Abstract
Genetic evidence suggests that conjugation of Small Ubiquitin-like Modifier proteins (SUMOs) plays an important role in kinetochore function, although the mechanism underlying these observations are poorly defined. we found that depletion of the SUMO protease SENP6 from HeLa cells causes chromosome misalignment, prolonged mitotic arrest and chromosome missegregation. Many inner kinetochore proteins (IKPs) were mis-localized in SENP6-depleted cells. This gross mislocalization of IKPs is due to proteolytic degradation of CENP-I and CENP-H via the SUMO targeted Ubiquitin Ligase (STUbL) pathway. Our findings show that SENP6 is a key regulator of inner kinetochore assembly that antagonizes the cellular STUbL pathway to protect IKPs from degradation during S phase. Here, we will briefly review the implications of our findings and present new data on how SUMOylation during S phase can control chromosome alignment in the subsequent metaphase.
Key words: SUMO, kinetochore, mitosis, SENP6, CENP-H, CENP-I
Introduction: The SUMO Pathway
Conjugation of Small Ubiquitin-like Modifier proteins (SUMOs) to cellular target proteins regulates many processes, including nucleo-cytoplasmic transport, mitosis, gene expression and DNA-metabolism.1 SUMOs undergo post-translational proteolytic processing, and mature SUMOs become attached to lysine residues within their target proteins through a conserved enzymatic cascade requiring the sequential action of an activating or E1 enzyme (Aos1/Uba2), a conjugating or E2 enzyme (Ubc9) and usually one of several ligases or E3 enzymes.2 SUMOylation is highly dynamic and is readily reversed by a single family of SUMO proteases.3,4 The same group of proteases both generates mature SUMOs by proteolytic processing (peptide bond cleavage) and removes SUMO from the conjugated substrates (isopeptide bond cleavage). Budding yeast has two SUMO proteases, Ulp1 and 2,5 while mammals have six genes encoding SUMO proteases, SENP1, 2, 3, 5, 6 and 7.4
Fungi have a single SUMO (Smt3p in budding yeast). Vertebrates possess three commonly expressed SUMOs: SUMO-2 and 3 are ∼95% identical to each other, and each shows ∼45% identity with SUMO-1.2 Due to their high sequence identity, SUMO-2 and 3 remain indistinguishable in most contexts, and can be collectively called SUMO-2/3. Most SUMOylation involves SUMO-2/3, since they are present at significantly higher concentrations than SUMO-1,6 and are more dynamic.7 Notably, SUMO-2/3 can also form conjugated SUMO chains.8 SUMO-1 does not typically form chains in vivo,8 and SUMO-1 conjugation is relatively more stable.9 Interestingly, SUMO-1 is dispensable for mouse development.10 All mammalian SENPs act on SUMO-2/3-conjugated species, while only SENP-1 and 2 act on SUMO-1-conjugated species.11 Chains of Smt3p or SUMO-2/3 are disassembled by a sub-family of SUMO proteases, which includes Ulp2p in yeast and SENP6 and SENP7 in vertebrates.12–16 These enzymes are collectively called SUMO chain editors.
SUMOylation can cause a variety of outcomes for any particular target protein, including changes in the target's sub-cellular localization, activity, protein-protein interactions and stability. Mechanistically, these changes often reflect the loss or acquisition of interaction surfaces upon conjugation of the SUMO group.1 Many cellular proteins contain SUMO-interacting motifs (SIMs) that allow low-affinity interactions with free or conjugated SUMOs.17–20 Some SIMs bind particular SUMO polypeptides selectively,19,21 allowing paralog-specific interactions. SIM-SUMO interactions frequently promote assembly of higher-order protein complexes.22,23
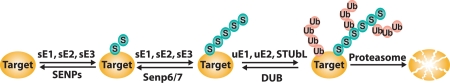
Recently, SIM-SUMO-based interactions were shown to be crucial for proteasomal degradation of some poly-SUMOylated targets through the action of SIM-containing ubiquitin ligases, dubbed as SUMO targeted ubiquitin ligases (STUbLs) (Fig. 1). The most studied STUbLs are Slx5/Slx8 in yeast and RNF4 in vertebrates.24–30 The biological roles of this degradation pathway are not yet fully understood; notably, SUMOylated species are often formed as intermediates in the assembly of large protein complexes,22,23,31 so it is possible STUbLs contribute to quality control mechanisms for disposal of mis-assembled or stalled intermediates.
Figure 1.
Schematic representation of STUbL degradation pathway. Enzymes of the SUMO conjugation pathway are indicated with s (sE1, sE2, sE3), while enzymes involved in ubiquitin conjugation are indicated with u (uE1, uE2) or as STUbL (E3 enzyme). SENPs are SUMO deconjugating enzymes, while DUBs are ubiquitin deconjugating enzymes.
Deletion of budding yeast Ulp2p disrupts mitotic progression and chromosome segregation.5,32 We tested whether depletion of the vertebrate SUMO chain editor SENP6 from HeLa cells could provoke analogous defects. We found that SENP6 depletion disrupted chromosome congression and caused prolonged delays in mitosis.33 These findings can be understood in the context of a novel role for SUMOylation in governing inner kinetochore assembly. Here, we present an overview of how SUMOylation controls kinetochore function within mitosis, as well as our findings regarding its distinct role in the regulation of inner kinetochore assembly.
SUMOylation during Mitosis: Outer Kinetochore and Inner Centromeric Proteins
Centromeres are chromatin domains on each chromosome that possess a specialized histone H3 variant, CEN-H3 (CENP-A in vertebrates and Cse4 in yeast).34 Kinetochores are large proteinaceous structures, which are assembled onto centromeres and which capture microtubules (MTs) to form the kinetochore fibers (k-fibers) that bi-orient the sister chromatids and allow their anaphase segregation to daughter cells.35 Kinetochores consist of around a hundred different proteins, arranged in multiple sub-complexes.36 Kinetochores have three other important roles: (1) Kinetochores nucleate MTs during early prometaphase that enhance polar MT capture;37 (2) Kinetochores generate a “wait anaphase” signal in the absence of proper MT attachment (bi-orientation).38 The pathway through which this signal is generated is called the Spindle Assembly Checkpoint (SAC); (3) Kinetochores generate the mechanochemical force required for anaphase separation of sister chromatids.39
When visualized by electron microscopy, vertebrate kinetochores show two electron dense regions (EDRs) separated by a translucent gap.40 The EDR proximal to the chromosome is known as the inner kinetochore, and the distal EDR is known as the outer kinetochore. Many of the enzymatic functions of kinetochores probably reside within the outer kinetochore, whereas the inner kinetochore may play a more structural role. Consistent with this idea, inner kinetochore components are epigenetically maintained in association with centromeres throughout the cell cycle while the outer kinetochore is reassembled prior to each mitosis.41,42 The fibrous corona is a region associated with the external face of the outer kinetochore that contains components of the SAC pathway and motor proteins. The inner centromeric region (ICR) is the domain of the centromeres that lies between the inner kinetochore plates.
In vertebrates, mitotic SUMOylation has been demonstrated for ICR, outer kinetochore and fibrous corona proteins (Table 1). Experiments in mitotic Xenopus egg extracts (XEEs) using GFP-SUMO-2/3 demonstrate considerable accumulation of SUMO-2/3 in the ICR region.43 ICR components that are confirmed SUMOylation targets include Topoisomerase II,44 poly(ADP-ribose) polymerase 1 (PARP1)45 and the chromosome passenger complex component Borealin.46 SUMOylation appears to be important for remodeling of attachments of Topoisomerase II to chromatin43 and for negative regulation of PARP1 activity against chromosomal substrates.45 The biological role of Borealin SUMOylation is not clear.46
Table 1.
SUMOylated kinetochore components
| Kinetochore proteins (gi number) | Amino acids (theoretical Mw kDa) | Number of SUMOylation concensus (aa positions) | Number of SIM motifs (aa positions) | SUMOylation status | Part of | Localization depends on | |
| Inner Kinetochore (CCAN or NAC-CAD) | CeNP-H (12597655) | 247 (29) | 1 (80–83) | 1 (195–198) | SUMOylated in S Phase | CeNP-H/I/K complex | CENP-A, CENP-T/W complex, CENP-M and CENP-N |
| CENP-I (41352697) | 756 (87) | None | 1 (309–312) | ||||
| Outer Kinnetochore (KMN Network) | Nuf2 (117968420) | 464 (54) | 3 (164–167, 334–337, 347–350) | 1 (15–18) | Ndc80 complex | CENP-A, CENP-C, Mis12 complex, KNL1, CENP-H/I/K complex (through CENP-H) | |
| Fibrous Corona | CENP-E (71061468) | 2701 (316) | 13 (637–640, 864–867, 1109–1112, 163–1166, 1698–1701, 1901–1904, 2051–2054, 2075–2078, 2183–2186, 2254–2257, 2453–2456, 2545–2548, 2592–2595) | 2 (2034–2037, 2307–2310) | SUMOylated in mitosis | None | KMN network |
| BubR1 (59814247) | 1050 (120) | 2 (4–7, 768–771) | 3 (242–245, 793–796, 860–863) |
The table lists known SUMO conjugation targets associated with the kinetochore. SUMO consensus motif (ΦK-X-E) or SIM motif (V/I-X-V/I-V/I, V/I-V/I-X-V/I) in each protein are listed. Φ denotes large hydrophobic amino acid, and X denotes any amino acid.
Outer kinetochore and fibrous corona proteins that are confirmed SUMOylation targets include BubR1 (a SAC component), Nuf2 (a member of the Hec1 MT binding complex) and CENP-E (a plus end-directed MT motor) (Table 1). In addition, CENP-E is a poly-SUMO-2/3 binding protein.47 Suppression of mitotic SUMOylation in HeLa cells by overexpression of SUMO protease SENP2 disrupts CENP-E targeting to kinetochores.47 Notably, mutation of SIMs within the C-terminal tail of CENP-E also blocks its recruitment to kinetochores, suggesting that its capacity to recognize poly-SUMOylated species is particularly important for its localization at this site. Nuf2 and BubR1 are implicated in targeting of CENP-E to kinetochores,48,49 so it is attractive to speculate that their SUMOylation may directly promote CENP-E recruitment through its SIM domains.47
Finally, a complex containing the nucleoporin RanBP2, SUMO-1-conjugated RanGAP1 and Ubc9 is recruited to outer kinetochores after initial attachments have formed between centrosome-nucleated MTs and kinetochores.50 RanGAP1 is the GTPase activating protein for Ran, a Ras-family GTPase with critical roles in interphase nuclear trafficking and in mitotic spindle assembly.51,52 Recruitment of this complex to kinetochores appears to be important for suppressing MT nucleation at kinetochores53 and for forming correct bi-oriented k-fibers.54,55 Interestingly, RanBP2 also possesses a domain with SUMO E3 activity, which has been implicated as a ligase for ICR substrates.46
SUMOylation of all known targets within the ICR, outer kinetochore and fibrous corona occurs within mitosis. While some of these proteins can be modified in other contexts,56 it is not clear whether SUMOylation at other points of the cell cycle can control their mitotic functions. While we have not performed an exhaustive search, it is notable that none of the ICR, outer kinetochore or fibrous corona proteins that we have tested to date appear to be substrates of SENP6, since they showed neither substantial increases in their SUMOylated populations nor were they destabilized after SENP6 depletion. It thus appears likely that SUMOylated targets within these structures are deconjugated by other proteases, and that SUMOylation may act in distinct ways during different phases of the cell cycle to control kinetochore function.
SUMOylation of CENP-H/I/K during S Phase
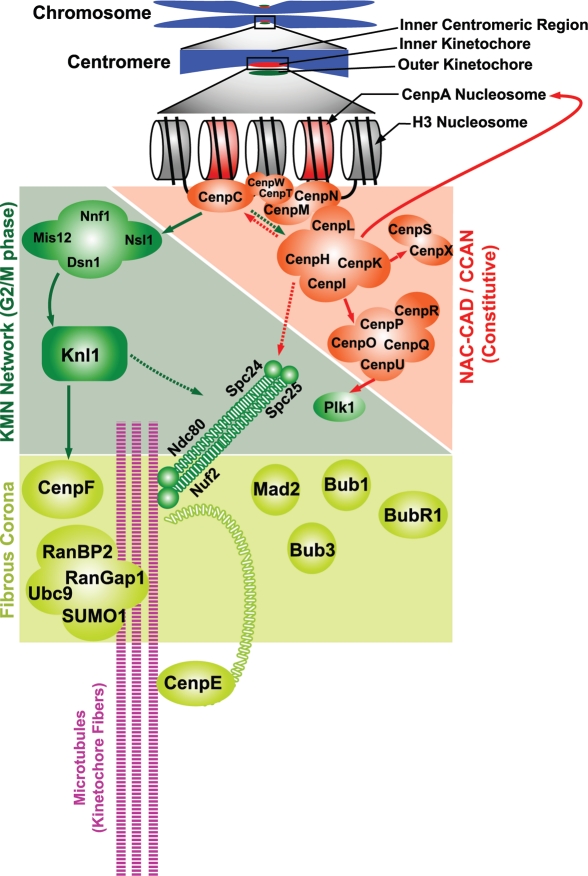
The inner kinetochore components that directly interact with CENP-A nucleosomes have been classified as the nucleosome-associated complex (NAC), and those that interact with CENP-A indirectly are known as CENP-A nucleosome-associated distal (CAD) complex;57 together, they are known as constitutive centromere-associated network (CCAN)42 (Supp. table 1 and Fig. 2). While CCAN components are essential for chromosome congression, spindle assembly and sister kinetochore bi-orientation, the biochemical functions of most individual CCAN proteins remain undefined. However, it is clear that they can play distinct roles in kinetochore assembly, and that multiple CCAN sub-complexes may be subject to SUMOylation: CENP-C is essential for recruitment of the outer kinetochore proteins.58 CENP-C interacts with the SUMO pathway genetically,59 and it can become SUMO conjugated in vitro,60 but the in vivo circumstances and molecular function of CENP-C SUMOylation remain obscure. The CENP-H/I/K complex is important for CENP-A loading in G1 phase,61,62 as well as for inner kinetochore assembly through recruitment of the CENP-O/P/Q/U/R63 and CENP-S/X64 complexes (Fig. 2). CENP-H/I/K has also been implicated in the control of MT plus end dynamics and flux at the kinetochore.65 As discussed below, we found that CENP-H/I/K components are important SUMO-2/3 conjugation substrates that are modified during S phase.
Figure 2.
Schematic representation of the genetic and molecular interactions of the different kinetochore proteins. Filled arrows indicate complete dependence and broken arrows indicate partial dependence.
We examined HeLa cells depleted of SENP6 by siRNA mediated knockdown33 to determine whether they would show mitotic defects, analogous to those observed in budding yeast lacking Ulp2p.5,32 While many aspects of spindle assembly appeared grossly normal, cells lacking SENP6 consistently showed an increase in pole-to-pole distance of around 15%, as well as a broader distribution of chromosome within the metaphase plate and increased distance between sister kinetochores.33 Moreover, although the bulk of chromosomes in cells lacking SENP6 congressed onto the metaphase plate, a few chromosomes typically remained unaligned and close to the poles. The unaligned kinetochores activated the SAC, prolonging mitosis for several hours, but the cells did not achieve complete chromosome congression despite this delay. The depleted cells frequently slipped out of mitosis with unaligned chromosomes, resulting in mis-segregation. We found only modest increase in the overall level of SUMOylated species in SENP6-depleted cells.13
To understand this phenotype, we systematically surveyed the abundance of representative members of all major kinetochore complexes. Most remarkably, while CENP-A and CENP-C were correctly localized, the CENP-H/I/K complex was absent from the kinetochore.33 We measured the levels of CENP-H and CENP-I and found the abundance of both the proteins was dramatically reduced after SENP6 depletion,33 suggesting that their absence was the result of destabilization. We hypothesized that one or more component of the CENP-H/I/K complex became polySUMOylated in SENP6-depleted cells, resulting in its STUbL-mediated degradation. One prediction of this hypothesis is that loss of both SENP6 and RNF4 would result in the accumulation of polySUMOylated forms of the targeted protein(s). Consistent with this prediction, RNF4 depletion restored the levels of CENP-H and -I in SENP6-depleted cells, and both proteins accumulated in highly SUMOylated species.33 These findings show that CENP-H/I/K complex members are bona fide SUMO-2/3 conjugation substrates in vivo.
Consequences of CENP-H/I/K Degradation after SENP6 Depletion
In surveying kinetochore components, we did not observe the wholesale degradation of other proteins in the manner that we had found for CENP-H and -I,33 indicating that the CENP-H/I/K may be unique among the kinetochore complexes as a target of RNF4-mediated degradation in the absence of SENP6. Previous reports have established that the CENP-H/I/K complex depends on the CENP-T/W complex and CENP-L, M and N for its localization at the kinetochore.57 In turn, the CENP-O/P/Q/U/R63 and CENP-S/X64 complexes require CENP-H/I/K for their localization. Loss of the CENP-H/I/K complex also results in reduced levels of some outer kinetochore complexes, including the Hec1 and Mis12 complexes.66 The CENP-O/P/Q/U/R complex recruits polo like kinase 1 (Plk1), which is responsible for phosphorylating many kinetochore proteins,63,67–70 suggesting that Plk1 localization should indirectly depend upon the CENP-H/I/K complex.
The pattern in which other kinetochore components were re-localized after SENP6 depletion was consistent with the previously described hierarchy of kinetochore protein recruitment. Although we found no degradation of CENP-O/P/Q/U/R complex members, CENP-O was absent from kinetochores, as would be expected in the absence of CENP-H/I/K. In turn, mis-localization of CENP-O/P/Q/U/R should disrupt Plk1 recruitment.68 We tested this prediction by examining the localization of Plk1 in cells depleted of SENP6, CENP-I and CENP-O (Fig. 3). We found that depletion of each of these proteins caused the loss of Plk1 from kinetochores, but not from centrosomes. Cells lacking either SENP6 or CENP-I frequently showed large cytoplasmic Plk1 aggregates in SENP-6 and CENP-I depleted cells [Fig. 3(ii) and (iii)] which are not present in CENP-O depleted cells [Fig. 3(iv)]. We often observed cytosolic CENP-O foci after SENP-6 knockdown with a similar appearance (data not shown), so it is possible that these aggregates reflect association of the mis-localized populations of CENP-O/P/Q/U/R complex and Plk1. Consistent with the correct recruitment of Plk1 to centrosomes, we did not find major defects in centrosomal maturation or MT nucleation (data not shown). Finally, the predicted absence of CENP-S/X complex from kinetochores after degradation of CENP-H/I/K may underlie the increase in intrakinetochore distance in metaphase SENP-6 depleted cells.64
Figure 3.
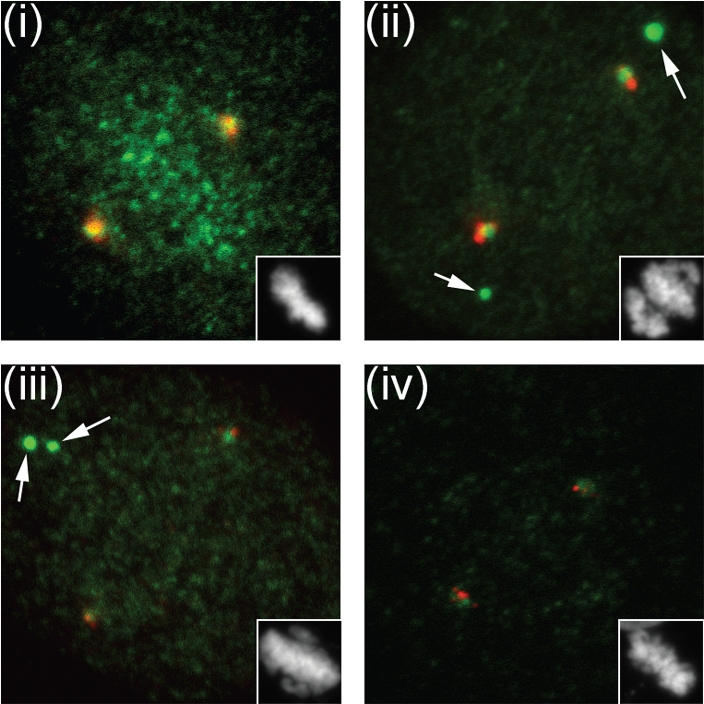
Plk localization is indirectly disrupted after SENP6 depletion. we treated HeLa cells with Control, SENP-6, CENP-I and CENP-O siRNA for 48 hours, and simultaneously synchronized the cells using a double thymidine block protocol, as described.3 Ten hours after release from the second thymidine block, the cells were preextracted, fixed in 4% paraformaldehyde, and stained for centrosomes (Centrin 2 antisera; red) and Plk1 (green) Images (i–iv) represents control, SENP-6, CENP-I and CENP-O knockdown respectively. SENP6 depletion in HeLa cells resulted in loss of punctuate Plk1 signal at the metaphase plate (i and ii), which we have confirmed as centromeric Plk1 (data not shown). Similar loss of Plk1 was also observed in case of CENP-I and CENP-O knockdown (iii and iv). The arrows indicate cytoplasmic Plk1 aggregates.
Quantitative immunofluorescence showed that the Hec1 complex was partially mis-localized in the absence of SENP6,33 consistent with its pattern of mis-localization after CENP-H/I/K complex depletion.66 Interestingly, we observed differential loading of Hec1 on the attached and unattached kinetochores within individual cells: Hec1 levels were reduced around 45% on the aligned kinetochores, whereas they were reduced around 75% on the unaligned kinetochores. This differential pattern was surprising because earlier reports had not found that Hec1 recruitment is sensitive to the attachment status of individual kinetochores.71,72 It is interesting to speculate that loss of CENP-H/I/K may lead to inconsistent loading of the Hec1 complex onto kinetochores. In this case, our observations would imply that kinetochores require a threshold level of Hec1 for efficient MT capture, and that kinetochores that do not meet this threshold may remain unaligned indefinitely. Mis12 is a member of another key outer kinetochore complex whose localization has been reported depend partially upon CENP-H/I/K.66 Mis12 showed decreased localization on kinetochores, particularly those of unattached chromosomes, although the extent of this decrease was somewhat less than we observed for Hec1.33 Notably, outer kinetochore components whose recruitment has been reported to be independent of CENP-H/I/K (CENP-E and -F66) did not show significant changes after SENP6 depletion.
Together, these patterns are consistent with the idea that CENP-H/I/K complex members are the key targets of SENP6 within the kinetochores, since the defects that result from SENP6 depletion closely mimic those caused by loss of CENP-H/I/K.
Implications for Kinetochore Assembly
Our observations do not clarify why the CENP-H/I/K complex is regulated through this elaborate mechanism that balances the activities of SENP6 with RNF4. Nor, indeed, do they reveal why CENP-H/I/K complex members are SUMOylated in the first place. It was striking that the maximum accumulation of these polySUMOylated species occurred during S phase in cells that were synchronized using a double thymidine block/release (DTB) protocol, rather than in mitosis.33 S phase is an interval of dynamic recruitment of CENP-H/I/K complex to constitutive inner kinetochore structures,73 and it is possible that maximal levels of their SUMOylation corresponds to the interval of inner kinetochore assembly. It is thus attractive to speculate that CENP-H and -I SUMOylation promotes assembly of inner kinetochores, in a manner that may be analogous to the role of SUMOylation in the assembly of other multi-protein complexes.22,23,31 In this context, it is notable that co-depletion of RNF4 substantially rescues chromosome congression defects observed in the absence of SENP6 alone. CENP-H and -I accumulate in highly SUMOylated forms under these circumstances,33 indicating that complete de-SUMOylation of CENP-H and -I cannot be essential for assembly of sufficiently functional inner kinetochores.
There are several possible ways in which CENP-H and -I SUMOylation could promote inner kinetochore assembly. First, SUMOylation may enhance formation of the CENP-H/I/K complex. As shown in Supp. Table 1, CENP-H contains both a SUMOylation consensus motif and SIM, whereas CENP-I and CENP-L have SIMs. Alternatively, SUMO moieties attached to the CENP-H/I/K complex could provide an interaction surface for CENP-H/I/K complex loading onto inner kinetochores or for recruitment of other SIM-containing kinetochore proteins (Suppl. Table 1). If de-SUMOylation were only achieved after inner kinetochore assembly were complete, persistent SUMOylation of assembly intermediates may allow their interactions with RNF4 and their degradation, thus disposing of stalled or superfluous assembly intermediates.
It is clearly possible that such regulated degradation of assembly intermediates could be a mechanism to prevent the formation of ectopic centromeres. Ectopic centromeres are extremely deleterious for genomic stability as they cause chromosome fragmentation during anaphase.74,75 CENP-A loading in extra-centromeric sites is very inefficient but not unprecedented,76–78 suggesting that its deposition is normally tightly controlled. The CENP-H/I/K complex enhances the loading of CENP-A containing nucleosomes;61 if unrestricted, such an activity could imaginably bypass normal restraints for CENP-A deposition at extra-centromeric sites. This model would predict tight temporal or spatial regulation of SENP6, RNF4 or both enzymes to prevent inappropriate deposition of centromeric components.
Implications for the SUMO Pathway
STUbLs mediate the targeted destruction of poly-SUMOylated substrates, while chain editing SUMO proteases mediate the deconjugation of the same species. Our studies show that the direct antagonism of these classes of enzymes can form a biologically significant network, regulating the abundance of their targets. It appears likely that this paradigm will also be applicable to other proteins whose degradation is controlled by RNF4, such as PML79,80 and Hypoxia-Inducible Factor-2α.81
SUMO-2/3 modification can be reversed by six SUMO proteases, which are distributed to different regions of the nucleus.4 Given that nuclear subdomains are extremely dynamic,82 any of these SENPs might in principle be able to deconjugate SUMO-2/3 chains from CENP-H/I, whereas it appears that only SENP6 actually fulfills this function in reality.33 The explanation for this exquisite specificity might lie in the enzymatic properties and the protein levels of individual SENPs. SENP6 shows a higher affinity for poly-SUMO chains than SENP2, and it is more active in cleavage of polySUMOylated substrates.14 An elevated affinity of SENP6 towards SUMO chains can be accomplished through its multiple SUMO interacting motifs, and RNF4 has been proposed to act preferentially on polySUMOylated substrates for the same reason.29 This model predicts that other SENPs cleave SUMOylated species with one or two SUMO moieties. When the chain length exceeds this level, SUMOylated species become high affinity targets of RNF4 and SENP6, which catalyze their destruction or reduce their level of SUMOylation below the threshold required for high affinity recognition. Since SENP7 also preferentially acts on SUMO-2/3 chains,14,15 it would be expected to function in a similar manner. However, SENP7 is much less abundant than SENP6 in HeLa cells (unpublished observation), which may explain its inability to compensate SENP6 deficiency in our experiments.
Obviously, it is also possible that SENP6 or RNF4 may have specificity for individual polySUMOylated target proteins. We have not observed concentration of SENP6 near centromeres13 or observed detectable amount of SENP6 in CENP-H/I immunoprecipitations (unpublished observation), so we currently have no evidence that such a mechanism is applicable in the case of the CENP-H/I/K complex.
Conclusions and Future Perspectives
Our findings provide the first case in which inner kinetochore assembly is regulated through post-translational modification. The use of SUMOylation in this context may be uniquely valuable, as it provides both a mechanism to facilitate complex assembly and a mechanism through which inappropriate structures can be destroyed. It will now be important to discover how this modification is used and what binding interfaces may be made or disrupted through SUMOylation. It will also be important to discover the E3 enzyme(s) that controls CENP-H/I/K complex modification, and to understand how this enzyme and SENP6 are temporally and spatially regulated during the process of kinetochore assembly. It appears likely to us that similar regulatory networks can be used in a variety of cellular contexts, and it will be fascinating to discover how they are adapted within different situations.
Acknowledgements
We would like to thank Wilma L. Lingle (Mayo Clinic Cancer Center, Rochester, Minnesota) for kindly providing the anti-Centrin2 antibody (Fig. 3).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12619
Supplementary Material
References
- 1.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 2.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 3.Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 7.Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 9.Zhu S, Goeres J, Sixt KM, Bekes M, Zhang XD, Salvesen GS, et al. Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1. Mol Cell. 2009;33:570–580. doi: 10.1016/j.molcel.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Mukhopadhyay D, Mathew S, Hasebe T, Heimeier RA, Azuma Y, et al. Identification and developmental expression of Xenopus laevis SUMO proteases. PLoS One. 2009;4:8462. doi: 10.1371/journal.pone.0008462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 13.Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, et al. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J Cell Biol. 2006;174:939–949. doi: 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lima CD, Reverter D. Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J Biol Chem. 2008;283:32045–32055. doi: 10.1074/jbc.M805655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen LN, Geoffroy MC, Jaffray EG, Hay RT. Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem J. 2009;421:223–230. doi: 10.1042/BJ20090246. [DOI] [PubMed] [Google Scholar]
- 16.Vertegaal AC. SUMO chains: polymeric signals. Biochem Soc Trans. 38:46–49. doi: 10.1042/BST0380046. [DOI] [PubMed] [Google Scholar]
- 17.Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 18.Minty A, Dumont X, Kaghad M, Caput D. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem. 2000;275:36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 19.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 20.Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 21.Zhu J, Zhu S, Guzzo CM, Ellis NA, Sung KS, Choi CY, et al. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J Biol Chem. 2008;283:29405–29415. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization and repression of sumoylated transcription factors. Mol Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 23.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekiyama N, Arita K, Ikeda Y, Hashiguchi K, Ariyoshi M, Tochio H, et al. Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45. Proteins. 78:1491–1502. doi: 10.1002/prot.22667. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Prelich G. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol Cell Biol. 2009;29:1694–1706. doi: 10.1128/MCB.01470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie Y, Rubenstein EM, Matt T, Hochstrasser M. SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev. doi: 10.1101/gad.1906510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol. 2009;10:564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 30.Hunter T, Sun H. Crosstalk between the SUMO and ubiquitin pathways. Ernst Schering Found Symp Proc. 2008:1–16. doi: 10.1007/2789_2008_098. [DOI] [PubMed] [Google Scholar]
- 31.Panse VG, Kressler D, Pauli A, Petfalski E, Gnadig M, Tollervey D, et al. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 2006;7:1311–1321. doi: 10.1111/j.1600-0854.2006.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strunnikov AV, Aravind L, Koonin EV. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics. 2001;158:95–107. doi: 10.1093/genetics/158.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukhopadhyay D, Arnaoutov A, Dasso M. The SUMO protease SENP6 is essential for inner kinetochore assembly. J Cell Biol. 188:681–692. doi: 10.1083/jcb.200909008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalal Y, Bui M. Down the rabbit hole of centromere assembly and dynamics. Curr Opin Cell Biol. doi: 10.1016/j.ceb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 36.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitamura E, Tanaka K, Komoto S, Kitamura Y, Antony C, Tanaka TU. Kinetochores generate micro-tubules with distal plus ends: their roles and limited lifetime in mitosis. Dev Cell. 18:248–259. doi: 10.1016/j.devcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nezi L, Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Curr Opin Cell Biol. 2009;21:785–795. doi: 10.1016/j.ceb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 41.Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 43.Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 2005;24:2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryu H,, Al-Ani G, Deckert K, Kirkpatrick D, Gygi SP, Dasso M, et al. PIASy mediates SUMO-2/3 conjugation of poly (ADP-ribose) polymerase1 (PARP1) on mitotic chromosomes. J Biol Chem. doi: 10.1074/jbc.M109.074583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein UR, Haindl M, Nigg EA, Muller S. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol Biol Cell. 2009;20:410–418. doi: 10.1091/mbc.E08-05-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell. 2008;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan GK, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu D, Ding X, Du J, Cai X, Huang Y, Ward T, et al. Human NUF2 interacts with centromere-associated protein E and is essential for a stable spindle microtubule-kinetochore attachment. J Biol Chem. 2007;282:21415–21424. doi: 10.1074/jbc.M609026200. [DOI] [PubMed] [Google Scholar]
- 50.Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14:611–617. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 51.Dasso M. Ran at kinetochores. Biochem Soc Trans. 2006;34:711–715. doi: 10.1042/BST0340711. [DOI] [PubMed] [Google Scholar]
- 52.Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- 53.Torosantucci L, De Luca M, Guarguaglini G, Lavia P, Degrassi F. Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol Biol Cell. 2008;19:1873–1882. doi: 10.1091/mbc.E07-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arnaoutov A, Dasso M. Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle. 2005;4:1161–1165. doi: 10.4161/cc.4.9.1979. [DOI] [PubMed] [Google Scholar]
- 55.Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, et al. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol. 2005;7:626–632. doi: 10.1038/ncb1263. [DOI] [PubMed] [Google Scholar]
- 56.Martin N, Schwamborn K, Schreiber V, Werner A, Guillier C, Zhang XD, et al. PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. EMBO J. 2009;28:3534–3548. doi: 10.1038/emboj.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 58.Kwon MS, Hori T, Okada M, Fukagawa T. CENP-C is involved in chromosome segregation, mitotic checkpoint function and kinetochore assembly. Mol Biol Cell. 2007;18:2155–2168. doi: 10.1091/mbc.E07-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fukagawa T, Regnier V, Ikemura T. Creation and characterization of temperature-sensitive CENP-C mutants in vertebrate cells. Nucleic Acids Res. 2001;29:3796–3803. doi: 10.1093/nar/29.18.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chung TL, Hsiao HH, Yeh YY, Shia HL, Chen YL, Liang PH, et al. In vitro modification of human centromere protein CENP-C fragments by small ubiquitin-like modifier (SUMO) protein: definitive identification of the modification sites by tandem mass spectrometry analysis of the isopeptides. J Biol Chem. 2004;279:39653–39662. doi: 10.1074/jbc.M405637200. [DOI] [PubMed] [Google Scholar]
- 61.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 62.Okada M, Okawa K, Isobe T, Fukagawa T. CENPH-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol Biol Cell. 2009;20:3986–3995. doi: 10.1091/mbc.E09-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hori T, Okada M, Maenaka K, Fukagawa T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell. 2008;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amano M, Suzuki A, Hori T, Backer C, Okawa K, Cheeseman IM, et al. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J Cell Biol. 2009;186:173–182. doi: 10.1083/jcb.200903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amaro AC, Samora CP, Holtackers R, Wang E, Kingston IJ, Alonso M, et al. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat Cell Biol. 12:319–329. doi: 10.1038/ncb2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol Biol Cell. 2008;19:587–594. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee KS, Oh DY, Kang YH, Park JE. Self-regulated mechanism of Plk1 localization to kinetochores: lessons from the Plk1-PBIP1 interaction. Cell Div. 2008;3:4. doi: 10.1186/1747-1028-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang YH, Park JE, Yu LR, Soung NK, Yun SM, Bang JK, et al. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol Cell. 2006;24:409–422. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 69.Wong OK, Fang G. Plx1 is the 3F3/2 kinase responsible for targeting spindle checkpoint proteins to kinetochores. J Cell Biol. 2005;170:709–719. doi: 10.1083/jcb.200502163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong OK, Fang G. Cdk1 phosphorylation of BubR1 controls spindle checkpoint arrest and Plk1-mediated formation of the 3F3/2 epitope. J Cell Biol. 2007;179:611–617. doi: 10.1083/jcb.200708044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, et al. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hori T, Haraguchi T, Hiraoka Y, Kimura H, Fukagawa T. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- 73.Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amor DJ, Choo KH. Neocentromeres: role in human disease, evolution and centromere study. Am J Hum Genet. 2002;71:695–714. doi: 10.1086/342730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: new insights into centromere structure, disease development and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakano M, Okamoto Y, Ohzeki J, Masumoto H. Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes. J Cell Sci. 2003;116:4021–4034. doi: 10.1242/jcs.00697. [DOI] [PubMed] [Google Scholar]
- 78.Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JY, Berns MW, et al. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci USA. 2009;106:15762–15767. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 80.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 81.van Hagen M, Overmeer RM, Abolvardi SS, Vertegaal AC. RNF4 and VHL regulate the proteasomal degradation of SUMO-conjugated Hypoxia-Inducible Factor-2alpha. Nucleic Acids Res. 38:1922–1931. doi: 10.1093/nar/gkp1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Misteli T. Physiological importance of RNA and protein mobility in the cell nucleus. Histochem Cell Biol. 2008;129:5–11. doi: 10.1007/s00418-007-0355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.