysics Resource of the Structural Biology Laboratory. This work was supported by the National Institutes of Health Intramural Research Program and by a grant to AMW from the Multiple Myeloma Research Foundation.
Footnotes
SUPPLEMENTAL DATA Supplemental Data includes Supplemental Experimental Methods; Supplemental References, 10 figures and one table.
ACCESSION NUMBERS Coordinates and structure factors of the Ube2g2:G2BR complex were deposited in the Protein Data Bank under accession code 3H8K.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arai R, Yoshikawa S, Murayama K, Imai Y, Takahashi R, Shirouzu M, Yokoyama S. Structure of human ubiquitin-conjugating enzyme E2 G2 (UBE2G2/UBC7) Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:330–334. doi: 10.1107/S1744309106009006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazirgan OA, Hampton RY. Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem. 2008;283:12797–12810. doi: 10.1074/jbc.M801122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- Briggman KB, Majumdar A, Coleman CS, Chau V, Tolman JR. NMR assignment of human ubiquitin conjugating enzyme Ubc7. J Biomol NMR. 2005;32(4):340. doi: 10.1007/s10858-005-1257-7. [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D, 3rd, Fukuda M, Ohta T, Klevit R. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci U S A. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci U S A. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Pickart CM. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J Biol Chem. 1990;265:21835–21842. [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Bonvin AM, Winkler GS, van Schaik FM, Timmers HT, Boelens R. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12:633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Weissman AM. A field guide to ubiquitylation. Cell Mol Life Sci. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Jensen JP, Weissman AM. Subcellular localization and ubiquitin-conjugating enzyme (E2) interactions of mammalian HECT family ubiquitin protein ligases. J Biol Chem. 1997;272:15085–15092. doi: 10.1074/jbc.272.24.15085. [DOI] [PubMed] [Google Scholar]
- Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- Kostova Z, Mariano J, Scholz S, Koenig C, Weissman AM. A Ubc7p binding domain in Cue1p activates endoplasmic reticulum-associated protein degradation. J Cell Sci. 2009;122:1374–1381. doi: 10.1242/jcs.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin Cell Dev Biol. 2007;18:770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick KL, Tsai YC, Yang Y, Weissman AM. RING fingers and relatives: determinators of protein fate. In: Mayer RJ, Ciechanover A, Rechsteiner M, editors. Ubiquitin and the Chemistry of Life. Weinhein: Wiley-VCH; 2006. pp. 44–104. [Google Scholar]
- Madura K, Dohmen RJ, Varshavsky A. N-recognin/Ubc2 interactions in the N-end rule pathway. J Biol Chem. 1993;268:12046–12054. [PubMed] [Google Scholar]
- Moraes TF, Edwards RA, McKenna S, Pastushok L, Xiao W, Glover JN, Ellison MJ. Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13. Nat Struct Biol. 2001;8:669–673. doi: 10.1038/90373. [DOI] [PubMed] [Google Scholar]
- Morito D, Hirao K, Oda Y, Hosokawa N, Tokunaga F, Cyr DM, Tanaka K, Iwai K, Nagata K. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRΔF508. Mol Biol Cell. 2008;19:1328–1336. doi: 10.1091/mbc.E07-06-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadav E, Shmueli A, Barr H, Gonen H, Ciechanover A, Reiss Y. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem Biophys Res Commun. 2003;303:91–97. doi: 10.1016/s0006-291x(03)00279-1. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Brodsky JL. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci U S A. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Pichler A, Knipscheer P, Saitoh H, Sixma TK, Melchior F. The RanBP2 SUMO E3 ligase is neither HECT- nor RING-type. Nat Struct Mol Biol. 2004;11:984–991. doi: 10.1038/nsmb834. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Kim S, Jaffray E, Song J, Chen Y, Hay RT. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat Struct Mol Biol. 2005;12:67–74. doi: 10.1038/nsmb878. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Mendoza A, Mariano JM, Zhou M, Kostova Z, Chen B, Veenstra T, Hewitt SM, Helman LJ, Khanna C, Weissman AM. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat Med. 2007;13:1504–1509. doi: 10.1038/nm1686. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Wu K, Chen A, Tan P, Pan ZQ. The Nedd8-conjugated ROC1-CUL1 core ubiquitin ligase utilizes Nedd8 charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. J Biol Chem. 2002;277:516–527. doi: 10.1074/jbc.M108008200. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Kikkert M, van Voorden S, Wiertz E, Rapoport TA. Inaugural Article: Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


