Abstract
Epstein-Barr virus (EBV) has been shown to encode at least 40 microRNAs (miRNAs), an important class of molecules that negatively regulate the expression of many genes through posttranscriptional mechanisms. Here, we have used real-time PCR assays to quantify the levels of EBV-encoded BHRF1 and BART miRNAs in latently infected cells and in cells induced into the lytic cycle. During latency, BHRF1 miRNAs were seen only in cells with detectable Cp- and/or Wp-initiated EBNA transcripts, while the BART miRNAs were expressed in all forms of latent infection. Surprisingly, levels of different BART miRNAs were found to vary up to 50-fold within a cell line. However, this variation could not be explained by differential miRNA turnover, as all EBV miRNAs appeared to be remarkably stable. Following entry into the virus lytic cycle, miR-BHRF1-2 and -1-3 were rapidly induced, coincident with the onset of lytic BHRF1 transcripts, while miR-BHRF1-1 expression was delayed until 48 h and correlated with the appearance of Cp/Wp-initiated EBNA transcripts. In contrast, levels of BART miRNAs were relatively unchanged during virus replication, despite dramatic increases in BART transcription. Finally, we show that BHRF1 and BART miRNAs were delayed relative to the induction of BHRF1 and BART transcripts in freshly infected primary B cell cultures. In summary, our data show that changes in BHRF1 and BART transcription are not necessarily reflected in altered miRNA levels, suggesting that miRNA maturation is a key step in regulating steady-state levels of EBV miRNAs.
Epstein-Barr virus (EBV), a B lymphotropic gammaherpesvirus with potent growth-transforming properties, is etiologically linked to a number of malignancies of lymphoid and epithelial cell origin, including Burkitt's lymphoma (BL), posttransplant lymphoproliferative disease (PTLD), and nasopharyngeal carcinoma (NPC) (52). As illustrated in Fig. 1A, these different tumor settings can be distinguished by alternative patterns of EBV latent gene expression. Thus, EBV-driven PTLD lesions and growth-transformed lymphoblastoid cell lines (LCLs) display a latency III form of infection, characterized by the expression of six EBV nuclear antigens transcribed from one of two alternative promoters (Wp and Cp), and three latent membrane proteins (50, 63); in addition, a recent study (36) reported that LCLs also weakly express the viral Bcl2 homologue BHRF1 as a latent antigen. In contrast, most BL tumor cell lines which retain the original BL tumor phenotype in vitro show a more restricted pattern of latent antigen expression (termed latency I), in which the Cp, Wp, and LMP promoters are silent and a single nuclear antigen EBNA1 is transcribed from a novel promoter, Qp (46, 54). However, a subset of BL lines display a third form of latency (termed Wp-restricted latency), in which Wp-initiated transcripts give rise to EBNA1, -3A, -3B, -3C, and BHRF1 (35, 36, 38). In addition to the above-mentioned latent antigens, two sets of RNAs are also expressed in all forms of EBV infection. These are the noncoding EBER RNAs (6, 42) and the BamHI A rightward transcripts (BARTs), a complex family of highly spliced transcripts originally identified in nasopharyngeal carcinoma (NPC) tumor cells, and whose protein-coding potential remains controversial (15, 16, 26, 53, 57).
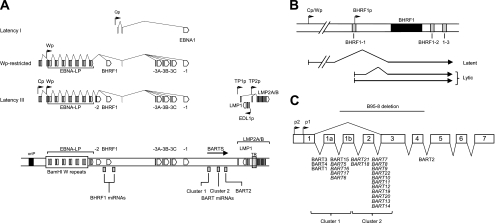
FIG. 1.
Schematic organization of the EBV genome and location of EBV miRNAs. (A) Three different forms of virus latent gene expression in BL cell lines and LCLs. The location of viral promoters and splice structures of viral transcripts are shown relative to a linear representation of the EBV genome. Conventional latency I BLs express a single latent antigen EBNA1 transcribed from a viral promoter in the BamHI Q region (Qp). Wp-restricted BL lines are characterized by the presence of an EBNA2-deleted EBV genome; these BLs express EBNA1, -3A, -3B, -3C, and -LP, along with BHRF1, all transcribed from the latency III BamHI W promoter (Wp), but in the absence of EBNA2 and the latent membrane proteins (LMPs). Growth-transformed LCLs express all six EBNAs and BHRF1, predominantly from transcripts initiated at the BamHI C promoter (Cp), along with the LMPs which are transcribed from separate promoters in the BamHI N region. EBERs (not shown) and BamHI A rightward transcripts (BARTs) are present in all three forms of latency. Also shown are the positions of the BHRF1 and BART miRNAs, the latent origin of replication (oriP), and the terminal repeat region (TR). (B) Detailed structure of BHRF1 transcripts and location of BHRF1-derived miRNAs. All three BHRF1 miRNAs may be generated either by processing of an intron present within the Cp/Wp-initiated primary EBNA transcript or by processing of the 5′ and 3′ untranslated regions within latent BHRF1 transcripts with the W2-Y1-Y2-BHRF1 structure. In contrast, lytic BHRF1 transcripts initiated from the alternative BHRF1p promoter encode only miR-BHRF1-2 and miR-BHRF1-3. (C) Detailed structure of the highly spliced BARTs and location of the BART-derived miRNAs (adapted from Edwards et al. [21]). The BART miRNAs form two clusters within the BART introns, with the exception of mirBART2, which lies further downstream. The prototype B95-8 EBV strain carries a 12-kb deletion in this region which removes several cluster 1 miRNAs and all cluster 2 miRNAs (shown in italics).
To date, most studies on the role of EBV in the pathogenesis of virus-associated malignancies have focused on the contributions of the latent gene products as signaling effector molecules or transcription factors (39). However, in recent years it has been shown that EBV also encodes an important class of molecules called microRNAs (miRNAs) (12, 30, 48). These small (∼19- to 24-nucleotide [nt]) noncoding RNAs negatively regulate gene expression through binding to specific sequences in the 3′ untranslated region (UTR) of target mRNAs, leading either to translation inhibition or to mRNA cleavage (22). Cellular miRNAs are thought to regulate expression of up to one-third of all human genes (43) and have been implicated in a diverse range of biological functions, including cell growth, apoptosis, and tumorigenesis (8, 11, 27). As illustrated in Fig. 1A, EBV encodes at least 40 miRNAs that map to two regions of the viral genome (12, 19, 30, 48, 65); the BHRF1 miRNAs are located immediately upstream and downstream of the BHRF1 open reading frame (Fig. 1B), while the BART miRNAs lie within the intronic regions of the BARTs (Fig. 1C). While these viral miRNAs appear to have been conserved during lymphocryptovirus evolution (12), to date there is little information regarding the possible functions of these miRNAs in the viral life cycle. Specific EBV miRNA targets which have been identified include the viral proteins LMP1 (miR-BART1-5p, -16-5p, and -17-5p), LMP2A (miR-BART22), and DNA polymerase (miR-BART2) and the cellular targets PUMA (miR-BART5) and CXCL-11 (miR-BHRF1-3) (9, 18, 44, 45, 60).
While the roles of the EBV miRNAs are still unresolved, we have a better understanding of the expression profiles of these molecules from studies of EBV-positive cell lines (12, 21, 48, 49, 61) and primary tumor material (19, 41, 65). Thus, it has been reported that the BHRF1 miRNAs are present in cells with a latency III pattern of antigen expression, suggesting that all three miRNAs are generated from Cp/Wp-initiated transcripts. High levels of miR-BHRF1-2 and miR-BHRF1-3 are also observed during EBV lytic replication, although in this context, BHRF1 and its associated miRNAs are transcribed from an alternative promoter, BHRF1p (47); as illustrated in Fig. 1B, this lytic BHRF1 transcript overlaps the miR-BHRF1-1 sequence and therefore can generate only the two downstream BHRF1 miRNAs (7). In the case of the BART miRNAs, previous reports have indicated that these species are present in all forms of latent infection (12, 48, 49), consistent with the view that they are processed from the ubiquitous BART transcripts but are preferentially expressed in EBV-positive epithelial and NPC tumor cells (19, 21). Interestingly, alternative splicing of these complex BART transcripts may affect BART miRNA expression, as suggested by the recent finding that splicing of exon 1 directly to exon 3 favored miRNA production (21).
Previous reports characterizing the expression of EBV miRNAs have relied upon Northern blotting, cloning, or arrays to detect miRNA expression (12, 48, 60, 65). These methods require large amounts of RNA, have limited sensitivity, and provide only qualitative data. To overcome these limitations, we have used real-time PCR to quantify the absolute levels of EBV miRNAs in EBV-infected cells. In parallel, we have investigated the relationship between expression of the BHRF1 and BART transcripts and the levels of BHRF1 and BART miRNAs in latent and productive infection. Our data confirm and extend the existing literature on EBV miRNA expression and demonstrate that changes in BHRF1 and BART transcription do not necessarily correlate with changes in miRNA abundance.
MATERIALS AND METHODS
Cell culture.
The BL lines used in this work included the standard EBV-positive latency I BL lines Dante-BL, Sav-BL, Ezema-BL, Akata-BL, Kem-BL, and MutuI-BL cl.59 (28, 31), the group III BL lines MutuIII-BL and GlorIII-BL, which have drifted in culture to a latency III form of infection, the Wp-restricted BL lines Oku-BL, Sal-BL, and Ava-BL (35), and latency I and Wp-restricted sublines derived from Awia-BL (37). The LCLs carrying natural EBV isolates included IM51.1, IM81.1, IM83.1, and IM93.1, established by spontaneous transformation from EBV-infected donors (10), and EH LCL1 and LCL2, generated by in vitro infection of EBV-naïve B cells with the recombinant 2089 EBV genome (20). In addition, the following reference cell lines served as standards for quantifying EBV transcripts: X50/7 LCL (62), CD+Oku LCL (35), the spontaneously permissive cell line Sal tr-LCL (containing 2% BZLF1-positive cells), and the EBV-positive nasopharyngeal cell line C666-1 (17). EBV-negative DG75 cells were included as a negative control in all experiments. AKBM is a derivative of Akata-BL stably transfected with a green fluorescent protein (GFP) reporter plasmid under the control of the early BMRF1 promoter (51). All cell lines were maintained in exponential growth in RPMI medium-10% selected fetal calf serum (Biosera) supplemented with 2 mM l-glutamine (Gibco) and penicillin-streptomycin (Sigma). Where indicated, RNA transcription was inhibited by the addition of 5 μg/ml actinomycin D (Sigma) to the cell culture medium, and cells were harvested for analysis at the indicated time points.
Reactivation of EBV in AKBM cells and isolation of cells in the lytic cycle.
Viral reactivation in AKBM cells was induced by cross-linking surface immunoglobulin (Ig) with 100 μg/ml goat anti-human IgG antibody (Cappel) as described previously (51). Cells were then transferred into fresh medium, and 2 × 106 cells were harvested at each indicated time point. To monitor the induction of the virus lytic cycle, an aliquot of cells was assayed by flow cytometry, either directly for GFP expression or after being stained with the BZLF1-specific monoclonal antibody (MAb) BZ.1, as described previously (51). To isolate productively infected cells, induced AKBM cells were sorted into GFP-positive and GFP-negative populations using a MoFlo cell sorter (Dako, North America). Where indicated, EBV genome replication was inhibited by the addition of 200 μg/ml acycloguanosine (Sigma) to the culture medium.
Measurement of EBV genome load.
Genomic DNA was extracted using a GenElute mammalian genomic DNA kit (Sigma). EBV genome load was determined by quantitative PCR (QPCR) using primer-probe combinations to amplify EBV DNA polymerase (BALF5) and the cellular beta 2-microglobulin sequences, as described previously (34).
Primary infection of B cells.
Primary B cells were isolated from peripheral blood samples using CD19 Dynabeads (Invitrogen) and subsequently exposed to preparations of recombinant 2089 EBV (20) at a multiplicity of infection (MOI) of 100 using published methods (56). Infected cells were cultured in fresh medium and harvested at the indicated time points.
Quantitative PCR analysis of EBV transcripts.
Total RNA was prepared using a Nucleospin RNA isolation kit (Machery-Nagel), treated with DNase I to remove residual DNA (DNA-free; Ambion), and reverse transcribed using gene-specific cDNA primers (10). Relative levels of Qp-, Cp-, and Wp-initiated transcripts, BARTs (exon 6-7 splice junction), lytic and latent BHRF1 transcripts, and immediate-early BZLF1 transcripts were determined by QPCR using published primers and TaqMan probes (10, 36). In addition, a new assay was designed to detect both latent and lytic BHRF1 transcripts as follows: cDNA primer 5′-TCTTGCTGCTAGCT-3′, forward primer 5′-CCCTCTTAATTACATTTGTGCCAGAT-3′, reverse primer 5′-TCCCGTATACACAGGGCTAACAGT-3′, and probe 5′-FAM (6-carboxyfluorescein)-TAGAGCAAGATGGCCTATTCAACAAGGGAGA-TAMRA (6-carboxytetramethylrhodamine)-3′. Two further assays were also designed to amplify across the exon 2-3 splice junction in conventional BART mRNAs (BART 2-3) or across the exon 1-3 splice junction in the alternative splice variant lacking exon 2 (BART 1-3) using the following primers and probes: common cDNA primer 5′-TCTAAAGTCATACGCCC-3′ (exon 3), BART 2-3 forward primer 5′-TCCACTTTGTGTTACAGGTCCG-3′ (exon 2-3 junction), BART 1-3 forward primer 5′-CTCTTCATGTGAGGTCCGGC-3′ (exon 1-3 junction), common reverse primer 5′-TGTGTCCGGTAAACGCCATA-3′ (exon 3), and BART probe 5′-FAM-CCACGGAGACTCGGACGTAGCCCTT-TAMRA-3′ (exon 3).
Relative quantification of gene expression was performed using an Applied Biosystems 7500 sequence detection system. Each PCR run included duplicate test cDNA samples and serial cDNA dilutions prepared from a suitable reference line (Akata-BL for Qp, X50/7 for Wp, CD+Oku LCL for Cp and latent BHRF1, Sal tr-LCL for BZLF1 and lytic BHRF1, and C666-1 for BARTs), which were used to construct relative standard curves. EBV gene expression data were normalized either to cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or to phosphoglycerate kinase (PGK) expression data quantified using commercially available assays (Applied Biosystems). Normalized data were expressed relative to the level for the appropriate reference line, which was assigned an arbitrary value of 1. Note that in the case of lytic transcripts, the data were further adjusted such that the final values were expressed relative to the level for a culture containing 100% BZLF1-positive cells.
Quantitative PCR analysis of EBV miRNAs.
All EBV miRNA sequences were obtained from the Sanger miRNA database, version 12 (http://microRNA.sanger.ac.uk/) (29). Real-time PCR assays to quantify EBV miRNAs were based on the stem-loop reverse transcriptase (RT) primer method described by Chen et al. (13). Stem-loop RT primers and primer-probe combinations for each target miRNA were either designed using the custom TaqMan small RNA assay service (miR-BART5, miR-BART7, miR-BART22, miR-BART13, and miR-BART2-5p) or obtained commercially (miR-BHRF1-1, miR-BHRF1-2, miR-BHRF1-3, miR-BART3, miR-BART4, miR-BART1-3p, and miR-BART15) (Applied Biosystems). For miRNA analysis, RNA was prepared using a mirVana miRNA isolation kit (Applied Biosystems) according to the manufacturer's instructions. Input RNA (10 ng) was reverse transcribed in a 20-μl reaction volume containing up to 7 RT stem-loop primers using a TaqMan microRNA reverse transcription kit (Applied Biosystems). QPCRs (in 20-μl reaction mixtures) were performed using an Applied Biosystems 7500 sequence detection system. In each PCR, duplicate aliquots of each test cDNA were run alongside serial cDNA dilutions prepared from a suitable reference line (X50/7 for miR-BHRF1 miRNAs, and C666-1 for miR-BART miRNAs), which were used to construct relative calibration curves. To convert from relative to absolute quantitation, target miRNAs were obtained as synthetic oligonucleotides (Eurogentec), pooled, and reverse transcribed as described above. Aliquots of cDNA corresponding to 108 to 102 miRNA copies were then subjected to PCR amplification and the data used to generate calibration curves from which the absolute number of EBV miRNA molecules in reference X50-7 and C666.1 cells could be determined. Expression data were normalized to the level for RNU48 as a loading control (Applied Biosystems).
Sequencing of BHRF1 miRNAs.
BHRF1 sequences spanning all three miRNAs were PCR amplified using the primers 5′-GGACTTGGGGTGATTTTCTATGT-3′ and 5′-CCCACACTCACCTCAGTTATTTC-3′ under the following conditions: 94°C for 5 min; 35 cycles of 94°C for 60 s, 56°C for 60 s, and 72°C for 2 min; and 72°C for 10 min. The 1,600-bp fragment was then gel purified and sequenced using an automated Applied Biosystems 3730 system (Functional Genomics and Proteomics Services, School of Biosciences, University of Birmingham, United Kingdom). The pre-miRNA secondary structure prediction through free energy minimization was calculated using the software program Mfold (http://mfold.rna.albany.edu/) (66).
RESULTS
Validation of miRNA PCR assays.
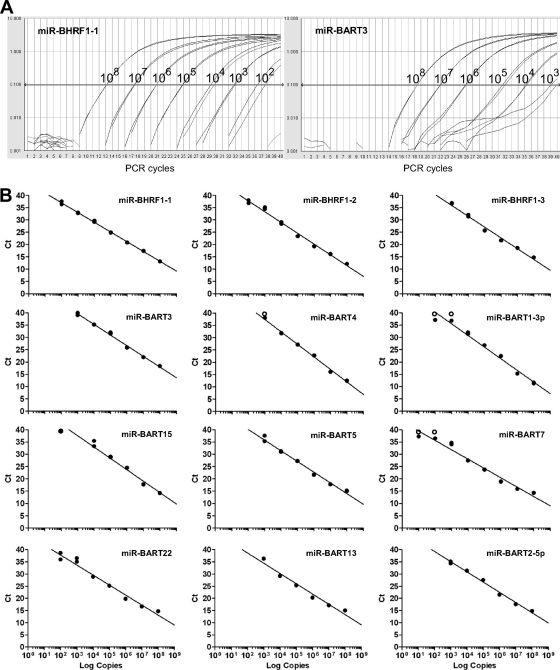
In the initial experiments, we validated the specificity and sensitivity of a panel of QPCR assays designed to quantify selected EBV miRNAs; these included all three miRNAs derived from the BHRF1 region, five miRNAs from BART cluster 1, three miRNAs from BART cluster 2, and miR-BART2-5p. We also optimized a multiplex cDNA synthesis method incorporating up to 7 miRNA-specific RT primers such that several miRNAs could be assayed from the same cDNA pool but without significantly reducing the sensitivity of detection (data not shown). Using cDNA prepared by this multiplex protocol, we then validated both the specificity and the sensitivity of each PCR assay in a series of control experiments. First, using RNA from six EBV-negative cell lines, we showed that all but two assays always gave negative PCR results; the exceptions were the assays for miR-BART3 and miR-BART4, in which these cell lines occasionally yielded extremely weak signals (data not shown; see Fig. 6B). Second, using cell mixtures containing serial dilutions of an EBV-positive LCL in an EBV-negative cell background, we found that all assays reproducibly detected a single EBV-infected cell (data not shown). Finally, we determined the absolute sensitivity of each assay by using cDNA prepared from serial dilutions of a pool of synthetic oligonucleotides containing all 12 target miRNAs. Data from representative amplification plots for serial dilutions of miR-BHRF1-1 and miR-BART3 are shown in Fig. 2A, while standard curves for all 12 assays are shown in Fig. 2B. All assays demonstrated an excellent correlation between cycle threshold (CT) values and RNA input over at least a 5-log range and had a lower detection limit of between 100 and 1,000 miRNA copies.
FIG. 2.
Sensitivity of QPCR assays for detecting EBV miRNAs. (A) cDNA samples containing known numbers of synthetic miRNAs (108 to 102 copies) were used to test the absolute sensitivity of each QPCR assay. Shown are representative PCR amplification plots for miR-BHRF1-1 and miR-BART3. (B) Standard curves obtained for all 12 miRNAs obtained by plotting the cycle threshold (CT) values against log input RNA copy number. Open circles indicate absence of a detectable signal (CT = 40).
BHRF1 transcription and miR-BHRF1 miRNA expression in EBV-positive B cell lines.
In the next series of experiments, we sought to explore the relationship between BHRF1 transcription and the levels of mature BHRF1 miRNAs in well-characterized EBV-positive B cell lines with different patterns of latent gene expression. These test lines included seven conventional latency I BL lines, five Wp-restricted BL lines, two latency III BL lines, and seven virus-transformed LCLs; note that EH LCL1 and EH LCL2 were generated from a recombinant prototype EBV strain which, like B95-8, lacks many of the BART miRNAs (Fig. 1C). We first verified that these cell lines showed the expected pattern of latent viral promoter usage by quantifying levels of Qp-, Cp-, and Wp-initiated EBNA transcripts (Fig. 3). Thus, latency I BL lines exclusively used Qp with only trace or undetectable levels of Cp/Wp activity. In contrast, the Wp-restricted BL lines were distinguished by high levels of Wp activity, usually in the absence of the other latent promoters. As expected, latency III BLs and LCLs showed variable levels of both Cp and Wp usage, with the exception of X50-7, which lacks Cp sequences (62). Furthermore, all test cell lines were tightly latent since immediate-early BZLF1 transcripts were either undetectable or present at very low levels, equivalent to fewer than 1% BZLF1-positive cells (Fig. 4A).
FIG. 3.
Expression of latent transcripts in EBV-positive B cell lines. Latency I-associated Qp transcripts and latency III-associated Cp- and Wp-initiated transcripts were quantified by QPCR in seven latency I BLs (Dante, Sav, Ezema, Akata, Kem, and Awia cl. a), five Wp-restricted BLs (Awia cl. k and m, Oku, Sal, and Ava), two latency III BLs (Mutu III and Glor III), and seven LCLs (X50-7, IM51, -81, -83, -93, EH LCL1, and EH LCL2); DG75 was included as a negative control. Data were normalized to cellular PGK levels and expressed relative to a suitable positive reference line, which was assigned an arbitrary value of 1. Error bars indicate standard deviations for replicate assays.
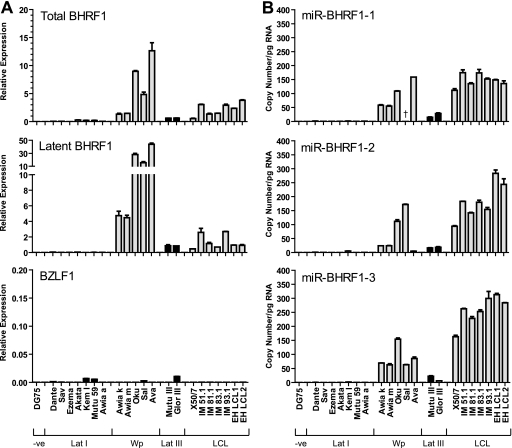
FIG. 4.
Expression of BHRF1 transcripts and BHRF1 miRNAs in EBV-positive B cell lines. (A) Total BHRF1 transcripts, Cp/Wp-initiated latent BHRF1 transcripts, and immediate-early BZLF1 transcripts were quantified by QPCR with the same cell lines as those shown in Fig. 3. BHRF1 values are expressed relative to the level for a reference LCL, while BZLF1 values are expressed relative to the level for a spontaneous LCL and adjusted so that a value of 1 is equivalent to 100% positive cells. (B) Expression of BHRF1 miRNAs in the same cell lines determined by QPCR. Data were normalized to RNU48 expression and expressed as absolute copy numbers per pg total RNA. Error bars indicate standard deviations for replicate assays. Note that Sal-BL is deleted for miR-BHRF1-1, as indicated by the dagger symbol.
Having validated our test lines, we next quantified BHRF1 transcripts using two different primer-probe combinations, one designed to detect both lytic and latent BHRF1 transcripts and the other designed to specifically amplify Cp/Wp-initiated latent BHRF1 transcripts. As shown by the data in Fig. 4A, the BHRF1 expression profiles were comparable for both assays and indicated that BHRF1 transcripts were detectable only in cells with Cp and/or Wp activity. Consistent with the very low BZLF1 signals, negligible levels of lytic BHRF1 transcripts were observed in these test lines (data not shown), thus excluding the possibility that a small population of productively infected cells were making a significant contribution to the total levels of BHRF1 transcripts. We then went on to measure the levels of miR-BHRF1 miRNAs in the same panel of cell lines using our newly developed QPCR assays. The data (Fig. 4B) show that BHRF1 miRNA expression varied widely, ranging from 0 to 5 copies per pg RNA in latency I BL lines to more than 300 copies per pg RNA in latency III LCLs. Interestingly, levels of the miR-BHRF1 miRNAs in the LCLs were at least as high as those in the Wp-restricted BL lines, even though the latter has much higher levels of BHRF1 transcripts. In most cases, we noted that all three miR-BHRF1 miRNAs were coordinately expressed within a cell line, with no more than 3-fold variation in absolute values between individual miRNAs. One exception was Sal-BL, which expressed abundant levels of mir-BHRF1-2 and mir-BHRF1-3 but is known to be deleted for miR-BHRF1-1 (35); a second exception was Ava-BL, where levels of miR-BHRF1-2 were unusually low compared to those of the other BHRF1 miRNAs.
BHRF1 miRNA sequence variation.
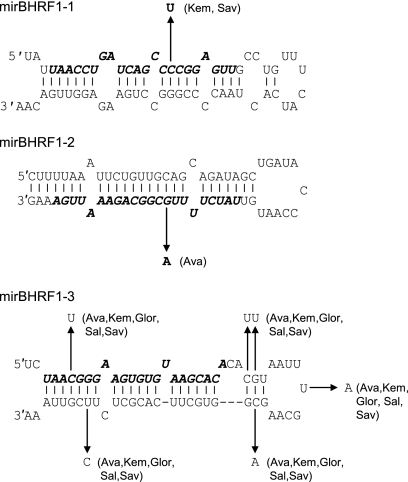
Since previous studies have revealed that sequence changes can interfere with miRNA processing (21, 49), we considered the possibility that EBV strain polymorphisms might account for some of the observed variation in BHRF1 miRNA levels. To this end, we determined the sequence of the BHRF1 pre-miRNAs in 12 EBV isolates and compared the sequences with the published B95-8 genome; the number and position of any sequence changes are summarized in Fig. 5. Overall, miR-BHRF1-1 and miR-BHRF1-2 sequences were highly conserved. In two cases (Kem-BL and Sav-BL), we found a single change in the mature miR-BHRF1-1 miRNA; the effect of this mutation on pre-miRNA processing or PCR detection is unclear, as the BHRF1 region is not transcribed in these latency I lines (Fig. 4A). One cell line, Ava-BL, contained a single polymorphism in the mature miR-BHRF1-2 sequence which disrupts a G-C base pair in the stem-loop. With Mfold used to calculate the change in minimum free energy, this G-to-A substitution is predicted to significantly destabilize the pre-miRNA hairpin loop structure (Fig. 5) and therefore may account for the low levels of miR-BHRF1-2 detected in this cell line (Fig. 4B). Finally, five cell lines contained six common nucleotide changes in the miR-BHRF1-3 pre-miRNA. Surprisingly, these changes did not decrease the minimum free energy structure of the BHRF1-3 pre-miRNA and are therefore unlikely to affect folding or processing.
FIG. 5.
Sequence variation in BHRF1 miRNAs. Shown is a summary of sequence changes seen in Sav-BL, Akata-BL, Kem-BL, Mutu-BL, Awia-BL, Oku-BL, Sal-BL, Ava-BL, Glor-BL, X50-7 LCL, and IM51 LCL. In each case, the prototype B95-8 sequence of the pre-miRNA is shown with the mature miRNA indicated in bold italicized text. Sequence changes are marked by the arrows, with the cell line indicated in brackets. Using Mfold to calculate the minimum free energy (ΔG) of the folded pre-miRNA, the C-to-U change in mir-BHRF1-1 is predicted to increase ΔG from −25.5 to −22.5 kcal/mol. The G-to-A substitution seen in miR-BHRF1-2 markedly increases ΔG from −27.1 to −20.5 kcal/mol. In the case of miR-BHRF1-3, the six nucleotide changes marginally decrease ΔG from −23.2 to −24.3 kcal/mol.
BART transcription and miR-BART miRNA expression in EBV-positive B cell lines.
In the next series of experiments, we examined the expression of the BART transcripts and derived miRNAs in the same panel of B cell lines (Fig. 6). Since particular BART splice variants may favor the production of BART miRNAs (21), we quantified BART transcription using two different PCR assays, the first specific for the conventional transcript containing exons 1, 2, and 3 (exon 2-3 assay) and the second specific for the exon 2-deleted variant (exon 1-3 assay). In contrast to BHRF1 transcripts, BART transcripts were detected across all forms of latency, albeit at highly variable levels (Fig. 6A); BART transcripts containing exon 2-3 and exon 1-3 splice junctions appeared to be coordinately expressed. Screening the same samples for the presence of nine representative BART miRNAs, we found levels ranging from fewer than 10 to more than 400 copies per pg RNA (Fig. 6B to D). Overall, it is clear that the B cell lines expressed lower levels of BART miRNAs than the epithelial cell line C666-1 (data included in Fig. 6B to D for comparison). Interestingly, we observed up to a 50-fold variation between different BART miRNAs within a given cell line. BART miRNA levels also varied widely between cell lines, but all cell lines showed the same hierarchy of BART miRNA expression, with miR-BART7 usually the most abundant and miR-BART13 the most scarce. While there was a good correlation between BART transcription and BART miRNAs in the BL lines, levels of BART transcripts were frequently lower in the LCLs despite the fact that BLs and LCLs expressed similar levels of BART miRNAs.
FIG. 6.
Expression of BART transcripts and mirBART miRNAs in EBV-positive B cell lines. (A) BART transcripts were quantified by QPCR using assays specific for either the exon 1-3 or the exon 2-3 splice variants in the same cell lines as those shown in Fig. 3. Data were normalized to PGK expression, and results are expressed relative to those for the EBV-positive epithelial tumor cell line C666-1. Panels B and C show expression of cluster 1 and cluster 2 BART miRNAs, respectively. Data were normalized to RNU48 expression and are expressed as copy numbers per pg input RNA. Error bars indicate standard deviations for replicate assays. Data points highlighted with daggers correspond to cell lines carrying a BART deletion. For comparison, copy numbers of BART miRNAs per pg C666-1 RNA are shown in parentheses in each histogram.
Stability of EBV miRNAs.
The above-mentioned findings highlighted both the dramatic differences in overall levels of viral miRNA expression between different cell lines and the wide variation in levels of individual miRNAs within a cell line. One possible explanation for this variability could be differences in the turnover of individual EBV miRNAs. To investigate this in more detail, we examined the half-life of selected miRNAs in three cell lines, X50/7 LCL, Sal-BL, and Ava-BL. Cells were cultured in the presence of the RNA synthesis inhibitor actinomycin D and then harvested for analysis at various time points after treatment. Residual levels of BHRF1 and BART transcripts and derived miRNAs were then monitored by QPCR (Fig. 7); as a control, we also assayed c-myc RNA since this transcript has been reported to have a very short half-life of around 12 min (58). Figure 7A shows the individual data points from X50-7 cells up to 24 h posttreatment, while Fig. 7B summarizes the data from all three cell lines. In the case of X50/7, c-myc RNA levels decreased rapidly to less than 10% after 4 h of exposure to actinomycin D, while the latent BHRF1 transcripts also decreased, but to a lesser extent. In contrast, the BHRF1 miRNAs were very stable, decreasing by less than 2-fold even after 24 h. We then measured levels of BART transcripts and three miRNAs (miR-BART4, miR-BART7, and miR-BART2-5p) previously shown to have markedly different steady-state levels. We found that both the BART transcripts and the BART miRNAs remained relatively unchanged in X50/7 up to 24 h (Fig. 7A). Similar data for the stability of the transcripts and miRNAs were obtained from Sal-BL and Ava-BL (Fig. 7B). Thus, it appears that EBV miRNAs are very stable, and therefore, the observed variation in miRNA levels cannot be explained by different rates of miRNA turnover.
FIG. 7.
Stability of EBV transcripts and miRNAs. (A) X50/7 cells were treated with 5 μg/ml actinomycin D, harvested at the indicated time points, and then analyzed for EBV transcripts and miRNAs. The left hand panel shows the residual expression of c-myc RNA, latent BHRF1 transcripts, and BHRF1 miRNAs, while the right hand panel shows the expression of BART (1-3) transcripts and selected BART miRNAs. (B) Summary of data from Sal-BL, Ava-BL, and X50/7. The histograms indicate the expression of the indicated transcripts and miRNAs measured at 12 h after the addition of actinomycin D. All data were normalized to RNA input.
Expression of EBV miRNAs during lytic replication.
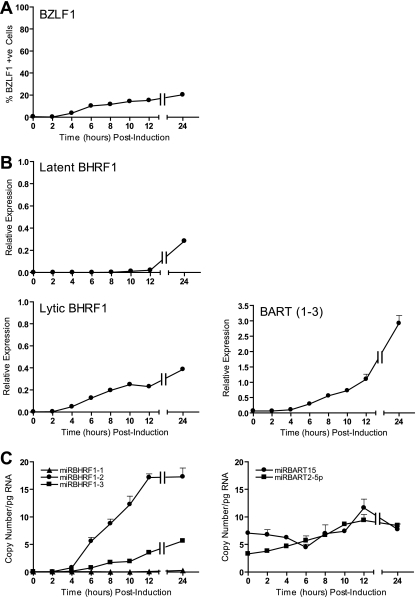
Since it has been reported that BHRF1 and BART transcripts are induced during productive infection (32, 47, 64), we next asked to what extent miR-BHRF1 and miR-BART miRNA levels might also be increased in the EBV lytic cycle. To this end, we exploited the AKBM cell system in which EBV-positive Akata-BL cells are stably transfected with a GFP reporter gene under the control of the EBV lytic cycle BMRF1 promoter; following Ig cross-linking, a significant proportion of cells enter into the lytic cycle and these cells can be identified by GFP expression. In the first instance, we monitored the production of BHRF1 and BART transcripts and derived miRNAs in AKBM cells at various time points up to 24 h postinduction. In the representative experiment whose results are shown in Fig. 8A, BZLF1-positive lytic cells were detected by flow cytometry within 4 h and increased to around 22% of the culture at 24 h. Lytic BHRF1 transcripts appeared with kinetics similar to those of BZLF1 (Fig. 8B) and coincided with marked increases in miR-BHRF1-2 and -1-3 miRNAs but not miR-BHRF1-1 (Fig. 8C); this is consistent with the idea that BHRF1 transcription is initiated from the lytic promoter BHRF1p, rather than the latent promoters Cp/Wp, at these early time points. In contrast, levels of BART-derived miRNAs (here illustrated with data for miR-BART15 and miR-BART2-5p) barely increased during the same 24-h period, despite the fact that BART transcript levels increased more than 45-fold (Fig. 8B and C).
FIG. 8.
Expression of EBV transcripts and miRNAs in lytic AKBM cultures at 24 h postinduction. (A) BZLF1-positive AKBM cells were enumerated at the indicated time points postinduction by staining with BZ.1 anti-BZLF1 antibody and then analyzed by flow cytometry. (B) BHRF1 transcripts and BARTs quantified by QPCR at the indicated time points. Latent BHRF1 values are expressed relative to the level for a reference LCL, while lytic BHRF1 and BZLF1 values are expressed such that a value of 1 is equivalent to 100% positive cells. (C) Expression of selected BHRF1 miRNAs and BART miRNAs determined by QPCR at the indicated time points. Data were normalized to RNU48 expression and are expressed as copy numbers per pg input RNA. Error bars indicate standard deviations for replicate assays.
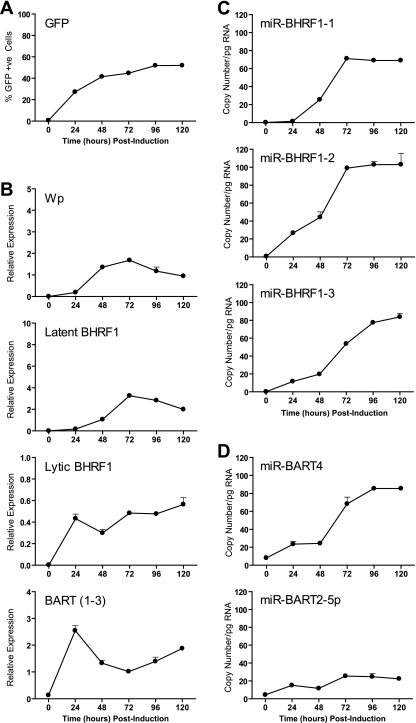
To examine whether the relatively small increases in miR-BART miRNA expression might be due to a delay in processing the BART transcripts, we repeated the experiment but now analyzed gene expression in the induced AKBM cells for up to 120 h postinduction. Lytic BHRF1 and BART transcripts showed levels of induction at 24 h similar to those observed in the previous experiments and thereafter remained relatively constant (Fig. 9B). However, we now observed that latent BHRF1 transcripts became detectable at later time points, coincident with induction of the latency III promoters Wp and Cp (Fig. 9B and data not shown) and leading to expression of all three miR-BHRF1 miRNAs (Fig. 9C). Consistent with our earlier findings, we again noted a rapid and dramatic increase in BART transcription accompanied by only modest 5- to 8-fold increases in BART miRNAs.
FIG. 9.
Extended time course showing expression of EBV transcripts and miRNAs in lytic AKBM cultures. (A) Lytic GFP-positive AKBM cells were enumerated at the indicated time points postinduction by flow cytometry. (B) Wp, latent BHRF1 and lytic BHRF1 transcripts, and BARTs (exon 1-3 splice form) were quantified by QPCR. Panels C and D show expression of selected BHRF1 and BART miRNAs, respectively, determined by QPCR. Error bars indicate standard deviations for replicate assays.
The above-mentioned findings suggested that BHRF1 miRNA production during virus replication is biphasic, with early lytic BHRF1 transcripts selectively generating miBHRF1-2 and miR-BHRF1-3 while delayed latent BHRF1 transcripts are processed to give all three miR-BHRF1 miRNAs. To investigate BHRF1 miRNA expression in more detail, we performed two additional experiments. In the first, we confirmed that the lytic and latent BHRF1 transcripts were present in the same cell population. Briefly, AKBM cells were induced into the lytic cycle as before and then sorted into GFP-positive (lytic) and GFP-negative (latent) populations at 48 h postinduction. We found that both latent and lytic BHRF1 transcripts, and all three miR-BHRF1 miRNAs, were greatly enriched in the GFP-positive fraction relative to the level for the GFP-negative population (data not shown).
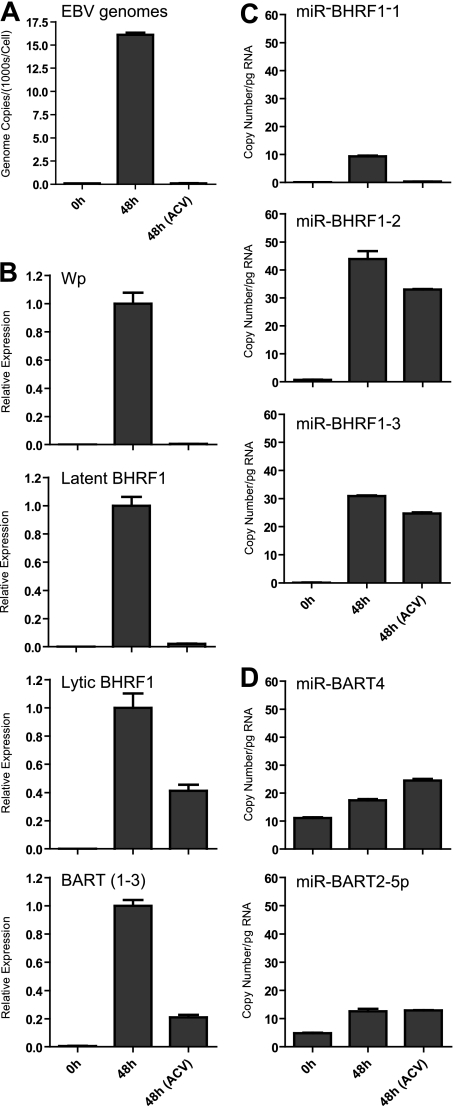
The appearance of latency III transcripts during the later stages of virus replication has been reported previously (64). We speculated that such transcripts might be expressed from the very high copy numbers of newly replicated EBV genomes, since such genomes are unmethylated and could therefore act as templates for Wp- and Cp-initiated transcription. To test this hypothesis, we induced AKBM cells in the presence of acyclovir (ACV) to block viral DNA replication. By QPCR, we confirmed that the average EBV genome load increased from around 50 genomes per cell to over 15,000 genomes per cell in induced control AKBM cells. The addition of ACV completely blocked this genome amplification (Fig. 10A) and also ablated induction of latency III transcripts and miR-BHRF1-1 miRNA (Fig. 10B and C), supporting the view that these transcripts are dependent on viral DNA synthesis. In contrast, ACV treatment only modestly reduced levels of lytic BHRF1 and BART transcripts (2-fold and 5-fold, respectively) (Fig. 10B) and made little difference to the expression of the two remaining miR-BHRF1 miRNAs or BART miRNAs (Fig. 10C and D).
FIG. 10.
Effect of ACV on expression of EBV transcripts and miRNAs in induced AKBM cultures. (A) EBV genome load quantified by QPCR at 48 h postinduction. Data were normalized to cellular beta-2-microblobulin copy numbers and are expressed as numbers of EBV genomes per cell. (B) Wp, latent and lytic BHRF1 transcripts and BARTs (exon 1-3 splice) quantified by QPCR at 48 h postinduction. Data are expressed relative to values seen in the uninhibited culture. Panels C and D show expression of selected BHRF1 and BART miRNAs, respectively, determined by QPCR. Data were normalized to RNU48 expression and are expressed as copy numbers per pg input RNA. Error bars indicate standard deviations for replicate assays.
In the final experiment, we asked whether the failure to significantly increase BART miRNA expression during virus replication was due to changes in BART transcript or BART miRNA stability. Briefly, AKBM cells were induced as before, and then, at 12 h postinduction, the cells were either cultured in normal medium or transferred to medium containing actinomycin D to block further RNA transcription. We found that the BART miRNAs had similarly long half-lives in lytically induced AKBM cultures and in the latently infected cell lines examined previously (data not shown), arguing that the weak induction of the BART miRNAs was not due to increased miRNA turnover during the lytic cycle.
Expression of EBV miRNAs during primary infection of B cells.
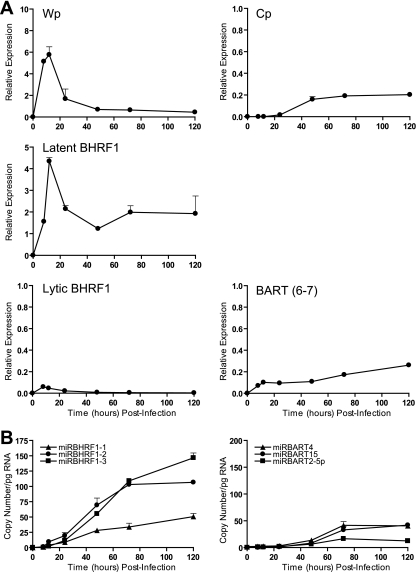
In the final set of experiments, we examined the expression of EBV miRNAs in newly infected primary B cells. On this point, the kinetics of EBV gene expression during primary B cell infection are well established. Thus, Wp is activated immediately postinfection (1, 56, 59), leading to the initial expression of EBNA2, EBNA-LP, and also BHRF1 (1, 2, 4, 36). Wp activity then falls after 24 h, as Cp activity becomes dominant, concomitant with the broadening of latent antigen expression to the full spectrum of EBNAs and LMPs. Primary B cells were therefore exposed to EBV preparations in vitro, cultured, and harvested for analysis at the indicated time points. Note that the recombinant 2089 EBV strain used in this work contains the same 12-kb BamHI A deletion as the prototype B95-8 strain (Fig. 1C); therefore, in this experiment it was necessary to detect the BART transcripts using an alternative primer-probe combination which specifically amplified sequences across the exon 6-7 splice junction. The data in Fig. 10A show the results of this BART6-7 assay, alongside those of standard assays used to measure Wp-initiated transcripts, Cp-initiated transcripts, and BHRF1 transcripts. As expected, Wp activity was already induced by 8 h postinfection, peaked at 12 h, and then fell as the level of Cp activity began to increase; latent BHRF1 expression was also robustly induced by 8 h (Fig. 10A). In contrast, the appearance of the BHRF1 miRNAs was significantly delayed relative to that of BHRF1 transcription, with weak signals first detected between 12 and 24 h, which then gradually increased throughout the experiment (Fig. 10B). Kinetic analysis of BART RNAs also showed a similar lag in the appearance of mature BART miRNAs relative to BART transcription (Fig. 10A and B).
DISCUSSION
EBV miRNAs have attracted a lot of interest in recent years because of their potential roles in B cell growth transformation and tumor pathogenesis. However, to date there is little information on the quantitative expression of these molecules in different types of infection. In the present work, we have validated a panel of real-time PCR assays to quantify the expression of three BHRF1 miRNAs and nine representative BART miRNAs (including all five BART miRNAs present in the B95-8 prototype EBV genome). In control experiments using synthetic miRNAs and control cell lines (Fig. 2), these assays were shown to be both specific and highly sensitive.
We initially focused on miRNA profiling of 21 EBV-positive lymphoid cell lines with well-defined forms of latent infection. While this issue has been addressed in earlier studies, data have been reported for only a very limited number of lymphoid cell lines, and these were not fully characterized in terms of virus gene expression (12, 19, 21, 48, 61). The present work shows that all three BHRF1 miRNAs are robustly expressed at comparable levels in latency III cell lines which use Cp/Wp to drive EBNA transcription. While this finding is consistent with earlier Northern blot-based studies (12, 61), our data also provide a number of novel findings. First, we demonstrate that the BHRF1 miRNAs are also abundantly expressed in a range of Wp-restricted BL lines (Fig. 4B). While the biological significance of this finding remains to be determined, we have previously reported that Wp-restricted BL lines are much more resistant to cell death stimuli than standard latency I BL lines (38). A number of recent reports (5, 24, 36) have focused on the role of BHRF1, EBNA-LP, and the EBNA3 proteins in providing a prosurvival signal in Wp-restricted BLs, but the current work now adds the BHRF1 miRNAs to the list of candidate genes which may mediate this phenotype.
Second, our data show for the first time that alterations in BHRF1 transcription do not necessarily correlate with changes in steady-state BHRF1 miRNA levels. This is best exemplified by the Wp-restricted BL lines, which on average expressed 10-fold-more-latent BHRF1 transcripts than LCLs but did not express higher levels of mature BHRF1 miRNAs (Fig. 4). These discordant results cannot be explained by differences in the expression of the miRNA processing machinery, since we found that the RNA levels of Drosha and Dicer, two enzymes involved in miRNA biogenesis (8), were relatively constant in a selection of cell lines tested (data not shown). Our data also argue that this variation is not due to differences in RNA turnover, since BHRF1 transcripts and BHRF1 miRNAs have similar half-lives in X50/7 LCL and two Wp-restricted BL lines, Sal-BL and Ava-BL (Fig. 7). We note that BHRF1 pre-miRNAs may potentially be generated either by processing of a large BHRF1-containing intron present in all primary Cp/Wp-initiated EBNA transcripts (61) or from the 5′ and 3′ UTRs present within latent BHRF1 transcripts. Our data appear to suggest that the second pathway may be less important for determining the levels of BHRF1 miRNAs in latent infection.
Our study also provides the first detailed analysis of BHRF miRNA sequence variation among different EBV-positive cell lines. Overall, we found that the miR-BHRF1-1 and miR-BHRF1-2 sequences were well conserved in 12 virus genomes tested. In contrast, several cell lines contained six changes in the miR-BHRF1-3 pre-miRNA sequence, yet remarkably these mutations did not appear to significantly affect miR-BHRF1-3 expression. These findings strongly support the view that BHRF1 miRNAs have been functionally conserved during EBV evolution. It should be noted that when sequence variation occurs in the mature miRNA sequence, we cannot readily distinguish whether the mutation affects pre-miRNA processing or PCR detection. However, our data (Fig. 4) suggest that the changes seen in miR-BHRF1-3 have little or no effect on PCR amplification.
Extending our studies to the BART region, our quantitative analysis confirms earlier reports that the BART miRNAs are readily detectable in all forms of virus latency, albeit at lower levels than in EBV-infected epithelial cell lines (12, 21, 48, 49), and also highlights the huge variation in overall BART miRNA levels between different cell lines. To some extent, this variation could be explained by differences in overall levels of BART transcription (Fig. 6). However, in sharp contrast to the coordinate expression of the BHRF1 miRNAs, different BART miRNAs were found to be expressed at dramatically different levels within a cell line, with miR-BART7, miR-BART4, and miR-BART15 frequently the most abundant and miR-BART2-5p and miR-BART13 consistently the least abundant (Fig. 6). While a similar hierarchy of miRNA expression has been noted in previous studies of B cells (49) and epithelial cells (19), it was unclear if this pattern reflected miRNA sequence polymorphisms or differences in miRNA processing and stability. We argue that these differences are unlikely to be related to sequence variation, since a number of studies have documented the high degree of sequence conservation of BART miRNAs in different EBV strains (12, 21); of the nine BART miRNAs examined here, none have previously been reported to carry polymorphisms in the mature miRNA sequence. Our data also suggest that this variation is not due to differences in the stability of individual BART miRNAs within a cell line, since all the BART miRNAs tested appeared to have half-lives in excess of 12 h, in common with cellular miRNAs (22). We conclude therefore that differences in steady-state BART miRNA levels are determined primarily by variations in the efficiency of miRNA maturation from the BART transcripts.
While this work was in progress, Pratt and coworkers (49) described a similar quantitative study of EBV miRNA expression in EBV-positive cell lines. While their conclusions are broadly similar to those of the present work, we have extended the analysis to include a larger collection of cell lines and examined a different panel of miRNAs. We also noted a number of discordant results between the two studies. Overall, we found that the absolute levels of BHRF1 and BART miRNAs were 5- to 20-fold higher than previously estimated (49), and therefore, it is more likely that they are present at physiologically significant levels. In addition, Pratt et al. claimed that miR-BHRF1-3 could be detected in a number of latency I BLs. One of these lines was Oku-BL, a tumor line established in our laboratory which we have previously shown to exhibit a Wp-restricted form of latent gene expression (35, 38). The present study clearly shows that expression of BHRF1 miRNAs in Oku-BL is associated with Wp usage (Fig. 3 and 4). Furthermore, our data support the conclusion that extremely low levels of BHRF1 miRNAs are occasionally seen in some latency I BLs but that these signals are always accompanied by similar trace levels of Wp- and Cp-initiated transcripts, suggesting that a small population of cells have drifted to latency III during passage in vitro. Our findings thus underline the importance of carefully validating the patterns of virus promoter usage before drawing conclusions on miRNA expression.
A number of previous studies have reported contradictory findings on EBV miRNA expression during virus replication (12, 49, 61). Our data clearly show that BHRF1 miRNAs are strongly induced in the lytic cycle, approaching levels seen in latency III LCLs. Our detailed kinetic studies indicate that both lytic and latent BHRF1 transcripts potentially contribute to this increase in BHRF1 miRNAs (Fig. 9); thus, the initial increases in miR-BHRF1-2 and miR-BHRF1-3 correlated with the induction of early lytic BHRF1 transcripts, while the expression of BHRF1-1 was delayed until after the induction of Cp/Wp-initiated transcripts (64) and dependent on new viral DNA replication (Fig. 10). In contrast, virus replication was associated with relatively small increases in BART miRNA levels (Fig. 8 and 9), in agreement with two earlier studies (49, 61). However, the latter finding is difficult to reconcile with the much larger increases in overall BART transcription during the lytic cycle. As the BART miRNAs are very stable in both the latent and the lytic cycles, we conclude that the BART transcripts which accumulate during replication are inefficiently processed into mature miRNAs. These observations raise the question as to the role of the BARTs in the context of virus replication. On this point, two open reading frames (RPMS1 and A73) have previously been identified within the BART transcripts, and a number of studies have proposed that these may give rise to functional proteins (53, 57). While there is no evidence to date to support the view that these protein products are naturally expressed in latently infected cells (3, 23, 25, 40), our data would suggest that such protein products might be detectable in productive infection where there are dramatically elevated levels of BART transcripts. Alternatively, the BARTs might have other roles, such as encoding a recently identified small nucleolar RNA, v-snoRNA1 (33).
In the final set of experiments, we looked at miRNA expression during the early events of B cell transformation. Interestingly, we found that EBV miRNA production was delayed with respect to the induction of BHRF1 and BART transcripts (Fig. 11). The lack of significant levels of EBV miRNAs at early time points, when EBNA2 and EBNA-LP are already abundantly expressed (1, 2), argues that the BHRF1 and BART miRNAs are not essential for the initiation of B cell growth transformation. On this point, a recent study (55) has demonstrated that the BHRF1 miRNAs protect B cells against apoptosis during the early stages of B cell infection; however, the earliest time point that was analyzed in that study was 5 days postinfection, by which time BHRF1 miRNAs are robustly detectable (Fig. 11). It should also be noted that the efficient transformation mediated by the prototype B95-8 virus clearly demonstrates that many of the BARTs are dispensable for transformation in vitro. On the question of the delayed miRNA production, it is tempting to speculate that BHRF1 miRNAs are only efficiently processed once the Wp-to-Cp switch has occurred between 24 and 48 h postinfection. Interestingly, BART transcripts can also be expressed from two alternative promoters (14), P1, which is expressed early postinfection, and P2, which is only activated after 48 h.
FIG. 11.
Expression of latent transcripts and EBV miRNAs in primary B cells infected with EBV in vitro. (A) Wp- and Cp-initiated transcripts, BHRF1 transcripts, and BARTs were quantified by QPCR at the indicated time points postinfection. Since the recombinant EBV used in these experiments carries the same BamHI A deletion as the B95-8 strain, BARTs were detected using a primer-probe combination which amplified across the exon 6-7 splice junction. (B) Expression of selected BHRF1 and BART miRNAs measured by QPCR. Data were normalized to RNU48 expression and are expressed as copy numbers per pg input RNA. Error bars indicate standard deviations for replicate assays.
In summary, our results extend the existing literature of the analysis of BHRF1 and BART miRNAs in lymphoid cell lines and during productive EBV infection. Our conclusion that changes in BHRF1 and BART transcription do not necessarily correlate with mature miRNA levels raises new questions regarding the regulation of EBV miRNA processing, which may have important implications for B cell growth transformation and virus replication.
Acknowledgments
This work was supported by funding from Cancer Research UK.
We thank Alan Rickinson for critical comments on the manuscript and Rachael Cartlidge, Deborah Croom-Carter, Emily Heath, Claire Shannon-Lowe, and Nikki Smith for providing cell lines.
Footnotes
Published ahead of print on 10 November 2010.
REFERENCES
- 1.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. [DOI] [PubMed] [Google Scholar]
- 2.Allday, M. J., D. H. Crawford, and B. E. Griffin. 1989. Epstein-Barr virus latent gene expression during the initiation of B cell immortalization. J. Gen. Virol. 70:1755-1764. [DOI] [PubMed] [Google Scholar]
- 3.Al-Mozaini, M., et al. 2009. Epstein-Barr virus BART gene expression. J. Gen. Virol. 90:307-316. [DOI] [PubMed] [Google Scholar]
- 4.Altmann, M., and W. Hammerschmidt. 2005. Epstein-Barr Virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. 3:e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderton, E., J. Yee, P. Smith, T. Crook, R. E. White, and M. J. Allday. 2008. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt's lymphoma. Oncogene 27:421-433. [DOI] [PubMed] [Google Scholar]
- 6.Arrand, J. R., and L. Rymo. 1982. Characterization of the major Epstein-Barr virus-specific RNA in Burkitt lymphoma-derived cells. J. Virol. 41:376-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Austin, P. J., E. Flemington, C. N. Yandava, J. L. Strominger, and S. H. Speck. 1988. Complex transcription of the Epstein-Barr virus BamHI fragment H rightward open reading frame 1 (BHRF1) in latently and lytically infected B lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 85:3678-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 9.Barth, S., et al. 2008. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 36:666-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell, A. I., et al. 2006. Analysis of Epstein-Barr virus latent gene expression in endemic Burkitt's lymphoma and nasopharyngeal carcinoma tumour cells by using quantitative real-time PCR assays. J. Gen. Virol. 87:2885-2890. [DOI] [PubMed] [Google Scholar]
- 11.Bushati, N., and S. M. Cohen. 2007. microRNA functions. Annu. Rev. Cell Dev. Biol. 23:175-205. [DOI] [PubMed] [Google Scholar]
- 12.Cai, X., et al. 2006. Epstein-Barr Virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, C., et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, H., et al. 2005. Regulation of expression of the Epstein-Barr virus BamHI-A rightward transcripts. J. Virol. 79:1724-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, H., P. Smith, R. F. Ambinder, and S. D. Hayward. 1999. Expression of Epstein-Barr virus BamHI-A rightward transcripts in latently infected B cells from peripheral blood. Blood 93:3026-3032. [PubMed] [Google Scholar]
- 16.Chen, H. L., et al. 1992. Transcription of BamHI-A region of the EBV genome in NPC tissues and B cells. Virology 191:193-201. [DOI] [PubMed] [Google Scholar]
- 17.Cheung, S. T., et al. 1999. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int. J. Cancer 83:121. [DOI] [PubMed] [Google Scholar]
- 18.Choy, E. Y., et al. 2008. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 205:2551-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cosmopoulos, K., et al. 2009. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J. Virol. 83:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards, R. H., A. R. Marquitz, and N. Raab-Traub. 2008. Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. J. Virol. 82:9094-9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabian, M. R., N. Sonenberg, and W. Filipowicz. 2010. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79:351-379. [DOI] [PubMed] [Google Scholar]
- 23.Fries, K. L., et al. 1997. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J. Virol. 71:2765-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garibal, J., et al. 2007. Truncated form of the Epstein-Barr virus protein EBNA-LP protects against caspase-dependent apoptosis by inhibiting protein phosphatase 2A. J. Virol. 81:7598-7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilligan, K., J. Rajadurai, and J. Lin. 1991. Expression of the Epstein-Barr virus BamHI A fragment in nasopharyngeal carcinoma: evidence for a viral protein expressed in vivo. J. Virol. 65:6252-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilligan, K., et al. 1990. Novel transcription from the Epstein-Barr virus terminal EcoRI fragment, DIJhet, in a nasopharyngeal carcinoma. J. Virol. 64:4948-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottwein, E., and B. R. Cullen. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3:375-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory, C. D., M. Rowe, and A. B. Rickinson. 1990. Different Epstein-Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt's lymphoma cell line. J. Gen. Virol. 71(Pt. 7):1481-1495. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths-Jones, S., H. K. Saini, S. van Dongen, and A. J. Enright. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36:D154-D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grundhoff, A., C. S. Sullivan, and D. Ganem. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habeshaw, G., Q. Y. Yao, A. I. Bell, D. Morton, and A. B. Rickinson. 1999. Epstein-Barr virus nuclear antigen 1 sequences in endemic and sporadic Burkitt's lymphoma reflect virus strains prevalent in different geographic areas. J. Virol. 73:965-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hummel, M., and E. Kieff. 1982. Mapping of polypeptides encoded by the Epstein-Barr virus genome in productive infection. Proc. Natl. Acad. Sci. U. S. A. 79:5698-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hutzinger, R., et al. 2009. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 5:e1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Junying, J., et al. 2003. Absence of Epstein-Barr virus DNA in the tumor cells of European hepatocellular carcinoma. Virology 306:236-243. [DOI] [PubMed] [Google Scholar]
- 35.Kelly, G., A. Bell, and A. Rickinson. 2002. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat. Med. 8:1098-1104. [DOI] [PubMed] [Google Scholar]
- 36.Kelly, G. L., et al. 2009. An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in burkitt lymphomagenesis: the Wp/BHRF1 link. PLoS Pathog. 5:e1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly, G. L., A. E. Milner, G. S. Baldwin, A. I. Bell, and A. B. Rickinson. 2006. Three restricted forms of Epstein-Barr virus latency counteracting apoptosis in c-myc-expressing Burkitt lymphoma cells. Proc. Natl. Acad. Sci. U. S. A. 103:14935-14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly, G. L., et al. 2005. Epstein-Barr virus nuclear antigen 2 (EBNA2) gene deletion is consistently linked with EBNA3A, -3B, and -3C expression in Burkitt's lymphoma cells and with increased resistance to apoptosis. J. Virol. 79:10709-10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kieff, E., and A. B. Rickinson. 2007. Epstein-Barr virus and its replication, p. 2511. In D. M. Knipe et al. (ed.), Fields virology, 5th ed. Lippincott-Raven, Philadelphia, PA.
- 40.Kienzle, N., et al. 1998. Identification of a cytotoxic T-lymphocyte response to the novel BARF0 protein of Epstein-Barr virus: a critical role for antigen expression. J. Virol. 72:6614-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim do, N., et al. 2007. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. J. Virol. 81:1033-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lerner, M. R., N. C. Andrews, G. Miller, and J. A. Steitz. 1981. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. U. S. A. 78:805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15-20. [DOI] [PubMed] [Google Scholar]
- 44.Lo, A. K., et al. 2007. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc. Natl. Acad. Sci. U. S. A. 104:16164-16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lung, R. W., et al. 2009. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia 11:1174-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nonkwelo, C., J. Skinner, A. Bell, A. Rickinson, and J. Sample. 1996. Transcription start sites downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J. Virol. 70:623-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearson, G. R., et al. 1987. Identification of an Epstein-Barr virus early gene encoding a second component of the restricted early antigen complex. Virology 160:151-161. [DOI] [PubMed] [Google Scholar]
- 48.Pfeffer, S., et al. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 49.Pratt, Z. L., M. Kuzembayeva, S. Sengupta, and B. Sugden. 2009. The microRNAs of Epstein-Barr Virus are expressed at dramatically differing levels among cell lines. Virology 386:387-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rea, D., et al. 1994. Epstein-Barr virus latent and replicative gene expression in post-transplant lymphoproliferative disorders and AIDS-related non-Hodgkin's lymphomas. French Study Group of Pathology for HIV-associated Tumors. Ann. Oncol. 5:113-116. [DOI] [PubMed] [Google Scholar]
- 51.Ressing, M. E., et al. 2005. Impaired transporter associated with antigen processing-dependent peptide transport during productive EBV infection. J. Immunol. 174:6829-6838. [DOI] [PubMed] [Google Scholar]
- 52.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe et al. (ed.), Fields virology, 5th ed., vol. II. Lippincott, Williams & Wilkins, Philadelphia, PA.
- 53.Sadler, R. H., and N. Raab-Traub. 1995. Structural analyses of the Epstein-Barr virus BamHI A transcripts. J. Virol. 69:1132-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaefer, B. C., J. L. Strominger, and S. H. Speck. 1995. Redefining the Epstein-Barr virus-encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc. Natl. Acad. Sci. U. S. A. 92:10565-10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seto E Fau-Moosmann, A., et al. 2010. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. Plos Pathog. 6:e1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shannon-Lowe, C., et al. 2005. Epstein-Barr virus-induced B-cell transformation: quantitating events from virus binding to cell outgrowth. J. Gen. Virol. 86:3009-3019. [DOI] [PubMed] [Google Scholar]
- 57.Smith, P. R., et al. 2000. Structure and coding content of CST (BART) family RNAs of Epstein-Barr virus. J. Virol. 74:3082-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swartwout, S. G., and A. J. Kinniburgh. 1989. c-myc RNA degradation in growing and differentiating cells: possible alternate pathways. Mol. Cell. Biol. 9:288-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tierney, R. J., et al. 2000. Methylation of transcription factor binding sites in the Epstein-Barr virus latent cycle promoter Wp coincides with promoter down-regulation during virus-induced B-cell transformation. J. Virol. 74:10468-10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia, T., et al. 2008. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 68:1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xing, L., and E. Kieff. 2007. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J. Virol. 81:9967-9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yandava, C. N., and S. H. Speck. 1992. Characterization of the deletion and rearrangement in the BamHI C region of the X50-7 Epstein-Barr virus genome, a mutant viral strain which exhibits constitutive BamHI W promoter activity. J. Virol. 66:5646-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young, L., et al. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 321:1080-1085. [DOI] [PubMed] [Google Scholar]
- 64.Yuan, J., E. Cahir-McFarland, B. Zhao, and E. Kieff. 2006. Virus and cell RNAs expressed during Epstein-Barr virus replication. J. Virol. 80:2548-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu, J. Y., et al. 2009. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 83:3333-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]