Abstract
Cooperative binding of transcription factors (TFs) to promoters and other regulatory regions is essential for precise gene expression. The classical model of cooperativity requires direct interactions between TFs, thus constraining the arrangement of TF sites in regulatory regions. Recent genomic and functional studies, however, demonstrate a great deal of flexibility in such arrangements with variable distances, numbers of sites, and identities of TF sites located in cis-regulatory regions. Such flexibility is inconsistent with cooperativity by direct interactions between TFs. Here, we demonstrate that strong cooperativity among noninteracting TFs can be achieved by their competition with nucleosomes. We find that the mechanism of nucleosome-mediated cooperativity is analogous to cooperativity in another multimolecular complex: hemoglobin. This surprising analogy provides deep insights, with parallels between the heterotropic regulation of hemoglobin (e.g., the Bohr effect) and the roles of nucleosome-positioning sequences and chromatin modifications in gene expression. Nucleosome-mediated cooperativity is consistent with several experimental studies, is equally applicable to repressors and activators, allows substantial flexibility in and modularity of regulatory regions, and provides a rationale for a broad range of genomic and evolutionary observations. Striking parallels between cooperativity in hemoglobin and in transcriptional regulation point to a general mechanism that can be used in various biological systems.
Keywords: protein–DNA interactions, promoter, enhancer, histone, Monod–Wyman–Changeux
In higher eukaryotes, cis-regulatory regions are 200 to 3,000 base pairs (bps) in length and may contain clusters of 3 to 50 TF binding sites (TFBSs) (1–4). The arrangement, identity, and affinity of the sites determine the function of the regulatory region. Cooperative binding of TFs to regulatory regions leads to highly cooperative gene activation and is essential for development (5) and other vital processes (6).
Cooperative binding is traditionally explained by protein–protein interactions among TFs (7, 8). While this mechanism finds support in bacterial and some eukaryotic systems (9), several functional and genomic observations are inconsistent with it. Cooperativity by protein–protein interactions (directly or via DNA looping) (7, 8, 10) can significantly constrain arrangements of TFBSs, allowing only those that provide the correct orientation, order, and distance between TFs. On the contrary, recent evolutionary analysis of Drosophila enhancers revealed massive turnover and rearrangements of TFBSs (11, 12). Furthermore, functional studies demonstrated that cis-regulatory regions could tolerate incorporation of new binding sites (promiscuity) and significant alterations in TFBS placement while retaining in vivo functionality (11, 13, 14). The few mechanisms that have been proposed to explain flexible arrangements of TFBSs and promiscuity are based on the idea of transcriptional synergy (i.e., cooperative recognition or simultaneous contact between TFs and some part of the transcription machinery) (13, 14), rather than cooperative binding of TFs to DNA.
An alternative mechanism of cooperativity considered here is based on synergistic binding of noninteracting TFs mediated by a nucleosome. The phenomenon of nucleosome-mediated cooperativity has been documented by a series of in vivo and in vitro experiments (6, 15–17), which demonstrated synergistic binding and gene activation by nonendogenous TFs (e.g., Gal4 and LexA) that occupied sites on nucleosomal DNA (Fig. 1). Such cooperativity requires only the DNA-binding domains of TFs, suggesting that it does not involve chromatin modification or direct protein–protein interactions (18). Experimental studies (16) and an earlier model (19) of synergistic binding to nucleosomal DNA considered only two close-by (∼30 bps) sites (Fig. 1B) that interact through an assisted unwrapping mechanism: binding of the first TF leads to partial unwrapping of nucleosomal DNA, thus making the site of the second TF more accessible (19).
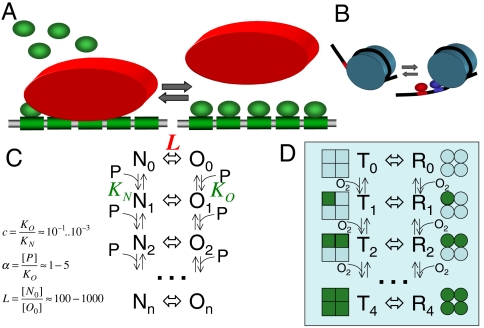
Fig. 1.
The model of nucleosome-mediated cooperativity. (A) DNA region containing an array of n sites (green boxes) that can be bound by a histone core (red oval), thus becoming nucleosomal DNA, or remain naked. In either the nucleosomal (N) or the open (O) state, the DNA can be bound by transcription factors (TFs, green ovals). Binding of TFs to the nucleosomal DNA is diminished as compared to naked DNA but is possible due to transient, partial unwinding of the DNA (shown in B) (36). The equilibrium of the system is fully characterized by the scheme in C. The states of the system: nucleosomal (Ni) and open (Oi), with i being the number of TFs bound. In this form, the nucleosome-TF system is identical to the Monod–Wyman–Changeux (MWC) model of cooperativity in hemoglobin (D). The N and O forms of the DNA correspond to the T and R states of hemoglobin; TF binding is equivalent to O2 binding (see Table S1). Like the MWC model, the nucleosome-TF system is determined by three dimensionless parameters: L, c and α (see text). (B) The model of Polach and Widom (19): synergistic binding by two TFs to nearby sites through partial unwinding of nucleosomal DNA. The mechanism requires the constant presence of a nucleosome and does not lead to nucleosome eviction. In our model, binding of multiple TFs can evict a nucleosome completely thus allowing more distant sites to interact, more sites to be involved and hence a higher Hill coefficient.
Here we introduce and analyze a different mechanism of nucleosome-mediated cooperativity. We show that competition between histones and TFs for a region of DNA that bears an array of TFBSs induces strongly cooperative binding of TFs and cooperative nucleosome eviction. We find this mechanism of cooperativity is identical to the Monod–Wyman–Changeux (MWC) model of cooperativity in hemoglobin (20). Using this analogy, we gain deeper insights into a range of phenomena such as the role of nucleosome-positioning sequences and histone modifications, low-affinity TFBS, and TFBS clustering (see Table S1). Finally, we review experimental evidences in support of our nucleosome-mediated mechanism (Table S2).
Presented mechanism of nucleosome-mediated cooperativity is sufficiently general, provides a high Hill coefficient (21), and leads to passive nucleosome eviction. These aspects distinguish it from assisted unwrapping (19), which requires that a nucleosome remains in place but cannot achieve a high Hill coefficient due to gradual unwrapping of nucleosomal DNA (see SI Text). Our framework, in essence, integrates DNA unwrapping as a mechanism of TF access to nucleosomal DNA, with the possibility of nucleosome eviction. The high energetic cost of nucleosome eviction and greater length of DNA that becomes accessible to TF binding upon eviction provide high cooperativity for TF binding. Note that our model does not consider active (ATP-dependent) nucleosome modification by recruited TFs. This important mechanism of chromatin remodeling may follow the initial cooperative binding of TFs and passive nucleosome eviction. Similarly, we do not consider the effect of nucleosome eviction on the positioning of neighboring nucleosomes (22, 23). A recent study (24) modeled the effect of two TFs binding their sites in nucleosomal DNA, demonstrating cooperative binding, which, however, do not exhibit a high Hill coefficient (SI Text, Table S3, and Fig. S3).
Proposed mechanism requires several TFBSs located within nucleosomal DNA (see refs. 7, 25, and 26 for examples). As such, it is more applicable to gene regulation in multicellular eukaryotes where the sites are shorter and more sites are clustered together to form a regulatory region than to yeast where one to two sites can be sufficient (27).
In general, the input of a regulatory module is an external stimulus (e.g., a ligand or activation of an upstream kinase) and the output is gene expression. Here, the input is a concentration of TFs activated by such stimuli and poised to bind their sites. The output is TF occupancy of a regulatory region, rather than gene expression, because expression can be a complex function of the occupancy. Our model is equally applicable to repressors and activators, providing cooperative binding irrespective of the effect of bound TFs on gene expression.
Results
The Model of Nucleosome-Mediated Cooperativity.
We consider interactions of TFs with a stable nucleosome, containing an array of n TFBSs within its DNA footprint (147 bps) (Fig. 1). This region of DNA can be in one of two states: the nucleosome (N) state and the open (O) state, in which histones are absent from the region. While histones limit access of other proteins to nucleosomal DNA, the nucleosome is highly dynamic, with DNA unwrapping and wrapping at a high rate, thus making nucleosomal DNA at least partially accessible to TFs (28, 29). TFBSs can be occupied by TFs in either the N or O state leading to 2n states labeled Ni and Oi (i = 0,1,…n), where i is the number of occupied sites. The equilibrium between the N and O states in the absence of bound TFs is characterized by the constant L = [N0]/[O0], where L≫1 for a stable nucleosome. The affinity of TFs for the sites depends on the state, with binding constants KN and KO (KO ≪ KN due to higher affinity in the open state). Suppression of TF binding to nucleosomal DNA is reflected by the free energy cost of DNA unwrapping required to accommodate one more TF (30) and is taken into account by parameter c ≡ KO/KN ≪ 1.
For simplicity of presentation, we assume all TFBSs to have the same affinity and experience the same suppression by the nucleosome. More complicated models that drop these assumptions and consider partial unwrapping are considered in SI Text but yield similar results.
TFs can be activated by different mechanisms: increased intranuclear concentration of a TF (e.g., by facilitating its nuclear localization or inhibiting degradation) or elevated TF affinity for its cognate sites (e.g., through a conformational change upon a modification or binding to a ligand). Both factors are taken into account by a dimensionless parameter α = [P]/KO, where [P] is the intranuclear concentration of the activated TF. In equilibrium, the system is fully determined by three dimensionless parameters: c, L, and α (Fig. 1). We study equilibrium properties, assuming that rapid exchange of TFs and histones leads to fast equilibration.
Cooperative Binding and Nucleosomal Occupancy.
The two quantities of primary biological interest are TF occupancy per site (Y) and the occupancy by the nucleosome (YN). Using statistical mechanics, and analogy to the MWC model, we obtain expressions for these quantities (see SI Text):
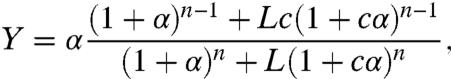 |
[1] |
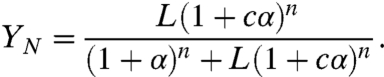 |
[2] |
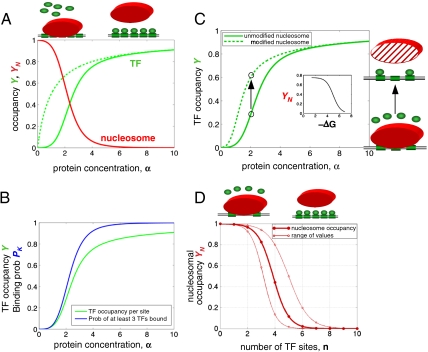
Fig. 2A presents TF and nucleosome occupancies as a function of TF concentration, computed using parameters inferred from experiments with α = 0–10, L ≈ 103, c ≈ 10-2 (see Materials and Methods). Strikingly, nucleosome-mediated cooperativity results in a sharp transition with a twofold increase in TF concentration leading to a more than eightfold increase in the occupancy. The nucleosome occupancy also changes cooperatively, dropping from about 65% to less than 10% due to a twofold change in TF concentration.
Fig. 2.
Nucleosome-mediated cooperativity and its implications. (A) Cooperative transition in the equilibrium TFBS occupancy, Y (solid green line), and nucleosome occupancy, YN (red line), as a function of TF concentration α (Eqs. 1 and 2, n = 6, L = 103, c = 10-3). Notice that nucleosome-mediated cooperativity leads to suppression of TF binding at a low concentration, as compared to noncooperative binding (dashed green line). (B) Probability of having at least 3 TFs bound P3 as a function of TF concentration (blue). Mean occupancy per site, Y, is shown for comparison (green). (C) The Bohr effect: attenuation of histone affinity for DNA, due to modifications or as a function of DNA sequence (modified—dashed line, unmodified—solid line), leads to a shift in TF-nucleosome competition and displacement of the nucleosome by TFs (arrow). This competition renders nucleosomal occupancy, YN, considerably responsive to small changes in nucleosome affinity (Inset), as demonstrated by the dependence of YN on -ΔG = kBT log(L/L′) (kcal/mol). (D) The effect of number of TF sites, n, on nucleosome stability, obtained for three concentrations of TF: α = 3, 5, 8 (lines from top to bottom). There is a critical number of sites (∼4–5) below which TFs are unable to displace a nucleosome and above which the nucleosome is unstable even at a lower concentration of TFs.
Complex cis-regulatory elements of higher eukaryotes may require several activating TFs (or several copies of the same TF) to be bound for initiation of gene expression (4, 31). To take this into account, we calculate another quantity, the probability of having at least k TF bound, Pk, as a measure of occupancy, which also shows a significant cooperativity (Fig. 2B and SI Text).
Importantly, as in the case of hemoglobin, the cooperativity stems from suppression of TF binding at low TF concentration (Fig. 2A, dotted vs. solid green lines). This behavior is distinct from cooperative binding enhanced by attractive protein–protein interactions. Cooperative suppression of TF binding by nucleosomes, however, could be advantageous in multicellular eukaryotes that contain an overwhelming number of spurious binding sites in their genomes (12, 27).
Surprisingly, our model suggests that nucleosome destabilization should have opposite effects on cooperativity and on TF binding. Factors that destabilize nucleosomes, [e.g., histone modification, poly(dA:dT), etc.] facilitate TF binding but make this binding less cooperative. Similarly, factors that stabilize nucleosomes suppress binding at a low TF concentration, making binding more cooperative. In Discussion, we review recent genomics findings that reveal signatures of nucleosome stabilization in human regulatory regions (26).
Analogy to Hemoglobin.
Strikingly, the system of TFs and a nucleosome is identical to the scheme of cooperativity in hemoglobin described by the classical MWC model (20) (see Fig. 1 C and D). Table S1 summarizes equivalent parameters and analogous phenomena between the two systems. The MWC model considers the equilibrium between two states of hemoglobin: the R state, which has a higher affinity for O2, and the low-affinity T state. In the absence of the O2, hemoglobin is mostly in the T state. Binding of O2 shifts the equilibrium toward the R state, making binding of successive oxygen molecules more likely and thus cooperative (Fig. 1D). The R and T states of hemoglobin correspond to the O and N states of the nucleosomal DNA, and the binding of O2 to hemoglobin domains corresponds to the binding of TFs to individual sites (Fig. 1 C and D and Table S1).
The analogy to the MWC model allows us to reveal features of the nucleosome–TF system that are essential for cooperativity (Table S1). The MWC system has a strong cooperativity as long as L is sufficiently large (L > 100) and c is sufficiently small (c < 0.1), i.e., the nucleosome is stable (in the absence of TFs) and significantly attenuates TF binding. These requirements are consistent with estimated parameters (see Materials and Methods), as well as with high stability (32, 33) and slow exchange (34, 35) of nucleosomes in regions depleted in TFBSs. In vitro studies of TF binding to nucleosomal DNA demonstrate the required attenuation of TF binding (36).
The Bohr Effect and Chromatin Modification.
We also use the analogy to hemoglobin to examine implications of sequence-specific nucleosome positioning, histone modifications, and other processes involved in gene regulation. These effects can be considered as allosteric heterotropic regulation of the nucleosome–TF system, analogous to heterotropic effectors of hemoglobin. A prototypical heterotropic allosteric regulation of hemoglobin is the Bohr effect: Lowering the pH decreases the affinity to oxygen, thus providing more oxygen to actively working muscles. The basis of the Bohr effect is the higher affinity of hydrogen ions to the T state. Thus, low pH stabilizes the T state, shifting the equilibrium away from the high-affinity R state. Other allosteric effectors of hemoglobin (e.g., DPG) act in a similar way: Binding hemoglobin in one state affects the R-T equilibrium and thus changes the affinity of hemoglobin to oxygen.
While heterotropic effectors of hemoglobin affect equilibrium between R and T states, effectors of the nucleosomes–TF system such as histone modifications and histone-binding proteins influence nucleosome stability, thus altering the balance between N and O states. Fig. 2C shows the manifestation of the Bohr effect in the TF-nucleosome system: Small changes in nucleosomal affinity (from L to L′) due to histone modifications can shift the balance in TF-nucleosome competition toward or away from the nucleosome. For example, a modification that reduces nucleosome stability by about ΔG = 1 kcal/mol (ΔG = kBT log(L/L′)) can lead to an 80% drop in nucleosome occupancy (Fig. 2C, Inset) and a concurrent rise of the TF occupancy (Fig. 2C). Similarly, nucleosome-positioning sequences have a similar effect: They alter nucleosome stability, thus shifting the occupancy curve. Competition between the nucleosome and TFs leads to amplification of the nucleosome-positioning sequence signal, i.e., small changes in histone affinity translate into significant changes in nucleosome occupancy (Fig. 2C, Inset).
Similarly, small changes in TF concentration can significantly reduce nucleosomal occupancy (Fig. 2A and Fig. S3C) in TFBS-rich regions. For example, activation of a tissue-specific TF can lead to selectively reduced chromatization and increased accessibility of tissue-specific regulatory regions (Fig. 3). This is in agreement with a recent genome-scale mapping of DNase I hypersensitive sites (DHS), which found that several loci exhibit tissue-specific DHS profile (37). Note that this mechanism of passive nucleosome eviction does not rely on recruitment of chromatin modification machinery, which may play a role in further destabilizing nucleosomes and expanding nucleosome-free regions.
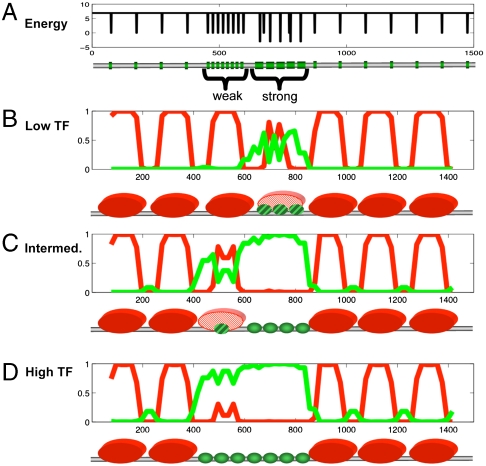
Fig. 3.
Cooperative binding to high- and low-affinity sites. The nucleosomal (red) and TF (green) occupancy profiles for a regulatory region that contains clusters of high- and low-affinity sites. (A) The binding energy profile: a cluster of 8 low-affinity sites and a cluster of 5 high-affinity sites located over the background of spurious low-affinity sites (27). The region contains seven stable, equally spaced nucleosomes with a liner of 50 bp. (B–D) Diagrams show nucleosomal occupancy YN (red), and TF cluster occupancy P3 for three values of TF concentration. While an intermediate TF concentration is sufficient to get high-affinity clusters nucleosome-free and TF-bound, a higher concentration is needed for low-affinity clusters. A combination of low- and high-affinity sites in a regulatory region can result in different responses to various TF concentrations. Notice that isolated low-affinity sites are unable to displace nucleosomes. Nucleosomes were assumed to be well-positioned by DNA sequence, and sliding was disregarded. The following parameters were used: c = 0.01; L = 1,000, αnon-site = 0.001; αhigh-affinity = 20; αlow-affinity = 1.
Critical Size of the TFBS Cluster.
Nucleosome-induced cooperativity, however, has some properties without counterparts in hemoglobin. For example, the number and the affinity of binding sites are constant in hemoglobin but vary in cis-regulatory regions. Fig. 2D presents nucleosomal occupancy as a function of the number, n, of TFBSs. As the number of TFBSs exceeds a certain critical value nc, nucleosomal occupancy drops sharply, manifesting another allosteric effect in the system. Our calculations show that the critical number of TFBSs is given by nc ≈ log(L)/ log(1 + α), yielding a narrow range nc = 3–6 that is not very sensitive to model parameters (see SI Text). This range of TFBSs per nucleosomal footprint is consistent with the recent characterization of Drosophila enhancers (1, 3) that contain about 20 TFBSs per 0.7–1 Kb (i.e., 4–6 sites per nucleosome) (2). Similarly, the cis-regulatory system of endo16 in sea urchins has about 50 sites localized within 2.3 Kb and grouped into seven clusters of five to 10 sites (4). Such passive eviction of nucleosomes could explain widespread depletion and rapid exchange of nucleosomes in TFBS-rich regions (33, 34, 38–40).
This cluster size is consistent with our recent information-theoretical estimates (27) of the minimal significant TFBS cluster. Nucleosomes-mediated cooperativity can suppress binding to widespread spurious sites that are unlikely to cluster, while providing binding to clusters of sites. An example on Fig. 3 demonstrates that clusters of five high-affinity (or eight low-affinity TFBSs) become occupied and nucleosome-free, while isolated sites remain unoccupied by TFs.
Our approach allows one to consider the contributions of low-affinity sites (9, 41) and mixtures of sites of different TFs (see SI Text). Fig. 3 illustrates how arrays of low-affinity and high-affinity sites in nucleosomal DNA respond differently to increasing levels of TFs.
Discussion
Below we discuss several experimental results that support the nucleosome-mediated mechanism of cooperativity, summarized in Table S2, and propose direct experimental tests of the mechanism. We believe that the proposed mechanism contributes to the complex interplay of nucleosome-positioning DNA sequences (23, 42–46), TFs interacting with each other (2) as well as with histones (47) and regulating gene expression (25, 41). The relative importance of different mechanisms of cooperativity (16) and nucleosome positioning (23) may vary for different DNA regions and different organisms (46). A recent study examined intrinsic and extrinsic nucleosome-positioning factors and demonstrated that taking into account TF-nucleosome competition improves the accuracy of nucleosome position predictions (23).
Most direct evidences of nucleosome-mediated cooperativity come from experimental studies that demonstrated cooperative binding of nonendogenous TFs without involvement of protein–protein interactions and for a range of up to 200 bps (15–17). Moreover, experiments with TFs lacking activation domains have shown that synergistic activation of gene expression is determined more by the number of TFBSs than by the interactions with general TFs, polymerase, or chromatin modification machinery (31, 48). Consistent with the nucleosome-induced mechanism, transcomplementation experiments on stripe 2 enhancers demonstrated that precise expression does not require special Bcd–Hb interactions and can be achieved by chimeric and nonendogenous (i.e., noninteracting) TFs (9). The range of nucleosome-induced cooperativity (∼150–200 bps) is also consistent with the modularity of the otd enhancer, which contains two 180-bp TFBS clusters, each able to provide the correct expression pattern (49).
Presented mechanism also ties together several observations in genomics. Passive nucleosome eviction can explain how low nucleosomal density is maintained on cis-regulatory regions and how sharp boundaries of such nucleosome-depleted regions are achieved. The critical number of sites nc = 3–5 (see above) required for the TF-induced nucleosome displacement is consistent with clustered arrangements of TFBSs and can explain why such clustering serves as a powerful criterion for bioinformatic identification of cis-regulatory regions (3, 50). Our mechanism suggests a possible role for low-affinity TFBSs, which are abundant (2, 51) and essential (9) in Drosophila in assisting high-affinity sites to displace nucleosomes. Cooperative nucleosome displacement by competing TF serves, along with sequence information (42–46, 52), to determine nucleosome positioning but, in contrast, can be tissue or condition specific.
The nucleosome-mediated mechanism allows significant flexibility in arrangements of TFBSs while retaining cooperativity, requiring only that a sufficient number of sites are located sufficiently deep inside a nucleosomal footprint. Widespread turnover of sites in enhancers (1–3) and the paucity of local protein–protein interactions (for example, in a crystal structure of interferon-beta enhanceosome) (53) are difficult to reconcile with the classical model of cooperativity by protein–protein interactions. The nucleosome-mediated mechanism can explain observed promiscuity of regulatory regions: Unrelated TFs can cooperate by evolving proximal TFBSs or by site duplication and divergence. A new TF can become a part of an existing assembly by acquiring TFBSs within an existing cluster, a fairly fast and widespread evolutionary process (12, 54). A classical model of cooperativity, in contrast, requires interacting TFs to evolve protein–protein interfaces used for interactions—a much slower evolutionary process.
The nucleosome-mediated mechanism, however, works only for TFBSs separated by at most 150 bps. This range can be increased by synergy of nearby nucleosomes (55) and spread much further through recruitment of chromatin modification machinery and positive feedback in this process (56).
The proposed mechanism makes concrete predictions that can be tested experimentally. First, it suggests a dependence of the cooperative effect and nucleosome occupancy on the number of TFBSs. Both the nucleosome occupancy and level of gene expression can be assayed directly for synthetic or natural promoters that contain TFBS clusters of different densities and spacings and that are also confirmed to be SWI/SNF-independent. Our model specifically predicts that cooperativity (as measured by the Hill coefficient) increases with the number of sites and is greater for sites located closer to the nucleosome center. Second, the cooperativity of TF binding is predicted to depend on nucleosome stability in a nontrivial fashion: Stabilization of the nucleosome (e.g., by strong nucleosome-positioning DNA sequences) is expected to make binding and expression more cooperative, while destabilization of the nucleosome [e.g., by poly(dA:dT) sequences (57, 58) or nucleosome eviction by a nonendogenous TF or polymerase] is expected to decrease cooperativity. This prediction can be used to distinguish our mechanism from the effect of direct protein–protein interactions that are expected to exhibit the opposite dependence on nucleosome stability. A recent study of human nucleosome positioning (26) provided evidences consistent with our predictions. First, it reports elevated in vivo nucleosomal occupancy of human regulatory sequences. This observation is consistent with high nucleosome stability required by nucleosome-mediated cooperativity. Second, this study demonstrates that such high nucleosomal occupancy is encoded in the DNA sequences of regulatory regions, and such sequences are depleted in nucleosome-repelling rigid poly(dA:dT). Again, the depletion of poly(dA:dT) supports the requirement of our model for stable nucleosome positioning. Moreover, poly(dA:dT) that flank TFBS clusters can further stabilize TF-competing nucleosomes by preventing their sliding. Removal of such sequences, we predict, will reduce the degree of cooperativity, while increasing the binding affinity for TFs.
Nucleosome-mediated cooperativity works particularly well for clusters of several TFBSs and thus may be utilized more by higher eukaryotes with more complex regulatory regions (4). Yeast, to the contrary, has simpler promoters with individual sites, rather than clusters carrying regulatory potential (27), and may rely less on nucleosome-mediated cooperativity. Consistent with this argument is the observation that yeast has a lower nucleosome occupancy and lower intrinsic nucleosome propensity of regulatory regions (44, 59).
In summary, we have shown how competition between a nucleosome and TFs can lead to cooperative binding of TFs and cooperative eviction of nucleosomes from regulatory regions. We have established and employed the analogy between this process and cooperativity in hemoglobin according to the MWC model. This analogy has allowed us to consider chromatin modification and nucleosome-positioning sequences as heterotropic allosteric effectors, similar to the Bohr effect. Most importantly, presented mechanism explains several observations in comparative and functional genomics that cannot be reconciled with the classical model of cooperativity via direct protein–protein interactions or a simplistic view of nucleosomes as suppressors of gene expression. Our study provides a mechanistic rationale for widespread flexibility in arrangement of binding sites, allowing highly evolvable regulatory regions. Finally, the analogy between cooperativity in hemoglobin and nucleosome-mediated cooperativity of TFs emphasizes a widespread MWC mechanism of cooperativity observed in a range of biological systems from protein folding (60) to receptor coupling in bacteria (61), ligand-gated ion channels, and enzymatic phosphorylation (62).
Materials and Methods
Approach.
We use a statistical mechanics approach to derive occupancy and other equilibrium properties of the system presented in Fig. 1 (see SI Text). The advantage of our approach is that it allows generalization for more complex cases considered in SI Text.
Estimation of Parameters from Experimental Data.
To estimate α, we take into account TF binding to both specific and nonspecific DNA, with binding constants K and KNS, respectively. Due to competition between specific and nonspecific DNA the occupancy of a single site is y = [P]/([P] + K(1 + [DNA]/KNS)) = α/1 + α, where α ≡ [P]/Keff, effective binding constant defined as Keff = K(1 + [DNA]/KNS) ≈ K[DNA]/KNS, and [DNA] is the concentration of TF-accessible nonspecific DNA. The dissociation constant of most eukaryotic TFs is in the range of K ≈ 1–10 nM, while known nonspecific binding constants are KNS ≈ 1–10 μM (63, 64). Using the length of genomic DNA and the measured copy number of TFs ([P] ≈ 500–5,000 in yeast and [P] ≈ 104–105 protein copies per nucleus in multicellular eukaryotes) (65) (BioNumbers database, http://bionumbers.hms.harvard.edu/) with the assumption of 90% chromatinization, we obtain the range of α ≈ 0.5–5.
We estimate L using in vivo nucleosome equilibrium occupancy f = [N]/([N] + [O]), yielding L = f/(1 - f). Although for stable nucleosomes, occupancy is very close to 1, the fraction of DNA covered by nucleosomes provides a lower bound for f and has a range of 0.9–0.99 (66), yielding L > 10–100. This constitutes a lower bound for f and L, because most nucleosome-free regions are maintained by competition with TFs or active chromatin modification.
Parameter c of the model reflects suppression of TF binding by a nucleosome while in the N state. Acting through steric hindrance, such suppression is not permanent and nucleosomal DNA becomes transiently exposed for TF binding (Fig. 1B) (36, 67). This suppression is equivalent to the experimentally measured equilibrium constant of site exposure, which depends on the location of the site with respect to the center of the nucleosome and has the range c = 2·10-2–10-5. Detailed treatment of partially unwrapped states (see SI Text) shows that they can be aggregated into a single N state.
Obtained estimated values of parameters are sufficient to provide nucleosome-mediated cooperativity. The mechanism of cooperativity is very robust, requiring only L≫1 and c ≪ 1 for the onset of cooperativity (Fig. S1).
Supplementary Material
Acknowledgments.
I am grateful to Mehran Kardar, Stanislav Shvartsman, Paul Wiggins, Jane Kondev, Ned Wingreen, Rob Phillips, George Benedek, and Ilya Ioschikhes for insightful discussions of this study; to John Stamatoyannopoulos for carefully reading of and commenting on the manuscript as well as providing several important suggestions; and to Jason Leith, Zeba Wunderlich, and Albert Liau for discussing and editing the manuscript. Part of this work took place at the Kavli Institute for Theoretical Physics, Santa Barbara, CA, and the Aspen Center for Physics, Aspen, CO. I acknowledge the support of the National Institutes of Health-funded National Center for Biomedical Computing, i2b2, Informatics for Integrating Biology and the Bedside, and National Cancer Institute-funded Physical Sciences in Oncology Center at MIT.
Footnotes
The author declares no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913805107/-/DCSupplemental.
References
- 1.Schroeder MD, et al. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:e271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papatsenko DA, et al. Extraction of functional binding sites from unique regulatory regions: The Drosophila early developmental enhancers. Genome Res. 2002;12:470–481. doi: 10.1101/gr.212502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman BP, et al. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 2004;5:R61. doi: 10.1186/gb-2004-5-9-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Amsterdam; Boston: Elsevier/Academic; 2006. [Google Scholar]
- 5.Lebrecht D, et al. Bicoid cooperative DNA binding is critical for embryonic patterning in Drosophila. Proc Natl Acad Sci USA. 2005;102:13176–13181. doi: 10.1073/pnas.0506462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pettersson M, Schaffner W. Synergistic activation of transcription by multiple binding sites for NF-kappa B even in absence of co-operative factor binding to DNA. J Mol Biol. 1990;214:373–380. doi: 10.1016/0022-2836(90)90187-q. [DOI] [PubMed] [Google Scholar]
- 7.Ptashne M, Gann A. Genes & Signals. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- 8.Bintu L, et al. Transcriptional regulation by the numbers: Models. Curr Opin Genet Dev. 2005;15:116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- 10.Saiz L, Vilar JM. DNA looping: The consequences and its control. Curr Opin Struct Biol. 2006;16:344–350. doi: 10.1016/j.sbi.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moses AM, et al. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- 14.Lin YS, Carey M, Ptashne M, Green MR. How different eukaryotic transcriptional activators can cooperate promiscuously. Nature. 1990;345:359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- 15.Miller JA, Widom J. Collaborative competition mechanism for gene activation in vivo. Mol Cell Biol. 2003;23:1623–1632. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vashee S, Melcher K, Ding WV, Johnston SA, Kodadek T. Evidence for two modes of cooperative DNA binding in vivo that do not involve direct protein-protein interactions. Curr Biol. 1998;8:452–458. doi: 10.1016/s0960-9822(98)70179-4. [DOI] [PubMed] [Google Scholar]
- 17.Adams CC, Workman JL. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol. 1995;15:1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morse RH. Getting into chromatin: How do transcription factors get past the histones? Biochem Cell Biol. 2003;81:101–112. doi: 10.1139/o03-039. [DOI] [PubMed] [Google Scholar]
- 19.Polach KJ, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- 20.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 21.Hill AV. The possible effects of the aggregation of the molecules of hæmoglobin on its dissociation curves. J Physiol. 1910;40:1–7. [Google Scholar]
- 22.Wasson T, Hartemink AJ. An ensemble model of competitive multi-factor binding of the genome. Genome Res. 2009;19:2101–2112. doi: 10.1101/gr.093450.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morozov AV, et al. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009;37:4707–4722. doi: 10.1093/nar/gkp475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raveh-Sadka T, Levo M, Segal E. Incorporating nucleosomes into thermodynamic models of transcription regulation. Genome Res. 2009;19:1480–1496. doi: 10.1101/gr.088260.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam FH, Steger DJ, O’Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tillo D, et al. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 2010;5:e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wunderlich Z, Mirny LA. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 2009;25:434–440. doi: 10.1016/j.tig.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Poirier MG, Bussiek M, Langowski J, Widom J. Spontaneous access to DNA target sites in folded chromatin fibers. J Mol Biol. 2008;379:772–786. doi: 10.1016/j.jmb.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 31.Oliviero S, Struhl K. Synergistic transcriptional enhancement does not depend on the number of acidic activation domains bound to the promoter. Proc Natl Acad Sci USA. 1991;88:224–228. doi: 10.1073/pnas.88.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 33.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 34.Dion MF, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 35.Phair RD, et al. Global nature of dynamic protein-chromatin interactions in vivo: Three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 37.John S, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 38.Sabo PJ, et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc Natl Acad Sci USA. 2004;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valouev A, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mavrich TN, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levine M. A systems view of Drosophila segmentation. Genome Biol. 2008;9:207.1–207.3. doi: 10.1186/gb-2008-9-2-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trifonov EN, Sussman JL. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci USA. 1980;77:3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ioshikhes I, Bolshoy A, Derenshteyn K, Borodovsky M, Trifonov EN. Nucleosome DNA sequence pattern revealed by multiple alignment of experimentally mapped sequences. J Mol Biol. 1996;262:129–139. doi: 10.1006/jmbi.1996.0503. [DOI] [PubMed] [Google Scholar]
- 44.Ioshikhes IP, Albert I, Zanton SJ, Pugh BF. Nucleosome positions predicted through comparative genomics. Nat Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- 45.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peckham HE, et al. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim HD, O’Shea EK. A quantitative model of transcription factor-activated gene expression. Nat Struct Mol Biol. 2008;15:1192–1198. doi: 10.1038/nsmb.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu HE, Kodadek T, Johnston SA. A single GAL4 dimer can maximally activate transcription under physiological conditions. Proc Natl AcadSci USA. 1995;92:7677–7680. doi: 10.1073/pnas.92.17.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao Q, Finkelstein R. Targeting gene expression to the head: the Drosophila orthodenticle gene is a direct target of the Bicoid morphogen. Development. 1998;125:4185–4193. doi: 10.1242/dev.125.21.4185. [DOI] [PubMed] [Google Scholar]
- 50.Sinha S, van Nimwegen E, Siggia ED. A probabilistic method to detect regulatory modules. Bioinformatics. 2003;19(Suppl 1):292i–301i. doi: 10.1093/bioinformatics/btg1040. [DOI] [PubMed] [Google Scholar]
- 51.Zhang C, et al. A clustering property of highly-degenerate transcription factor binding sites in the mammalian genome. Nucleic Acids Res. 2006;34:2238–2246. doi: 10.1093/nar/gkl248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 53.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berg J, Willmann S, Lassig M. Adaptive evolution of transcription factor binding sites. BMC Evol Biol. 2004;4:42.1–42.12. doi: 10.1186/1471-2148-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat Struct Mol Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 56.Sneppen K, Micheelsen MA, Dodd IB. Ultrasensitive gene regulation by positive feedback loops in nucleosome modification. Mol Syst Biol. 2008;4:182.1–182.5. doi: 10.1038/msb.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tillo D, Hughes TR. G + C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442.1–442.13. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kay MS, Baldwin RL. Alternative models for describing the acid unfolding of the apomyoglobin folding intermediate. Biochemistry. 1998;37:7859–7868. doi: 10.1021/bi9802061. [DOI] [PubMed] [Google Scholar]
- 61.Skoge ML, Endres RG, Wingreen NS. Receptor-receptor coupling in bacterial chemotaxis: Evidence for strongly coupled clusters. Biophys J. 2006;90:4317–4326. doi: 10.1529/biophysj.105.079905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 63.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 64.Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR. Comparative binding of p53 to its promoter and DNA recognition elements. J Mol Biol. 2005;348:589–596. doi: 10.1016/j.jmb.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 65.Chen SC, Zhao T, Gordon GJ, Murphy RF. Automated image analysis of protein localization in budding yeast. Bioinformatics. 2007;23(13):i66–71. doi: 10.1093/bioinformatics/btm206. [DOI] [PubMed] [Google Scholar]
- 66.Sabo PJ, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 67.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.