Abstract
The catalytic A1 subunit of cholera toxin (CTA1) is an ADP-ribosyltransferase with three distinct subdomains: CTA11 forms the catalytic core of the toxin; CTA12 is an extended linker between CTA11 and CTA13; and CTA13 is a compact globular region. CTA1 crosses the endoplasmic reticulum (ER) membrane to enter the cytosol where it initiates a cytopathic effect. Toxin translocation involves ER-associated degradation (ERAD), a quality control system that exports misfolded proteins from the ER to the cytosol. At the physiological temperature of 37°C, the free CTA1 subunit is in a partially unfolded conformation that triggers its ERAD-mediated translocation to the cytosol. Thus, the temperature sensitivity of CTA1 structure is an important determinant of its function. Here, we examined the contribution of CTA1 subdomain structure to the thermal unfolding of CTA1. Biophysical measurements demonstrated that the CTA11 subdomain is thermally unstable, and that the CTA12 subdomain provides a degree of conformational stability to CTA11. The CTA13 subdomain does not affect the overall stability of CTA1, but the thermal unfolding of CTA1 appears to begin with a local loss of structure in the CTA13 subdomain: glycerol and acidic pH both inhibited the thermal disordering of full-length CTA1 but not the disordering of a CTA1 construct lacking the A13 subdomain. These observations provide mechanistic insight regarding the thermal unfolding of CTA1, an event which facilitates its subsequent translocation to the cytosol.
Cholera toxin (CT)1 is an AB protein toxin that contains an enzymatic A moiety and a cell-binding B moiety (1, 2). Many plant and bacterial toxins share this general structural organization. In the case of CT, the B subunit is a homopentameric ring-like structure that binds to GM1 gangliosides on the eukaryotic plasma membrane. The CTA subunit is initially produced as a single polypeptide of 240 amino acid residues, and it is then proteolytically cleaved at residue 192 to generate a disulfide-linked A1/A2 heterodimer. CTA1 is a 22 kDa ADP-ribosyltransferase with three distinct subdomains: CTA11 (residues 1 to 132) is the globular, compact catalytic core of the toxin; CTA12 (residues 133 to 161) wraps around the A11 subdomain and forms an extended bridge which links CTA11 with CTA13; and CTA13 (residues 162 to 192) is a globular region with many hydrophobic residues and the cysteine residue which covalently tethers CTA1 to CTA2 (3, 4). The 5.5 kDa CTA2 subunit consists of an α-helix that abuts the CTA1 subunit and an elongated C-terminal tail that sits within the central cavity of the B pentamer (5). CTA2 maintains numerous non-covalent interactions with CTB, thereby connecting the cell-binding and catalytic components of the toxin.
CT travels as an intact AB holotoxin from the cell surface to the endoplasmic reticulum (ER) (6). The disulfide bond linking CTA1 to CTA2 is reduced in the ER (7, 8), which then permits the chaperone-dependent dissociation of CTA1 from CTA2/CTB5 (9). CTA1 is kept in a folded conformation when associated with other components of the CT holotoxin (10–12), but it shifts to a partially unfolded state when released from CTA2/CTB5 at physiological temperature (11). This conformational change identifies the dissociated CTA1 subunit as a misfolded protein for processing by the quality control mechanism of ER-associated degradation (ERAD) (13). ERAD is a normal cellular process that recognizes misfolded or misassembled proteins in the ER and exports them to the cytosol for degradation by the ubiquitin-proteasome system (14). CTA1 evades ubiquitin-dependent proteasomal degradation after its translocation to the cytosol because the toxin has a dearth of lysine residues for ubiquitin conjugation (11, 15, 16). An interaction with ADP-ribosylation factors and possibly other cytosolic proteins allows the translocated pool of CTA1 to attain an ordered, active conformation which could further help it resist proteasomal degradation (11, 17, 18). The refolded CTA1 subunit subsequently modifies its Gsα target to elicit a cytopathic effect. Other AB toxins such as ricin and Shiga toxin also exploit ERAD for A chain translocation to the cytosol (19).
The thermal instability of the isolated CTA1 subunit plays an essential role in toxin translocation to the cytosol. However, the relationship between CTA1 subdomain structure and CTA1 thermal instability has not been examined. In addition, the process of CTA1 thermal unfolding remains uncharacterized. To address these issues, we analyzed the structural characteristics of CTA1, a CTA1 construct lacking most of the A13 subdomain (CTA11–168), and a CTA1 construct lacking both the A12 and A13 subdomains (CTA11–133). Our data indicate that the A11 subdomain alone is thermally unstable, that the A12 subdomain provides a degree of conformational stability to CTA11, and that the CTA13 subdomain does not affect overall toxin stability. Our data also suggest that the thermal unfolding of CTA1 begins with a localized loss of structure in the A13 subdomain. These results provide insight into the molecular mechanism of CTA1 unfolding, a key event for ERAD-mediated toxin translocation to the cytosol and cellular intoxication.
EXPERIMENTAL PROCEDURES
Structural studies
The temperature-dependent unfolding of CTA1 was studied with a J-810 spectrofluoropolarimeter (Jasco Corp., Tokyo, Japan) and a Jasco PFD-425S Peltier temperature controller. Experiments were performed with a 4-mm optical path-length rectangular quartz cuvette. The protein concentrations of His-tagged CTA1 constructs were 70 µg in 220 µL of 20 mM sodium borate buffer (pH 7.0) containing 100 mM NaCl. Protein concentration of the commercially available CTA1/CTA2 heterodimer (Calbiochem, La Jolla, CA) was 34 µg in 200 µL of 20 mM sodium phosphate buffer (pH 6.0 or 7.5) containing 150 mM NaCl and, to reduce the heterodimer, 10 mM β-mercaptoethanol. Thermal unfolding was carried out in the temperature range of 18–55°C, and samples were allowed to equilibrate for 4 min at each temperature before measurement. For fluorescence spectra, CTA1 tryptophan residues were excited at 290 nm and the fluorescence emission was measured between 300 and 400 nm. Circular dichroism (CD) spectra were recorded from 190 to 320 nm, which covers both near-UV and far-UV range, and were averaged from 5 scans. The same sample was used for all measurements to eliminate possible errors resulting from sample-to-sample variability. For CD measurements, the observed ellipticity was converted to mean residue molar ellipticity, [θ], in units of degrees×cm2×dmol−1 using
| (1) |
where θobs is the measured ellipticity in millidegrees, c is the molar concentration of the protein, nres is the number of amino acid residues in the protein, and l is the optical path-length in millimeters.
The thermal profiles of conformational changes in the protein, as determined by CD and fluorescence spectroscopy and plotted in Figures 2–5, were fitted with simulated curves using the following equation (22):
| (2) |
where X is the ellipticity or the fluorescence measured at a given temperature, fL is the fraction of amino acids representing the native conformation at low temperature, XL and XH are limiting values of X at low and high temperatures, respectively. The parameter, fL, is given by
| (3) |
where the temperature dependence of the free energy of unfolding (ΔG) is described by
| (4) |
T is the absolute temperature, Tm is the transition temperature (i.e., the midpoint of transition between folded and unfolded states), ΔH is the apparent enthalpy of unfolding, and ΔC is the heat capacity of unfolding. The parameter ΔC = 0.39 kcal mol−1K−1 was used, consistent with our earlier data (11). Values of ΔH that were used to obtain the best fit between experimental and simulated curves are listed in Supporting Information Table S1. The locations of CTA1 tryptophan and tyrosine residues, which impact fluorescence and near-UV CD measurements, are shown in Supporting Information Figure S1. Replicate experiments with CTA1 and CTA11–168 indicated that the Tm values did not deviate by more than 1°C.
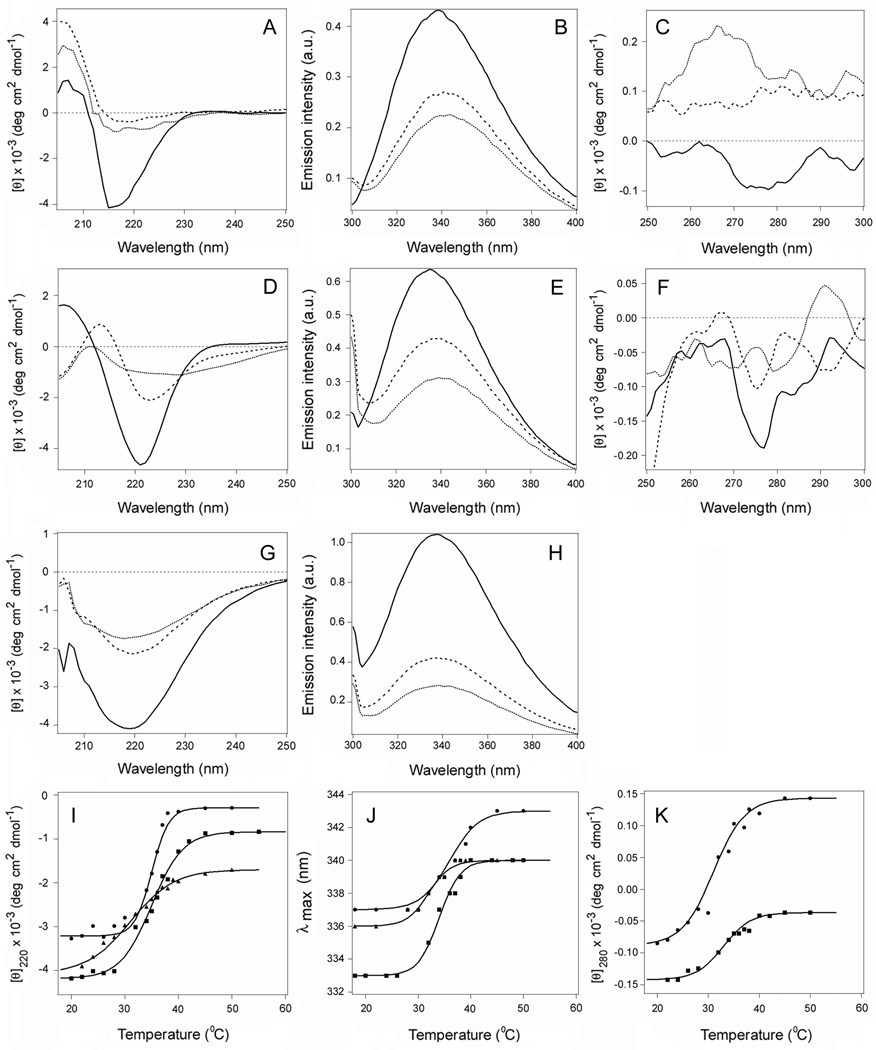
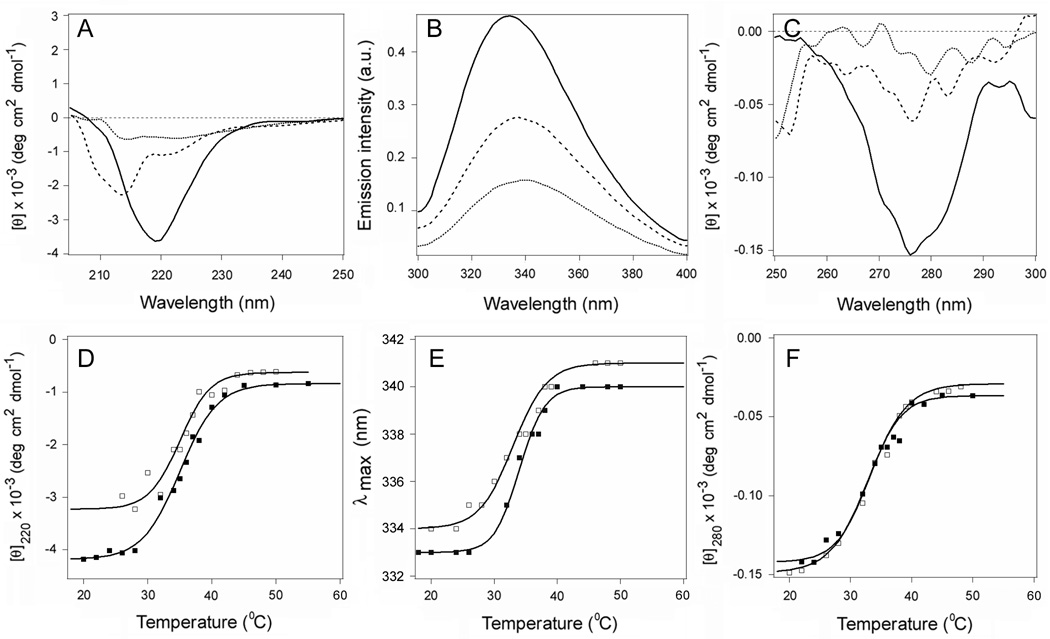
Figure 2.
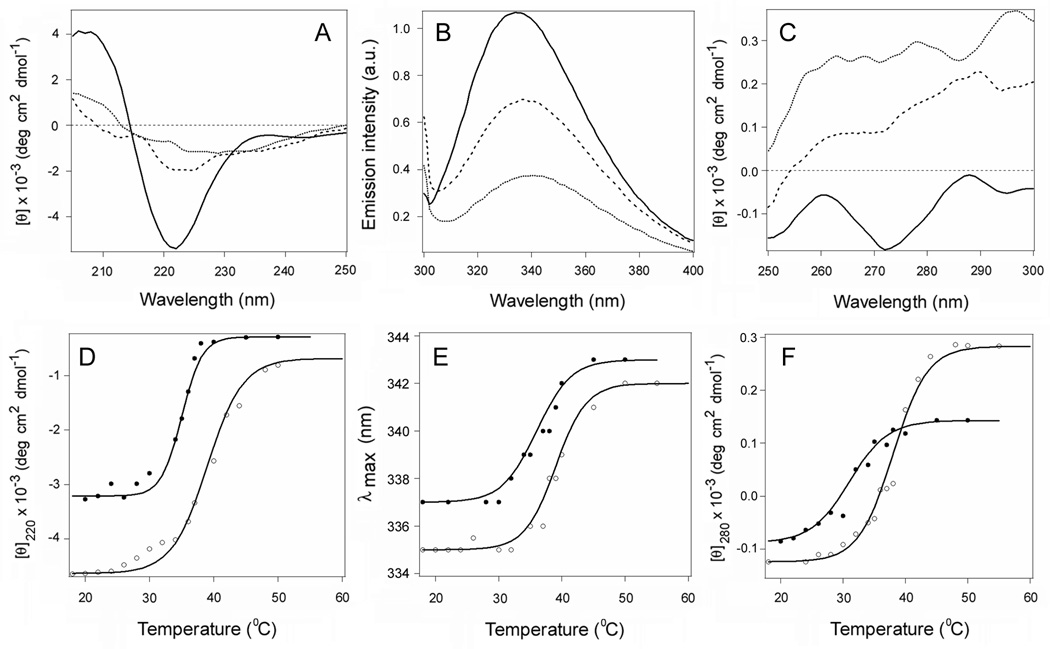
Temperature-induced unfolding of CTA1, CTA11–168, and CTA11–133. (A–H): The thermal denaturation of CTA1 (A–C), CTA11–168 (D–F), or CTA11–133 (G–H) was monitored by far-UV CD (A, D, G), fluorescence spectroscopy (B, E, H), and near-UV CD (C, F). Spectra recorded at 18°C (solid line), 37°C (dashed line), and 50°C (dotted line) are shown. The full spectral profiles obtained over a temperature range from 18°C to 55°C are shown in Supporting Information Figure S3. (I–K): Thermal unfolding profiles for CTA1 (circles), CTA11–168 (squares), and CTA11–133 (triangles) were derived from data collected during the change in temperature from 18°C to 55°C. The experimental data points were fitted with theoretical curves simulated through Eq. 2–4, using the Tm and ΔH parameters summarized in Tables 1 and S1. (I): For far-UV CD, the mean residue molar ellipticities at 220 nm ([θ]220) were plotted as a function of temperature. (J): For fluorescence spectroscopy, the maximum emission wavelengths were plotted as a function of temperature. (K): For near-UV CD analysis, the mean residue molar ellipticities at 280 nm ([θ]280) were plotted as a function of temperature.
Figure 5.
Temperature-induced unfolding of CTA11–168 at acidic pH. (A–C): The thermal denaturation of CTA11–168 pH 6.0 buffer was monitored by far-UV CD (A), fluorescence spectroscopy (B), and near-UV CD (C). Spectra recorded at 18°C (solid line), 37°C (dashed line), and 50°C (dotted line) are shown. The full spectral profiles obtained over a temperature range from 18°C to 55°C are shown in Supporting Information Figure S9. (D–F): Thermal unfolding profiles for CTA11–168 in pH 7.0 buffer (closed squares) or pH 6.0 buffer (open squares) were derived from the raw data presented in Supporting Information Figures S3D–F and S9, respectively. The experimental data points were fitted with theoretical curves simulated through Eq. 2–4, using the Tm and ΔH parameters summarized in Tables 1 and S1. (D): For far-UV CD, the mean residue molar ellipticities at 220 nm ([θ]220) were plotted as a function of temperature. (E): For fluorescence spectroscopy, the maximum emission wavelengths were plotted as a function of temperature. (F): For near-UV CD analysis, the mean residue molar ellipticities at 280 nm ([θ]280) were plotted as a function of temperature.
CTA1 constructs
The construction and purification of His-tagged CTA1 and CTA11–168 were previously described (20, 21). A clone expressing His-tagged CTA11–133 was made by PCR amplification from the full-length His-tagged CTA1 template in pT7CTA1h6 (21) with a T7 vector primer and TCAGAATTCCCAAAATGAACTCGATACC primer that appended an EcoRI site encoding Asn-Ser after residue Gly-133. An XbaI-EcoRI digest of this PCR fragment was cloned back into pT7CTA1h6 in place of the full-length CTA1 coding sequence. The predicted DNA sequence was confirmed. CTA11–133 encodes a protein ending with Gly-133 followed by the NSHHHHHH tag and was purified as previously described (20, 21). Although the presence of the His6 tag could affect the absolute Tm value and its pH dependence, each CTA1 construct had the same C-terminal His6 tag so experimental differences between them should not reflect the presence of the tag. The CTA1 preparations were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and visualized with Coomassie staining in order to confirm the purity of the protein preparations (Supporting Information Fig. S2).
In order to facilitate protein refolding, the purified toxins were dialyzed against five changes of 500 mL of 20 mM sodium borate buffer (pH 7.0) containing 100 mM NaCl before experimental use. The first step was for 2 h at 4°C in 6 M urea buffer; the second step was for 2 h at 4°C in 2 M urea buffer; the third step was for 2 h at 4°C in 1 M urea buffer; the fourth step was for 2 h at 4°C in urea-free buffer; and the last step was again for 2 h at 4°C in urea-free buffer. Slide-a-lyzer 3500 MWCO dialysis cups (Pierce, Rockford, IL) were used for the procedure. After dialysis, the His-tagged toxins were used immediately.
RESULTS
Organization and stability of the CTA1 subdomains
The subdomain structure of CTA1 is presented in Figure 1. The C-terminal A13 subdomain of CTA1 was originally thought to serve as the trigger for ERAD-mediated toxin translocation to the cytosol, but a CTA11–169 construct2 lacking most of the A13 subdomain could still move from the ER to the cytosol (21). This observation was consistent with a model of toxin translocation in which the ERAD system is activated by a global loss of structure in the isolated CTA1 subunit (11, 20, 21). We accordingly predicted that the CTA11–168 construct would, like the full-length construct, be thermally unstable. Biophysical measurements involving far-UV CD, fluorescence spectroscopy, and near-UV CD were conducted on CTA1 and CTA11–168 to test this prediction (Fig. 2 and Supporting Information Fig. S3). The loss of toxin structure with increasing temperature occurred for both full-length CTA1 (Figs. 2A–C) and CTA11–168 (Figs. 2D–F). Temperature-dependent changes to the toxin secondary structure were characterized with a Tm of 36°C for CTA1 and a Tm of 35°C CTA11–168; temperature dependencies of the peak fluorescence wavelength (λmax) were characterized with a Tm of 35.5°C for CTA1 and a Tm of 35°C for CTA11–168; and temperature-induced changes to the toxin tertiary structure were characterized with a Tm of 32°C for CTA1 and a Tm of 32.5°C for CTA11–168 (Table 1). The similar transition temperatures of CTA1 and CTA11–168 suggested that truncation of the CTA13 subdomain had little effect on the thermal stability of CTA1.
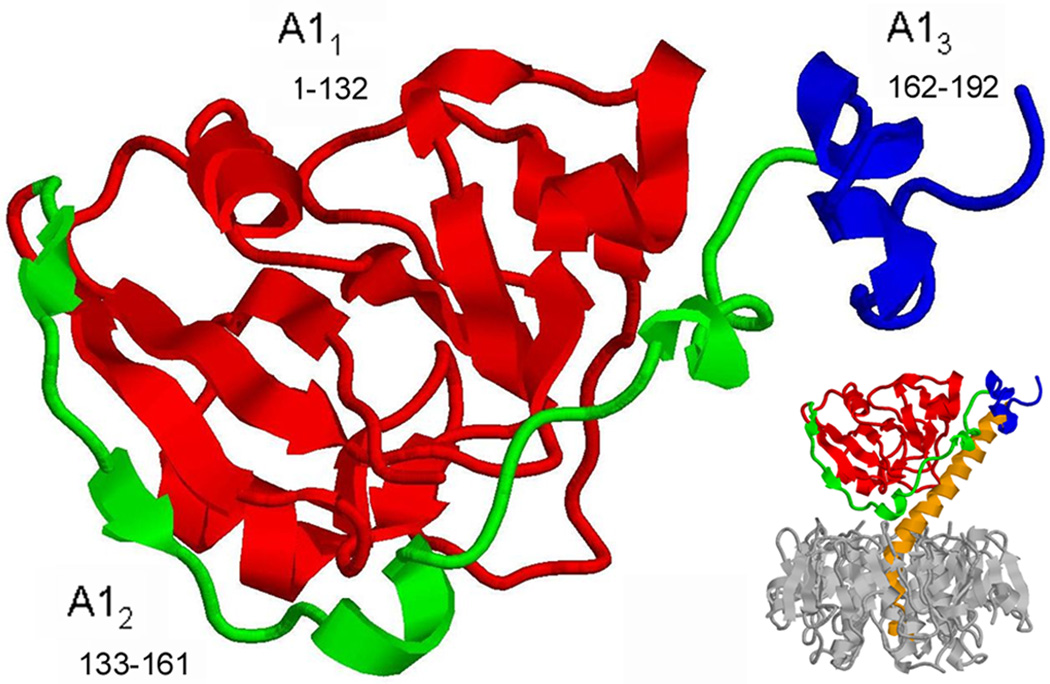
Figure 1.
Ribbon diagram of the CTA1 subunit. The presented structural model of the isolated CTA1 subunit is based upon the X-ray crystal structure of CT as determined by O'Neal et al. (Protein Data Bank entry 1S5F) (4). The subdomains, as described by Zhang et al. (3), are highlighted in different colors: the A11 subdomain is in red, the A12 subdomain is in green, and the A13 subdomain is in blue. Inset shows the orientation of CTA1 within the holotoxin, with CTA2 shown in orange and the B pentamer in grey. The images were generated using RasMol.
Table 1.
Thermal stabilities of CTA1 and the CTA1 deletion constructs.a
| Tm (°C) | ||||
|---|---|---|---|---|
| far-UV CD | λmax | near-UV CD | ||
| CTA1 | 36 | 35.5 | 32 | |
| CTA1 pH 6.0 | 39 | 37 | 38 | |
| CTA11–168 | 35 | 35 | 32.5 | |
| CTA11–168 + glycerol | 35 | 33 | 33 | |
| CTA11–168 pH 6.0 | 35 | 34 | 32 | |
| CTA11–133 | 31 | 32 | -- | |
It should be noted that the CD spectra of CTA11–168 exhibited a significantly weaker ππ* transition intensity between 200 and 210 nm and red-shifted nπ* bands compared to the full-length CTA1 molecule (Supporting Information Fig. S3). A relatively stronger ππ* signal is interpreted in terms of more twisted β-strands (23), and red-shifted nπ* bands indicate less polar environment (24). Our data therefore suggest that truncation of the CTA13 subdomain unwinds the twisted β-strands in the molecule, possibly accompanied with a tightening of its tertiary structure. The latter would cause a blue shift in tryptophan fluorescence spectra, which is indeed confirmed by the Trp fluorescence data which indicates a 3–4 nm blue-shifted spectra of CTA11–168 compared to those of CTA1 (Figs. 2B,E,J).
The near-UV CD spectra of CTA1 in the 250–300 nm region are generated by Tyr and Trp residues: CTA1 has no intramolecular disulfide bonds, and Phe side chains generate weaker bands at lower wavelengths (24, 25). Full-length CTA1 demonstrates positive bands around 260 and 290 nm, most likely due to the Lb transitions of both Tyr and Trp residues, intervened with a broad negative band around 275 nm that can be assigned to the La transitions (Fig. 2C and Supporting Information Fig. S3C) (25). In the spectrum of CTA11–168, the negative (La) band becomes more pronounced (Fig. 2F and Supporting Information Fig. S3F), which may reflect conformational changes and the microenvironment of the aromatic amino acid side chains.
To determine the contribution of the A12 subdomain to CTA1 stability, additional biophysical measurements were performed on a CTA11–133 construct which lacked both the A12 and A13 subdomains (Figs. 2G–H and Supporting Information Figs. S3G–H). Near-UV CD measurements could not be accurately recorded for this construct. However, far-UV CD and fluorescence spectroscopy clearly demonstrated that CTA11–133 was less stable than either full-length CTA1 or CTA11–168 (Figs. 2I–J). CTA11–133 displayed a secondary structure Tm of 31°C, and the temperature dependence of its λmax exhibited a Tm of 32°C (Table 1). These Tm values were, respectively, 5°C and 3.5°C lower than the corresponding Tm values for full-length CTA1. Thus, the A12 subdomain provides a degree of conformational stability to the catalytic A11 subdomain of CTA1. In addition, the CD spectra of CTA11–133 showed even weaker ππ* bands in comparison to CTA11–168 (Supporting Information Fig. S3). This may indicate a further relaxation of the β-strand twist in CTA11–133 and a larger spectral contribution from its α-helices.
CTA1 unfolding begins at the C-terminal subdomain
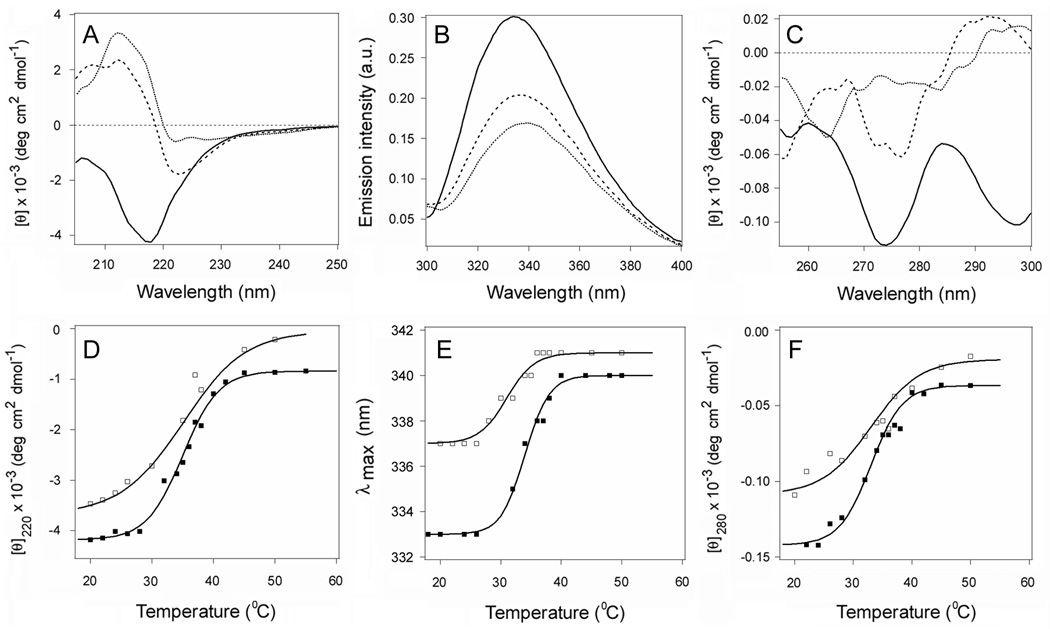
NMR measurements have shown that the C-terminus of CTA1 is partially unfolded even at 15°C (17). This suggests the overall loss of CTA1 structure that occurs with increasing temperature may begin with further unfolding of the A13 subdomain. To examine this possibility, we monitored the thermal unfolding of CTA11–168 in the presence of 10% glycerol (Fig. 3 and Supporting Information Fig. S4). Previous work has shown that glycerol blocks the thermal disordering of tertiary structure in the full-length CTA1 subunit (20). If the temperature-induced unfolding of CTA1 was initiated at its C-terminus, then glycerol would likely prevent the further loss of structure in the CTA13 subdomain. Truncation of the CTA13 subdomain would consequently eliminate the stabilizing effect of glycerol on the overall structure of CTA1. Consistent with this prediction, we found that CTA11–168 exhibited similar Tm values in either the absence or presence of 10% glycerol as determined by near-UV, far-UV CD, and fluorescence techniques (Table 1). The secondary and tertiary structural stability of CTA11–168 was therefore unaffected by glycerol treatment. In contrast, as determined by near-UV CD, exposure to glycerol resulted in a 5.5°C increase in the tertiary structure Tm for full-length CTA1 (20). It thus appears that the stabilizing effect of glycerol on CTA1 structure is mediated through an effect on the A13 subdomain. This observation further suggests that unfolding of the C-terminus precedes the global loss of CTA1 tertiary structure.
Figure 3.
Temperature-induced unfolding of CTA11–168 in the presence of 10% glycerol. (A–C): The thermal denaturation of CTA11–168 in the presence of 10% glycerol was monitored by far-UV CD (A), fluorescence spectroscopy (B), and near-UV CD (C). Spectra recorded at 18°C (solid line), 37°C (dashed line), and 50°C (dotted line) are shown. The full spectral profiles obtained over a temperature range from 18°C to 55°C are shown in Supporting Information Figure S4. (D–F): Thermal unfolding profiles for CTA11–168 in the absence (closed squares) or presence (open squares) of 10% glycerol were derived from the raw data presented in Supporting Information Figures S3D–F and S4, respectively. The experimental data points were fitted with theoretical curves simulated through Eq. 2–4, using the Tm and ΔH parameters summarized in Tables 1 and S1. (D): For far-UV CD, the mean residue molar ellipticities at 220 nm ([θ]220) were plotted as a function of temperature. (E): For fluorescence spectroscopy, the maximum emission wavelengths were plotted as a function of temperature. (F): For near-UV CD analysis, the mean residue molar ellipticities at 280 nm ([θ]280) were plotted as a function of temperature.
Acid-induced stabilization of CTA1
To further examine the processive unfolding of CTA1, we used a second experimental condition to stabilize the toxin structure. It has been reported that acidic pH prevents the thermal unfolding of ricin A chain (26). Furthermore, intoxication with either ricin or Shiga toxin is blocked at acidic pH because of an apparent inhibition of toxin export from the ER to the cytosol (27–30). Although the molecular basis of this effect was not determined, we hypothesized that acidic pH had a stabilizing effect on A chain structure which consequently disrupted toxin-ERAD interactions and A chain translocation to the cytosol. We therefore predicted that acidic pH would inhibit the thermal unfolding of CTA1.
CD and fluorescence spectroscopy were used to examine the effect of acidic pH on temperature-induced changes to the structure of CTA1 (Fig. 4 and Supporting Information Fig. S5). At pH 6.0, the thermal unfolding of CTA1 was characterized with a secondary structure Tm of 39°C, a λmax Tm of 37°C, and a tertiary structure Tm of 38°C (Table 1). Thus, at acidic pH the Tm for CTA1 tertiary structure shifted to a higher temperature by 6°C. The thermal disordering of tertiary structure in a reduced CTA1/CTA2 heterodimer was also inhibited at pH 6.0 (Supporting Information Fig. S6, Table S2). Furthermore, cells bathed at acidic pH could deliver the CT holotoxin from the cell surface to the ER but could not export reduced CTA1 from the ER to the cytosol (Supporting Information Figs. S7–S8). These results, which were previously observed for glycerol-treated cells (20), strongly suggest that ERAD-mediated toxin translocation is triggered by a loss of CTA1 tertiary structure.
Figure 4.
Temperature-induced unfolding of CTA1 at acidic pH. (A–C): The thermal denaturation of CTA1 in pH 6.0 buffer was monitored by far-UV CD (A), fluorescence spectroscopy (B), and near-UV CD (C). Spectra recorded at 18°C (solid line), 37°C (dashed line), and 50°C (dotted line) are shown. The full spectral profiles obtained over a temperature range from 18°C to 55°C are shown in Supporting Information Figure S5. (D–F): Thermal unfolding profiles for CTA1 in pH 7.0 buffer (closed circles) or pH 6.0 buffer (open circles) were derived from the raw data presented in Supporting Information Figures S3A–C and S5, respectively. The experimental data points were fitted with theoretical curves simulated through Eq. 2–4, using the Tm and ΔH parameters summarized in Tables 1 and S1. (D): For far-UV CD, the mean residue molar ellipticities at 220 nm ([θ]220) were plotted as a function of temperature. (E): For fluorescence spectroscopy, the maximum emission wavelengths were plotted as a function of temperature. (F): For near-UV CD analysis, the mean residue molar ellipticities at 280 nm ([θ]280) were plotted as a function of temperature.
Acid-induced stabilization of CTA1 involves an effect on the C-terminal subdomain
Although acidic pH inhibited the thermal disordering of tertiary structure in full-length CTA1, it did not alter the thermal stability of CTA11–168 (Fig. 5 and Supporting Information Fig. S9). Both the tertiary and secondary structures of CTA11–168 exhibited nearly identical Tm values at pH 7.0 and pH 6.0, contrasting with the 6°C increase in tertiary structure Tm for full-length CTA1 at pH 6.0 as measured by near-UV CD (Table 1). The stabilizing effect of acidic pH on CTA1 tertiary structure thus appeared to involve the C-terminal A13 subdomain. This observation was consistent with the data derived from glycerol-treated toxin samples and again suggested that the thermal unfolding of CTA1 begins with a local loss of structure in the A13 subdomain.
DISCUSSION
In order to access the host cell cytosol, CTA1 undergoes what has been described as an order-disorder-order transition (17). The ordered CTA1 subunit travels from the cell surface to the ER as part of an intact CT holotoxin. In the ER, chaperone-assisted dissociation of reduced CTA1 from CTA2/CTB5 releases the structural constraints on CTA1 unfolding and allows CTA1 to assume a disordered conformation which triggers its ERAD-mediated export to the cytosol (9, 11, 20). The translocated pool of CTA1 then interacts with cytosolic ADP-ribosylation factors and possibly other host factors to regain an active, ordered conformation (11, 17). In this work, we examined the molecular details of CTA1 unfolding and identified the contribution of CTA1 subdomains to thermal disordering of the toxin.
A previous NMR study reported that a CTA1 construct lacking the C-terminal A13 subdomain retained the same core structure found in the full-length toxin (17). Likewise, CTA11–168/169 retained some enzymatic and biological activity (21). CTA11–168 also retained the same thermal stability as full-length CTA1 (Fig. 2, Table 1), which explains why the CTA11–169 construct was in a translocation-competent state (21). However, our data indicated that CTA11–133 was less stable than full-length CTA1: truncation of both the A12 and A13 subdomains reduced the secondary structure Tm by 5°C and the λmax Tm by 3.5°C (Fig. 2, Table 1). This destabilization was not seen when only the A13 subdomain was removed from CTA1. Accordingly, we conclude that the A12 subdomain, which wraps around one face of the A11 subdomain, provides a degree of conformational stability to the catalytic core of the toxin.
As previously reported (11, 20), the isolated CTA1 subunit was in a partially unfolded conformation at the physiological temperature of 37°C. The thermal disordering of CTA1 tertiary structure preceded the thermal perturbation of CTA1 secondary structure under control conditions. However, the thermal disordering of CTA1 tertiary structure was shifted to higher temperatures and occurred in tandem with the temperature-induced loss of CTA1 secondary structure when toxin samples were exposed to 10% glycerol (20) or pH 6.0 buffer (Fig. 4, Fig. S6, Table 1, Table S2). These effects were recorded with full-length CTA1 but not CTA11–168: neither glycerol nor acidic pH altered the thermal unfolding of CTA11–168 (Fig. 3, Fig. 5, Table 1). Collectively, these data indicated that the stabilizing effect of glycerol and acidic pH on CTA1 structure is mediated through an effect on the A13 subdomain. As an inhibition of unfolding in the A13 subdomain prevented the temperature-induced loss of structure in other regions of CTA1, it appears that the thermal unfolding of CTA1 begins with a localized loss of structure in the C-terminal domain. This event occurs after CTA1 dissociation from the holotoxin and may result, in part, from the loss of potentially stabilizing contacts between the A13 subdomain and CTA2.
The A13 subdomain itself was not required for toxin translocation to the cytosol (21). ERAD thus recognizes an unfolded region of CTA1 in the A11 and/or A12 subdomain(s). However, unfolding of the CTA13 subdomain preceded the loss of structure in other regions of CTA1. Forced stabilization of the CTA13 subdomain accordingly inhibited thermal unfolding of the CTA11 and CTA12 subdomains. This, in turn, blocked toxin-ERAD interactions and toxin translocation to the cytosol. These effects were first noted with glycerol (20) and were confirmed here with acidic pH, a second independent method of stabilizing CTA1 through an effect on the A13 subdomain.
We could not directly correlate the acid-induced inhibition of CTA1 translocation to an acid-induced inhibition of CT intoxication. Previous work has shown that experimentally induced alkalinization of the endosomes does not inhibit CT intoxication of cultured cells, but the effect of acidic conditions on CT intoxication was not examined (31). Here, initial control experiments found that cells incubated in acidified medium do not generate elevated levels of intracellular cAMP in response to treatment with forskolin, an agonist of adenlyate cyclase (data not shown). Since the ADP-ribosylation of Gsα by CTA1 leads to the activation of adenylate cyclase and high levels of intracellular cAMP, the acid-induced inhibition of adenylate cyclase activity prevented us from assessing the effect of acidic pH on CT intoxication. However, previous studies have shown that a block of toxin export from the ER to the cytosol will have a corresponding block on cellular intoxication (20, 32, 33).
Technical limitations also prevented us from monitoring the ER-to-cytosol translocation of CTA11–168 in cells exposed to 10% glycerol or acidic pH. Our data demonstrate that glycerol and acidic pH do not inhibit the thermal unfolding of CTA11–168, so glycerol and acidic pH would not be expected to block the ERAD-mediated translocation of CTA11–168. These potential experiments require a CT holotoxin containing the truncated CTA11–168 construct, but the cysteine residue linking CTA1 to CTA2/CTB5 is missing from CTA11–168. Thus, it is not possible to incorporate CTA11–168 into a CT holotoxin.
Thermal instability in the catalytic CTA1 subunit appears to identify the toxin as a misfolded protein for ERAD-mediated translocation to the cytosol. To elucidate the molecular details of this process, we have used glycerol and acidic pH as experimental conditions to alter the thermal unfolding of various CTA1 constructs. These conditions would not be found in the ER, but they were instead used to disrupt the normal biological process of CTA1 unfolding. With this approach we have provided mechanistic insight into the molecular mechanism of CTA1 unfolding, a key event for ERAD-mediated toxin translocation to the cytosol and cellular intoxication.
We previously suggested that the thermal stabilization of CTA1 may represent a novel therapeutic strategy (20). Here, we have more precisely identified the C-terminal A13 subdomain as a specific site where thermal unfolding of the CTA1 molecule is initiated. Specific stabilization of the A13 subdomain thus represents a possible therapeutic target. Inhibition of unfolding in the A13 subdomain prevents the temperature-induced loss of structure in other regions of CTA1. The stabilized conformation of CTA1 is not recognized as an ERAD substrate, which consequently prevents toxin translocation to the cytosol and productive intoxication. Thus, it should be possible to block the in vivo activity of CT with structure-based inhibitors of A13 unfolding.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health grants R03 AI067987 to K. Teter, R01 AI073783 to K. Teter, and R01 AI031940 to R.K. Holmes.
Abbreviations: CT, cholera toxin; CTA1, the catalytic A1 subunit of CT; CD, circular dichroism; ER, endoplasmic reticulum; ERAD, ER-associated degradation; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; Tm, transition temperature; λmax, peak fluorescence wavelength
In previous work (21), two variants of the A13 deletion construct were generated: His-tagged CTA11–168 was placed in a prokaryotic expression vector for protein purification, and CTA11–169 was placed in a eukaryotic expression vector for direct, plasmid-borne expression in the ER of transfected cultured cells.
SUPPORTING INFORMATION AVAILABLE
A table of ΔH values, a table of unfolding data for reduced CTA1/CTA2, locations of CTA1 tryptophan and tyrosine residues in ribbon diagrams and space-filling models, assessment of protein purification by SDS-PAGE, the full series of His6-tagged CTA1 biophysical measurements from 18°C to 55°C, the thermal unfolding of a reduced CTA1/CTA2 heterodimer at pH 6.0 and 7.5, and experiments demonstrating the cellular effects resulting from acid-induced stabilization to the tertiary structure of CTA1 are provided as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.De Haan L, Hirst TR. Cholera toxin: a paradigm for multi-functional engagement of cellular mechanisms (Review) Mol. Membr. Biol. 2004;21:77–92. doi: 10.1080/09687680410001663267. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez J, Holmgren J. Cholera toxin structure, gene regulation and pathophysiological and immunological aspects. Cell. Mol. Life Sci. 2008;65:1347–1360. doi: 10.1007/s00018-008-7496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang RG, Scott DL, Westbrook ML, Nance S, Spangler BD, Shipley GG, Westbrook EM. The three-dimensional crystal structure of cholera toxin. J. Mol. Biol. 1995;251:563–573. doi: 10.1006/jmbi.1995.0456. [DOI] [PubMed] [Google Scholar]
- 4.O'Neal CJ, Jobling MG, Holmes RK, Hol WG. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science. 2005;309:1093–1096. doi: 10.1126/science.1113398. [DOI] [PubMed] [Google Scholar]
- 5.O'Neal CJ, Amaya EI, Jobling MG, Holmes RK, Hol WG. Crystal structures of an intrinsically active cholera toxin mutant yield insight into the toxin activation mechanism. Biochemistry. 2004;43:3772–3782. doi: 10.1021/bi0360152. [DOI] [PubMed] [Google Scholar]
- 6.Lencer WI, Tsai B. The intracellular voyage of cholera toxin: going retro. Trends Biochem. Sci. 2003;28:639–645. doi: 10.1016/j.tibs.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Majoul I, Ferrari D, Soling HD. Reduction of protein disulfide bonds in an oxidizing environment. The disulfide bridge of cholera toxin A-subunit is reduced in the endoplasmic reticulum. FEBS Lett. 1997;401:104–108. doi: 10.1016/s0014-5793(96)01447-0. [DOI] [PubMed] [Google Scholar]
- 8.Orlandi PA. Protein-disulfide isomerase-mediated reduction of the A subunit of cholera toxin in a human intestinal cell line. J. Biol. Chem. 1997;272:4591–4599. [PubMed] [Google Scholar]
- 9.Tsai B, Rodighiero C, Lencer WI, Rapoport TA. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- 10.Goins B, Freire E. Thermal stability and intersubunit interactions of cholera toxin in solution and in association with its cell-surface receptor ganglioside GM1. Biochemistry. 1988;27:2046–2052. doi: 10.1021/bi00406a035. [DOI] [PubMed] [Google Scholar]
- 11.Pande AH, Scaglione P, Taylor M, Nemec KN, Tuthill S, Moe D, Holmes RK, Tatulian SA, Teter K. Conformational instability of the cholera toxin A1 polypeptide. J. Mol. Biol. 2007;374:1114–1128. doi: 10.1016/j.jmb.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surewicz WK, Leddy JJ, Mantsch HH. Structure, stability, and receptor interaction of cholera toxin as studied by Fourier-transform infrared spectroscopy. Biochemistry. 1990;29:8106–8111. doi: 10.1021/bi00487a017. [DOI] [PubMed] [Google Scholar]
- 13.Teter K, Allyn RL, Jobling MG, Holmes RK. Transfer of the cholera toxin A1 polypeptide from the endoplasmic reticulum to the cytosol is a rapid process facilitated by the endoplasmic reticulum-associated degradation pathway. Infect. Immun. 2002;70:6166–6171. doi: 10.1128/IAI.70.11.6166-6171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazes B, Read RJ. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry. 1997;36:11051–11054. doi: 10.1021/bi971383p. [DOI] [PubMed] [Google Scholar]
- 16.Rodighiero C, Tsai B, Rapoport TA, Lencer WI. Role of ubiquitination in retro-translocation of cholera toxin and escape of cytosolic degradation. EMBO Rep. 2002;3:1222–1227. doi: 10.1093/embo-reports/kvf239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ampapathi RS, Creath AL, Lou DI, Craft JW, Jr, Blanke SR, Legge GB. Order-disorder-order transitions mediate the activation of cholera toxin. J. Mol. Biol. 2008;377:748–760. doi: 10.1016/j.jmb.2007.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wernick NL, De Luca H, Kam WR, Lencer WI. N-terminal extension of the cholera toxin A1-chain causes rapid degradation after retrotranslocation from endoplasmic reticulum to cytosol. J. Biol. Chem. 2010;285:6145–6152. doi: 10.1074/jbc.M109.062067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lord JM, Roberts LM, Lencer WI. Entry of protein toxins into mammalian cells by crossing the endoplasmic reticulum membrane: co-opting basic mechanisms of endoplasmic reticulum-associated degradation. Curr. Top. Microbiol. Immunol. 2005;300:149–168. doi: 10.1007/3-540-28007-3_7. [DOI] [PubMed] [Google Scholar]
- 20.Massey S, Banerjee T, Pande AH, Taylor M, Tatulian SA, Teter K. Stabilization of the tertiary structure of the cholera toxin A1 subunit inhibits toxin dislocation and cellular intoxication. J. Mol. Biol. 2009;393:1083–1096. doi: 10.1016/j.jmb.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teter K, Jobling MG, Sentz D, Holmes RK. The cholera toxin A13 subdomain is essential for interaction with ADP-ribosylation factor 6 and full toxic activity but is not required for translocation from the endoplasmic reticulum to the cytosol. Infect. Immun. 2006;74:2259–2267. doi: 10.1128/IAI.74.4.2259-2267.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavigne P, Crump MP, Gagne SM, Hodges RS, Kay CM, Sykes BD. Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper. J. Mol. Biol. 1998;281:165–181. doi: 10.1006/jmbi.1998.1914. [DOI] [PubMed] [Google Scholar]
- 23.Woody RW. Theory of circular dichroism of proteins. In: Fasman GD, editor. Circular Dichroism and the Conformational Analysis of Biomolecules. New York and London: Plenum Press; 1996. pp. 25–67. [Google Scholar]
- 24.Sreerama N, Woody RW. Circular dichroism of peptides and proteins. In: Berova N, Nakanishi K, Woody RW, editors. Circular Dichroism: Principles and Applications. Hoboken, NJ: John Wiley & Sons, Inc.; 2000. pp. 601–620. [Google Scholar]
- 25.Woody RW, Dunker AK. Aromatic and cysteine side-chain circular dichroism in proteins. In: Fasman GD, editor. Circular Dichroism and the Conformational Analysis of Biomolecules. New York and London: Plenum Press; 1996. pp. 109–157. [Google Scholar]
- 26.Argent RH, Parrott AM, Day PJ, Roberts LM, Stockley PG, Lord JM, Radford SE. Ribosome-mediated folding of partially unfolded ricin A-chain. J. Biol. Chem. 2000;275:9263–9269. doi: 10.1074/jbc.275.13.9263. [DOI] [PubMed] [Google Scholar]
- 27.Sandvig K, Olsnes S. Entry of the toxic proteins abrin, modeccin, ricin, and diphtheria toxin into cells. II. Effect of pH, metabolic inhibitors, and ionophores and evidence for toxin penetration from endocytotic vesicles. J. Biol. Chem. 1982;257:7504–7513. [PubMed] [Google Scholar]
- 28.Sandvig K, Olsnes S, Brown JE, Petersen OW, van Deurs B. Endocytosis from coated pits of Shiga toxin: a glycolipid-binding protein from Shigella dysenteriae 1. J. Cell Biol. 1989;108:1331–1343. doi: 10.1083/jcb.108.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eiklid K, Olsnes S. Entry of Shigella dysenteriae toxin into HeLa cells. Infect. Immun. 1983;42:771–777. doi: 10.1128/iai.42.2.771-777.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wesche J, Rapak A, Olsnes S. Dependence of ricin toxicity on translocation of the toxin A-chain from the endoplasmic reticulum to the cytosol. J. Biol. Chem. 1999;274:34443–34449. doi: 10.1074/jbc.274.48.34443. [DOI] [PubMed] [Google Scholar]
- 31.Lencer WI, Strohmeier G, Moe S, Carlson SL, Constable CT, Madara JL. Signal transduction by cholera toxin: processing in vesicular compartments does not require acidification. Am. J. Physiol. 1995;269:G548–G557. doi: 10.1152/ajpgi.1995.269.4.G548. [DOI] [PubMed] [Google Scholar]
- 32.Teter K, Holmes RK. Inhibition of endoplasmic reticulum-associated degradation in CHO cells resistant to cholera toxin, Pseudomonas aeruginosa exotoxin A, and ricin. Infect. Immun. 2002;70:6172–6179. doi: 10.1128/IAI.70.11.6172-6179.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernardi KM, Forster ML, Lencer WI, Tsai B. Derlin-1 facilitates the retro-translocation of cholera toxin. Mol. Biol. Cell. 2008;19:877–884. doi: 10.1091/mbc.E07-08-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.