Abstract
The Phospho.ELM resource (http://phospho.elm.eu.org) is a relational database designed to store in vivo and in vitro phosphorylation data extracted from the scientific literature and phosphoproteomic analyses. The resource has been actively developed for more than 7 years and currently comprises 42 574 serine, threonine and tyrosine non-redundant phosphorylation sites. Several new features have been implemented, such as structural disorder/order and accessibility information and a conservation score. Additionally, the conservation of the phosphosites can now be visualized directly on the multiple sequence alignment used for the score calculation. Finally, special emphasis has been put on linking to external resources such as interaction networks and other databases.
INTRODUCTION
Over the past few years, many advances have been made in mass spectrometry techniques and protein enrichment strategies that have significantly improved the detection efficiency of phosphorylated proteins (1,2). Consequently, steadily increasing numbers of phosphorylated peptides are being reported from mouse and human cell lines as well as tissue samples (3,4).
However, the knowledge of the phosphorylated sites per se is neither sufficient to identify how signals are propagated into cells nor adequate to define the complexity of the intracellular networks. To fully appreciate the relevance of phosphoproteomic approaches it is essential to gain additional knowledge about the biological conditions under which the phosphorylation occurs, to identify the enzymes (kinases and phosphatases) that switch ‘on and off’ their substrates, and to understand the functional consequences that these modification events have on cellular processes.
Amino acid phosphorylation is probably the most abundant of the intracellular post-translational protein modifications used to regulate the state of eukaryotic cells, with estimates ranging up to 500 000 phosphorylation sites in the human proteome (5). Is this vast number plausible? It is considered that cell regulatory systems exhibit the property of robustness, but that this vital property cannot be achieved without system complexity (6). Complexity is therefore inevitable and unavoidable, yet it is probable that it has so far been systematically underestimated. However, there are now indications that we are at the dawn of a new and more realistic era in our approaches to signaling research (7). More and more authors are highlighting the importance of factors such as cooperativity, networking, redundancy and decision-making by in-complex molecular switching as we move away from overly linear pathway-based descriptions of cellular systems (8–14). In this context, the efforts to deploy large scale phosphoproteomics to map cellular networks (e.g. (15–17)) can be seen as indispensable to the process of covering more of the signaling network space.
An important aspect to consider is the evolutionary conservation of the phospho-residues. Due to their crucial role in regulating protein function, one could expect phosphorylation events to be conserved among species. However, phosphorylation motifs are short, strongly dependent on the surrounding context (18) and often reside in unstructured and rapidly-evolving regions (19), hence they have been difficult to trace evolutionarily and mixed conclusions have been reported (20,21). Lack of data has limited the possibility for this kind of analysis; only recently has phosphoproteomic data been available from different model organisms, thereby enabling comparative studies of the evolutionary and functional dynamics of reversible phosphorylation across eukaryotes (20). A significant, though not so surprising, observation is that phosphorylated residues are significantly more conserved than equivalent but non-phosphorylated ones (10,22,23).
Since the future knowledge and exploitation of reversible phosphorylation relies on the accessibility of the data, it is of fundamental importance to develop and maintain public repositories to facilitate data retrieval for both wet lab scientists and computational biologists. In this article we describe the content and the more recent features of Phospho.ELM, a manually curated web-based resource dedicated to eukaryotic phosphorylation sites.
THE Phospho.ELM RESOURCE AND ITS USAGE
The core structure of the database has been retained (24,25) and extended, while new features have been implemented to improve data retrieval and presentation. In addition to a much larger data set, information for the phosphorylated residue, i.e. a conservation score (CS) and the surface accessibility score (either calculated or predicted), have also been included in the update.
The Phospho.ELM data can be accessed by a user-friendly web interface, directly via URL, or programmatically via a XML/Soap Web Service. The user can query the database by keyword or sequence identifier [from UniProt (26) or Ensembl (27)] to get information about single proteins/substrates, or by kinase name to retrieve all phosphorylated substrates of a particular kinase. It is also possible to restrict the query to different taxonomy groups. Table 1 lists all available options for querying the database.
Table 1.
Examples of different search options for retrieving data stored in the Phospho.ELM resource
| Phospho.ELM data retrieval methods | |
|---|---|
| Input Query | Result |
| Access via WEB interface: http://phospho.elm.eu.org/ | |
| Protein name (keyword) | Retrieval of phosphorylation sites identified in sequences of the same substrate for multiple species |
| UniProt or Ensembl ACC | Retrieval of phosphorylation sites of a specific sequence |
| Kinase name | Retrieval of phosphorylation sites recognized by the specified kinase |
| Access via url: input prefix: http://phospho.elm.eu.org/ | |
| byAccession/src_human.html | Retrieval of the human ‘src’ (UniProt ACC P12931) |
| byAccession/P12931.html | |
| byAccession/P12931,P07948.html | Retrieval of all the phosphorylation sites of two proteins (UniProt ACC P12931 and P07948) |
| byAccession/P12931,P07948.csv | Retrieval of a plain output of the phosphorylation sites of two proteins (UniProt ACC P12931 and P07948) |
| byDomain/CBL_SH2.html | Retrieval of phosphorylation sites which bind to the phospho-binding domain CBL_SH2 |
| P12931.fasta | Retrieval of the protein sequence stored in the Phospho.ELM database |
| Access via PhosphoBlast: http://phospho.elm.eu.org/pELMBlastSearch.html | |
| Uniprot AC or text sequence | Retrieval of phosphorylation sites that are conserved in related proteins (whether orthologues or paralogues) |
| Access via Web service: http://phospho.elm.eu.org/webservice/phosphoELMdb.wsdl | |
| pELMdbws = phosphoELMdbLocator().getphosphoELMdb() | For retrieving phosphorylation sites recognized by the selected kinase (e.g. ALK) |
| kinaseName = ‘ALK' | |
| req = getInstancesByKinaseTextSearchRequestMsg() | |
| req._QueryText = kinaseName | |
| result = pELMdbws.getInstancesByKinaseTextSearch(req) | |
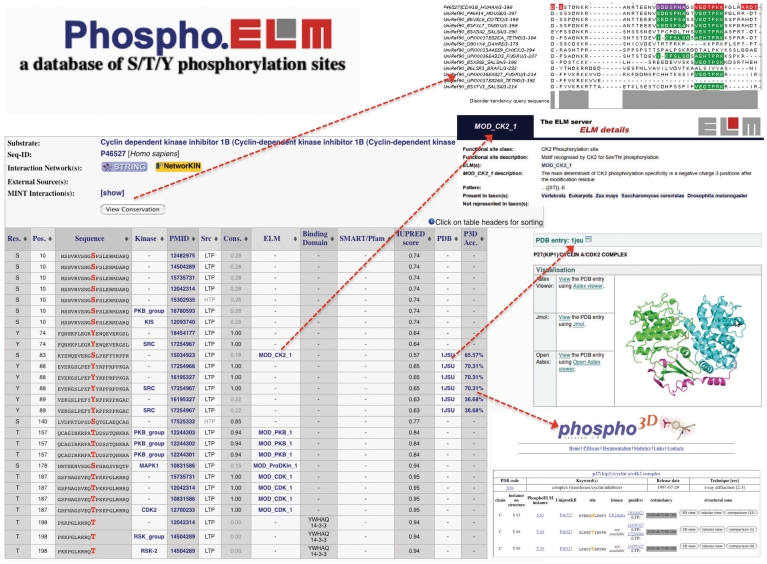
The results page displays all phosphoproteins and phosphorylation sites (instances) meeting the searching criteria; the results tables can be sorted on any column, aiding inspection of the data according to different criteria, such as: the residue number and code, the PubMed references, the sequence (±10) surrounding the phospho-residue and, when available, the upstream kinases as well as the phosphopeptide-binding domains, such as the SH2, 14-3-3 or PTB domains (these data have been annotated for 1250 phosphosites). Furthermore, links to matching kinase recognition motif entries stored in the ELM database (28) is provided when available. An example Phospho.ELM output is shown in Figure 1.
Figure 1.
Output example of a Phospho.ELM search using the Cyclin dependent kinase inhibitor 1B (UniProt P46527) as query. The results table contains: the phosphorylated residue and its position; surrounding sequence; kinase responsible for the phosphorylation; literature reference; type of source (HTP/LTP); conservation score; link to ELM database; annotation of domain which binds to the phosphorylated residue; protein domain identified by SMART or Pfam; a disorder score calculated by IUPRED; link to PDB structure; and accessibility score calculated by Phospho3D. The conservation of the instance and the multiple sequence alignment that was used to calculate the CS can be inspected using the JALVIEW plugin (top right). Furthermore links to Phospho3D and the respective ELM entry are shown at the bottom right.
The kinases are currently known for only ∼12% of the curated instances. It should be mentioned that this percentage has decreased since the last Phospho.ELM publication (25); this data reflects the current limitation of both experimental and computational methods in assigning the kinase recognizing a given phosphorylation site. Since this information is relevant for gaining insights into the regulation of cellular processes, we provide direct links to NetworKIN (29), a database of predicted kinase–substrate relations.
In addition, we encourage our users to explore the links to other resources that integrate information on signaling networks or protein–protein interactions, such as MINT (30) and STRING (31), to expand their knowledge of the phosphoprotein substrates.
The entire Phospho.ELM data set can be freely downloaded in a tab-delimited format at: http://phospho.elm.eu.org/dataset.html
DATA SET
The current release of the Phospho.ELM data set (version 9.0) contains more than 42 500 non-redundant instances of phosphorylated residues in more than 11 000 different protein sequences (3370 tyrosine, 31 754 serine and 7449 threonine residues).
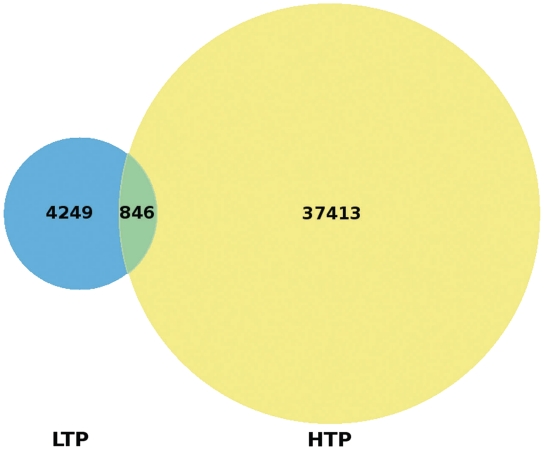
For each phosphosite we report whether the phosphorylation evidence has been identified by small-scale analyses (low-throughput, LTP) and/or by large-scale experiments (high-throughput, HTP), which mainly apply MS techniques. The Venn diagram in Figure 2 shows the remarkably small overlap between the LTP and HTP phospho-instances.
Figure 2.
Venn diagram comparing the sources of Phospho.ELM instances. A total of 4249 instances have been obtained exclusively by LTP experiments and 37 413 instances solely by HTP assays while 846 instances were confirmed by both HTP and LTP analyses.
The majority of the protein instances from Phospho. ELM are vertebrate (mostly Homo sapiens (62%) and Mus musculus (16%)) though 22% are from other species, mainly Drosophila melanogaster (13%) and Caenorhabditis elegans (7%).
In total, more than 300 different kinases have been annotated and a document providing additional information about all kinases annotated in Phospho.ELM can be found at http://phospho.elm.eu.org/kinases.html.
CONSERVATION SCORE
In order to improve the biological understanding of a particular site and thereby indirectly providing the users with additional evidence, we have added information about sequence conservation. This will help researchers to better assess the reliability of the identified sites, especially for those derived from proteomic analyses. For each instance, we have calculated the conservation score (CS) as described in Chica et al. (32).
The conservation of the phosphorylation sites in the database has been calculated using a tree-based approach specifically developed for assessing the conservation of short linear protein motifs (32), also accessible as individual service at http://conscore.embl.de. The method takes into account the presence/absence of the phosphorylated serine, threonine or tyrosine in relation to the global sequence conservation and gives a value between 0 and 1, where 1 indicates conservation in all the homologous sequences at a certain distance from the query sequence, and 0 corresponds to absence of conservation.
The CS of an instance is an estimation of the persistence of a phosphosite during the divergent evolution of a homologous protein sequence set. In a protein-centered view a high CS, together with other contextual information such as high residue accessibility, represents cumulative evidence for the biological relevance of such a site. This is particularly useful when analyzing HTP sites that might be phosphorylated in vitro but not in vivo. The distribution of the CS of manually annotated instances is indeed significantly (P-value <2.2e-16) skewed towards 1, in comparison to that of the instances coming from HTP experiments (Figure 3).
Figure 3.
Distribution of the conservation scores for LTP and HTP instances in the Phospho.ELM database. The CS varies between 0 and 1, where 1 represents the highest conservation. The two distributions differ according to the Kolmogorov-Smirnov test with P-value <2.2e-16, with the LTP sites being more conserved.
In a protein interaction network context, the CS can be used to suggest evolutionarily stable protein interactions as well as taxa-specific interactions that might have been gained during evolution as regulatory circuits are changed and modulated.
Alignments between the phosphoprotein and the corresponding homologous sequences are available for close inspection of the conservation of the phosphosites of interest in different species (Figure 1, button ‘view conservation’). To this end, the alignment editor Jalview (33) (http://www.jalview.org) has been embedded as a JAVA plugin in the HTML output. Here, known instances are highlighted in different colors according to the phospho-residues (light green for phosphotyrosine, purple for phosphoserine and red for phosphothreonine), while the conservation of the corresponding peptides in the aligned sequences are displayed as dark green columns.
We urge users to look at these alignments, particularly if the CS is low, since there are several factors unrelated to the evolution of the protein sequence, e.g. sequencing errors in not well studied genomes, which could artificially diminish the score even if the site itself is quite conserved across different species.
STRUCTURAL INFORMATION
Phosphorylation sites are often found in intrinsically disordered regions of proteins (34), which usually cannot be experimentally determined by X-ray crystallography. However, in a number of cases, they lie on globular domains whose sequence can confidently be mapped onto X-ray determined structures.
For the latter sites, accessibility to the solvent can be calculated. Currently we have been able to assign an accessibility value to 3% of all the sites in Phospho.ELM (1281 of 42 574 instances) and we anticipate that this number will increase in parallel with the increase in solved structures. These data are particularly relevant for bioinformaticians who develop computational methods to predict kinase substrates. Because of the transient nature of phosphorylation events, phosphorylation sites tend to lie on the surface of proteins. Many studies [see Via et al. (35) for a summary] have shown that the substrate specificity is not only dependent on the primary sequence of the motif hosting the phosphorylation site, but also on its structural conformation.
The correspondence between a Phospho.ELM sequence and an X-ray Protein Data Bank (PDB) structure (36) is based on sequence alignment using at least 98% global sequence identity and 100% identity at the phosphorylation site. When more than one PDB structure corresponded to a single Phospho.ELM sequence, one with the lowest resolution was retained. Whenever a site can be mapped onto a PDB structure, its solvent accessibility (SA in Å2) is taken from DSSP (37) and the corresponding percentage is obtained by normalizing the SA to the phospho-residue accessibility maximum value [as determined in (38)].
Furthermore, we have also integrated protein surface accessibility data and links to structural data (when available) obtained from the Phospho3D database (39). For details on the structure, one can follow the link to PDBe (40) (http://www.ebi.ac.uk/pdbe).
The accessibility data, however, should be interpreted in the context of the structure. For example, a low accessibility value (of ∼18%) is reported in the Phospho.ELM entry for the human Src (UniProt P12931) tyrosine 530, which is a well known substrate of the CSK kinase. This is due to the fact that the structure used to calculate the accessibility has been determined in a closed Src conformation, where the phosphorylated tail binds to the SH2 domain. In general, when evaluating the SA of an instance, the user is advised to be aware of the instance molecular context. Note that in most cases, the best resolution structures are not in the phosphorylated conformation (i.e. with the phosphate moiety attached) or, as in the Src example, they are in a phosphorylated but closed, inactive conformation. In particular, if a site becomes available to its cognate kinase only as a consequence of a conformational change, this might be reflected in a (transiently) low accessibility value.
In the great majority of the cases (∼97%), either an X-ray reference structure is not available for a Phospho.ELM sequence or a structure can be found but the phosphorylation site falls in an unresolved (disordered) region of the structure. In these cases, we provide the users with predicted accessibility values. The SA predictions were carried out using the real-SPINE integrated system of neural networks (41).
The accessibility score is shown as the last column in the HTML output (Figure 1) and, when provided, it is linked to the Phospho3D resource (http://arianna.bio.uniroma1.it/phospho3d). The user is encouraged to investigate this link to gain more insight into the structural features of the particular protein including a 3D representation of the instance as well as a comparison of all PDB entries available for that instance. The PDB entry that was used to calculate the score, is listed in the second last column of the HTML output and is linked to the PDBe resource at the EBI where different viewers are available for closer inspection of the site.
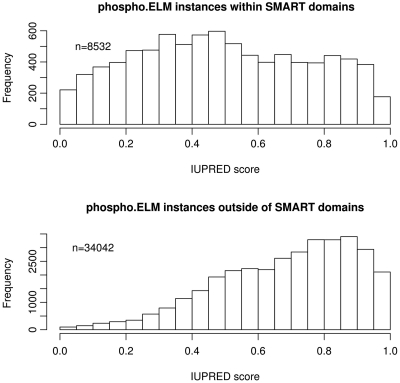
In addition to providing more evidence about the structural properties of the region surrounding the phosphosites, we determine if the sites reside within domains annotated in the SMART resource (42). Furthermore, for each phosphosite, a probabilistic score was calculated ranging from 0 (complete order) to 1 (complete disorder) using the IUPRED intrinsic order–disorder predictor (43). The IUPRED algorithm uses the parameter ‘long’ and a window of 21 residues for smoothing the score. In the HTML output table, IUPRED scores below 0.5 (predicted ordered) are colored in grey while IUPRED scores above 0.5 (predicted disordered) are colored in black. Figure 4 shows the distributions of IUPRED scores for instances that reside either within (upper panel) or outside (lower panel) known SMART domains. Outside the known domains, the sites are strongly skewed to the native disorder values, reaffirming the earlier analyses (34). These curves may help in understanding the nature of cell regulation as they imply that most protein phosphorylation explicitly modulates protein–protein interactions in dynamic regulatory systems, rather than through allosteric regulation of the shape of the modified protein, although this is clearly an important function of the less abundant in-domain sites.
Figure 4.
Histograms of IUPRED Score of Phospho.ELM instances within and outside of known domains. Instances with an IUPRED score above 0.5 are predicted to be in a region of polypeptide sequence that is intrinsically disordered (i.e. cannot fold into a stable native structure). Instances that reside outside globular domains have a tendency towards higher IUPRED scores (disordered, lower panel) whereas the scores of instances within domains are more evenly distributed (upper panel). Note that sites mapping outside the known domains are predicted to be predominantly in natively disordered polypeptides.
CONCLUSION
A few years ago it was estimated that more than 100 000 phosphorylation sites might exist in the human proteome (44). Recently this number has been corrected upwards to more than 500 000 sites (5). This newer value implies an average of approximately 25 sites ‘per protein’ yet it seems quite plausible, given the low LTP/HTP overlap in Figure 2.
Other bioinformatics resources also incorporate substantial phosphorylation data: more general ones are UniProt (26), HPRD (45), and PhosphoSitePlus (46). The latter provides mainly phosphorylation sites from Vertebrata, but also includes data from other PTMs such as acetylation, methylation, ubiquitination, and O-glycosylation. Both PHOSIDA (47) and phosphoPEP (48) are specialized in annotation of large-scale experiments; phosphoGRID for Saccharomyces cerevisiae (49), virPTM for viruses (50) and P3DB for various plants (51) are devoted to specific species. Additional information on phosphorylation resources can be found at the Phospho.ELM link page (http://phospho.embl.de/links.html) and at the GPS compendium of computational resources for protein phosphorylation (52) (http://gps.biocuckoo.org/links.php). In addition, a collection of phosphorylation databases and predictors has been recently published in a review by Via et al. (35). Though we intend to incorporate the most relevant large-scale analyses, we consider our main effort should be on the collection of manually curated phosphorylation sites and related information derived from small-scale experiments on various model organisms. In the near future, we plan to integrate more information on phosphorylation motifs and protein kinase specificity, such as the kinase docking motifs (53). In order to provide an up-to-date and comprehensive resource, we encourage our users to participate in the curation of the Phospho.ELM resource by submitting their own data (http://phospho.elm.eu.org/submit.html).
FUNDING
Novo Nordisk Foundation Center for Protein Research. Funding for open access charge: EMBL, Heidelberg, Germany.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to acknowledge all the Phospho.ELM users who, by reporting missing sites or sending us their data sets, have contributed to improving the database. Many thanks to Leonardo Briganti (MINT database) and Florian Gnad (PHOSIDA database) for technical support, and to Alessandro Barbato for running SA predictions. We are grateful to Norman Davey and Kim Van Roey for critical reading of the article.
REFERENCES
- 1.Cantin GT, Yi W, Lu B, Park SK, Xu T, Lee JD, Yates JR. Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J. Proteome Res. 2008;7:1346–1351. doi: 10.1021/pr0705441. [DOI] [PubMed] [Google Scholar]
- 2.Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 3.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics. 2004;3:1093–1101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Lemeer S, Heck AJ. The phosphoproteomics data explosion. Curr. Opin. Chem. Biol. 2009;13:414–420. doi: 10.1016/j.cbpa.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 6.Kitano H. A robustness-based approach to systems-oriented drug design. Nat. Rev. Drug Discov. 2007;6:202–210. doi: 10.1038/nrd2195. [DOI] [PubMed] [Google Scholar]
- 7.Feller SM. The dawn of a new era in cell signalling research. Cell Commun. Signal. 2010;8:7. doi: 10.1186/1478-811X-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitty A. Cooperativity and biological complexity. Nat. Chem. Biol. 2008;4:435–439. doi: 10.1038/nchembio0808-435. [DOI] [PubMed] [Google Scholar]
- 9.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan CS, Bodenmiller B, Pasculescu A, Jovanovic M, Hengartner MO, Jorgensen C, Bader GD, Aebersold R, Pawson T, Linding R. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci. Signal. 2009;2:ra39. doi: 10.1126/scisignal.2000316. [DOI] [PubMed] [Google Scholar]
- 11.Gibson TJ. Cell regulation: determined to signal discrete cooperation. Trends Biochem. Sci. 2009;34:471–482. doi: 10.1016/j.tibs.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Mayer BJ, Blinov ML, Loew LM. Molecular machines or pleiomorphic ensembles: signaling complexes revisited. J. Biol. 2009;8:81. doi: 10.1186/jbiol185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jorgensen C, Linding R. Simplistic pathways or complex networks? Curr. Opin. Genet. Dev. 2010;20:15–22. doi: 10.1016/j.gde.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preisinger C, von Kriegsheim A, Matallanas D, Kolch W. Proteomics and phosphoproteomics for the mapping of cellular signalling networks. Proteomics. 2008;8:4402–4415. doi: 10.1002/pmic.200800136. [DOI] [PubMed] [Google Scholar]
- 16.de la Fuente van Bentem S, Mentzen WI, de la Fuente A, Hirt H. Towards functional phosphoproteomics by mapping differential phosphorylation events in signaling networks. Proteomics. 2008;8:4453–4465. doi: 10.1002/pmic.200800175. [DOI] [PubMed] [Google Scholar]
- 17.Morandell S, Stasyk T, Skvortsov S, Ascher S, Huber LA. Quantitative proteomics and phosphoproteomics reveal novel insights into complexity and dynamics of the EGFR signaling network. Proteomics. 2008;8:4383–4401. doi: 10.1002/pmic.200800204. [DOI] [PubMed] [Google Scholar]
- 18.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK. Evolutionary rate heterogeneity in proteins with long disordered regions. J. Mol. Evol. 2002;55:104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- 20.Boekhorst J, van Breukelen B, Heck AJ, Snel B. Comparative phosphoproteomics reveals evolutionary and functional conservation of phosphorylation across eukaryotes. Genome Biol. 2008;9:R144. doi: 10.1186/gb-2008-9-10-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lienhard GE. Non-functional phosphorylations? Trends Biochem. Sci. 2008;33:351–352. doi: 10.1016/j.tibs.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Tan CS, Pasculescu A, Lim WA, Pawson T, Bader GD, Linding R. Positive selection of tyrosine loss in metazoan evolution. Science. 2009;325:1686–1688. doi: 10.1126/science.1174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diella F, Cameron S, Gemund C, Linding R, Via A, Kuster B, Sicheritz-Ponten T, Blom N, Gibson TJ. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: a database of phosphorylation sites–update 2008. Nucleic Acids Res. 2008;36:D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl’s 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould CM, Diella F, Via A, Puntervoll P, Gemund C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linding R, Jensen LJ, Pasculescu A, Olhovsky M, Colwill K, Bork P, Yaffe MB, Pawson T. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 2008;36:D695–D699. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chica C, Labarga A, Gould CM, Lopez R, Gibson TJ. A tree-based conservation scoring method for short linear motifs in multiple alignments of protein sequences. BMC Bioinformatics. 2008;9:229. doi: 10.1186/1471-2105-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Via A, Diella F, Helmer-Chitterich M, Gibson TJ. From sequence to structural analysis in protein phosphorylation motifs. Frontiers in Bioscience. 2011;16:1261–1275. doi: 10.2741/3787. [DOI] [PubMed] [Google Scholar]
- 36.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 38.Miller S, Janin J, Lesk AM, Chothia C. Interior and surface of monomeric proteins. J. Mol. Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 39.Zanzoni A, Ausiello G, Via A, Gherardini PF, Helmer-Citterich M. Phospho3D: a database of three-dimensional structures of protein phosphorylation sites. Nucleic Acids Res. 2007;35:D229–D231. doi: 10.1093/nar/gkl922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Velankar S, Best C, Beuth B, Boutselakis CH, Cobley N, Sousa Da Silva AW, Dimitropoulos D, Golovin A, Hirshberg M, John M, et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2010;38:D308–D317. doi: 10.1093/nar/gkp916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dor O, Zhou Y. Real-SPINE: an integrated system of neural networks for real-value prediction of protein structural properties. Proteins. 2007;68:76–81. doi: 10.1002/prot.21408. [DOI] [PubMed] [Google Scholar]
- 42.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakiewicz RD, Comb MJ. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J. Biol. Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 45.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 47.Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R. PhosphoPep–a database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 2008;26:1339–1340. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stark C, Su TC, Breitkreutz A, Lourenco P, Dahabieh M, Breitkreutz BJ, Tyers M, Sadowski I. PhosphoGRID: a database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database. 2010 doi: 10.1093/database/bap026. 2010, doi:10.1093/database/bap026; [28 January 2010, Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz D, Church GM. Collection and motif-based prediction of phosphorylation sites in human viruses. Sci. Signal. 2010;3 doi: 10.1126/scisignal.2001099. rs2. [DOI] [PubMed] [Google Scholar]
- 51.Gao J, Agrawal GK, Thelen JJ, Xu D. P3DB: a plant protein phosphorylation database. Nucleic Acids Res. 2009;37:D960–D962. doi: 10.1093/nar/gkn733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue Y, Gao X, Cao J, Liu Z, Jin C, Wen L, Yao X, Ren J. A summary of computational resources for protein phosphorylation. Curr. Protein Pept. Sci. 2010;11:485–496. doi: 10.2174/138920310791824138. [DOI] [PubMed] [Google Scholar]
- 53.Remenyi A, Good MC, Lim WA. Docking interactions in protein kinase and phosphatase networks. Curr. Opin. Struct. Biol. 2006;16:676–685. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]