Abstract
Variation in DNA sequence contributes to individual differences in quantitative traits, but in humans the specific sequence variants are known for very few traits. We characterized variation in gene expression in cells from individuals belonging to three major population groups. This quantitative phenotype differs significantly between European-derived and Asian-derived populations for 1,097 of 4,197 genes tested. For the phenotypes with the strongest evidence of cis determinants, most of the variation is due to allele frequency differences at cis-linked regulators. The results show that specific genetic variation among populations contributes appreciably to differences in gene expression phenotypes. Populations differ in prevalence of many complex genetic diseases, such as diabetes and cardiovascular disease. As some of these are probably influenced by the level of gene expression, our results suggest that allele frequency differences at regulatory polymorphisms also account for some population differences in prevalence of complex diseases.
The expression levels of genes determine the distinctive characteristics of cells. Recent studies have shown that gene expression levels in humans differ not only among cell types within an individual but also among individuals1,2. This observation led to analysis of gene expression as a phenotype and to the identification of polymorphic genetic variants that influence individual differences in expression level3–8. However, these studies of the genetics of human gene expression have been restricted to individuals from one European-derived sample, the families collected by the Centre d’Etude du Polymorphisme Humain (CEPH). Differences between populations in gene expression phenotypes have not been characterized. We present an analysis of such differences.
Much of the recognized genetic variation among populations is in DNA polymorphisms with no known functional significance. On the other hand, some allele frequency differences between populations have highly significant phenotypic consequences. Among the best-established are the differences in allele frequencies for mendelian genetic diseases. The marked population differences in prevalence of these qualitative phenotypes (such as cystic fibrosis9 and Tay-Sachs disease10) are entirely due to differences in frequencies of the mutant alleles. However, genetic differences among populations in quantitative phenotypes are potentially just as important functionally.
Here we extend the comparative genetic analysis of population differences from qualitative phenotypes to a particular quantitative phenotype, the expression level of genes. The choice of gene expression as a phenotype provides a large set of comparable traits, all measured at the same time in each individual. Our goals are to determine what proportion of gene expression phenotypes differs significantly between populations and to what extent the phenotypic differences are attributable to specific genetic polymorphisms. We find that at least 25% of the gene expression phenotypes differ significantly between the major populations studied, and specific genetic variation (in allele frequency) accounts for the difference in the most significant instances among the phenotypes that are cis regulated.
We measured the expression of genes in Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines from three populations that are part of the samples from the International HapMap Project11. These include 60 European-derived individuals from the Utah pedigrees of the Centre d’Etude du Polymorphisme Humain (CEU), 41 Han Chinese in Beijing (CHB) and 41 Japanese in Tokyo (JPT).
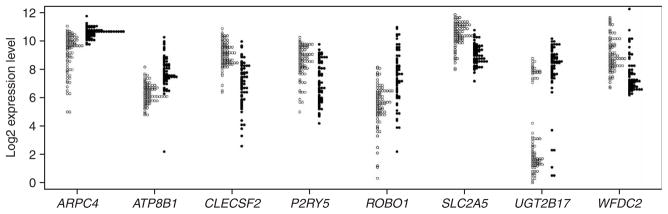
We used the Affymetrix Genome Focus Array that contains ~8,500 annotated human genes to measure expression of genes in the 142 individuals from the three populations. We focused on 4,197 genes that are expressed in lymphoblastoid cell lines. There were 939 genes whose expression was significantly different by the t test (P < 10−5; Pc < 0.05 after Šidák correction12) between the CEU and CHB samples and 756 genes that differed between the CEU and JPT samples. In contrast, there were only 27 genes whose expression differed significantly (P < 10−5) between the CHB and JPT samples. Because the mean expression levels of most genes are similar between the CHB and JPT samples, we combined the samples as ‘CHB+JPT’ for subsequent analysis, as did the International HapMap Consortium11. At P < 10−5, there were 1,097 genes that differed between CEU and the combined CHB+JPT samples (Supplementary Table 1 online). Figure 1 shows eight of the gene expression phenotypes with the largest differences between the CEU and CHB+JPT samples. Even when the mean expression differed significantly between populations, the magnitude of the difference was quite small for most genes, and the area of overlap was large. Table 1 describes the 35 genes whose mean expression differs by twofold or more between the CEU and CHB+JPT samples.
Figure 1.
Gene expression in CHB+JPT (open circles) and CEU (filled circles) for eight genes that differ in mean expression level between the populations (N = 82 for CHB+JPT, and N = 60 for CEU). Additional information is found in Table 1.
Table 1.
Thirty-five genes with greater than twofold difference in mean expression between CEU and CHB+JPT samples
| Gene symbol | Gene name | Ratio of CEU expression level to CHB+JPT expression level (95% c.i.)b | P (t test) versus |
|---|---|---|---|
| UGT2B17a | UDP glucuronosyltransferase 2 family, polypeptide B17 | 22.3 (16.8, 29.5) | 1.03 × 10−18 |
| ROBO1a | Roundabout, axon guidance receptor, homolog 1 | 4.0 (3.6, 4.5) | 7.10 × 10−10 |
| ATP8B1a | ATPase, class I, type 8B, member 1 | 2.9 (2.8, 3.0) | 6.87 × 10−14 |
| ARPC4a | Actin-related protein 2/3 complex, subunit 4, 20 kDa | 2.7 (2.6, 2.8) | 7.07 × 10−15 |
| DPYSL2 | Dihydropyrimidinase-like 2 | 2.6 (2.6, 2.6) | 9.78 × 10−31 |
| RGS20 | Regulator of G-protein signaling 20 | 2.3 (2.2, 2.5) | 6.77 × 10−8 |
| FCER2 | FC fragment of IGE, low affinity II, receptor for CD23A | 2.3 (2.2, 2.3) | 8.53 × 10−15 |
| MEIS2 | MEIS1, myeloid ecotropic viral integration site 1 homolog 2 | 2.2 (2.1, 2.3) | 1.52 × 10−9 |
| COPG | Coatomer protein complex, subunit gamma | 2.2 (2.1, 2.2) | 1.05 × 10−14 |
| HSPB1 | Heat shock protein 1 | 2.1 (2.1, 2.2) | 2.15 × 10−14 |
| TCF7 | Transcription factor 7 | 2.0 (2.0, 2.0) | 4.94 × 10−14 |
| HDGFRP3 | Hepatoma-derived growth factor, related protein 3 | 2.0 (2.0, 2.1) | 3.51 × 10−11 |
| STXBP2 | Syntaxin binding protein 2 | 2.0 (1.9, 2.0) | 2.95 × 10−13 |
| D21S2056E | EST | 2.0 (1.9, 2.0) | 1.19 × 10−11 |
| MAN2A1 | Mannosidase, alpha, class 2A, member 1 | −2.0 (−2.0, −2.0) | 1.01 × 10−12 |
| STS | Steroid sulfatase, arylsulfatase C, isozyme S | −2.0 (−1,9, −2.0) | 8.48 × 10−11 |
| CTSS | Cathepsin S | −2.0 (−1.9, −2.0) | 7.52 × 10−18 |
| PDE4B | Phosphodiesterase 4B, CAMP-specific | −2.1 (−2.1, −2.2) | 4.70 × 10−15 |
| OCIL | C-type lectin domain family 2, member D | −2.1 (−2.1, −2.2) | 6.66 × 10−10 |
| TMPRSS3 | Serine protease TADG12 | −2.1 (−2.0, −2.2) | 8.02 × 10−7 |
| NFIL3 | Nuclear factor, interleukin 3–regulated | −2.1 (−2.1, −2.1) | 5.85 × 10−17 |
| LEF1 | Lymphoid enhancer-binding factor 1 | −2.1 (−2.0, −2.2) | 1.07 × 10−6 |
| DNAJB9 | DNAJ (HSP40) homolog, subfamily B, member 9 | −2.1 (−2.0, −2.1) | 9.11 × 10−15 |
| ARL7 | ADP-ribosylation factor-like 4C | −2.1 (−2.0, −2.1) | 1.72 × 10−11 |
| PTPRO | Protein tyrosine phosphatase, receptor type, O | −2.1 (−2.0, −2.1) | 1.35 × 10−13 |
| ADM | Adrenomedullin | −2.2 (−2.1, −2.3) | 2.92 × 10−7 |
| IGF1 | Insulin-like growth factor 1 (somatomedin C) | −2.2 (−2.1, −2.2) | 1.81 × 10−14 |
| PSPHL | Phosphoserine phosphatase | −2.3 (−2.1, −2.6) | 8.09 × 10−6 |
| CD38 | CD38 antigen (P45) | −2.4 (−2.4, −2.5) | 4.47 × 10−12 |
| SEC10L1 | Exocyst complex component 5 | −2.5 (−2.4, −2.6) | 1.97 × 10−11 |
| WFDC2 a | WAP four-disulfide core domain 2 | −2.6 (−2.5, −2.8) | 8.68 × 10−10 |
| P2RY5 a | Purinergic receptor P2Y, G protein–coupled, 5 | −2.6 (−2.5, −2.8) | 1.19 × 10−8 |
| SLC2A5 a | Solute carrier family 2 (facilitated glucose/fructose transporter), member 5 | −2.6 (−2.5, −2.7) | 1.24 × 10−18 |
| NK4 | Interleukin 32 | −2.6 (−2.4, −2.9) | 6.32 × 10−6 |
| CLECSF2 a | C-type lectin domain family 2, member b | −4.3 (−4.1, −4.6) | 2.65 × 10−14 |
Distribution of expression levels for these eight genes is shown in Figure 1.
Negative ratio indicates that mean expression is higher in CHB+JPT.
The gene with the greatest difference between the CEU and CHB+JPT samples was UGT2B17; its mean expression in the CEU individuals was 22 times higher than in the CHB+JPT samples. In both populations, there is a polymorphism for deletion of this gene13. Homozygotes for the deletion are more common in CHB+JPT than in the CEU samples14, accounting for the lower average expression of this gene in CHB+JPT (Fig. 1).
We considered it essential to replicate the marked similarity of the Asian-derived populations and their distinctness from the CEU. We followed up the initial findings with an analysis of 24 samples from the Han Chinese of Los Angeles (CHLA) who are part of the Human Variation Panel15. Among the 35 genes in Table 1, only one (3%) differed significantly (P < 0.05) between the CHLA and the CHB+JPT samples, but 32 (91%) differed significantly between CHLA and CEU.
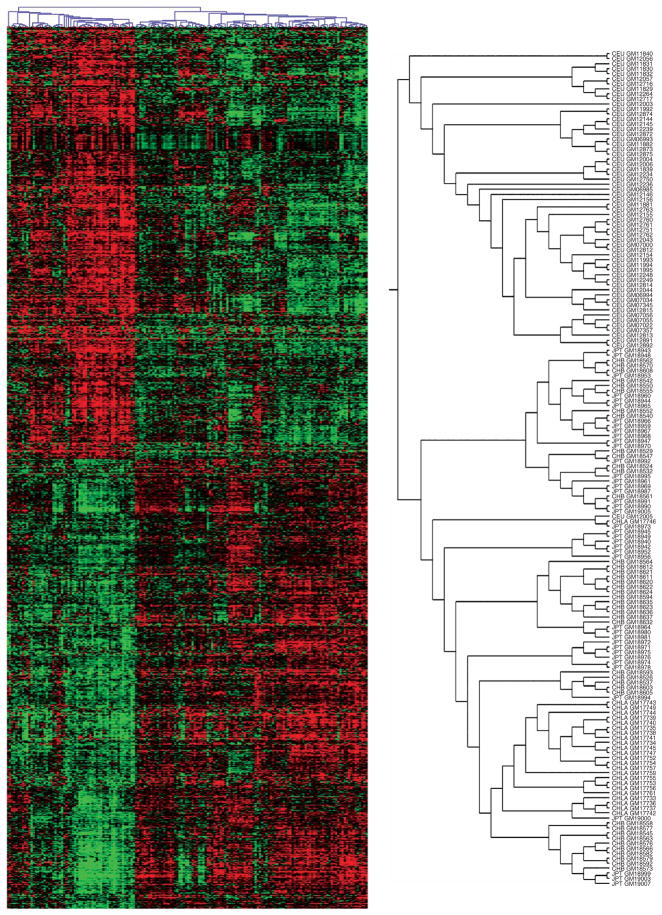
To investigate the population differences in a multilocus fashion, we carried out cluster analysis16 and grouped the samples from 60 CEU, 41 CHB, 41 JPT and 24 CHLA by similarity of expression level for the 1,097 genes that are differentially expressed between the HapMap CEU and CHB+JPT samples. We expected that the CHB and JPT would cluster together, separately from the CEU. However, we were most interested in how the CHLA samples would be grouped, as they were not used in identifying the 1,097 genes. There were two main clusters (Fig. 2): one consisted entirely of CEU individuals (59 of the 60 CEU), and the other consisted of all the Asian-derived individuals (82 CHB+JPT and 24 CHLA individuals) plus one CEU. Thus, the samples from the Han Chinese of Los Angeles were much more similar in expression profile to the HapMap CHB+JPT samples than to the HapMap CEU samples. This confirms that there is a characteristic expression pattern that the CHLA samples share with the CHB+JPT. The CHLA samples were collected separately from the CHB and JPT samples; therefore, the expression differences between the European- and Asian-derived samples are not an artifact of how the cells were processed.
Figure 2.
Results of cluster analysis. The 166 individuals are represented by columns, and the 1,097 genes of the main analysis are represented by rows. For each gene, expression level for each individual is indicated by color; intensity of red is proportional to degree of expression above the mean, and intensity of green is proportional to degree of expression below the mean. The analysis grouped the individuals into two main distinguishable groups (see enlarged tree diagram at right). One group consists of 59 CEU samples, and the other consists of the 82 CHB+JPT samples, the 24 CHLA samples and 1 CEU sample.
Our second goal was to determine to what extent the expression-phenotype differences are associated with, and possibly attributable to, specific genetic differences. A large catalog of population differences at the DNA level is available (in the form of SNP frequencies11) for the same HapMap samples we studied at the expression level. We did the analysis in two steps. First, we carried out genome-wide association (GWA) analysis with these SNPs for each of the 1,097 phenotypes to localize the genetic determinants of variation in gene expression. We did this analysis in CEU and CHB+JPT. Then we compared the results for the two samples in order to identify the genetic differences that might explain expression differences between the populations.
We carried out the GWA analysis with the SNP markers as follows. For each expression phenotype, we tested ~2 million SNPs for association by linear regression of expression level on SNP genotype (coded 0, 1, 2). To adjust for the large number of tests, we set the significance level at nominal P = 2.5 × 10−8 (Pc = 0.05 after Šidák correction), which is conservative. Among 1,097 phenotypes tested, we would expect approximately 55 (0.05 × 1,097) to appear significant by chance. We found 104 phenotypes that showed significant association with one or more markers in the CEU samples: 10 phenotypes with ‘cis’ association and 94 with ‘trans’ association. In the CHB+JPT samples, we found 89 phenotypes with significant association: 23 with cis association and 66 with trans association. We have operationally defined a cis-regulated gene by the presence of significant association with SNP(s) in the region 500 kb upstream of the start of the transcript to 500 kb downstream of the 3′ end. (This definition allows for linkage disequilibrium between a marker and the actual regulatory variants, and for long-range cis regulators.) Among the findings for either population alone, we expect some to be false positive results as indicated above. However, when we found the identical marker (among 2 million) to be significantly associated with the same expression phenotype in both populations, we considered the result very unlikely to be a false positive; instead, it is likely to be the ‘true’ regulatory variant, or in very strong linkage disequilibrium with a regulatory variant.
The most direct comparison between CEU and CHB+JPT, with respect to regulatory differences at the DNA level, is possible when a gene expression phenotype is associated with the same SNP in both populations. We restricted our attention to the 11 phenotypes of this kind, where the SNPs were the most highly significant in both populations. (In our data, these SNPs were all in cis to the expressed gene.) For these phenotypes, the association was either significant at P < 2.5 × 10−8 in both populations or significant at approximately P < 2.5 × 10−8 in one population and somewhat less significant in the other (Table 2).
Table 2.
Contribution (R2) of SNP genotypes to differences in mean expression between populations for 11 cis-regulated gene expression phenotypes
| Gene | P value (t test) | CEU (log2 expression) | CHB+JPT (log2 expression) | cis SNP | P value for association (CEU) | P value for association (CHB+JPT)a | More highly expressed allele | Frequency of high allele (CEU) | Frequency of high allele (CHB+JPT) | R2 due to genotype variationb | R2 due to population- specific genotype effectsc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UGT2B17 | 1.03 ± 10−18 | 7.9 | 3.4 | rs3100645 | 3.3 ± 10−12 | 6.5 ± 10−37 | A | 0.68 | 0.15 | 0.81 | 0.05 |
| POMZP3 | 6.94 ± 10−6 | 7.3 | 6.6 | rs2005354 | 9.7 ± 10−22 | 1.9 ± 10−14 | G | 0.28 | 0.06 | 0.75 | 0.00 |
| PEX6 | 1.33 ± 10−6 | 7.3 | 6.7 | rs2395943 | 4.3 ± 10−15 | 1.4 ± 10−7 | G | 0.59 | 0.36 | 0.52 | 0.05 |
| PSPHL | 8.09 ± 10−6 | 7.2 | 8.4 | rs11982736 | 5.8 ± 10−13 | 2.3 ± 10−9 | A | 0.17 | 0.45 | 0.51 | 0.02 |
| CSTB | 9.79 ± 10−9 | 11.6 | 11.1 | rs2838386 | 1.2 ± 10−7 | 5.5 ± 10−10 | A | 0.63 | 0.19 | 0.50 | 0.01 |
| DNAJD1 | 5.82 ± 10−7 | 10.9 | 11.2 | rs2281778 | 2.5 ± 10−9 | 3.4 ± 10−7 | C | 0.76 | 0.90 | 0.43 | 0.08 |
| AP3S2 | 1.50 ± 10−11 | 9.7 | 9.4 | rs4932265 | 1.9 ± 10−10 | 5.8 ± 10−7 | T | 0.29 | 0.16 | 0.39 | 0.18 |
| HSD17B12 | 3.10 ± 10−6 | 11.2 | 11.5 | rs1061810 | 2.7 ± 10−11 | 3.4 ± 10−9 | T | 0.76 | 0.80 | 0.39 | 0.12 |
| NUBP2 | 6.14 ± 10−11 | 8.6 | 8.1 | rs1065663 | 3.2 ± 10−5 | 1.3 ± 10−8 | A | 0.81 | 0.69 | 0.32 | 0.16 |
| B4GALT1 | 1.14 ± 10−6 | 8.9 | 8.3 | rs10511909 | 4.7 ± 10−10 | NAd | G | 0.17 | 0 | 0.22 | 0.04 |
| TPP2 | 2.81 ± 10−9 | 9.4 | 8.7 | rs1887355 | 4.1 ± 10−6 | 3.3 ± 10−8 | G | 0.56 | 0.69 | 0.18 | 0.31 |
See Methods for calculation of R2. ‘High allele’ refers to the more highly expressed allele.
To allow for possible effects of heterogeneity between CHB and JPT, we used membership in these two groups as a covariate in calculating the P value for association in CHB+JPT.
Component 1; assumes same genotype effects for each population but not necessarily the same genotype frequencies.
Component 2; measures the additional variation explained by population-specific genotype effects, after adjusting for inclusion of component 1. See Methods.
Monomorphic (C allele) in CHB+JPT sample. In CHB+JPT the closest SNP (rs10124479) is associated with the expression level of B4GALT1 (P = 1.04 × 10−11); however, this SNP is not significantly associated with expression in CEU. For analysis, we treated the associated allele of rs10124479 in CHB+JPT as equivalent to the G allele of rs10511909 in CEU.
As the same cis markers are associated with the expression phenotypes in both populations, we assumed that the actual cis regulators were the same in both populations. At this point, however, we did not know whether the mean differences between populations were due mainly to different SNP genotype frequencies or to different mean expression levels for the same SNP genotypes (‘population-specific genotype effects’).
We used nested linear models for gene expression level to partition the overall expression variation sequentially into three components: (i) the effect of genotype variation, allowing for population differences in genotype frequencies, but not for population-specific genotype effects; (ii) additional variation explained by population-specific genotype effects and (iii) additional variation explained by departures from genetic additivity (dominance). The contributions of these components were represented by the fraction R2 of the total sum of squares (see Methods).
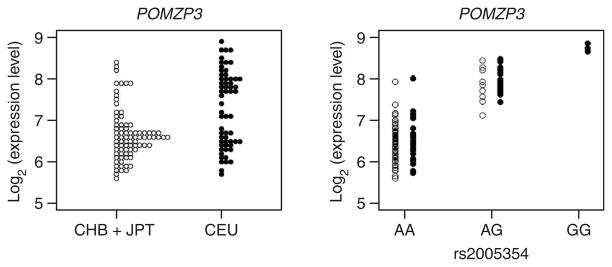
Except for one gene (TPP2), the highly significant expression differences between CEU and CHB+JPT were due to differences in genotype (allele) frequency much more than to population-specific genotype effects (Table 2). For five of the genes, the genotype frequency difference accounted for 50% or more of the variation. For example, the G allele of SNP rs2005354 was associated with higher expression of POMZP3 in both populations (Fig. 3). However, the frequency of the G allele was appreciably greater in CEU (0.28) than in CHB+JPT (0.06), with corresponding differences in genotype frequencies. The result is that the mean expression level was higher in CEU (7.3) than in CHB+JPT (6.6). Additional examples are shown in Table 2. We assumed that in these cases the SNP was itself a regulator of gene expression or was in strong linkage disequilibrium with a regulator. For most of these genes, therefore, there was little evidence that the regulators themselves differed. Instead, different frequencies of allelic forms of the regulator accounted for the population differences in expression levels for these expression phenotypes. (We did not find large contributions from dominance for any of the expression phenotypes in Table 2; the largest R2 for this component was only 0.08, for UGT2B17).
Figure 3.
The population difference in expression of POMZP3 is accounted for by the allele frequency difference at the very closely linked SNP rs2005354. The left panel shows the distribution of expression level in the same format as in Figure 1. The right panel shows the expression level separately for individuals with each genotype of the SNP.
In addition to the variation analyzed above, some variation in expression phenotypes between populations can probably be attributed to different regulatory mechanisms. For four phenotypes, we found significant cis association in the CHB+JPT sample but not in the CEU sample (Supplementary Table 2 online). We note, however, that the CHB+JPT sample size is larger (n = 82) than the CEU sample (n = 60), so it is possible that the corresponding cis effects exist in CEU but were not strong enough to be detected with the smaller sample size.
There were four additional phenotypes with significant evidence for trans regulators in both populations, but all four mapped to different genomic regions in the two populations (Supplementary Table 3 online). In several cases, the results were highly significant even after correction for multiple testing. This evidence for different locations suggests that different regulatory mechanisms may account for the variation in expression levels between populations. Nevertheless, we recognize that the genetic analysis for trans regulators has been much less conclusive than for cis regulators, and the apparent differences in location of regulators may be due to association findings that are false positives.
What can we conclude about the relationship between DNA sequence variation and variation in expression phenotype? Our previous studies3,4 showed that expression variation within the CEPH Utah sample is associated with polymorphic variation at the DNA level (that is, with SNPs). Here we have found that 1,097 expression phenotypes (~25% of those tested) differ significantly between the populations studied. Because so many phenotypes differ, when we combine them for analysis, we are able to classify individuals very accurately (as in Fig. 2). However, our primary interest is not in classification but rather in accounting for the expression differences that we found between the populations and in the implications of this finding.
We found that the difference in expression for a set of phenotypes is accounted for by a simple aspect of population genetics. There are marked between-population differences in allele frequencies of the same SNPs that are associated with within-population regulation of expression. In the 11 phenotypes we investigated in detail, these allele frequency differences explain 18%–81% of the total variation in expression level. For five phenotypes, allele frequency differences at SNPs associated with the regulators account for more than half the total variation in expression. In other words, the population differences in these expression phenotypes are largely attributable to frequency differences at the DNA sequence level. Similar results have been found for differences between two strains of Drosophila melanogaster17
In our analysis, we tested a large set of quantitative phenotypes. By our very stringent criteria, we identified specific genetic polymorphisms strongly associated with the differences between human populations in at least a dozen of these phenotypes (Table 2 and Supplementary Tables 1 and 2). Our approach yields a large collection of comparable measurements, consisting of gene expression phenotypes that can be examined simultaneously and compared among individuals. There are a few other polymorphisms that seem to account for population differences in quantitative traits: these include several examples for skin color (for a review, see ref. 18), which has also been attributed to at least one SNP variant19. Unlike expression profiles, however, these quantitative traits do not lend themselves to analysis as a ‘collection,’ and very few can be confidently associated with a specific genetic polymorphism. A collection of differences in gene expression therefore provides a distinctive way to approach the subject of genetically determined population differences.
The findings for gene expression are relevant for understanding the genetics of disease susceptibility—in particular, susceptibility to complex genetic diseases. In discussions of the genetics of complex disease, it is has been noted20 that variants in coding regions of candidate genes do not account for a large proportion of disease susceptibility. A reasonable conclusion is that variation in gene expression is responsible instead. There are well-known population differences in the prevalence of complex genetic diseases such as hypertension and type 2 diabetes mellitus. Our results suggest that genetically determined differences in gene expression contribute to these population differences. Analysis of variation in gene expression will enhance understanding of both the underlying genetics and the population differences observed in complex genetic diseases.
METHODS
Study participants and expression phenotyping
Lymphoblastoid cell lines for 60 HapMap CEU, 41 HapMap CHB, 41 HapMap JPT and 24 Han Chinese of Los Angeles (CHLA) were obtained from Coriell Cell Repositories and grown to a density of 5 × 105 cells/ml in RPMI 1640 with 15% FBS (vol/vol), 100 units penicillin/ml, 100 μg streptomycin sulfate/ml and 1% L-glutamine (wt/vol). Several CHB and JPT samples from the HapMap collection were excluded because cell lines were not available at the time of the study. Total RNA was extracted with the RNeasy Mini-Kit (Qiagen) and hybridized onto Affymetrix Genome Focus arrays according to the manufacturer’s protocol. The growing and processing of the HapMap cell lines was randomized by population group to eliminate batch effects that may contribute to apparent population differences in gene expression; the CHLA cells were studied later and were grown and processed at one time.
Expression arrays were analyzed using the Affymetrix MAS 5.0 software. The expression intensity was scaled to 500 and log2 transformed. The 4,197 genes that were called ‘Present’ in at least 80% of the samples in one population were used for further analysis.
Significance tests
The significance of the difference between sample means was first tested by the t test. To assess the effect of possible departures from the assumptions for the parametric test, we compared the results with those from the nonparametric Wilcoxon rank-sum test and found very similar results. With the Wilcoxon test, there are 1,104 genes that are significantly different between the CHB+JPT and CEU samples at P < 10−5. More than 90% of the genes that are significant by the t test are also significant by Wilcoxon test. For several randomly selected phenotypes, we calculated empirical P values by a permutation test. The empirical P values and those from the t test did not differ appreciably. We conclude that the t test is a satisfactory test for significant differences between CHB+JPT and CEU.
Cluster analysis
The pairwise similarity of all 166 subjects was calculated as the Pearson correlation coefficient of the expression levels of the 1,097 genes that were found to be differentially expressed between the CEU and CHB+JPT samples. The individuals were then grouped by hierarchical clustering using the average linkage method, as implemented in MultiExperiment Viewer.
Genome-wide association analysis
Log2 of expression level as the dependent variable was regressed on SNP genotype (coded 0, 1, and 2). Genotypes from release 19 of the International HapMap Project were used. All markers with minor allele frequency >5% were included. Analysis was carried out separately for the CEU and CHB+JPT samples. Correction for multiple testing was performed by the Šidák procedure for 2,050,366 markers in the CHB+JPT samples and 2,246,676 markers in the CEU samples. The corrected P value of 0.05 corresponds to a nominal P value of ~2.5 × 10−8.
Contributions to total sum of squares of expression variation
With nested multiple regression models, the total sums of squares of expression variation can be decomposed sequentially into contributing sums of squares. Each sum of squares measures the additional part of the total variation accounted for when one predictor variable is added to the model, given that the previous predictors are already included. Thus the contribution of each predictor is measured after adjusting for inclusion of previous predictors. In our data, which contain unequal numbers in the genotype categories, different ordering of predictors gives slightly different results.
We present the contributions from three components.
Genotype variation. This component assumes an additive (linear) model, with slope and intercept the same in both populations; only the frequencies of the three genotypes may vary among populations.
Population-specific genotype effects (after adjusting for (i)). This component assumes a different additive (linear) model for each population (that is, populations may differ with respect to intercept, slope or both).
Dominance (after adjusting for (i) and (ii)). This component relaxes the additive assumption in (i) and (ii) and allows for an alternative genetic model (for example, dominance); effectively allows for different mean expression for each of the six groups defined by genotype and population.
This analysis was carried out in SAS for each of the expression phenotypes in Table 2. We report component (i) as ‘R2 due to genotype variation’ and component (ii) as ‘additional R2 due to population-specific genotype effects’. We did not find large contributions from (iii) for any of the expression phenotypes in Table 2.
Supplementary Material
Acknowledgments
We thank T. Weber for carrying out the microarray hybridizations, V. Mancuso for processing samples and data analysis and H. H. Kazazian and K. Ewens for comments on the manuscript. This work was supported by US National Institutes of Health grants (to R.S.S, V.G.C) and by the W.W. Smith Chair (V.G.C.).
Footnotes
Accession codes. Gene Expression Omnibus (GEO): GSE5859.
URLs. Human Variation Panel: http://ccr.coriell.org/nigms/cells/humdiv.html. MultiExperiment Viewer: http://www.tm4.org. Information on HapMap SNP markers can be found at http://www.hapmap.org.
Note: Supplementary information is available on the Nature Genetics website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 2.Cheung VG, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 3.Cheung VG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monks SA, et al. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 8.Pastinen T, et al. A survey of genetic and epigenetic variation affecting human gene expression. Physiol Genomics. 2004;16:184–193. doi: 10.1152/physiolgenomics.00163.2003. [DOI] [PubMed] [Google Scholar]
- 9.Tsui LC. Mutations and sequence variations detected in the cystic fibrosis transmembrane conductance regulator (CFTR) gene: a report from the Cystic Fibrosis Genetic Analysis Consortium. Hum Mutat. 1992;1:197–203. doi: 10.1002/humu.1380010304. [DOI] [PubMed] [Google Scholar]
- 10.Paw BH, Tieu PT, Kaback MM, Lim J, Neufeld EF. Frequency of three Hex A mutant alleles among Jewish and non-Jewish carriers identified in a Tay-Sachs screening program. Am J Hum Genet. 1990;47:698–705. [PMC free article] [PubMed] [Google Scholar]
- 11.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westfall PH, Young SS. Resampling-Based Multiple Testing. John Wiley & Sons; New York: 1992. [Google Scholar]
- 13.Wilson W, III, et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics. 2004;84:707–714. doi: 10.1016/j.ygeno.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 14.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 15.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 16.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osada N, Kohn MH, Wu CI. Genomic inferences of the cis-regulatory nucleotide polymorphisms underlying gene expression differences between Drosophila melanogaster mating races. Mol Biol Evol. 2006;23:1585–1591. doi: 10.1093/molbev/msl023. [DOI] [PubMed] [Google Scholar]
- 18.McEvoy B, Beleza S, Shriver MD. The genetic architecture of normal variation in human pigmentation: An evolutionary perspective and model. Hum Mol Genet. 2006;15:R176–R181. doi: 10.1093/hmg/ddl217. [DOI] [PubMed] [Google Scholar]
- 19.Lamason RL, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 20.Knight JC. Regulatory polymorphisms underlying complex disease traits. J Mol Med. 2005;83:97–109. doi: 10.1007/s00109-004-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.