Abstract
Intellectual disability (ID), also referred to as mental retardation (MR), is frequently the result of genetic mutation. Where ID is present together with additional clinical symptoms or physical anomalies, there is often sufficient information available for the diagnosing physician to identify a known syndrome, which may then educe the identification of the causative defect. However, where co-morbid features are absent, narrowing down a specific gene can only be done by ‘brute force’ using the latest molecular genetic techniques. Here we attempt to provide a systematic review of genetic causes of cases of ID where no other symptoms or co-morbid features are present, or non-syndromic ID. We attempt to summarize commonalities between the genes and the molecular pathways of their encoded proteins. Since ID is a common feature of autism, and conversely autistic features are frequently present in individuals with ID, we also look at possible overlaps in genetic etiology with non-syndromic ID.
Keywords: Intellectual disability, Non-syndromic, Genetic basis, Convergent pathways
Introduction
Definition and prevalence of intellectual disability
Intellectual disability (ID) is a common neurodevelopmental disorder that is characterized by an intelligence quotient (IQ) of 70 or below, and deficits in at least two behaviors related to adaptive functioning diagnosed by 18 years of age (American Psychiatric Association 2000). The prevalence of ID is between 1% and 3% (Roeleveld et al. 1997; Leonard and Wen 2002) and is present in every social class and culture (Leonard and Wen 2002). Despite its universal occurrence, there tends to be higher prevalence of ID in areas of lower socioeconomic status and developing countries, particularly for mild cases (Drews et al. 1995; Roeleveld et al. 1997; Durkin et al. 1998; Durkin 2002; Emerson 2007). It has been suggested that this discrepancy is likely due to environmental factors (Roeleveld et al. 1997; Durkin et al. 1998; Emerson 2007).
Approximately 30% more males are diagnosed with ID than females (American Psychiatric Association 2000; McLaren and Bryson 1987). However, despite a higher ratio of males to females among milder cases of ID, the ratio decreases as IQ decreases (American Psychiatric Association 2000; McLaren and Bryson 1987). Some studies suggest that severe ID may be more prevalent among females (Katusic et al. 1996; Bradley et al. 2002), however these studies were performed in quite specific communities, and may not necessarily be generalizable to other regions.
Classification of ID by IQ and syndromic vs. non-syndromic
ID is divided into 5 categories based on IQ: Mild, moderate, severe, profound and unable to classify (DSM IV). However, epidemiological studies often use a simplified classification, grouping their subjects into mild ID (IQ50-70) and severe ID (IQ<50) (Ropers and Hamel 2005). While the prevalence of severe ID is relatively stable, the prevalence of mild ID is variable and often depends heavily on external environmental factors such as level of maternal education, access to education/opportunity and access to healthcare (Leonard and Wen 2002; Drews et al. 1995; Roeleveld et al. 1997). Study design, age of subjects, and the catchment population for the studies may also contribute to the variability seen across mild ID prevalence studies (Leonard and Wen 2002; Drews et al. 1995; Roeleveld et al. 1997).
In addition to categorization by severity/IQ level, ID can also be grouped into syndromic intellectual disability (S-ID) and non-syndromic intellectual disability (NS-ID). In S-ID, patients present with one or multiple clinical features or co-morbidities in addition to ID. While S-ID has a clear definition, there is debate over the classification of NS-ID. Traditionally, NS-ID has been defined by the presence of intellectual disability as the sole clinical feature. However, it has been a challenge to rule out the presence of more subtle neurological anomalies and psychiatric disorders in these patients, as they may be less apparent, or difficult to diagnose due to the cognitive impairment. Additionally, symptoms of some syndromes may be so subtle that they are extremely difficult to diagnose unless the features are looked for specifically in the context of a known genetic defect previously associated with these features (Ropers 2006). Thus the distinction between S-ID and NS-ID is often blurred.
Causes of ID
ID can be caused by environmental and/or genetic factors. However, for up to 60% of cases, there is no identifiable cause (Rauch et al. 2006). Environmental exposure to certain teratogens, viruses or radiation can cause ID, as can severe head trauma or injury causing lack of oxygen to the brain. While these factors explain some cases of NS-ID, it is also important to consider genetic etiology.
Genetic causes of ID are thought to be present in 25–50% of cases, although this number increases proportionally with severity (McLaren and Bryson 1987). Chromosomal abnormalities have been reported in ID, with a broad range of prevalence, and many different types of aberrations have been identified (Rauch et al. 2006). Autosomal trisomies that are compatible with human viability and aneuploidies of the X-chromosome almost always result in some degree of ID as part of a syndrome, as illustrated by trisomy 21, or Down Syndrome—the most common genetic form of ID (Rauch et al. 2006). Additionally, pathogenic copy number variants (CNV) have been found to be associated with ID in a large number of studies, and will likely contribute to the discovery of many ID causing genes in the future (Ropers and Hamel 2005; Zahir and Friedman 2007).
Over the past 15 years many single gene causes of NS-ID have been identified. Many of these NS-ID genes may also cause S-ID, autism or other neurodevelopmental phenotypes, making it likely that other genetic modifiers or environmental factors may be involved in disease etiology. It also stresses the importance of detailed genotype/phenotype comparisons, which are often difficult to elucidate. There is also a possibility that some instances of NS-ID are multifactorial, with more than one gene contributing to disease in an individual, however this has not been well studied. Most known NS-ID genes are on the X-chromosome, however the number of autosomal genes associated with NS-ID is growing rapidly (Chelly et al. 2006).
Relevance of studying NS-ID genetics
Lionel Penrose was quoted as wishing to see each ID individual “as an integral part of the human race in its struggle for evolution and survival, unwittingly yielding information of the greatest value in the progressive understanding of the biological structure of the whole group” (from Berg 1998). As Penrose implied, the identification of the biological causes of NS-ID is necessary to our understanding of human cognition and intellect. Because NS-ID presents with intellectual impairment as the only feature, genes that cause it are likely related to the processes of learning and memory. These processes are fundamental to our understanding of the formation of normal intellectual capabilities, and in particular how intellect develops from a neurological perspective. Additionally, finding genes that cause NS-ID might help us to decipher relevant pathways that are involved in neurological development. Understanding these pathways may aid us in treating or relieving symptoms of NS-ID in certain cases. Knowledge of pathways involved in NS-ID will also make it easier to select candidate genes to analyze in research and clinically based studies. Understanding the genetics of a complex disease like NS-ID is also relevant to genetic counseling in families with affected individuals, particularly where consanguinity is involved (Modell and Darr 2002).
Identifying genes that cause NS-ID: methodology and obstacles
Homozygosity mapping
Given the innate heterogeneity of the NS-ID phenotypes, it is of little surprise that its genetics are equally as complex. It is thought that rare variants are likely responsible for most cases of NS-ID, and this has been the focus of NS-ID genetics for some time, although association studies are still used to identify polymorphisms that potentially contribute to the phenotype (e.g. SNAP25; Gosso et al. 2008). It is likely that many rare variants across many genes result in the same phenotype. Under the rare variants model of causation, large consanguineous families are particularly useful. Rare mutations, which may be identified in such families, can provide us with information about the types of etiological aberrations in NS-ID as well as the genes and relevant pathways that may be essential for normal neuronal functioning.
Our increasing ability to perform high throughput analyses of genotype using microarrays has been a significant contributor to the increased discovery rate of NS-ID genes over the past 10 years (Lugtenberg et al. 2007). All of the genes identified for autosomal recessive NS-ID thus far have been identified through microarray technology combined with homozygosity mapping using large consanguineous families. In these studies, large multiplex families are obtained and detailed family histories are taken. Affected family members are typically assessed for additional clinical phenotypes that may suggest S-ID, and are screened for more commonly known causes such as fragile-X mutations and gross chromosomal anomalies. DNA from the blood of affected family members, along with one or two unaffected family members, is analyzed using microarray technology and analysis software.
This method allows for a fast screen of individuals genotypes, and allows the researcher to identify regions of the genome that are homozygous for the same alleles among all affected individuals. Typically (or at least ideally), a single long stretch of DNA (often over 3 Mb), is identified, and the disease causing mutation would lie within this region. Use of consanguineous multiplex families allows the identification of these linkage regions with relative ease. The regions are then confirmed by microsatellite analysis, and sequencing of genes within the region will often lead to the identification of a disease gene.
Consanguinity is marriage between closely related individuals, and is common in many countries around the world, particularly in the Middle East and Asia. In Pakistan—a country of 173 million people—consanguineous unions make up 62.7% of marriages, and ~80% of which are between first cousins, according to a national census (Hussain and Bittles 1998). Other countries that also have high prevalence of consanguinity include Iran, where the overall prevalence is 40%, and India where prevalence ranges from 16% to 33% depending on the region (Hussain and Bittles 2000; Najmabadi et al. 2007).
Multiply affected consanguineous families have been integral in determining autosomal recessive causes of disease. The children of consanguineous individuals will have more homozygous DNA than the offspring of an outbred marriage. This leads to an increased likelihood of rare, recessive disease-causing variants being inherited from both parents. This is known as autozygosity or homozygosity-by-descent (HBD), which occurs when a rare allelic variant is passed down to offspring from a common ancestor via both maternal and paternal lineages. In populations where consanguinity is prevalent, there is a significant increase in infant morbidity and mortality (Modell and Darr 2002; Gustavson 2005; Khlat and Khoury 1991). One study showed that, in consanguineous populations, there were 10 times more recessively inherited congenital conditions, many of which resulted in early death (reported from the Birmingham Birth Study, as reviewed in Modell and Darr 2002).
Advantages and disadvantages of homozygosity mapping
There are several reasons why homozygosity mapping has been a successful approach for identifying rare genetic variants. Intuitively, the co-segregation of large stretches of homozygous alleles only in affected family members in a consanguineous family increases the likelihood that the region contains a gene that is relevant to the phenotype, and statistics in the literature support this concept (Lander and Botstein 1987). The method has been successfully utilized to identify recessive causes of many diseases such as Joubert syndrome, Charcot-Marie-Tooth syndrome, syndromic deafness, and oligodontia (Noor et al. 2008; Bolino et al. 2000; Senderek et al. 2003; Verpy et al. 2001; Noor et al. 2009).
A major drawback is the scarcity of families in outbred western populations that are suitable for this method. Also, a lack of appropriate clinical assessment tools and infrastructure may pose problems in some of the countries where consanguineous families are more common. Aside from the lack of large pedigrees, there are further obstacles in identifying the gene of interest. Homozygosity mapping studies, just like linkage studies, often identify very large regions in which the disease gene may be found. Some of these regions contain hundreds of genes, and selecting relevant candidate genes can be problematic. Often, there is no clear candidate gene, or there are many genes in the region with no known function and thus selection of suitable candidate genes is tricky. Using Next Generation Sequencing (NGS) might be an effective way to overcome this, however innate variation within the genome will likely result in the identification of multiple genetic aberrations in several genes that need to be ruled out as disease genes. For instance, in one of the few reports so far on identification of a recessive gene (DHODH, for Miller syndrome) through exome sequencing, many variants were identified, however, after filtering for known SNPs, just 9 candidate genes remained (Ng et al. 2010). For application in consanguineous families, this approach should be simpler, as one would expect that the exome sequence data would also be able to indicate regions of the genome containing large stretches of homozygosity, so the candidate genes could be narrowed down further to such a region.
Sequencing candidate genes
The sequencing of candidate genes has previously been employed in the identification of many XLMR genes. The X-chromosome has been a target in many studies looking for causes of NS-ID because of the high male to female ratio in the NS-ID population. The result is that most of the known NS-ID genes are X-linked. Often, large cohorts of individuals with ID are collected and target genes that have a suspected role in development or brain function are sequenced across the cohort. Recently, a re-sequencing of the X-chromosome exome in a large cohort has led to the identification of several novel NS-ID genes (Tarpey et al. 2009).
Typical genetic mapping strategies for autosomal dominant disorders have been unsuccessful for NS-ID, due to genetic heterogeneity and lack of suitable multiplex families, as procreation of affected individuals is unlikely. Thus, autosomal dominant NS-ID is likely to be sporadic, resulting from de novo mutations. Sequencing candidate genes may be the best approach for identifying autosomal dominant causes of NS-ID. SYNGAP1, STXBP1, and SHANK3 were all identified as autosomal dominant causes of NS-ID using candidate gene sequencing (Hamdan et al. 2009a, b; Michaud et al. 2009 Abstract).
While this approach can be effective, it may also require much work, and will frequently be unsuccessful if the level of genetic heterogeneity is as high as anticipated. However, as our knowledge of biological pathways involved in ID grows, our ability to select probable candidates will increase, and this strategy may become more plausible. Additionally, with improved technology such as NGS, techniques, sequencing entire exomes for causes of ID will become a less laborious and more productive screening method. NGS is an emerging method that produces large amounts of sequencing data quickly, at relatively low cost. With this technology, it is possible to sequence through large linkage regions, which can contain hundreds of genes, with much less labor and in shorter time than traditional sequencing. As well as application to critical linkage regions, NGS can be applied to HBD regions, or whole genome sequence analysis. It can also be applied in a more directed (and thus more economical) approach, by whole exome sequencing. This involves sequencing just the coding portion of the genome—approximately 1% of our genetic material. This method may be useful for identifying genes when no linkage or susceptibility region is known, as most disease causing mutations are likely to occur in exons.
Although NGS and exome sequencing present us with many opportunities for high-throughput analysis of whole genomes, some issues with the method have arisen. One major concern is that there are likely to be many false positives—there are thousands of common variants in the human genome, many of which have not been well documented. This is exemplified in the Tarpey et al. (Tarpey et al. 2009) re-sequencing of the X-chromosome exome. In this particular case, although standard rather than next-generation sequencing was used, Tarpey et al. (Tarpey et al. 2009) screened virtually the entire X-chromosome exome in a large cohort for genes causing ID. It was found that truncation of 1% or more of X-chromosome genes may still be compatible with normal phenotype (Tarpey et al. 2009). There are many examples of nonsense mutations that occur in the healthy population, where the resulting haploinsufficiency is presumably compensated for by other means (Tarpey et al. 2009). This demonstrates the need for caution when interpreting results of large quantities of sequencing data, such as that obtained by NGS and exome sequencing studies. For instance, in the whole genome sequencing of a patient with Charcot-Marie-Tooth neuropathy, ~9,000 synonymous and ~9,000 non-synonymous coding changes were identified, including 121 nonsense mutations (Lupski et al. 2010). (Also see comment in previous section on identification of the DHODH gene for Miller syndrome through exome sequencing; Ng et al. 2010).
Characterization of chromosomal aberrations
Characterization of chromosomal aberrations by breakpoint analysis has long been used as a method to identify autosomal dominant disease causing genes. Determining the exact location of the breakpoints and study of disrupted genes has led to the discovery of several candidate genes for non-syndromic autosomal dominant ID (NS-ADID). DOCK8, MBD5, CDH15 and KIRREL3 were all identified at chromosomal breakpoints of either gross chromosomal abnormalities or CNVs (Griggs et al. 2008; Bhalla et al. 2008; Wagenstaller et al. 2007). For MBD5, CDH15 and KIRREL3, further screening of these genes in cohorts of ID patients led to the identification of several mutations, indicating the utility of this method in identifying disease genes (Bhalla et al. 2008; Wagenstaller et al. 2007). X-linked NS-ID genes have also been found using this method. TSPAN7 was identified at the breakpoint of a translocation in one individual, and then screening of an NS-ID cohort led to the identification of mutations in TSPAN7 in 2/33 families (Zemni et al. 2000).
Subtelomeric rearrangements and copy number variants
Copy number variants (CNV), which in the past few years have been found to contribute significantly to common genetic variation, also confer susceptibility to numerous diseases. Although CNVs cover approximately 12% of the entire genome in the normal population, rare CNVs have been associated with various neuropsychiatric disorders such as schizophrenia and autism (Redon et al. 2006; Marshall et al. 2008; Stefansson et al. 2008), and have also been implicated in ID (Zahir and Friedman 2007; Knight et al. 1999)
Submicroscopic subtelomeric rearrangements have been long implicated in the etiology of NS-ID. These rearrangements include deletions as well as balanced translocations and other chromosomal aberrations that are unable to be seen under the microscope. Subtelomeric regions are frequently a focus for analysis of chromosomal rearrangements, as the density of genes in this region is greater than in the rest of the genome (Saccone et al. 1992), and about half of all segmental aneusomies involve subtelomeric and terminal regions of chromosomes (Biesecker 2002). Subtelomeric rearrangements have been shown to be a significant cause of ID in many chromosomal studies and are thought to be responsible for 3–6% of cases (Knight et al. 1999; Ledbetter and Martin 2007). In a 2002 meta-analysis of studies of subtelomeric abnormalities in ID cohorts, it was confirmed that this type of genetic aberration is present in ~6% of cases, and that ~50% of them are inherited (Biesecker 2002). These finding were quite exciting because of the large number of cases with subtelomeric abnormalities, and indicated that analysis of subtelomeric regions could identify the molecular cause of ID in many idiopathic individuals, as well as leading to more effective clinical testing. With the advent of CGH microarrays, identification of submicroscopic subtelomeric rearrangements became more robust and more clinically feasible (Stankiewicz and Beaudet 2007).
Currently, deletions in subtelomeric regions are also picked up by CNV analysis—a type of analysis that has become essential for determining the etiology of NS-ID. With the increasing availability and sensitivity of microarray technology, pathogenic CNVs have been consistently detected in 10–15% of individuals with ID across many studies (Fan et al. 2007; Koolen et al. 2009; McMullan et al. 2009) (as reviewed by Zahir and Friedman 2007). These CNVs include both rare de novo, as well as rare inherited mutations, which are still of unknown significance (but may yet be important, since rare inherited subtelomeric rearrangements have also been seen frequently; McMullan et al. 2009).
As a demonstration of the successful application of CNVs for identifying novel NS-ID genes, very recently, SHANK2 was identified as an NS-ID and autism disease gene, through the identification of CNVs that deleted SHANK2 coding regions in two affected individuals. Subsequent screening by sequencing in large cohorts identified further NS-ID and autism patients with SHANK2 mutations (Berkel et al. 2010). Additionally, in the largest study of its kind to date, SHANK2, SYNGAP1 and ILRAPL1 CNVs were identified in autism probands, illustrating the importance of CNV analysis and the identification of genetic overlap between autism and NS-ID (Pinto et al. 2010). CNV analysis will clearly play an important role in the identification of further ID-related genes. A great number of pathogenic CNVs have been identified in NS-ID in recent years, and mapping these breakpoints will likely be a successful method in determining autosomal dominant as well as X-linked causes of ID, just as mapping breakpoints of cytogenetic abnormalities has in the past (Ropers and Hamel 2005).
Non-syndromic X-linked intellectual disability (or mental retardation; NS-XLMR)
The X-chromosome has historically been the most thoroughly studied chromosome with regard to NS-ID due to the high male to female ratio. There are approximately 40 genes known to cause NS-ID, and ~80% of these reside on the X-chromosome. Some of these genes cause both S-ID and NS-ID, depending on the mutation, or may even vary within families, possibly modulated by additional factors. In the case of ATRX (MIM: 300032) the same mutation in the same family led to all family members having characteristic facial features, except for one who had NS-ID (Guerrini et al. 2000). This gene has been shown to cause several other ID syndromes, for which different mutations have been identified (Yntema et al. 2002; Gibbons et al. 2003; Howard et al. 2004).
Several other genes that classically cause syndromes may also cause NS-XLMR. MECP2 (MIM: 300005), which causes Rett syndrome, has been identified in a number of NS-ID cases (Orrico et al. 2000; Couvert et al. 2001; Dotti et al. 2002). Previously thought to be lethal in males, an MECP2 missense mutation was found to cause severe intellectual disability in males, and a much milder phenotype in females (Dotti et al. 2002). These findings suggest that there may be a quantifiable genotype/phenotype correlation for certain mutations. Notably, in Rett syndrome, some studies have demonstrated a genotype/phenotype correlation in terms of severity, as well as for specific phenotypic measures (Bebbington et al. 2008; Ham et al. 2005).
ARX (MIM: 300382) is one of the most frequently mutated genes in XLMR. It encodes a transcription factor, responsible for both gene repression and activation, that is essential for normal development of the CNS. Mutations in this gene are responsible for causing 7 distinct but overlapping ID-related phenotypes, including NS-ID (Friocourt et al. 2006). One mutation in this gene, a 24bp in-frame insertion leading to a lengthening of the polyalanine tract, is found to be associated with at least four of these phenotypes (Sherr 2003; Gecz et al. 2006). The reason for this striking pleiotropy is currently unknown but could be essential for understanding the role ARX in ID.
In all of these cases, where mutations of the same gene cause a variable phenotype, it may be important to assess if there are other factors are involved in phenotype expression. Although the gene clearly causes the disease phenotype, genetic background, epigenetic factors and other modifier genes or environmental factors could alter how the phenotype manifests. It might be useful to analyze these families’ genomes at much higher resolution in order to reveal other genetic factors that contribute to the heterogeneous phenotype.
A number of X-linked genes cause NS-ID as the predominant phenotype, and several of these are relatively common in comparison to most NS-ID genes, which have typically been identified in only one individual or family. For instance, mutations in JARID1C (MIM: 314690) are relatively common in NS-ID. It is likely involved in REST-mediated transcriptional regulation and chromatin remodeling (Tahiliani et al. 2007; Christensen et al. 2007). Other X-linked genes that regulate gene expression such as PQBP1 (MIM: 300463), MECP2, ATRX, and several zinc finger genes are less common causes of XLMR (Ropers 2006; Gecz et al. 2009). SCL6A8 (MIM: 300036) is another relatively common cause of XLMR (Salomons et al. 2001; Hahn et al. 2002; Clark et al. 2006). It encodes a creatine transporter, and mutations result in creatine deficiency (Hahn et al. 2002). Creatine deficiency has been suggested to be a relatively common cause of NS-ID (Lion-Francois et al. 2006). Despite the biochemical effects of mutations in SLC6A8, it is commonly accepted to be an NS-ID gene, as the outward phenotypic manifestation of mutations is NS-ID.
As previously mentioned, the X-chromosome has been highly scrutinized in the search for genes for NS-ID (McLaren and Bryson 1987). However, reviews of X-linked genes in ID have postulated that genes on the X-chromosome likely account for only 10–12% of the genes involved in genetic cases of ID (Ropers and Hamel 2005; Mandel and Chelly 2004; Kleefstra and Hamel 2005). Therefore genes on the X-chromosome appear to account for some, but not all, of the increased male prevalence (the X-chromosome contains ~4% of the genes in the genome). It is probable that X-linked genes are not the sole reason for gender differences, and that other factors, both genetic and non-genetic, might influence the sex ratio. These could include environmental influences or increased penetrance of certain autosomal mutations in males.
Non-syndromic autosomal dominant intellectual disability (NS-ADID)
Autosomal inheritance is a plausible mechanism for many NS-ID cases, and in recent years it has become the subject of intensive study. In contrast to XLMR genes, which contain both missense and nonsense mutations relatively frequently, the NS autosomal dominant and recessive genes identified so far appear to have mostly truncating mutations (see Table 1). For reasons discussed previously, only a few genes have been found that cause NS-ADID. MIM uses the acronym MRD for loci for “mental retardation, autosomal dominant”. For MRD1 (MIM 156200), the methyl binding domain 5 gene, MBD5 (MIM 611472) on 2q23.1 was identified by SNP microarray analysis, indicating a 200Kb de novo deletion, removing at least 6 exons of the gene in a female proband with sandal-toe and epilepsy but no facial dysmorphic features (Wagenstaller et al. 2007). An additional 4 S-ID probands with MBD5 missense mutations were also identified in this study.
Table 1.
Details of known NS-ID genes, as well as S-ID genes where allelic variants cause NS-ID, including the types of mutations found to cause disease and mode of inheritance
| Gene Name | OMIM Ref | X-linked/Autosomal (MR/ID) | Cytogenetic Banda | Phenotypic Expression* | Mutation Type | Reference |
|---|---|---|---|---|---|---|
| ACSL4 | 300157 | X-linked (MRX63 & 68) | Xq22.3 | Males: MR | Missense, splice site | (Meloni et al. 2002) |
| Females: Variable | ||||||
| AFF2/FMR2 | 300806 | X-linked | Xq28 | Dominantb | 5′ GGC repeat amplification leading to methylation | (Gecz et al. 1996) |
| AGTR2 | 300034 | X-linked (MRX88) | Xq23 | Recessive | Missense, Truncating | (Vervoort et al. 2002) |
| AP1S2 | 300629 | X-linked (MRX59) | Xp22.2 | Recessive | Truncating | (Tarpey et al. 2006) |
| ARHGEF6 | 300267 | X-linked (MRX46) | Xq26.3 | Recessive | Splice site | (Kutsche et al. 2000) |
| ARX | 300382 | X-linked | Xp21.3 | Males: MR; Females: Low-normal IQ | In frame insertions/deletions, missense | (Bienvenu et al. 2002; Stromme et al. 2002a, b; Troester et al. 2007) |
| ATRX | 300032 | X-linked | Xq21.1 | Recessive | Missense | (Yntema et al. 2002) |
| BRWD3 | 300553 | X-linked (MRX93) | Xq21.1 | Recessive | Missense, frame-shift | (Field et al. 2007) |
| CASK | 300172 | X-linked | Xp11.4 | Recessive | Missense | (Tarpey et al. 2009; Hackett et al. 2009) |
| CC2D1A | 610055 | Autosomal (MRT3) | 19p13.12 | Recessive | Truncating | (Basel-Vanagaite et al. 2006) |
| CDH15 | 114019 | Autosomal (MRD3) | 16q24.3 | Dominant | Missense | (Bhalla et al. 2008) |
| CRBN | 609262 | Autosomal (MRT2) | 3p26.3 | Recessive | Truncating | (Higgins et al. 2004) |
| DLG3 | 300189 | X-linked (MRX90) | Xq13.1 | Males: MR | Truncating | (Tarpey et al. 2004) |
| Females: Variable | ||||||
| DOCK8 | 611432 | Autosomal (MRD2) | 9p24.3 | Dominant | Deletion, translocation | (Griggs et al. 2008) |
| FGD1 | 305400 | X-linked | Xp11.22 | Recessive | Missense | (Lebel et al. 2002) |
| FTSJ1 | 300499 | X-linked (MRX9 & 44) | Xp11.23 | Recessive | Truncating, missense | (Freude et al. 2004) |
| GDI1 | 300104 | X-linked (MRX41 & 48) | Xq28 | Dominantb | Truncating, missense | (D’Adamo et al. 1998; (Bienvenu et al. 1998) |
| GRIK2 | 138244 | Autosomal (MRT6) | 6q21 | Recessive | Truncating | (Motazacker et al. 2007) |
| HUWE1 | 300697 | X-linked (MRXS-Turner & MRX17) | Xp11.22 | Recessive | Missense, microduplication | (Froyen et al. 2008) |
| IL1RAPL1 | 300206 | X-linked (MRX21) | Xp21.3–p21.2 | Recessive | Truncating, deletion | (Carrie et al. 1999; Nawara et al. 2008) |
| JARID1C (KDM5C) | 314690 | X-linked | Xp11.22 | Recessive | Truncating, missense | (Jensen et al. 2005) |
| KIRREL3 | 607761 | Autosomal (MRD4) | 11q24.2 | Dominant | Missense | (Bhalla et al. 2008) |
| MAGT1 | 300715 | X-linked (MRX95) | Xq21.1 | Dominant | Missense | (Molinari et al. 2008) |
| MBD5 | 611472 | Autosomal (MRD1) | 2q23.1 | Dominant | Deletion, missense | (Wagenstaller et al. 2007) |
| MECP2 | 300005 | X-linked (MRXS13) | Xq28 | Dominant | Truncating, missense, deletion | (Orrico et al. 2000) |
| NLGN4 | 300427 | X-linked (AUTSX2) | Xp22.31–p22.32 | Recessive | Truncating | (Laumonnier et al. 2004) |
| OPHN1 | 300127 | X-linked | Xq12 | Recessive | Truncating, deletion | (Billuart et al. 1998; Philip et al. 2003) |
| PAK3 | 300142 | X-linked (MRX30) | Xq22.3 | Recessive | Truncating, missense, splice site | (Allen et al. 1998; Rejab et al. 2008) |
| PQBP1 | 300463 | X-linked | Xp11.23 | Recessive | Missense | (Kalscheuer et al. 2003) |
| PRSS12 | 606709 | Autosomal (MRT1) | 4q26 | Recessive | Truncating | (Molinari et al. 2002) |
| PTCHD1 | X-linked | Recessive | Deletion | (Noor et al. in press) | ||
| RPS6KA3 | 300075 | X-linked (MRX19) | Xp22.12 | Recessive | Missense | (Merienne et al. 1999) |
| SHANK2 | 603290 | Autosomal | 11q13.3–13.4 | Dominant | CNV deletion, missense | (Berkel et al. 2010) |
| SHROOM4 | 300579 | X-linked | Xp11.22 | Dominant | Translocation; missense | (Stocco dos Santos et al. 2003; Hagens et al. 2006) |
| SLC6A8 | 300036 | X-linked | Xq28 | Dominantb | Truncating, missense | (Salomons et al. 2001; Hahn et al. 2002) |
| STXBP1 | 602926 | Autosomal | 9q34.11 | Dominant | Deletion, nonsense | (Hamdan et al. 2009a, b) |
| SYNGAP1 | 603384 | Autosomal (MRD5) | 6p21.32 | Dominant | Truncating | (Hamdan et al. 2009a, b) |
| SYP | 313475 | X-linked | Xp11.23 | Recessive | Truncating, missense | (Tarpey et al. 2009) |
| TSPAN7 | 300096 | X-linked (MRX58) | Xp11.4 | Recessive | Truncating, missense | (Zemni et al. 2000) |
| TRAPPC9 | 611966 | Autosomal (MRT13) | 8q24.3 | Recessive | Truncating | (Mir et al. 2009 |
| TUSC3 | 601385 | Autosomal (MRT7) | 8p22 | Recessive | Truncating | (Garshasbi et al. 2008; Molinari et al. 2008) |
| UPF3B | 300298 | X-linked (MRXS14) | Xq24 | Recessive | Truncating | (Tarpey et al. 2007) |
| ZNF41 | 314995 | X-linked (MRX89) | Xp11.3 | Variable | Missense, splice site | (Shoichet et al. 2003) |
| ZNF674 | 300573 | X-linked (MRX92) | Xp11.3 | Recessive | Truncating | (Lugtenberg et al. 2006 |
| ZNF711 | 314990 | X-linked | Xq21.1 | Recessive | Truncating | (Tarpey et al. 2009) |
| ZNF81 | 314998 | X-linked (MRX45) | Xp11.23 | Recessive | Missense | (Kleefstra et al. 2004) |
*In X-linked genes, male and female values reflect the effects of one variant
aCytoband reported in UCSC Genome Browser
bless severe in females
The gene for MRD2, on 9p24, dedicator of cytokinesis 8 (DOCK8; MIM 611432) was identified in 2 unrelated patients, by mapping breakpoints of a deletion and translocation respectively (Griggs et al. 2008).
Combining the mapping of translocations and CNV breakpoints with candidate gene sequencing has been an important method for identifying ADID genes. The MRD3 and MRD4 genes, CDH15 (MIM: 114019) and KIRREL3 (MIM: 607761) respectively, both were mapped to the chromosomal breakpoint of an individual with a balanced t(11;16)(q24.2;q24) translocation presenting with severe ID and several other dysmorphisms (Bhalla et al. 2008). The two genes, both encoding putative cell-adhesion proteins, were then sequenced in 600 ID patients, and several missense mutations not found in the general population were identified—4 in CDH15 and 3 in KIRREL3 (Bhalla et al. 2008). Some of the patients had additional clinical features, while others were NS-ID cases. This confirms the utility of chromosomal breakpoints for identifying candidate genes for ADID. It is also possible that additive effects of mutations in more than one ID gene may be involved in some instances, however evidence to support this is lacking, and additive gene effects are poorly understood. CDH15 is a cadherin gene that is involved in intercellular adhesion and is strongly expressed in cerebellum (Bhalla et al. 2008). KIRREL3 encodes a protein of unknown function, although it has been shown to co-localize and interact with CASK, an NS-ID-associated synaptic protein, and is expressed in adult and fetal brain (Bhalla et al. 2008). It also shares structural aspects with IL1RAPL1, one of the well-established X-linked NS-ID genes. Both contain several IG-like domains, an IGC2 domain, and a transmembrane region.
SYNGAP1 (MIM: 603384; MRD5), encoding SynGAP, a component of the NMDA-receptor (NMDAR) complex, was initially screened because mouse models carrying heterozygous mutations in Syngap were observed to have impaired learning and synaptic plasticity, along with defects in LTP (Hamdan et al. 2009a, b; Kim et al. 2003; Komiyama et al. 2002). The homozygous mutation in Syngap is post-natally lethal, suggesting that SYNGAP1 may be a plausible candidate for ADID in humans (Kim et al. 2003). Sequencing of the gene in a cohort of NS-ID probands led to the discovery of 3 unrelated individuals with heterozygous mutations in SYNGAP1 (Hamdan et al. 2009a, b). Two were nonsense and one was a frame shift leading to truncation (Hamdan et al. 2009a, b). NMDARs play a role in glutamate-activated excitation of postsynaptic neurons, and have been implicated in memory formation and synaptic plasticity. This gene has also been implicated in autism in a large-scale CNV analysis in which one proband was found to have a CNV loss overlapping the entire gene (Pinto et al. 2010).
Additionally, mutations in SHANK2 (MIM: 603290) have been reported in cases of autism and NS-ID. Four unrelated individuals with NS-ID and 7 unrelated individuals with autism have either CNV deletions or sequence mutations in SHANK2 (Berkel et al. 2010). One of the NS-ID patients had a de novo CNV deletion overlapping exon 7 of the gene, and the other three all have inherited missense mutations, one of which is also found in an unrelated autism proband (Berkel et al. 2010). Two of the individuals presenting with NS-ID in this study also show autistic features, but do not meet the autism diagnosis criteria. Mutations in SHANK2 on 11q13 were identified by CNV analysis with subsequent gene sequencing in a large autism and NS-ID cohort (Berkel et al. 2010). SHANK proteins are scaffolding proteins that are highly abundant at the post-synaptic density. Recent work has shown that HOMER and SHANK form a mesh-like matrix that creates a framework for structure and protein assembly at the post-synaptic density, and may be important for synaptic plasticity (Hayashi et al. 2009).
Non-syndromic autosomal recessive intellectual disability (NS-ARID)
MIM uses the acronym MRT for “mental retardation, autosomal recessive”. To date, only 6 MRT genes have been published. Only two of the NS-ARID genes published to date have been identified in more than one family. The first of these to be identified was TUSC3 (MIM: 601385), for MRT7, which encodes a protein that is likely involved in catalyzing the transfer of a 14-sugar oligosaccharide from dolichol to nascent protein, an essential step in N-linked protein glycosylation (Molinari et al. 2008; Garshasbi et al. 2008). A recent study has also demonstrated that TUSC3 is necessary for Mg2+ regulation, and knockdown of this gene causes decreased total and free intracellular Mg2+ in human cell lines, as well as arrested or abnormal development in zebrafish embryos (Zhou and Clapham 2009).
The second NS-ARID gene to be identified in more than one family is TRAPPC9 (MIM: 611966), for MRT13, which encodes a protein called NIBP. TRAPPC9 mutations have been found in 4 unrelated families from different parts of the world (Mir et al. 2009; Philippe et al. 2009; Mochida et al. 2009). NIBP directly interacts with NIK and IKKβ, which results in the activation the NF-κB pathway (Hu et al. 2005). It has been shown to be involved in axonal outgrowth in vitro, and may be involved in neuronal cell survival (Hu et al. 2005). Interestingly, the same truncating allelic variant segregates in two families from two different countries (Mir et al. 2009; Mochida et al. 2009). It is possible that this represents a historic variant passed down through many generations, and may represent a relatively common cause of NS-ARID, although this requires further investigation. It is also possible that the two families are distantly related although this is thought unlikely due to the distance between Pakistan and Israel, and the limited transmigration of the populations involved.
All other published NS-ARID genes have been identified in only one family. PRSS12 (MIM: 606709), also known as neurotrypsin, was the first of such genes to be identified (Molinari et al. 2002). It encodes a trypsin-like serine protease, which is expressed in the embryo, and is likely involved in synapse maturation and neural plasticity (Molinari et al. 2002; Gschwend et al. 1997; Wolfer et al. 2001). It functions in the proteolytic cleavage of agrin at the synapse, which requires postsynaptic NMDAR activation (Matsumoto-Miyai et al. 2009; Stephan et al. 2008; Reif et al. 2007). The result of mutation in these individuals is moderate to severe intellectual disability (Molinari et al. 2002).
CRBN (MIM: 609262) encodes the ATP-dependant Lon protease cereblon that is directly involved in assembly and surface expression of large-conductance Ca2+-activated K+ channels, which function in the control of neuronal excitability and transmitter release (Higgins et al. 2004; Jo et al. 2005). Mutations in CRBN appear to disturb the development of large-conductance Ca2+-activated K+ channels, which causes increased intracellular Ca2+ sensitivity and results in faster activation, and slower deactivation kinetics (Higgins et al. 2008). Nonsense mutations in this gene result in mild ID.
CC2D1A (MIM: 610055) codes for a protein that is a calcium-regulated transcriptional repressor and is a putative candidate for regulation of the NF-κB pathway (Basel-Vanagaite et al. 2006; Matsuda et al. 2003). CC2D1A, also known as Freud-1 (five prime repressor under dual repression binding protein-1), is also believed to regulate transcriptional repression of the serotonin 1A receptor gene HTR1A, and the dopamine receptor DRD2 gene (Ou et al. 2003; Rogaeva et al. 2007).
Nonsense mutations in GRIK2 (MIM: 138244) also cause NS-ARID (Motazacker et al. 2007). GRIK2 encodes a protein called GLuR6, which is a subunit of a kainate receptor (KAR). KARs are ionotropic glutamate receptors which respond to the excitatory neurotransmitter l-glutamate, similar to NMDA or AMPA receptors. They are highly expressed in the brain, particularly in the hippocampal mossy fibers, where GLuR6 has been found to modulate long-term potentiation (LTP) in mouse models (Bortolotto et al. 1999; Contractor et al. 2001). GLuR6 knockout mice show decreased LTP in mossy fibers; a phenotype which could be rescued by application of low levels of K+ indicating that KARs induce LTP via depolarization of the pre-synaptic terminal (Contractor et al. 2001; Schmitz et al. 2003). LTP in the hippocampus has been implicated as a mechanism for memory formation and learning (Bliss and Collingridge 1993; Fedulov et al. 2007).
Although there are only 6 known genes that segregate with NS-ARID, a further 8 loci have been identified through HBD mapping which are likely to lead to more gene discoveries in the near future (Najmabadi et al. 2007; Garshasbi et al. 2009 Abstract; Uyguner et al. 2007; Rafiq et al. 2010).
Three additional genes have been suggested as NS-ARID genes and reported at recent genetics meetings, but have yet to be published. Two of these genes were found to have missense mutations, which would represent the first instances of missense mutations causing autosomal NS-ID (Najmabadi et al. 2009 Abstract; Moheb et al. 2009 Abstract). One of these is the ZNF526 gene, which encodes a C2H2 zinc finger protein that is expressed in the brain (Moheb et al. 2009 Abstract). This mutation has been identified in two Iranian families. The other gene with missense mutations is ST3GAL3 (MIM: 606494), which codes for a glycosyl tranferase that catalyzes the transfer of sialic acid to galactose-containing substrates, and has been identified in two Iranian families (Najmabadi et al. 2009 Abstract; Grahn et al. 2002). Two more unrelated families have been mapped to the same locus, 1p34 (Najmabadi et al. 2009 Abstract). Finally, two unrelated families were found to have nonsense and frame shift mutations in ZC3H14, which is a recently described CCCH-type zinc finger gene (Garshasbi et al. 2009 Abstract; Leung et al. 2009). These three genes, if validated, will be interesting because all of them have been identified in more than one family.
See Table 1 for a summary of NS-ID genes.
Synthesizing our knowledge: the search for common pathways
As our knowledge of genes involved in ID expands and the number of genes we identify increases, common pathways are emerging. If a number of common pathways for ID can be confirmed, then, even without knowing which gene is involved in an individuals ID, we may be able to develop tests for specific biochemical markers that can indicate whether the level of activity of certain pathways is deficient. It is already well established that synaptic proteins are involved in memory and learning, and have been implicated in ID. See Table 2 for a summary of functions and domains of known NS-ID genes, and Table 3 for a summary of known protein interactions.
Table 2.
NS-ID genes and their protein products and functions Information about subcellular localization and protein domains were found using HPRD (www.hprd.org); SMART (http://smart.embl-heidelberg.de) and the NCBI gene database (http://www.ncbi.nlm.nih.gov)
| Gene Name | NS or Sa | Linked to Autism | Protein Product | Function | Subcellular localization (HPRD) | Protein Domains (SMART; HPRD; Bold-NCBI gene) |
|---|---|---|---|---|---|---|
| ACSL4 | NS | N | Acyl-CoA synthetase long-chain family member 4 | Fatty acid metabolism | Microsome; Mitochondia | 1 TM |
| AFF2/FMR2 | NS | Y | Fragile X mental retardation 2 | DNA binding protein: Potential activator of transcription | Nucleus | AF4 homology |
| AGTR2 | NS/S | N | Angiotensin II receptor, type 2 | G-protein-coupled receptor for Angiotensin II; mediator of programmed cell death | Plasma membrane | 7 TM |
| AP1S2 | NS/S | N | Adaptor-related protein complex 1 sigma 2 subunit | Complex involved in clathrin recruitment and sorting signal recognition. Synaptic vesicles/neurotransmitter release | Golgi apparatus | Clat Adaptor S |
| ARHGEF6 | NS | N | Rac/Cdc42 guanine nucleotide exchange factor 6 | GEF for Rac and Cdc42 | Cytoplasm | CH; SH3; RhoGEF; PH; CC |
| ARX | NS/S | Y | Aristaless related homeobox | Transcriptional regulation during development | Nucleus | HOX; CC |
| ATRX | NS/S | N | Transcriptional regulator ATRX | Chromatin remodeling | Nucleus | RING; CC; HELICc; DEXDc; SNF2_N |
| BRWD3 | NS/S | N | Bromo domain and WD repeat domain-containing protein 3 | JAK/STAT signaling in drosophila; Putative Chromatin Modifier | Nucleus | 8x WD40; 2x BROMO |
| CASK | NS/S | N | Calcium/calmodulin-dependent serine protein kinase | Kinase and scaffolding at synapses; MAGUK family protein | Synaptic junction, plasma membrane; Nucleus; Cytoplasm | S_TKc; 2x L27; PDZ; SH3; GuKc; |
| CC2D1A | NS | N | Coiled-coil and C2 domain containing 1A | Transcriptional regulator of neurotransmitter receptors; NF-kB pathway activator | Nucleus; Cytoplasm | 2x CC; C2; 4x DM14; |
| CDH15 | NS/S | N | Cadherin 15 | Ca2+ dependant intercellular adhesion protein | Plasma membrane | SP; 5x CA; 1x TM; Cadherin_C |
| CRBN | NS | N | Cereblon | Expression of potassium channels | Plasma membrane; cytoplasm | LON |
| DLG3 | NS | N | Synapse-associated protein 102 | Organization and scaffolding at post-synaptic density; MAGUK family protein | Synaptic junction, plasma membrane; endoplasmic reticulum; cytoplasm | 3x PDZ; SH3; GuKc |
| DOCK8 | N | Dedicator of cytokinesis 8 | Potential guanine nucleotide exchange factor (GEF); putative organizer of filamentous actin | Plasma membrane; cytoplasm; nucleus | Ded_cyto; 2x CC; C2; DUF3398 | |
| FGD1 | NS/S | N | Faciogenital dysplasia protein | GEF for Cdc42 | Cytoplasm; cytoskeleton | RhoGEF; 2x PH; FYVE; DH |
| FTSJ1 | NS | N | FtsJ homolog 1 | Processing and modification of rRNA | Nucleolus | AdoMet_MTases |
| GDI1 | NS | N | GDP dissociation inhibitor 1 | Inhibitor of Rab GTPases | Cytoplasm | NABD_Rossmann |
| GRIK2 | NS | Y | Glutamate receptor, ionotropic,, kainate 2 | Subunit of synaptic glutamate receptor (KARs) | Plasma membrane-synapse | SP; 4x TM; Lig_chan; PBPe; ANF_receptor; Periplasmic_Binding_Protein_Type_1 |
| HUWE1 | NS/S | N | HECT, UBA and WWE domain containing 1 | Ubiquitin E3 ligase; protein ubiquitination pathway | Nucleus; cytoplasm | DUF908; DUF913; UBA; WWE; UIM; HECTc; 2x CC |
| IL1RAPL1 | NS | Y | Interleukin 1 receptor accessory protein-like 1 | Part of the interleukin 1 receptor family; neuronal calcium-regulated vesicle release and dendrite differentiation | Plasma membrane | SP; 2x IG; IGc2; 1x TM; TIR |
| JARID1C/KDM5C | NS | Y | Jumonji, AT rich interactive domain 1C | Transcriptional regulation and chromatin remodeling | Nucleus | JmjN; BRIGHT; 2x PHD; JmjC; CC; Zf-C5HC2; PLU-1 |
| KIRREL3 | NS/S | N | kin of IRRE like 3 | Unknown: Potentially involved in synaptogenesis | Plasma membrane; cytoplasm; extracellular | SP; 3x IG; IGc2; 1x TM |
| MAGT1 | NS | N | Magnesium transporter 1 | Cellular Mg2+ uptake; oligosaccharide transferase; N-glycosylation | ER membrane | 5x TM; OST3_OST6; TRX_family |
| MBD5 | NS/S | N | methyl-CpG binding domain protein 5 | Putative role in transcriptional regulation | Nucleus | PWWP; MBD |
| MECP2 | NS/S | Y | Methyl CpG binding protein 2 | Methyl binding to control transcription | Nucleus | MBD; 2x AT Hook |
| NLGN4X | NS | Y | X-linked neuroligin 4 | Synapse adhesion protein | Plasma membrane-synapse | SP; 2x TM; PnbA; esterase_lipase |
| OPHN1 | NS/S | N | Oligophrenin 1 | Rho-GTPase activating protein | Cytoplasm | CC; PH; RhoGAP |
| PAK3 | NS | N | p21-activated kinase 3 | Downstream effector of Rho-GTPases | Cytoplasm | PBD; S_TKc |
| PQBP1 | NS/S | N | Polyglutamine binding protein 1 | Transcriptional activation | Nucleus; cytoplasm; nucleolus | WW |
| PRSS12 | NS | N | Neurotrypsin | Synaptic protease, cleaves agrin; synaptic plasticity | Extracelluar | SP; KR; 4x SR; Tryp_SPc |
| PTCHD1 | NS | Y | Patched domain 1 | Putative Hedgehog receptor | Plasma membrane | 12x TM; Ptc; MMPL |
| RPS6KA3 | NS/S | N | Ribosomal protein S6 kinase, 90kDa, polypeptide 3 | Ras/Map/ERK signaling pathway | Cytoplasm | 2x S_TKc; S_TK_X |
| SHANK2 | NS | Y | SH3 and multiple ankyrin repeat domains 2 | Scaffolding and cell adhesion protein; Synaptic plasticity | Dendrite; cytoplasm; post-synaptic density | 6x ANK; SH3; PDZ; SAM |
| SHROOM4 | NS/S | N | Shroom family member 4 | Cytoskeletal architecture | Cytoplasm | PDZ; 3x CC; ASD2 |
| SLC6A8 | NS | N | Solute carrier family 6 member 8 | Creatine transporter | Plasma membrane | 12x TM; SNF |
| STXBP1 | NS | N | Syntaxin-binding protein 1 | Synaptic vesicle docking and fusion; neurotransmission | Plasma membrane-synapse; cytoplasm | SEC1 |
| SYNGAP1 | NS | N | Synaptic Ras GTPase activating protein 1 | Part of the NMDA receptor complex; Negative regulation of Ras/Map/ERK pathway | Plasma membrane-synapse | PH; C2; RasGAP; CC; DUF3498 |
| SYP | NS/S | N | Synaptophysin | Synaptic vesicle protein | Synapse/vesicle membrane | 4x TM; MARVEL |
| TSPAN7 | NS/S | N | Tetraspanin 7 | Unknown: Transmembrane protein potentially involved in synapse maturation | Plasma membrane/synapse | Tetraspanin/4x TM |
| TRAPPC9 | NS | N | NIK- and IKKB-binding protein | Neuronal NF-kB signalling, vesicular transport | Golgi; ER; Cytoplasm | Trs120 |
| TUSC3 | NS | N | Tumor suppressor candidate 3 | Mg2+ uptake, oligosaccharide transferase; N-glycosylation | ER membrane | 5x TM; OST3_OST6 |
| UPF3B | NS/S | Y | UPF3 regulator of nonsense transcripts homolog B | mRNA nuclear export and surveillance | Nucleus; Cytoplasm; Nucleolus | SMG4_UPF3; 2x CC; 5x NLS |
| ZNF41 | NS | N | Zinc finger protein 41 | Putative repressor of transcription | Nucleus | KRAB; 17x ZnF_C2H2 |
| ZNF674 | NS | N | Zinc finger protein 674 | Putative repressor of transcription | Nucleus | KRAB; 11x ZnF_C2H2; |
| ZNF711 | NS | N | Zinc finger protein 711 | Unknown: Putative activator of transcription | Nucleus | Zfx_Zfy; 12x ZnF_C2H2; |
| ZNF81 | NS | N | Zinc finger protein 81 | Putative repressor of transcription | Nucleus | KRAB; 12x ZnF_C2H2; |
aS represents syndromic, NS represents nonsyndromic
CH calponin homology; PH pleckstrin homology; SH3 Src homology 3; CC coiled-coil; TM transmembrane; RhoGEF Rho guanine exchange factor; MBD Methyl CpG Binding; HOX homeodomain; DEXDc DEAD-like helicase superfamily; HELICc helicase superfamily c-terminal domain; S_TKc serine/threonine kinase catalytic domain; GuKc Guanylate kinase homologues; C2 Protein kinase C conserved region 2; CA Cadherin; SP signal peptide; Cadherin_C cadherin cytoplasmic region; DUF domain of unknown function; Ded_cyto dedicator of cytokinesis; DH DBL-homology; AdoMet_MTases SAM methyltranferase; NADB_Rossmann Rossmann-fold NAD(P)(+)-binding proteins; UBA Ubiquitin associated domain; UIM Ubiquitin interacting motif; Lig_chan-Glu_bd Ligated ion channel L-glutamate- and glycine-binding site; IG Immunoglobulin domain; IGc2 Immunoglobulin C-2 type; TIR Toll-interleukin resistance 1; JmjN jumonji; PHD plant homeodomain; PLU-1 PLU-like protein; KR Kringle; SR scavenger receptor Cys-rich; SNF sodium:neurotransmitter symporter family; ZnF Zinc finger; KRAB Kruppel associated box; SAM Sterile alpha motif; PnbA Carboxylesterase type B; zf- C5HC2 C5HC2 zinc finger; PBD P21-Rho-binding domain; Tryp_SPc Trypsin-like serine protease; S_TK_X Extension to Ser/Thr-type protein kinases; RasGAP Ras-GTPase Activating Domain; ASD2 Apx/Shroom domain ASD2; ANK Ankyrin repeats; NLS nuclear localization signal
Table 3.
NS-ID Gene Interactions Interactions were determined using 4 databases: BioGRID (www.thebiogrid.org), HPRD (www.hprd.org), String (www.string-db.org) and IntAct (http://www.ebi.ac.uk/intact). Interactions from all 4 databases are recorded for each gene. For each interaction identified on these websites there is a link to the appropriate publication for that interaction. In BioGRID, the publication can be found by clicking on the “publication” link. For HPRD the publication can be found by clicking on the “experiment type” next to each interaction. For String, clicking on the “experiments” link and then on the “details” for the interaction brings up the published abstract. For IntAct, the EBI identification number has been provided for each protein, and this can be queried on the IntAct database to find the publication information. Interactants not found in these databases have been cited independently. *Interactions have been identified in animal studies
| Gene Name | Interacting proteins (IntAct; STRING; BioGRID; HPRD) |
|---|---|
| ACSL4 | SPG20 (EBI-2643801) |
| AFF2/FMR2 | GRB2 (EBI-1964238); NCK1 (EBI-1968993); PLCG1 (EBI-1971165); MAPK14 (EBI-1959498) |
| AGTR2 | AGT; AGTRAP; ACE; MTUS1; TIMP3 (EBI-1749967); GNAI2*; GNAI3*; ZBTB16; PIK3CB; ERBB3 |
| AP1S2 | AP1G2 (EBI-516314); AP1G1 (EBI-516318); AP1S1 (EBI-1073310); GGA3 |
| ARHGEF6 | GIT1; PKLR; BMPR1B; PAK1-3; ARHGEF7; SMAD1-3; TGFBR1; TGFBR2; PARVB; CAPNS1; CDC42; EPHB2; YWHAG; SH2D1A; ADAM15; |
| ARX | None known |
| ATRX | DAXX (EBI-371424); NEK1 (EBI-695931); PTN (EBI-731356); HDAC1; HDAC4; RAD51; ATN1; FAM190B (EBI-731347); EZH2; EIF4A2 (EBI-1069160); MECP2*; SMC1A; CBX5; PTPN4; H3F3A; H3F3B; KIAA1377; LUC7L2; KIAA1128 |
| BRWD3 | UBXD7 (EBI-2009865) |
| CASK | LIN7A; DLG1; TBR1 (EBI-1216781); KCNJ12 (EBI-704591); APBA1; PARK2; RPH3A (EBI-1216824); F11R; ATP2B4; SDC1-4; DLG4; TSPYL2; ID1 (EBI-1215556); KNCJ4 (EBI-706129); NPHS1; CNTNAP2; CNTNAP4; HGS; SPATA2; UIMC1 (EBI-2515432); LIN7C; EPB41 (EBI-1219323); C16ORF70; CD2AP; FCHSD2 (EBI-1215794); LIN7B; EPS8; GRIK2; RAB3A; CASK; KCNJ2 (EBI-706107); DLG1; CADM1; NRXN1-3; DFNB31; CASKIN1; TANC1; GRIN2A; KCNA4; GLS2; NF1; HTR2C; ARHGEF7; KIRREL3 (Bhalla et al. 2008) |
| CC2D1A | CHMP4A; CHMP4C |
| CDH15 | CTNNA1; CTNNB1; JUP; GNA12; BOC; ARVCF; CDON; CDH9; CDH7 |
| CRBN | DDB1* (EBI-2559059); CSN6 (EBI-2510262); CUL4A |
| DLG3 | GRIN2A; GRIN2B; EXOC4; KNCJ12 (EBI-704591); APC; PTK2B; DLG4; SYNGAP1; GDA; CRIPT; GRIN2C; SSCR2*; GUCY1A2; NLGN2; ATP2B4; NLGN1; GRIK2, DLGAP1; NLGN3; ATP2B2; GRIK5; SEMA4C; EXOC7; KRT85 (EBI-1085330); KRT34 (EBI-1077921); KRT35; KRT31 (EBI-1082542); ERBB4 (EBI-80454); CAMK2A (EBI-1068669); CUL2 (EBI-1081586); ANXA1 (EBI-1077519); S100A3 (EBI-1081429); EXOC3; HIST1H2BC; KLHDC3 (EBI-1081381); KRT82 (EBI-1084718); LRP2; CALM1; CNKSR2; KIF1B; MAPK12; KCNA4; SCN4A; ABCA1 (EBI-784253); LPHN1; DLG1; LIN7A; GRIN1; DLGAP4; CACNG2; SCN5A; GLS2; KCNJ2; DLGAP3; GRIK5; PAEP; SCN4A; IDUA (EBI-737171); INSM1 (EBI-737174); MORN2 (EBI-737180); PPP1R14A* (EBI-776423); DLGAP2* (EBI-389360); MDH1 (EBI-735148); SF3B3 (EBI-737189); LRFN2* (EBI-877173); PLK2 (EBI-735154); RBP5 (EBI-737168); TRIM41 (EBI-737192); HAP1 (EBI-732554); SCN2A (EBI-737186); RAB31 (EBI-737183); MCM2 (EBI-732557); B3GNT8 (EBI-737165); |
| DOCK8 | CDC42; RAC1; RHOJ; RHOQ |
| FGD1 | CDC42; ELMO1; ABP1; CTTN; RHOA; RHOG; RHOU; RHOC; RAC2; IFNB1 |
| FTSJ1 | DMWD; FBL; NOP58; |
| GDI1 | RAB1B; RAB6A; RAB3B; RAB9A; RAB27A; PSEN1; RHOH; CDC42; RAC2; RHOA; RABAC1; EPB41; SPOP |
| GRIK2 | DLG4; SDCBP; PICK1; DLG1; GRIK5; GRIP1; GRIA1; GRIA2; GRID1; DLG3; CDH2; CTNNB1; CASK; GRIK2; GRIK4; CTNND1; PRKAA1; LIN7B; GRID2; GOLM1 (EBI-736856); GPAA1 (EBI-736877); LRSAM1 (EBI-737066); MLF1 (EBI-735133) |
| HUWE1 | CDKN2A (EBI-625921); UBL4A (EBI-2515855); FAF2 (EBI-2009967); UBE2E3 (EBI-2339614); GIYD2 (EBI-2372444); P53 EBI-626140); VCPIP1 (EBI-2513235); USP7 (EBI-2513133); MYC (EBI-1237540); UBXN1 (EBI-2010040); CDC6; USP49 (EBI-2512828); USP50; SMAD2; CCL1; RNF11; SMAD9; MCL1; ATM |
| IL1RAPL1 | NCS-1/FREQ; DLG4 (Pavlowsky et al. 2010); CFTR (EBI-1171098) |
| JARID1C/KDM5C | HIST2H3A; SMAD3; RNF2, MAX, REST NCOR1, HDAC1, HDAC2, E2F6, RING1, CBX3; |
| KIRREL3 | CASK (Bhalla et al. 2008) |
| MAGT1 | None known |
| MBD5 | None known |
| MECP2 | SIN3A; LBR; HDAC1; PRPF40A; SPI1; PRPF40B; SKI; DNMT; CDKL5; RBPJ; YBX1; CBX5; SMARCA2; SMARCB1; ATRX*; RCOR1; GTF2B; SUV39H3; HIST2H3A; SIN3B; NCOR1; SMARCE1 |
| NLGN4X | DLG4; DLGAP2 |
| OPHN1 | RHOA; RAC1; CDC42 |
| PAK3 | RAC1; RAF1; ARHGEF6; ARHGEF7; SYN1; PXN; MYO6; GIT2; NCK1; CDC42 |
| PQBP1 | POU3F2; POLR2A; EEF1A1 (EBI-730579) ATXN1; MED31 (EBI-730576); SF3A2; TXNL4A; WBP11 (EBI-956742); RIF1 (EBI-732938); C14ORF1 (EBI-735456); RAB8A (EBI-737426); C1ORF103 |
| PRSS12 | None known |
| PTCHD1 | None known |
| RPS6KA3 | MAPT; MAPK1; MAPK3; PLD1; BAD; HIST3H3; CREB1; FGF2; NR4A1; SYT3; KRT18; CREBBP; MAPK14; PEA15; IGF1; PDPK1; GSTK1 (EBI-1079890); CSNK2B (EBI-1371784) |
| SHANK2 | DLGAP1-4; DNM2; ARHGEF7; CTTN; SSTR2*; DLG4; DYNLL1; DYNLL2; MYO5A; LPHN1; LPHN2; BAI2; SLC9A3; BAIAP2; NCK1 (EBI-1968244); PLCG1 (EBI-1971189); PIK3R1 (EBI-1969713); PPP1R14A* (EBI-776423); SRC (EBI-1960646); GRB2 (EBI-1963670); GRIN2B* (EBI-770442); GRIN1 (EBI-396959); HOMER/Ves1 (Hayashi et al. 2009); CRK (EBI-1959944) |
| SHROOM4 | MYO6; MYO1A; MYO1C; MYO9B; CORO1A; CD2AP |
| SLC6A8 | CD59 |
| STXBP1 | STX1A; STX1B; STX2-4; STX5* SYTL4; STX1B2; SNAP25; ABPA1/Mint1; APBA2*; DLG4*; TUBB2A; TUBA4A; MAPT; HGS; DOC2A; PLD1; CDK5R1; NEFH; CDK5; USO1; PRKCA; PRKCB1; PRKCG |
| SYNGAP1 | ULK1; DLG4; DLG3; ULK2; CAMK2A; MPDZ; DLG3; DLG4; GRIN2A; GRIN2B; GRIN1; TRIP6; PDGFRB; KDR |
| SYP | GRB2; VAMP2; SIAH1; SIAH2; AP1G1; STX1A; EPOR |
| TSPAN7 | KPTN; FYTTD1 |
| TRAPPC9 | TRAPP complex; IKBKB; MAP3K14 |
| TUSC3 | PPP1CA |
| UPF3B | UPF1 (EBI-536644); UPF2 (EBI-374193); RBM8A; NCBP1 (EBI-1776148); HBB; USP21 (EBI-2512177); EIF6; EIF4A3 (EBI-464796); UPF3A; TTC19 (EBI-374205); MCRS1 (EBI-374202); EIF4G1 (EBI-464801); ITGB3BP (EBI-732256) |
| ZNF41 | SMAD2 |
| ZNF674 | None known |
| ZNF711 | PHF8 (Kleine-Kohlbrecher et al. 2010) |
| ZNF81 | None known |
Ionotropic glutamate receptors and excitatory synapses
Ionotropic glutamate receptors have long been suspected to be involved in the etiology of neuropsychiatric disease. Several examples of mutations in glutamate activated receptors and their downstream effectors are present in NS-ID (see Fig. 1). The MRT6 gene GRIK2 encodes GLuR6, which is a subunit of a Kainate receptor (KAR). Likewise, the MRD5 gene SYNGAP1 encodes SynGAP—a GTPase activating protein that is part of the NMDA receptor (NMDAR) complex, binding to the NR2B subunit (Kim et al. 2005). The NMDAR is a well-characterized ionotropic glutamate receptor. SynGAP is a negative regulator of NMDAR mediated ERK activation and causes inhibition of the Ras/ERK pathway (Kim et al. 2005). Over-expression of SynGAP has also been shown to down regulate GLuR1, a subunit of AMPA receptors (AMPAR), a class of ionotropic glutamate receptors which are regulated by the Ras/ERK pathway (Kim et al. 2005; Rumbaugh et al. 2006). Likewise, Syngap knockout mice implicate SynGAP in the regulation of LTP and AMPAR expression (Komiyama et al. 2002).
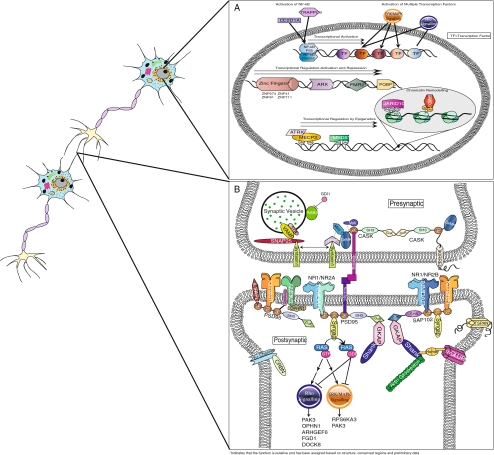
Fig. 1.
a The Nucleus: Many ID genes are involved in transcriptional regulation. Several of these genes encode transcription factors, such as the zinc fingers, PQBP1, ARX and FMR2. CC2D1A and TRAPPC9 activate the NF-κB transcription factor. Epigenetic regulation can also be seen in NS-ID genes. Mutations in genes encoding methyl-binding proteins like MeCP2, ATRX and MBD5, as well as chromatin remodeling proteins like JARID1C and BRWD3, result in ID. b The Glutamate Excitatory Synapse: The pre-synaptic portion of the excitatory synapse is where synaptic vesicles containing the neurotransmitter glutamate are exocytosed. The NS-ID-associated scaffolding protein CASK functions here, as do various other proteins involved in neurotransmitter release that have been implicated in NS-ID or autism, such as SNAP25, STXBP1, SYP, GDI1 and NRXN1. Postsynaptic excitatory synapses contain up to three types of ionotropic glutamate receptors as well as metabotropic glutamate receptors. In these receptors and their complex protein interactions, we find many proteins that are involved in NS-ID. Mutations in genes coding for subunits of ionotropic glutamate receptors, such as GLuR6 (Kainate receptor subunit) and GRIA3 (AMPAR subunit), cause MR. Additionally, many proteins in the postsynaptic density (PSD) including scaffolding and adhesion proteins (SAP102, SHANK3, NLGN4) have also been implied in the genetics of NS-ID and autism. CRBN is involved in regulating the expression of Ca2+ dependant K+ channels (BKCa, encoded by KCNMA1), and resulting ionic currents, at the synapse. IL1RAPL1, a relatively common genetic cause of NS-MR and autism is present in the PSD, as are OPHN1 and SYNGAP, which is a cause of autosomal dominant NS-ID. SYNGAP activates RAS, which leads to several signal transduction pathways resulting in transcriptional activation, including the ERK/MAP and RHO pathways, each of which have downstream effectors that are coded for other NS-ID related genes (Rho: PAK3, OPHN1, ARHGEF6, FGD1, DOCK8; ERK/MAPK: RPS6KA3, PAK3). A number of other autism or ID-associated proteins are also believed to function at the synapse (DPP6, DPP10, PCDH9, SLC6A8, PRSS12), but are not shown in this representation (Molinari et al. 2002; Hahn et al. 2002; Marshall et al. 2008; Hynes et al. 2009)
SAP102 (MRX90), which is part of the membrane-associated guanylate kinase (MAGUK) protein family and the product of DLG3, is also part of the NMDAR complex (Tarpey et al. 2004). MAGUKs are scaffolding proteins involved in the clustering, targeting and anchoring of ionotropic glutamate receptors in the excitatory postsynaptic density (Gardoni 2008). SAP102 directly interacts with the NR2B and NR2A subunits of the NMDAR and is likely to have a role in the clustering and targeting of these receptors (Muller et al. 1996). The protein is expressed at excitatory synapses, particularly during early brain development (Sans et al. 2000). When knocked down, SAP102 has been found to decrease AMPAR and NMDAR excitatory postsynaptic currents (EPSC), while over expression increases EPSC (Elias et al. 2008). Another MAGUK family protein gene, CASK, is frequently mutated in NS-ID as well (Tarpey et al. 2009; Hackett et al. 2009). It is a synaptic scaffolding protein, and acts as an Mg2+-independent neurexin kinase, and interacts with many other families of proteins at cellular junctions (Hata et al. 1996; Mukherjee et al. 2008). It also directly interacts with GLuR6 (Coussen et al. 2002).
Mutations in IL1RAPL1, a gene with several known mutations in NS-ID and autism, result in the incorrect localization of the MAGUK family protein PSD-95 (DLG4), which is important for organization and function of NMDA receptors, ion channels and other signaling proteins (Carrie et al. 1999; Gardoni 2008; Kim and Sheng 2004; Pavlowsky et al. 2010). IL1RAPL1 has been shown to interact with PSD-95, and knockout of this gene decreased the post-synaptic density (PSD) and the localization of PSD-95 at excitatory synapses. Loss of IL1RAPL1 also results in a decrease of activity in the JNK pathway, which led to decreased phosphorylation of PSD-95 (Pavlowsky et al. 2010). It has also been shown to be important for the formation of excitatory synapses in vivo (Pavlowsky et al. 2010). PSD-95 directly interacts with several known NS-ID associated proteins including: CASK, SynGAP, GLuR6 and neuroligins (Kim and Sheng 2004).
Three other synaptic proteins, NLGN4, SHANK2 and SHANK3, have also been identified as causes of NS-ID and/or autism. NLGN4 is a protein that acts in neuronal cell adhesion. It is an important element in postsynaptic differentiation, forming complexes with β-neurexins and PSD-95 (Ichtchenko et al. 1995; Irie et al. 1997; Scheiffele et al. 2000). NLGN4 is linked to glutamatergic postsynaptic proteins and neuroligin/neurexin complexes appear to be sufficient for synaptogenesis (Graf et al. 2009). Interestingly, heterozygous translocations and CNVs disrupting neurexin 1 (NRXN1), which interacts with NLGN4 at the synapse, have been associated with autism (Kim et al. 2008; Autism Genome Project Consortium et al. 2007), and homozygous mutation of NRXN1 causes the S-ID disorder Pitt-Hopkins-like syndrome-2 (PTHSL2; Zweier et al. 2009). NRXN1 also interacts with the NS-ID causing scaffolding protein CASK, as discussed earlier in this review (Hata et al. 1996). SHANK2 and SHANK3 encode scaffolding proteins present at the post-synaptic density and in dendrites. They are important for scaffolding in the post-synaptic density—connecting ion channels, neurotransmitter receptors and other membrane proteins to the actin cytoskeleton—and act as a structural framework at this site (Boeckers et al. 2002; Hayashi et al. 2009). They are also likely to play a role in neuronal plasticity (Boeckers et al. 2002; Hayashi et al. 2009).
OPHN1 encodes an activity-dependant protein that interacts with AMPARs and is essential for their stabilization, thus playing a significant role in synaptic maturation and plasticity (Nadif Kasri et al. 2009). Interestingly, GRIA3, which causes S-ID is a subunit of AMPAR. The S-ID phenotype caused by GRIA3 is variable, but one case presented with only ID and aesthenic body habitus (Wu et al. 2007; Bonnet et al. 2009). Due to the small number of mutations found in this gene, it is difficult to discern if the ID in certain individuals is actually syndromic. The finding that ID can be caused by mutations in all three classes of ionotropic glutamate receptors suggests that the activity and regulation of glutamatergic synapses is essential for normal cognition. This is not surprising, as LTP and LTD often originate from these synapses, and are processes that are thought to be important for synaptic plasticity, memory and learning (Malenka and Bear 2004).
Other synaptic and neuronal proteins in NS-ID
Activity downstream of ionotropic glutamate receptors is also important in understanding the etiology of NS-ID. PRSS12 encodes the protein neurotrypsin, a synaptic protease that is activated by the NMDA receptor (Molinari et al. 2002; Matsumoto-Miyai et al. 2009). It acts in the synapse to cleave agrin, which is present at neuromuscular junctions (NMJ) and in the CNS, but appears to have distinct functions at each location (Stephan et al. 2008; Bezakova and Ruegg 2003). Neurotrypsin-dependent agrin cleavage appears to be an essential process for the formation of dendritic filopodia in the hippocampus, and is dependent on LTP generated by NMDA and AMPA receptors (Matsumoto-Miyai et al. 2009). Dendritic filopodia are the precursors for new dendritic spines in activity dependant synaptogensis (Jontes and Smith 2000). With so many NS-ID genes coding for proteins that are involved in organizing, signaling and downstream effects of ionotropic glutamate receptors, it is likely that excitatory synapses are very important in the normal development of intellectual function. CRBN is another NS-ID gene that codes for a synaptic protease (Higgins et al. 2004). Although its function is not as well characterized as PRSS12, it is known to contain a LON-Protease domain. It, however, is better known to exist in neurons and assist in coordinating the expression function of Ca2+-dependant K+ channels (Higgins et al. 2008).
Cell adhesion
Earlier in this review, NLGN4 was discussed as a neuronal cell adhesion molecule involved in NS-ID etiology. Cell adhesion molecules are critical for the maintenance of synaptic structure and neuronal plasticity (Sudhof 2008). Not surprisingly, several ID genes are involved in cell adhesion. CDH15 is a cadherin gene localized in the brain and skeletal muscle (Bhalla et al. 2008). Mutations of this gene in individuals with ID were found to decrease cell adhesion by greater than 80% (Bhalla et al. 2008). PCDH19, a protocadherin, has also been implicated in epilepsy with mental retardation limited to females (EFMR) (Dibbens et al. 2008; Hynes et al. 2009). Additionally, it has been postulated that TSPAN7 is involved in a complex of ß-integrins, which are involved in cell–cell and cell–matrix interactions (Zemni et al. 2000).
The RHO pathway
Rho GTPases are also a common pathway in NS-XLMR (see Tables 2 and 3). OPHN1, PAK3, ARHGEF6 and FGD1 all encode proteins that are involved in cellular signaling through Rho GTPases or downstream effects (Ramakers 2002). OPHN1 encodes oligophrenin 1, which has been found to stimulate the GTPase activity of RhoA, Rac1 and Cdc42 (Billuart et al. 1998). PAK3, a serine/threonine protease that plays a role in regulating the actin cytoskeleton, is induced by Rac and Cdc42 (Allen et al. 1998; Manser et al. 1995; Daniels and Bokoch 1999). ARHGEF6 encodes a guanine nucleotide exchange factor (GEF) for Rac1 and Cdc42 that interacts with PAK family proteins (Manser et al. 1995; Daniels and Bokoch 1999). FGD1 is also a GEF for Cdc42 (Zheng et al. 1996). All of these gene products interact in the Rho signaling pathway, suggesting that events triggered by this pathway, such as vesicular trafficking in dendritic spines and cytoskeleton organization, may be important processes in cognition and that, when disturbed, cause ID.
Synaptic vesicle trafficking and exocytosis
Several NS-ID genes are involved in neurotransmitter release by exocytosis (see Fig. 1). This process has been extensively studied over the years and has a number of key components. The SNARE complex is essential for neurotransmitter release. Reconstitution experiments have shown that SNARE alone is sufficient for membrane fusion (Weber et al. 1998). SNAP25 (MIM: 600322), a subunit of plasma membrane SNARE complexes (t-SNARE) has been associated with ID through SNP analysis (Gosso et al. 2008). Studies of common variants in SNAP25 have shown significant association with the extremes of IQ (Gosso et al. 2008). This is one of the few examples of common variants playing a role in intellectual disability.
STXBP1 (Munc18-1) (MIM: 602926) binds to syntaxin-1, the other subunit of the t-SNARE complex, and there is evidence that it has a variety of functions in exocytosis including vesicle priming, SNARE assembly, localization of syntaxin-1 in the plasma membrane, as well as regulatory effects (As reviewed by Rizo and Rosenmund 2008). Truncating mutations in STXBP1 have been reported in two cases of NS-ID, and in ID with non-syndromic epilepsy (Hamdan et al. 2009a, b).
Additionally, SYP, which encodes synaptophysin, an integral membrane protein found in transport vesicles in the brain, also causes NS-ID (Tarpey et al. 2009). It interacts with synaptobrevin (VAMP2) and is involved in the regulation, sorting and distribution of synaptobrevin in neurons, but the molecular mechanism by which this occurs is unknown (Bonanomi et al. 2007). Synaptobrevin is the essential component of the vesicle SNARE complex (v-SNARE) (Rizo and Rosenmund 2008). Four mutations have been found in SYP in individuals with NS-ID and ID with epilepsy (Tarpey et al. 2009).
The Rab3 family of small GTPases is also known to be important in membrane trafficking and the release of neurotransmitters. GDI1 encodes a GDP-dissociation inhibitor, which causes GTPases Rab3a and Rab3b to remain inactive by maintaining it in their GDP-bound form (D’adamo et al. 1998; Bienvenu et al. 1998). Rab3 GTPase isoforms A, B and C are localized to synaptic vesicles, and function to regulate neurotransmitter release by regulating the SNARE complex (Fischer von Mollard et al. 1994; Geppert et al. 1997; Gonzalez and Scheller 1999). Interestingly, the scaffolding protein CASK interacts with rabphilin3a, an upstream effector of Rab3a (Zhang et al. 2001). Rabphilin3a maintains Rab3a in its active, GTP-bound state, so disruptions in the rabphilin3a complex with CASK may have a deleterious impact on its interaction with Rab3a (Geppert et al. 1997). It is possible that this leads to the disruption of synaptic vesicle exocytosis, but requires further investigation. Another Rab family gene, Rab39, was found to be mutated in two families with ID and variable other phenotypes including autistic features, macrocephaly and epilepsy (Giannandrea et al. 2010). This suggests that Rab family proteins are important for normal cognitive development and other Rab genes may be considered as candidates for ID.
The ERK/MAPK pathway
The ERK/MAPK pathway is a signaling cascade that responds to growth factors. This pathway is particularly interesting because it is required for certain types of synaptic plasticity (Zhu et al. 2002; Thomas and Huganir 2004). Genes coding for ERK/MAPK pathway proteins and regulators have also been found to cause NS-ID. SynGAP, discussed earlier in this review, has been shown to negatively regulate the ERK/MAPK pathway. When SynGAP activity was depleted by RNAi, ERK activation by NMDAR became sustained (Kim et al. 2005). Conversely, over-expression of SynGAP in cultured neurons diminishes ERK activation (Rumbaugh et al. 2006). These results suggest that SynGAP is necessary for terminating ERK activation thus negatively regulates excitatory synaptic transmission and AMPAR cell surface expression (Kim et al. 2005; Rumbaugh et al. 2006). RPS6KA3 is another NS-ID gene that is involved in ERK/MAPK signaling (Merienne et al. 1999). RPS6KA3 encodes Ribosomal S6 Kinase (RSK) 2, which is a downstream effector of the ERK signaling pathway. ERK activation of RSK2 causes phosphorylation of SHANK3, and appears to be required for normal functioning of AMPARs (Thomas and Huganir 2004). These findings suggest that the ERK/MAPK pathway is an important pathway for normal cognitive development.
Zinc finger proteins
A number of zinc finger proteins, autosomal and X-linked, have been implicated in NS-ID. Both missense and nonsense mutations cause NS-ID in these genes. ZNF41, ZNF81 and ZNF674 are members of a cluster of 7 highly related zinc-finger protein genes on the X-chromosome, and are thought to be involved in transcriptional regulation (Lugtenberg et al. 2006). These zinc finger proteins are all part of the Kruppel-associated box (KRAB) family, which is the largest class of zinc finger proteins (Urrutia 2003). KRAB family zinc finger genes tend to occur in clusters in the genome. The largest such cluster is present on 19q13 (Rousseau-Merck et al. 2002), and contains more than half of the known KRAB domain zinc finger proteins. KRAB family zinc finger proteins have been shown to contribute to transcriptional repression by binding to co-repressors (Witzgall et al. 1994; Margolin et al. 1994) They have been found to function in the formation of hp-1 heterochromatin via complex formation with Kap-1 and SetDB1, a histone 3 lysine 9 (H3-K9) methylase (Schultz et al. 2002). During mouse embryogenesis, KRAB domain proteins also cause irreversible gene silencing through methylation of promoter sequences (Wiznerowicz et al. 2007). It is also notable that KRAB family zinc finger proteins are only present in higher vertebrates such as mammals, birds and amphibians, but not in fish or lower eukaryotes, and it has been postulated that they may have evolved to play an important role in functions such as the nervous or immune systems (Urrutia 2003).
The gene ZNF711 is also implicated in NS-ID and is located on the X-chromosome (Tarpey et al. 2009). Currently the function is unknown but its sequence and domain structure bears similarity to other zinc fingers involved in transcriptional activation (Tarpey et al. 2009). ZC3H14, another zinc finger gene putatively involved in NS-ID (Garshasbi et al. 2009 Abstract), contains a polyadenosine RNA binding domain, and co-localizes with the splicing factor SC35, suggesting a potential role for it in mRNA processing, but further investigation is required to support this claim (Leung et al. 2009).
Since these genes are involved in NS-ID, it is possible that their protein products target the regulation of specific neuronal genes that are involved in cognitive development, learning or memory formation, resulting in an NS-ID phenotype. Future studies that detail the genes or chromosomal regions that are influenced by zinc finger proteins will result in a clearer picture of the mechanism by which mutations in zinc finger proteins cause NS-ID. Numerous other NS-ID genes also appear to be involved in transcriptional regulation (Gecz et al. 2009).
Transcriptional regulation and chromatin remodeling
In addition to the zinc finger proteins, several other transcriptional regulators are implicated in NS-ID. Regulation of transcription appears to be a common theme among NS-ID genes. CC2D1A, an autosomal gene, is responsible for repression of transcription of several serotonin and dopamine receptor genes, and is a putative activator of the NF-κB transcription factor (Basel-Vanagaite et al. 2006). Likewise, TRAPPC9 is an activator of the NF-κB pathway (Hu et al. 2005). NF-κB up-regulates the expression of many neuronal genes, as well as genes of the immune system. Multiple studies have implicated NF-κB in long-term memory formation (Albensi and Mattson 2000; Kassed et al. 2002; Meffert et al. 2003). Additionally, evidence suggests that NF-κB is induced in the hippocampus by group I metabotropic glutamate receptors (GpI-mGLuRs; O’Riordan et al. 2006).
ARX is a homeobox-containing gene that is part of the Aristaless-related gene family. This is a family of transcription factors that are required for various essential events during vertebrate embryogenesis, including CNS development (Meijlink et al. 1999). Based on the experimental data and gene structure it has been speculated that ARX regulates transcription by both gene activation and suppression (as reviewed by Friocourt et al. 2006). Many mutations have been identified in ARX causing intellectual disability. Diseases caused by mutations in this gene include: West syndrome (Stromme et al. 2002a), Partington syndrome (Stromme et al. 2002b), XLAG (Kitamura et al. 2002), XLMR (Bienvenu et al. 2002), Proud Syndrome (Kato et al. 2004), and various forms of epilepsy (Stromme et al. 2002a, b; Scheffer et al. 2002). There seems to be a genotype/phenotype correlation between the location and nature of the mutation and the severity of the phenotype. Loss of function mutations such as truncations and mutations occurring in the highly conserved homeobox domain tend to result in more severe disease including brain malformations as is seen in Proud Syndrome and XLAG (Sherr 2003; Gecz et al. 2006). This gene is one of the most frequently mutated in XLMR. Strikingly, one mutation, a 24-bp in frame duplication that expands the 12-polyalanine tract to 20, is found in >5% of individuals with XLMR (Bienvenu et al. 2002; Stromme et al. 2002a, b; Friocourt et al. 2006). This mutation is also found in individuals with other ARX related S-ID, emphasizing the pleiotropy observed with ARX mutations (Gecz et al. 2006; Friocourt et al. 2006). The activity of this transcription factor is clearly essential for normal cognitive development, and further study of the genes regulated by ARX will be important for our understanding of NS-ID.
Epigenetic regulation is also an important mechanism by which transcription is regulated. MECP2 and ATRX, two genes that function in epigenetic regulation, have been linked to NS-ID etiology. MECP2 encodes the methyl CpG binding protein 2 (MeCP2), which is believed to act as a transcriptional modulator, capable of repressing or activating genes through long-range chromatin re-organization through binding to methylated CpG DNA (see Gonzales and LaSalle 2010 for review). MeCP2 also interacts with ATRX, and in Mecp2 null mice ATRX localization in neurons is disturbed (Nan et al. 2007). Mutations in ATRX cause changes to DNA methylation at several specific highly repeated sequences during development, and MeCP2 mutations cause a loss of transcriptional regulation through binding methylated CpG-containing DNA (Dragich et al. 2000; Gibbons et al. 2000). Interestingly, these are both genes in which mutations cause variable phenotypes. MECP2 mutations cause Rett syndrome, MRXS13, LUBS X-linked ID syndrome, autism and NS-ID, and ATRX mutations cause alpha-thalassemia/MR syndrome, MR-hypotonic facies syndrome, alpha-thalassemia myelodysplasia syndrome and NS-ID.
Chromatin remodeling, a mechanism that is essential for both gene expression and the maintenance of chromatin structure, also appears to play a role in NS-ID etiology. JARIDIC, also known as KMD5C, is one of the more common genes related to X-linked intellectual disability with over twenty mutations known in XLMR patients (Tzschach et al. 2006; Tahiliani et al. 2007). It is a histone demethylase containing a PHD-finger domain that is characteristic of zinc finger proteins and specifically demethylates di- and tri-methylated histone 3 lysine 4 residues (H3K4me2/me3) (Cloos et al. 2008; Iwase et al. 2007; Tahiliani et al. 2007; Christensen et al. 2007). Trimethylation at this residue is extremely important for transcriptional regulation and chromatin structure. It is likely involved in REST-mediated transcriptional repression. It has been shown to regulate the expression of several REST-mediated genes, as well as regulate the H3K4me2/H3K4me3 levels at their promoters (Tahiliani et al. 2007). JARID1C has many allelic variants causing disease and has been linked with NS-ID, S-ID and autism (Jensen et al. 2005; Abidi et al. 2008; Adegbola et al. 2008).
Two other PHD-domain genes are mutated in different forms of ID. Mutations in PHF6 (MIM: 300414) on Xq26.3 result in Borjeson-Forssman-Lehmann syndrome (BFLS). This is a form of S-ID presenting with severe ID, epilepsy, obesity, endocrine defects and several dysmorphic features (Brun et al. 1974). Mutations in PHF8 (MIM: 300560) on Xp11.2 cause ID with characteristic facial features and cleft lip/palette—a form of S-ID also known as Siderius X-linked mental retardation syndrome. The gene functions as an H3K9me1/me2 demethylase and contains a PHD finger that binds to H3K9me2/me3 (Kleine-Kohlbrecher et al. 2010). It has recently been shown that PHF8 interacts with ZNF711, and that these two protein products co-localize at PHF8 target genes in the nucleus, resulting in transcriptional activation (Kleine-Kohlbrecher et al. 2010). Interestingly, JARID1C is one of these target genes (Kleine-Kohlbrecher et al. 2010). This study shows that PHF8, ZNF711, and JARID1C are involved in a common pathway of transcriptional regulation and chromatin remodeling, providing further evidence that these functions are important for normal cognitive development. It also illustrates the value of identifying pathways involved in transcriptional regulation, for the study of ID.
MBD5 (methyl binding domain 5) has been proposed as an autosomal dominant candidate for ID. Mutations in this gene result in variable phenotypes, ranging from syndromic to apparently non-syndromic ID (Wagenstaller et al. 2007). Although not much is known about the function of this gene in humans, it is a methyl-CpG binding protein and may, like other methyl-binding proteins, function in regulation of transcription. It is also notable that BRWD3, a gene found in both NS-ID and S-ID contains a bromodomain—a conserved protein motif that recognizes acetylated lysine residues, and is found in chromatin associated proteins and histone acetyltransferases (Field et al. 2007; Sanchez and Zhou 2009). Although there is currently no experimental data to confirm that BRWD3 is a chromatin modifying protein, it is a possibility based on its motif structure (Field et al. 2007). Further work on both of these gene products will provide evidence of whether or not they are important for chromatin remodeling and regulation of transcription.
Because of the variable phenotypes resulting from mutations in this class of genes, it seems likely that chromatin maintenance and remodeling play a crucial role in normal cognitive development. It is possible that different mutations might lead to varying levels of protein function, and that function might be retained or disturbed differentially by different mutations. In all of these genes, multiple allelic variants are involved and several forms of S-ID as well as NS-ID may result from mutation.
These are only some examples of potential mechanisms by which NS-ID is developed. As we learn more about genes involved in NS-ID, more common biological pathways will become apparent, and will lead us to an understanding of how it is acquired, and what can be done to prevent it or alleviate it in genetic cases.
Connecting ID and autism
CNVs, or structural variation within the genome, appear to contribute significantly to the etiology of both ID and autism. It is often necessary to look at these diagnoses together, as there is significant overlap between them. ID is present in ~67% of individuals with autism (Fombonne 2003). Additionally, in a study performed on an ID population, 28% met the criteria for an autism diagnosis on the ADI-R scale and only half of these had been previously diagnosed (Bryson et al. 2008). Similar studies have been replicated in the past, showing that within ID populations, the prevalence of autism is 8–20%, and that many more individuals with severe ID meet criteria for Autism Spectrum Disorder (ASD; Bryson et al. 2008; de Bildt et al. 2005; Stromme and Diseth 2000; Nordin and Gillberg 1996; Deb and Prasad 1994; Wing and Gould 1979).
ID and autism have multiple overlapping phenotypic domains. The three major phenotypes that characterize autism—language abnormalities, social deficits and stereotypies—can often be seen to varying degrees in ID individuals. Individuals with ID often display stereotypies, which tend to become more pronounced and often self-injurious, as IQ decreases (Symons et al. 2005). Studies have found that 30–60% of individuals with ID display some form of stereotypy (Bodfish et al. 1995; Bodfish et al. 2000; Goldman et al. 2009). Language deficits are often particularly severe in individuals with severe and profound ID.
Many ID syndromes have an incidence of autism that is significantly higher than the incidence for the general population. For example, A current review of the literature shows that 25–47% of individuals with fragile X syndrome, 5–10% of individuals with Downs syndrome, and 16–48% of individuals with tuberous sclerosis (TSC) have a concomitant autism/PDD diagnosis, compared to 0.3–0.6% in the general population (Fombonne 2003, Molloy et al. 2009). Other ID syndromes that have high incidences of concordant autism include Angelman syndrome, Rett syndrome, Joubert Syndrome and Cohen syndrome.
Although autism and ID constitute two separate diagnoses, the overlap cannot be ignored. These overlaps are difficult to quantify genetically due to the heterogeneity of both conditions, and the apparent contribution of rare genetic variants to both diseases. However, there are several genes that appear to be causative for both conditions. The Neuroligin 4 (NLGN4) gene has been linked to autism by several studies (Laumonnier et al. 2004; Marshall et al. 2008; Jamain et al. 2003). However, in 2004, Laumonnier et al. identified a family containing individuals with NS-ID, with or without ASD, segregating with a NLGN4 mutation (2004). More recently, a truncating mutation was found in SHANK3 in an individual with NS-ID (Michaud et al. 2009 Abstract). SHANK3 has also been found to cause autism in several studies (Durand et al. 2007; Marshall et al. 2008).
PTCHD1 is another X-linked gene that has been implicated in autism and NS-ID. A CNV, which deletes that copy of PTCHD1 entirely, causes NS-ID in one family (Noor et al., in press). Another CNV, which results in a loss of the first exon and upstream region of PTCHD1, results in autism in another family (Noor et al., in press). IL1RAPL1, which was initially identified as a cause of NS-ID, and has been shown to cause NS-ID in several individuals, has also been implicated in autism (Carrie et al. 1999; Marshall et al. 2008; Piton et al. 2008; Bhat et al. 2008). Similarly, a missense mutation in the NS-ID gene JARID1C was found in an autistic individual (Adegbola et al. 2008). Most recently, a de novo CNV deletion overlapping SYNGAP1 was identified in a female autism proband (Pinto et al. 2010). These genetic links are of much interest, particularly due to the strong phenotypic overlap seen in NS-ID and autism. These common genes will be an important factor in teasing out which biochemical processes are disturbed in different forms of developmental delay, and why a particular mutation in an individual might lead to one condition rather than the other.
It is also interesting to note that polymorphisms in GRIK2, one of the NS-ARID genes, have been associated with autism in several studies (Jamain et al. 2002; Shuang et al. 2004; Kim et al. 2007). One study found GRIK2 to be in linkage disequilibrium in an autistic population (Jamain et al. 2002). They found a SNP that causes an amino acid change and shows enhanced maternal transmission to be present in 8% of their autistic population, but in only 4% of the control population (Jamain et al. 2002). They propose that GRIK2 polymorphisms may be associated with an increased susceptibility to autism. Two additional independent studies from different populations have shown similar results (Shuang et al. 2004; Kim et al. 2007). However, the studies of GRIK2 in autism indicate a dominant model of inheritance, whereas inheritance of GRIK2 in NS-ID is recessive. It is unlikely that the GRIK2 polymorphisms alone cause autism, but they may contribute to the overall ASD susceptibility. As nonsense mutations in GRIK2 are responsible for the ARID, perhaps missense changes contribute to other neuropsychiatric disorders such as autism.
Potential therapies
For some NS-ID cases caused by single gene mutations, there is the potential for treatment using various categories of gene therapies. Some of these gene therapies are currently under investigation in animal models and humans and have shown promising results for ID-related phenotypes in mouse models.
Transgenic mouse models have been used to assess the effects of re-introducing functional MECP2 (the gene that causes Rett syndrome and some cases of NS-ID) into MECP2 deficient mice with some success. MECP2 null mice have been generated with transgenes that can be conditionally induced to express MECP2. Therefore, the gene may be expressed at experimental time points, which allows for an assessment of pharmacological relevance of re-introducing endogenous MECP2 or introducing ectopic MECP2 (Luikenhuis et al. 2004; Giacometti et al. 2007; Guy et al. 2007; Jugloff et al. 2008). Certain neurological and behavioral phenotypes of these mice were rescued with induction of the transgene, which shows that the damage done by lack of MECP2 is not completely irreversible, and that ID phenotypes can be rescued to some extent (Luikenhuis et al. 2004; Giacometti et al. 2007; Guy et al. 2007; Jugloff et al. 2008). Mice lacking MECP2 have a Rett-like phenotype, although presumably, similar phenotype rescue methods could be used to target NS-ID genes. One of the main issues with applying this strategy in humans is the problem of how to deliver functional copies of the gene to relevant tissues and cells without causing over-expression, which itself may have adverse effects.
A method that has been applied to the treatment of neuromuscular disorders, and may be of potential use for NS-ID, is exon skipping using antisense oligonucleotides (AO) or siRNA (Arnett et al. 2009). This therapy allows the exon containing the truncating mutation to be skipped so that the rest of the gene is transcribed (Arnett et al. 2009). The AOs are delivered using viral vectors. Major issues with this method include immunogenicity to the foreign virus and DNA (Arnett et al. 2009). Additionally, this method has only been assessed for effectiveness in muscle tissues. One of the main issues here would be to identify a vector that could transverse the blood-brain barrier. However, if this could be overcome, it could be an effective way to overcome nonsense-mediated RNA decay of ID genes, and as a consequence restore some function to NS-ID proteins.
Another attractive therapy, and perhaps the one with the most empirical support in humans is aminoglycoside-mediated suppression of nonsense mutations. Aminoglycosides, more generally known for their antibiotic properties, can cause read-through of nonsense mutations, so that translation would not be arrested by the mutation, and potentially full length protein may be produced. The use of aminoglycosides has been proposed and has been relatively successful for the read-through of several disease causing genotypes, including those found in MECP2 that cause Rett syndrome (Brendel et al. 2009). In HeLa cells transfected with common Rett syndrome disease variants of the MECP2 gene, read-through after treatment with aminoglycosides was between 10% and 21% depending on the variant (Brendel et al. 2009). Similar results have been achieved for other disease-causing genes. In studies of cystic fibrosis (CF) in humans, individuals with nonsense mutations in the CFTR gene had aminoglycoside (gentamicin) drops administered to their nasal pathway and showed some restored function of CFTR in 90% of patients (Wilschanski et al. 2003).
Although this method has had relatively encouraging results, there are some issues with it. There is evidence to suggest that naturally occurring stop codons lead to efficient termination due to their context, whereas premature stop codons may be more susceptible to read-through due to their location (Kerem 2004). This notion is further supported by the fact that certain mutations respond more efficiently to aminoglycosides than others (Kerem 2004). However, it is still possible that issues may arise with pseudogenes or other genes with nonsense mutations that have not resulted in a phenotype. There is potential for “junk” DNA to be transcribed and then compete for active sites with normal, functional proteins. Additionally, evidence has been found in CFTR nonsense mutation cell lines that the level of nonsense-mediated decay (NMD) has an effect on the efficiency of aminogylcosides (Linde et al. 2007). This makes sense, as NMD would result in there being less available transcript to read through. Other potential issues include nephrotoxicity and ototoxicity that occurs with aminoglycoside usage (Nagai and Takano 2004). However, this therapy should not be ruled out for potential usage in some monogenic NS-ID cases.
Overall, the fact that, for some therapies, there is a demonstrable improvement in the condition of mice with features of ID suggests that damage to the nervous system may be reversible in some types of ID. This has provided significant hope for potential treatment of ID and improving functional behaviors in individuals with ID. While this is very encouraging, more information is required concerning the genes and biological pathways involved in ID, in order to progress towards the goal of medicinal treatment of human ID.
Discussion
As more genes are being identified that cause NS-ID, certain pathways are emerging as central contributors to normal cognition, and researchers are beginning to fit pieces of the puzzle together. The hope is that this will result in greater accuracy in the selection of new candidate disease genes, as well as an enhanced efficiency for genetic diagnostics for NS-ID. This will also be essential to the next stages in the study of intellectual disabilities, as we move from identifying monogenic causes of NS-ID towards identifying more complicated genetic mechanisms, as well as identifying all the molecular complexes and pathways (and the ways in which they interact with each other) that are involved in human neurological development and cognitive functioning. For example, genes involved in neurotransmitter trafficking and release are very good candidates, as are genes that encode regulators of excitatory synapses. Based on the analysis of functions of genes involved in NS-ID, it is apparent that excitatory synapses play an integral role in the basis of cognition. Additional evidence of this is that memory and learning have been linked to LTP and LTD at these synapses, and thus both organization and communication at these locations are likely to affect cognition.
Additionally, transcription regulators such as zinc finger proteins and chromatin modifiers play a significant role in NS-ID etiology, although many of these genes cause variable NS-ID phenotypes. The location and nature of mutations within these genes may contribute to the phenotypic manifestation of syndromic versus non-syndromic ID. Learning more about the classes and functions of genes activated by these transcription regulators would improve our understanding of how mutations in these NS-ID genes exert an effect at the molecular level, as well as the phenotypic consequences. The same is also true for the signaling pathways, such as Rho and ERK/MAPK, implicated in NS-ID, where research into the downstream effects of mutations in these genes will assist our knowledge of the role of these pathways in normal neurodevelopment and cognition.
Interestingly, a recent study was conducted that analyzed SNP association across functional protein groups and found that genetic variations in synaptic heterotrimeric G-proteins are associated with cognitive function (Ruano et al. 2010). Although none of these genes had been implicated in intellectual disabilities previously, the approach suggests that, in addition to the biological pathways, specific classes of genes may also be fundamental for cognitive development.
The genetic causes of NS-ID are rapidly being uncovered and will be essential for genetic counseling, diagnostics and treatments in the future. The knowledge of genes that cause NS-ID will have an impact on how cases are treated in a clinical setting. Microarrays for mutation screening of known disease genes, as well as exome and whole genome sequencing, will likely become essential tools both for clinical diagnostic purposes and research. As microarray and next generation sequencing technology becomes less expensive and more accessible, identification of new genes for NS-ID will accelerate rapidly. This will inevitably feedback into clinical diagnostics and genetic counseling, with the addition of many more genes to the current catalogue of NS-ID genes. This, in turn, will further aid our understanding of the molecular pathways and processes involved in neurodevelopment and cognition.
Acknowledgements
LK is supported by an Ontario Graduate Scholarship. JBV is a National Alliance for Research on Schizophrenia and Depression Independent Investigator. We thank the anonymous peer-reviewers, whose comments and suggestions have helped to improve this review.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abidi FE, Holloway L, Moore CA, Weaver DD, Simensen RJ, Stevenson RE, Rogers RC, Schwartz CE. Mutations in JARID1C are associated with X-linked mental retardation, short stature and hyperreflexia. J Med Genet. 2008;45:787–793. doi: 10.1136/jmg.2008.058990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adegbola A, Gao H, Sommer S, Browning M. A novel mutation in JARID1C/SMCX in a patient with Autism Spectrum Disorder (ASD) Am J Med Genet A. 2008;146A:505–511. doi: 10.1002/ajmg.a.32142. [DOI] [PubMed] [Google Scholar]
- Albensi BC, Mattson MP. Evidence for the involvement of TNF and NF-kappaB in hippocampal synaptic plasticity. Synapse. 2000;35:151–159. doi: 10.1002/(SICI)1098-2396(200002)35:2<151::AID-SYN8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, Mulley JC, Walsh CA. PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and statistical manual of mental disorders, Text Revision. 4. Washington: American Psychiatric Association; 2000. [Google Scholar]
- Arnett AL, Chamberlain JR, Chamberlain JS. Therapy for neuromuscular disorders. Curr Opin Genet Dev. 2009;19:290–297. doi: 10.1016/j.gde.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Autism Genome Project Consortium. Szatmari P, Paterson AD, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Attia R, Yahav M, et al. The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive non-syndromic mental retardation. J Med Genet. 2006;43:203–210. doi: 10.1136/jmg.2005.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebbington A, Anderson A, Ravine D, et al. Investigating genotype-phenotype relationships in Rett Syndrome using an international data set. Neurology. 2008;70:868–875. doi: 10.1212/01.wnl.0000304752.50773.ec. [DOI] [PubMed] [Google Scholar]
- Berg JM. Lionel Sharples Penrose (1898–1972) aspects of the man and his works, with particular reference to his undertakings in the fields of intellectual disability and mental disorder. J Intellect Disabil Res. 1998;42:104–111. doi: 10.1046/j.1365-2788.1998.00119.x. [DOI] [PubMed] [Google Scholar]
- Berkel S, Marshall CR, Weiss B, et al. Mutations in the SHANK2 synaptic scaffolding gene in Autism Spectrum Disorder and mental retardation. Nat Genet. 2010. [DOI] [PubMed]
- Bezakova G, Ruegg MA. New insights into the roles of agrin. Nat Rev Mol Cell Biol. 2003;4:295–308. doi: 10.1038/nrm1074. [DOI] [PubMed] [Google Scholar]
- Bhalla K, Luo Y, Buchan T, et al. Alterations in CDH15 and KIRREL3 in patients with mild to severe intellectual disability. Am J Hum Genet. 2008;83:703–713. doi: 10.1016/j.ajhg.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat SS, Ladd S, Grass F, Spence JE, Brasington CK, Simensen RJ, Schwartz CE, Dupont BR, Stevenson RE, Srivastava AK. Disruption of the IL1RAPL1 gene associated with a pericentromeric inversion of the X chromosome in a patient with mental retardation and autism. Clin Genet. 2008;73:94–96. doi: 10.1111/j.1399-0004.2007.00920.x. [DOI] [PubMed] [Google Scholar]
- Bienvenu T, des Portes V, Saint Martin A, et al. Non-specific X-linked semidominant mental retardation by mutations in a rab GDP-dissociation inhibitor. Hum Mol Genet. 1998;7:1311–1315. doi: 10.1093/hmg/7.8.1311. [DOI] [PubMed] [Google Scholar]
- Bienvenu T, Poirier K, Friocourt G, et al. ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum Mol Genet. 2002;11:981–991. doi: 10.1093/hmg/11.8.981. [DOI] [PubMed] [Google Scholar]
- Biesecker LG. The end of the beginning of chromosome ends. Am J Med Genet. 2002;107:263–266. doi: 10.1002/ajmg.10160. [DOI] [PubMed] [Google Scholar]
- Billuart P, Bienvenu T, Ronce N, et al. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;392:923–926. doi: 10.1038/31940. [DOI] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Bodfish JW, Crawford TW, Powell SB, Parker DE, Golden RN, Lewis MH. Compulsions in adults with mental retardation: prevalence, phenomenology, and comorbidity with stereotypy and self-injury. Am J Ment Retard. 1995;100:183–192. [PubMed] [Google Scholar]
- Bodfish JW, Symons FJ, Parker DE, Lewis MH. Varieties of repetitive behavior in autism: comparisons to mental retardation. J Autism Dev Disord. 2000;30:237–243. doi: 10.1023/A:1005596502855. [DOI] [PubMed] [Google Scholar]
- Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED. ProSAP/Shank proteins—a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem. 2002;81:903–910. doi: 10.1046/j.1471-4159.2002.00931.x. [DOI] [PubMed] [Google Scholar]
- Bolino A, Muglia M, Conforti FL, et al. Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat Genet. 2000;25:17–19. doi: 10.1038/75542. [DOI] [PubMed] [Google Scholar]
- Bonanomi D, Rusconi L, Colombo CA, Benfenati F, Valtorta F. Synaptophysin I selectively specifies the exocytic pathway of synaptobrevin 2/VAMP2. Biochem J. 2007;404:525–534. doi: 10.1042/BJ20061907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet C, Leheup B, Beri M, Philippe C, Gregoire MJ, Jonveaux P. Aberrant GRIA3 transcripts with multi-exon duplications in a family with X-linked mental retardation. Am J Med Genet A. 2009;149A:1280–1289. doi: 10.1002/ajmg.a.32858. [DOI] [PubMed] [Google Scholar]
- Bortolotto ZA, Clarke VR, Delany CM, et al. Kainate receptors are involved in synaptic plasticity. Nature. 1999;402:297–301. doi: 10.1038/46290. [DOI] [PubMed] [Google Scholar]
- Bradley EA, Thompson A, Bryson SE. Mental retardation in teenagers: prevalence data from the Niagara Region, Ontario. Can J Psychiatry. 2002;47:652–659. doi: 10.1177/070674370204700707. [DOI] [PubMed] [Google Scholar]
- Brendel C, Klahold E, Gartner J, Huppke P. Suppression of nonsense mutations in Rett Syndrome by aminoglycoside antibiotics. Pediatr Res. 2009;65:520–523. doi: 10.1203/PDR.0b013e31819d9ebc. [DOI] [PubMed] [Google Scholar]
- Brun A, Borjeson M, Forssman H. An inherited syndrome with mental deficiency and endocrine disorder. A patho-anatomical study. J Ment Defic Res. 1974;18:317–325. doi: 10.1111/j.1365-2788.1974.tb01243.x. [DOI] [PubMed] [Google Scholar]
- Bryson SE, Bradley EA, Thompson A, Wainwright A. Prevalence of autism among adolescents with intellectual disabilities. Can J Psychiatry. 2008;53:449–459. doi: 10.1177/070674370805300710. [DOI] [PubMed] [Google Scholar]
- Carrie A, Jun L, Bienvenu T, et al. A new member of the IL-1 receptor family highly expressed in hippocampus and involved in X-linked mental retardation. Nat Genet. 1999;23:25–31. doi: 10.1038/12623. [DOI] [PubMed] [Google Scholar]
- Chelly J, Khelfaoui M, Francis F, Cherif B, Bienvenu T. Genetics and pathophysiology of mental retardation. Eur J Hum Genet. 2006;14:701–713. doi: 10.1038/sj.ejhg.5201595. [DOI] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Clark AJ, Rosenberg EH, Almeida LS, Wood TC, Jakobs C, Stevenson RE, Schwartz CE, Salomons GS. X-linked creatine transporter (SLC6A8) mutations in about 1% of males with mental retardation of unknown etiology. Hum Genet. 2006;119:604–610. doi: 10.1007/s00439-006-0162-9. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Swanson G, Heinemann SF. Kainate receptors are involved in short- and long-term plasticity at mossy fiber synapses in the hippocampus. Neuron. 2001;29:209–216. doi: 10.1016/S0896-6273(01)00191-X. [DOI] [PubMed] [Google Scholar]
- Coussen F, Normand E, Marchal C, Costet P, Choquet D, Lambert M, Mege RM, Mulle C. Recruitment of the kainate receptor subunit glutamate receptor 6 by cadherin/catenin complexes. J Neurosci. 2002;22:6426–6436. doi: 10.1523/JNEUROSCI.22-15-06426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvert P, Bienvenu T, Aquaviva C, et al. MECP2 is highly mutated in X-linked mental retardation. Hum Mol Genet. 2001;10:941–946. doi: 10.1093/hmg/10.9.941. [DOI] [PubMed] [Google Scholar]
- D’Adamo P, Menegon A, Lo Nigro C, et al. Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat Genet. 1998;19:134–139. doi: 10.1038/487. [DOI] [PubMed] [Google Scholar]
- Daniels RH, Bokoch GM. P21-activated protein kinase: a crucial component of morphological signaling? Trends Biochem Sci. 1999;24:350–355. doi: 10.1016/S0968-0004(99)01442-5. [DOI] [PubMed] [Google Scholar]
- de Bildt A, Sytema S, Kraijer D, Minderaa R. Prevalence of pervasive developmental disorders in children and adolescents with mental retardation. J Child Psychol Psychiatry. 2005;46:275–286. doi: 10.1111/j.1469-7610.2004.00346.x. [DOI] [PubMed] [Google Scholar]
- Deb S, Prasad KB. The prevalence of autistic disorder among children with a learning disability. Br J Psychiatry. 1994;165:395–399. doi: 10.1192/bjp.165.3.395. [DOI] [PubMed] [Google Scholar]
- Dibbens LM, Tarpey PS, Hynes K, et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet. 2008;40:776–781. doi: 10.1038/ng.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti MT, Orrico A, De Stefano N, Battisti C, Sicurelli F, Severi S, Lam CW, Galli L, Sorrentino V, Federico A. A Rett Syndrome MECP2 mutation that causes mental retardation in men. Neurology. 2002;58:226–230. doi: 10.1212/wnl.58.2.226. [DOI] [PubMed] [Google Scholar]
- Dragich J, Houwink-Manville I, Schanen C. Rett Syndrome: a surprising result of mutation in MECP2. Hum Mol Genet. 2000;9:2365–2375. doi: 10.1093/hmg/9.16.2365. [DOI] [PubMed] [Google Scholar]
- Drews CD, Yeargin-Allsopp M, Decoufle P, Murphy CC. Variation in the influence of selected sociodemographic risk factors for mental retardation. Am J Public Health. 1995;85:329–334. doi: 10.2105/AJPH.85.3.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with Autism Spectrum Disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin M. The epidemiology of developmental disabilities in low-income countries. Ment Retard Dev Disabil Res Rev. 2002;8:206–211. doi: 10.1002/mrdd.10039. [DOI] [PubMed] [Google Scholar]
- Durkin MS, Hasan ZM, Hasan KZ. Prevalence and correlates of mental retardation among children in Karachi, Pakistan. Am J Epidemiol. 1998;147:281–288. doi: 10.1093/oxfordjournals.aje.a009448. [DOI] [PubMed] [Google Scholar]
- Elias GM, Elias LA, Apostolides PF, Kriegstein AR, Nicoll RA. Differential trafficking of AMPA and NMDA receptors by SAP102 and PSD-95 underlies synapse development. Proc Natl Acad Sci USA. 2008;105:20953–20958. doi: 10.1073/pnas.0811025106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson E. Poverty and people with intellectual disabilities. Ment Retard Dev Disabil Res Rev. 2007;13:107–113. doi: 10.1002/mrdd.20144. [DOI] [PubMed] [Google Scholar]
- Fan YS, Jayakar P, Zhu H, Barbouth D, Sacharow S, Morales A, Carver V, Benke P, Mundy P, Elsas LJ. Detection of pathogenic gene copy number variations in patients with mental retardation by genomewide oligonucleotide array comparative genomic hybridization. Hum Mutat. 2007;28:1124–1132. doi: 10.1002/humu.20581. [DOI] [PubMed] [Google Scholar]
- Fedulov V, Rex CS, Simmons DA, Palmer L, Gall CM, Lynch G. Evidence that long-term potentiation occurs within individual hippocampal synapses during learning. J Neurosci. 2007;27:8031–8039. doi: 10.1523/JNEUROSCI.2003-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field M, Tarpey PS, Smith R, et al. Mutations in the BRWD3 gene cause X-linked mental retardation associated with macrocephaly. Am J Hum Genet. 2007;81:367–374. doi: 10.1086/520677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stahl B, Khokhlatchev A, Sudhof TC, Jahn R. Rab3C is a synaptic vesicle protein that dissociates from synaptic vesicles after stimulation of exocytosis. J Biol Chem. 1994;269:10971–10974. [PubMed] [Google Scholar]
- Fombonne E. Epidemiological surveys of autism and other pervasive developmental disorders: an update. J Autism Dev Disord. 2003;33:365–382. doi: 10.1023/A:1025054610557. [DOI] [PubMed] [Google Scholar]
- Freude K, Hoffmann K, Jensen LR, et al. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am J Hum Genet. 2004;75:305–309. doi: 10.1086/422507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friocourt G, Poirier K, Rakic S, Parnavelas JG, Chelly J. The role of ARX in cortical development. Eur J Neurosci. 2006;23:869–876. doi: 10.1111/j.1460-9568.2006.04629.x. [DOI] [PubMed] [Google Scholar]
- Froyen G, Corbett M, Vandewalle J, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet. 2008;82:432–443. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardoni F. MAGUK proteins: new targets for pharmacological intervention in the glutamatergic synapse. Eur J Pharmacol. 2008;585:147–152. doi: 10.1016/j.ejphar.2008.01.048. [DOI] [PubMed] [Google Scholar]
- Garshasbi M, Hadavi V, Habibi H, Kahrizi K, Kariminejad R, Behjati F, Tzschach A, Najmabadi H, Ropers HH, Kuss AW. A defect in the TUSC3 gene is associated with autosomal recessive mental retardation. Am J Hum Genet. 2008;82:1158–1164. doi: 10.1016/j.ajhg.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garshasbi M, Kahrizi K, Tzschach A, et al. ZC3H14 mutations cosegregate with Non-Syndromic Autosomal Recessive Mental Retardation (NS-ARMR) in two Iranian families. American Society of Human Genetics Meeting. 2009:190.
- Gecz J, Gedeon AK, Sutherland GR, Mulley JC. Identification of the Gene FMR2, associated with FRAXE mental retardation. Nat Genet. 1996;13:105–108. doi: 10.1038/ng0596-105. [DOI] [PubMed] [Google Scholar]
- Gecz J, Cloosterman D, Partington M. ARX: a gene for all seasons. Curr Opin Genet Dev. 2006;16:308–316. doi: 10.1016/j.gde.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Gecz J, Shoubridge C, Corbett M. The genetic landscape of intellectual disability arising from chromosome X. Trends Genet. 2009;25:308–316. doi: 10.1016/j.tig.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Geppert M, Goda Y, Stevens CF, Sudhof TC. The small GTP-binding protein Rab3A regulates a late step in synaptic vesicle fusion. Nature. 1997;387:810–814. doi: 10.1038/42954. [DOI] [PubMed] [Google Scholar]
- Giacometti E, Luikenhuis S, Beard C, Jaenisch R. Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proc Natl Acad Sci USA. 2007;104:1931–1936. doi: 10.1073/pnas.0610593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannandrea M, Bianchi V, Mignogna ML, et al. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. Am J Hum Genet. 2010;86:185–195. doi: 10.1016/j.ajhg.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, McDowell TL, Raman S, O’Rourke DM, Garrick D, Ayyub H, Higgs DR. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Pellagatti A, Garrick D, Wood WG, Malik N, Ayyub H, Langford C, Boultwood J, Wainscoat JS, Higgs DR. Identification of acquired somatic mutations in the gene encoding chromatin-remodeling factor ATRX in the Alpha-Thalassemia Myelodysplasia Syndrome (ATMDS) Nat Genet. 2003;34:446–449. doi: 10.1038/ng1213. [DOI] [PubMed] [Google Scholar]
- Goldman S, Wang C, Salgado MW, Greene PE, Kim M, Rapin I. Motor stereotypies in children with autism and other developmental disorders. Dev Med Child Neurol. 2009;51:30–38. doi: 10.1111/j.1469-8749.2008.03178.x. [DOI] [PubMed] [Google Scholar]
- Gonzales ML, LaSalle JM. The role of MeCP2 in brain development and neurodevelopmental disorders. Curr Psychiatry Rep. 2010;12:127–134. doi: 10.1007/s11920-010-0097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez L, Jr, Scheller RH. Regulation of membrane trafficking: structural insights from a rab/effector complex. Cell. 1999;96:755–758. doi: 10.1016/S0092-8674(00)80585-1. [DOI] [PubMed] [Google Scholar]
- Gosso MF, de Geus EJ, Polderman TJ, Boomsma DI, Heutink P, Posthuma D. Common variants underlying cognitive ability: further evidence for association between the SNAP-25 gene and cognition using a family-based study in two independent Dutch cohorts. Genes Brain Behav. 2008;7:355–364. doi: 10.1111/j.1601-183X.2007.00359.x. [DOI] [PubMed] [Google Scholar]
- Graf ER, Daniels RW, Burgess RW, Schwarz TL, DiAntonio A. Rab3 dynamically controls protein composition at active zones. Neuron. 2009;64:663–677. doi: 10.1016/j.neuron.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahn A, Barkhordar GS, Larson G. Cloning and sequencing of nineteen transcript isoforms of the human alpha2, 3-sialyltransferase gene, ST3Gal III; its genomic organisation and expression in human tissues. Glycoconj J. 2002;19:197–210. doi: 10.1023/A:1024253808424. [DOI] [PubMed] [Google Scholar]
- Griggs BL, Ladd S, Saul RA, DuPont BR, Srivastava AK. Dedicator of cytokinesis 8 is disrupted in two patients with mental retardation and developmental disabilities. Genomics. 2008;91:195–202. doi: 10.1016/j.ygeno.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwend TP, Krueger SR, Kozlov SV, Wolfer DP, Sonderegger P. Neurotrypsin, a novel multidomain serine protease expressed in the nervous system. Mol Cell Neurosci. 1997;9:207–219. doi: 10.1006/mcne.1997.0616. [DOI] [PubMed] [Google Scholar]
- Guerrini R, Shanahan JL, Carrozzo R, Bonanni P, Higgs DR, Gibbons RJ. A nonsense mutation of the ATRX gene causing mild mental retardation and epilepsy. Ann Neurol. 2000;47:117–121. doi: 10.1002/1531-8249(200001)47:1<117::AID-ANA20>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Gustavson KH. Prevalence and aetiology of congenital birth defects, infant mortality and mental retardation in Lahore, Pakistan: a prospective cohort study. Acta Paediatr. 2005;94:769–774. doi: 10.1080/08035250410025636. [DOI] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett Syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett A, Tarpey PS, Licata A, et al. CASK mutations are frequent in males and cause X-linked nystagmus and variable XLMR phenotypes. Eur J Hum Genet. 2009. [DOI] [PMC free article] [PubMed]
- Hagens O, Dubos A, Abidi F, et al. Disruptions of the novel KIAA1202 gene are associated with X-linked mental retardation. Hum Genet. 2006;118:578–590. doi: 10.1007/s00439-005-0072-2. [DOI] [PubMed] [Google Scholar]
- Hahn KA, Salomons GS, Tackels-Horne D, et al. X-linked mental retardation with seizures and carrier manifestations is caused by a mutation in the creatine-transporter gene (SLC6A8) located in Xq28. Am J Hum Genet. 2002;70:1349–1356. doi: 10.1086/340092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham AL, Kumar A, Deeter R, Schanen NC. Does genotype predict phenotype in Rett Syndrome? J Child Neurol. 2005;20:768–778. doi: 10.1177/08830738050200091301. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Spiegelman D, et al. Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N Engl J Med. 2009a;360:599–605. doi: 10.1056/NEJMoa0805392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Piton A, Gauthier J, et al. De novo STXBP1 mutations in mental retardation and nonsyndromic epilepsy. Ann Neurol. 2009b;65:748–753. doi: 10.1002/ana.21625. [DOI] [PubMed] [Google Scholar]
- Hata Y, Butz S, Sudhof TC. CASK: a novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J Neurosci. 1996;16:2488–2494. doi: 10.1523/JNEUROSCI.16-08-02488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MK, Tang C, Verpelli C, Narayanan R, Stearns MH, Xu RM, Li H, Sala C, Hayashi Y. The postsynaptic density proteins homer and shank form a polymeric network structure. Cell. 2009;137:159–171. doi: 10.1016/j.cell.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JJ, Pucilowska J, Lombardi RQ, Rooney JP. A mutation in a novel ATP-dependent lon protease gene in a kindred with mild mental retardation. Neurology. 2004;63:1927–1931. doi: 10.1212/01.wnl.0000146196.01316.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JJ, Hao J, Kosofsky BE, Rajadhyaksha AM. Dysregulation of large-conductance Ca2+-activated K+ channel expression in nonsyndromal mental retardation due to a cereblon p.R419X mutation. Neurogenetics. 2008;9:219–223. doi: 10.1007/s10048-008-0128-2. [DOI] [PubMed] [Google Scholar]
- Howard MT, Malik N, Anderson CB, Voskuil JL, Atkins JF, Gibbons RJ. Attenuation of an amino-terminal premature stop codon mutation in the ATRX gene by an alternative mode of translational initiation. J Med Genet. 2004;41:951–956. doi: 10.1136/jmg.2004.020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WH, Pendergast JS, Mo XM, et al. NIBP, a novel NIK and IKK(beta)-binding protein that enhances NF-(kappa)B activation. J Biol Chem. 2005;280:29233–29241. doi: 10.1074/jbc.M501670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain R, Bittles AH. The prevalence and demographic characteristics of consanguineous marriages in Pakistan. J Biosoc Sci. 1998;30:261–275. doi: 10.1017/S0021932098002612. [DOI] [PubMed] [Google Scholar]
- Hussain R, Bittles AH. Sociodemographic correlates of consanguineous marriage in the Muslim population of India. J Biosoc Sci. 2000;32:433–442. doi: 10.1017/S0021932000004338. [DOI] [PubMed] [Google Scholar]
- Hynes K, Tarpey P, Dibbens LM, et al. Epilepsy and mental retardation limited to females with PCDH19 mutations can present de novo or in single generation families. J Med Genet. 2009. [DOI] [PubMed]
- Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, Moomaw C, Sudhof TC. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Jamain S, Betancur C, Quach H, Philippe A, Fellous M, Giros B, Gillberg C, Leboyer M, Bourgeron T, Paris Autism Research International Sibpair (PARIS) Study Linkage and association of the glutamate receptor 6 gene with autism. Mol Psychiatry. 2002;7:302–310. doi: 10.1038/sj.mp.4000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LR, Amende M, Gurok U, et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Lee KH, Song S, Jung YK, Park CS. Identification and functional characterization of cereblon as a binding protein for large-conductance calcium-activated potassium channel in rat brain. J Neurochem. 2005;94:1212–1224. doi: 10.1111/j.1471-4159.2005.03344.x. [DOI] [PubMed] [Google Scholar]
- Jontes JD, Smith SJ. Filopodia, spines, and the generation of synaptic diversity. Neuron. 2000;27:11–14. doi: 10.1016/S0896-6273(00)00003-9. [DOI] [PubMed] [Google Scholar]
- Jugloff DG, Vandamme K, Logan R, Visanji NP, Brotchie JM, Eubanks JH. Targeted delivery of an Mecp2 transgene to forebrain neurons improves the behavior of female Mecp2-deficient mice. Hum Mol Genet. 2008;17:1386–1396. doi: 10.1093/hmg/ddn026. [DOI] [PubMed] [Google Scholar]
- Kalscheuer VM, Freude K, Musante L, et al. Mutations in the polyglutamine binding protein 1 gene cause X-linked mental retardation. Nat Genet. 2003;35:313–315. doi: 10.1038/ng1264. [DOI] [PubMed] [Google Scholar]
- Kassed CA, Willing AE, Garbuzova-Davis S, Sanberg PR, Pennypacker KR. Lack of NF-kappaB p50 exacerbates degeneration of hippocampal neurons after chemical exposure and impairs learning. Exp Neurol. 2002;176:277–288. doi: 10.1006/exnr.2002.7967. [DOI] [PubMed] [Google Scholar]
- Kato M, Das S, Petras K, et al. Mutations of ARX are associated with striking pleiotropy and consistent genotype-phenotype correlation. Hum Mutat. 2004;23:147–159. doi: 10.1002/humu.10310. [DOI] [PubMed] [Google Scholar]
- Katusic SK, Colligan RC, Beard CM, O’Fallon WM, Bergstralh EJ, Jacobsen SJ, Kurland LT. Mental retardation in a birth cohort, 1976–1980, Rochester, Minnesota. Am J Ment Retard. 1996;100:335–344. [PubMed] [Google Scholar]
- Kerem E. Pharmacologic therapy for stop mutations: how much CFTR activity is enough? Curr Opin Pulm Med. 2004;10:547–552. doi: 10.1097/01.mcp.0000141247.22078.46. [DOI] [PubMed] [Google Scholar]
- Khlat M, Khoury M. Inbreeding and diseases: demographic, genetic, and epidemiologic perspectives. Epidemiol Rev. 1991;13:28–41. doi: 10.1093/oxfordjournals.epirev.a036072. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee HK, Takamiya K, Huganir RL. The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity. J Neurosci. 2003;23:1119–1124. doi: 10.1523/JNEUROSCI.23-04-01119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Dunah AW, Wang YT, Sheng M. Differential roles of NR2A- and NR2B-containing NMDA receptors in ras-ERK signaling and AMPA receptor trafficking. Neuron. 2005;46:745–760. doi: 10.1016/j.neuron.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Kim SA, Kim JH, Park M, Cho IH, Yoo HJ. Family-based association study between GRIK2 polymorphisms and Autism Spectrum Disorders in the Korean Trios. Neurosci Res. 2007;58:332–335. doi: 10.1016/j.neures.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Yanazawa M, Sugiyama N, et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- Kleefstra T, Hamel BC. X-linked mental retardation: further lumping, splitting and emerging phenotypes. Clin Genet. 2005;67:451–467. doi: 10.1111/j.1399-0004.2005.00434.x. [DOI] [PubMed] [Google Scholar]
- Kleefstra T, Yntema HG, Oudakker AR, et al. Zinc finger 81 (ZNF81) mutations associated with X-linked mental retardation. J Med Genet. 2004;41:394–399. doi: 10.1136/jmg.2003.016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Christensen J, Vandamme J, et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38:165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Regan R, Nicod A, Horsley SW, Kearney L, Homfray T, Winter RM, Bolton P, Flint J. Subtle chromosomal rearrangements in children with unexplained mental retardation. Lancet. 1999;354:1676–1681. doi: 10.1016/S0140-6736(99)03070-6. [DOI] [PubMed] [Google Scholar]
- Komiyama NH, Watabe AM, Carlisle HJ, et al. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci. 2002;22:9721–9732. doi: 10.1523/JNEUROSCI.22-22-09721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen DA, Pfundt R, de Leeuw N, et al. Genomic microarrays in mental retardation: a practical workflow for diagnostic applications. Hum Mutat. 2009;30:283–292. doi: 10.1002/humu.20883. [DOI] [PubMed] [Google Scholar]
- Kutsche K, Yntema H, Brandt A, et al. Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for rho GTPases, in patients with X-linked mental retardation. Nat Genet. 2000;26:247–250. doi: 10.1038/80002. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel RR, May M, Pouls S, Lubs HA, Stevenson RE, Schwartz CE. Non-syndromic X-linked mental retardation associated with a missense mutation (P312L) in the FGD1 gene. Clin Genet. 2002;61:139–145. doi: 10.1034/j.1399-0004.2002.610209.x. [DOI] [PubMed] [Google Scholar]
- Ledbetter DH, Martin CL. Cryptic telomere imbalance: a 15-year update. Am J Med Genet C Semin Med Genet. 2007;145C:327–334. doi: 10.1002/ajmg.c.30149. [DOI] [PubMed] [Google Scholar]
- Leonard H, Wen X. The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev. 2002;8:117–134. doi: 10.1002/mrdd.10031. [DOI] [PubMed] [Google Scholar]
- Leung SW, Apponi LH, Cornejo OE, Kitchen CM, Valentini SR, Pavlath GK, Dunham CM, Corbett AH. Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA binding protein. Gene. 2009;439:71–78. doi: 10.1016/j.gene.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde L, Boelz S, Nissim-Rafinia M, et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest. 2007;117:683–692. doi: 10.1172/JCI28523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lion-Francois L, Cheillan D, Pitelet G, et al. High frequency of creatine deficiency syndromes in patients with unexplained mental retardation. Neurology. 2006;67:1713–1714. doi: 10.1212/01.wnl.0000239153.39710.81. [DOI] [PubMed] [Google Scholar]
- Lugtenberg D, Yntema HG, Banning MJ, et al. ZNF674: a new Kruppel-associated box-containing zinc-finger gene involved in nonsyndromic X-linked mental retardation. Am J Hum Genet. 2006;78:265–278. doi: 10.1086/500306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg D, Veltman JA, van Bokhoven H. High-resolution genomic microarrays for X-linked mental retardation. Genet Med. 2007;9:560–565. doi: 10.1097/GIM.0b013e318149e647. [DOI] [PubMed] [Google Scholar]
- Luikenhuis S, Giacometti E, Beard CF, Jaenisch R. Expression of MeCP2 in postmitotic neurons rescues rett syndrome in mice. Proc Natl Acad Sci USA. 2004;101:6033–6038. doi: 10.1073/pnas.0401626101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Mandel JL, Chelly J. Monogenic X-linked mental retardation: is it as frequent as currently estimated? The paradox of the ARX (Aristaless X) mutations. Eur J Hum Genet. 2004;12:689–693. doi: 10.1038/sj.ejhg.5201247. [DOI] [PubMed] [Google Scholar]
- Manser E, Chong C, Zhao ZS, Leung T, Michael G, Hall C, Lim L. Molecular cloning of a new member of the p21-Cdc42/rac-activated kinase (PAK) family. J Biol Chem. 1995;270:25070–25078. doi: 10.1074/jbc.270.42.25070. [DOI] [PubMed] [Google Scholar]
- Margolin JF, Friedman JR, Meyer WK, Vissing H, Thiesen HJ, Rauscher FJ., 3rd Kruppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci USA. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda A, Suzuki Y, Honda G, et al. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003;22:3307–3318. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- Matsumoto-Miyai K, Sokolowska E, Zurlinden A, et al. Coincident pre- and postsynaptic activation induces dendritic filopodia via neurotrypsin-dependent agrin cleavage. Cell. 2009;136:1161–1171. doi: 10.1016/j.cell.2009.02.034. [DOI] [PubMed] [Google Scholar]
- McLaren J, Bryson SE. Review of recent epidemiological studies of mental retardation: prevalence, associated disorders, and etiology. Am J Ment Retard. 1987;92:243–254. [PubMed] [Google Scholar]
- McMullan DJ, Bonin M, Hehir-Kwa JY, et al. Molecular karyotyping of patients with unexplained mental retardation by SNP arrays: a multicenter study. Hum Mutat. 2009;30:1082–1092. doi: 10.1002/humu.21015. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Meijlink F, Beverdam A, Brouwer A, Oosterveen TC, Berge DT. Vertebrate aristaless-related genes. Int J Dev Biol. 1999;43:651–663. [PubMed] [Google Scholar]
- Meloni I, Vitelli F, Pucci L, et al. Alport syndrome and mental retardation: clinical and genetic dissection of the contiguous gene deletion syndrome in Xq22.3 (ATS-MR) J Med Genet. 2002;39:359–365. doi: 10.1136/jmg.39.5.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merienne K, Jacquot S, Pannetier S, Zeniou M, Bankier A, Gecz J, Mandel JL, Mulley J, Sassone-Corsi P, Hanauer A. A missense mutation in RPS6KA3 (RSK2) responsible for non-specific mental retardation. Nat Genet. 1999;22:13–14. doi: 10.1038/8719. [DOI] [PubMed] [Google Scholar]
- Michaud J, Hamdan F, Gauthier J, et al. Large scale re-sequencing of autosomal synaptic genes reveals a high rate of de novo deleterious mutations in non-syndromic mental retardation. American Society of Human Genetics Meeting. 2009:2130.
- Mir A, Kaufman L, Noor A, et al. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-beta-binding protein, in nonsyndromic autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:909–915. doi: 10.1016/j.ajhg.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida GH, Mahajnah M, Hill AD, Basel-Vanagaite L, Gleason D, Hill RS, Bodell A, Crosier M, Straussberg R, Walsh CA. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet. 2009;85:897–902. doi: 10.1016/j.ajhg.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell B, Darr A. Science and society: genetic counselling and customary consanguineous marriage. Nat Rev Genet. 2002;3:225–229. doi: 10.1038/nrg754. [DOI] [PubMed] [Google Scholar]
- Moheb LA, Jensen LR, Garshasbi M, et al. Two independent mutations in the ZNF526 gene are associated with non-syndromic autosomal recessive mental retardation. American Society of Human Genetics Meeting. 2009:2078.
- Molinari F, Rio M, Meskenaite V, et al. Truncating neurotrypsin mutation in autosomal recessive nonsyndromic mental retardation. Science. 2002;298:1779–1781. doi: 10.1126/science.1076521. [DOI] [PubMed] [Google Scholar]
- Molinari F, Foulquier F, Tarpey PS, et al. Oligosaccharyltransferase-subunit mutations in nonsyndromic mental retardation. Am J Hum Genet. 2008;82:1150–1157. doi: 10.1016/j.ajhg.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy CA, Murray DS, Kinsman A, Castillo H, Mitchell T, Hickey FJ, Patterson B. Differences in the clinical presentation of trisomy 21 with and without autism. J Intellect Disabil Res. 2009;53:143–151. doi: 10.1111/j.1365-2788.2008.01138.x. [DOI] [PubMed] [Google Scholar]
- Motazacker MM, Rost BR, Hucho T, et al. A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am J Hum Genet. 2007;81:792–798. doi: 10.1086/521275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Sharma M, Urlaub H, Bourenkov GP, Jahn R, Sudhof TC, Wahl MC. CASK functions as a Mg2+-independent neurexin kinase. Cell. 2008;133:328–339. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller BM, Kistner U, Kindler S, et al. SAP102, a novel postsynaptic protein that interacts with NMDA receptor complexes in vivo. Neuron. 1996;17:255–265. doi: 10.1016/S0896-6273(00)80157-9. [DOI] [PubMed] [Google Scholar]
- Nadif Kasri N, Nakano-Kobayashi A, Malinow R, Li B, Van Aelst L. The rho-linked mental retardation protein oligophrenin-1 controls synapse maturation and plasticity by stabilizing AMPA receptors. Genes Dev. 2009;23:1289–1302. doi: 10.1101/gad.1783809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai J, Takano M. Molecular aspects of renal handling of aminoglycosides and strategies for preventing the nephrotoxicity. Drug Metab Pharmacokinet. 2004;19:159–170. doi: 10.2133/dmpk.19.159. [DOI] [PubMed] [Google Scholar]
- Najmabadi H, Garshasbi M, Bahman I, et al. Homozygosity mapping in 4 unrelated Iranian families with autosomal recessive mental retardation identifies overlapping linkage intervals on chromosome 1p34: a frequent cause of ARMR? American Society of Human Genetics Meeting. 2009:2019.
- Najmabadi H, Motazacker MM, Garshasbi M, et al. Homozygosity mapping in consanguineous families reveals extreme heterogeneity of non-syndromic autosomal recessive mental retardation and identifies 8 novel gene loci. Hum Genet. 2007;121:43–48. doi: 10.1007/s00439-006-0292-0. [DOI] [PubMed] [Google Scholar]
- Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, Kriaucionis S, Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci USA. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawara M, Klapecki J, Borg K, Jurek M, Moreno S, Tryfon J, Bal J, Chelly J, Mazurczak T. Novel mutation of IL1RAPL1 gene in a nonspecific X-linked mental retardation (MRX) family. Am J Med Genet A. 2008;146A:3167–3172. doi: 10.1002/ajmg.a.32613. [DOI] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Windpassinger C, Patel M, et al. CC2D2A, encoding a coiled-coil and C2 domain protein, causes autosomal-recessive mental retardation with retinitis pigmentosa. Am J Hum Genet. 2008;82:1011–1018. doi: 10.1016/j.ajhg.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Windpassinger C, Vitcu I, Orlic M, Rafiq MA, Khalid M, Malik MN, Ayub M, Alman B, Vincent JB. Oligodontia is caused by mutation in LTBP3, the gene encoding latent TGF-beta binding protein 3. Am J Hum Genet. 2009;84:519–523. doi: 10.1016/j.ajhg.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Whibley A, Marshall CR, Gianakopoulos PJ, Piton A, Carson AR, et al. Disruption at the PTCHD1 locus on Xp22.11 in autism spectrum disorder and intellectual disability. Science: Translational Medicine (in press). [DOI] [PMC free article] [PubMed]
- Nordin V, Gillberg C. Autism spectrum disorders in children with physical or mental disability or both. I: clinical and epidemiological aspects. Dev Med Child Neurol. 1996;38:297–313. doi: 10.1111/j.1469-8749.1996.tb12096.x. [DOI] [PubMed] [Google Scholar]
- O’Riordan KJ, Huang IC, Pizzi M, et al. Regulation of nuclear factor kappaB in the hippocampus by group I metabotropic glutamate receptors. J Neurosci. 2006;26:4870–4879. doi: 10.1523/JNEUROSCI.4527-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrico A, Lam C, Galli L, Dotti MT, Hayek G, Tong SF, Poon PM, Zappella M, Federico A, Sorrentino V. MECP2 mutation in male patients with non-specific X-linked mental retardation. FEBS Lett. 2000;481:285–288. doi: 10.1016/S0014-5793(00)01994-3. [DOI] [PubMed] [Google Scholar]
- Ou XM, Lemonde S, Jafar-Nejad H, Bown CD, Goto A, Rogaeva A, Albert PR. Freud-1: a neuronal calcium-regulated repressor of the 5-HT1A receptor gene. J Neurosci. 2003;23:7415–7425. doi: 10.1523/JNEUROSCI.23-19-07415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlowsky A, Gianfelice A, Pallotto M, et al. A postsynaptic signaling pathway that may account for the cognitive defect due to IL1RAPL1 mutation. Curr Biol. 2010;20:103–115. doi: 10.1016/j.cub.2009.12.030. [DOI] [PubMed] [Google Scholar]
- Philip N, Chabrol B, Lossi AM, Cardoso C, Guerrini R, Dobyns WB, Raybaud C, Villard L. Mutations in the oligophrenin-1 gene (OPHN1) cause X linked congenital cerebellar hypoplasia. J Med Genet. 2003;40:441–446. doi: 10.1136/jmg.40.6.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe O, Rio M, Carioux A, et al. Combination of linkage mapping and microarray-expression analysis identifies NF-kappaB signaling defect as a cause of autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:903–908. doi: 10.1016/j.ajhg.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- Piton A, Michaud JL, Peng H, et al. Mutations in the calcium-related gene IL1RAPL1 are associated with autism. Hum Mol Genet. 2008;17:3965–3974. doi: 10.1093/hmg/ddn300. [DOI] [PubMed] [Google Scholar]
- Rafiq MA, Ansar M, Marshall CR, et al. Mapping of three novel loci for non-syndromic autosomal recessive mental retardation (NS-ARMR) in consanguineous families from Pakistan. Clin Genet. 2010; [Epub ahead of print]. [DOI] [PubMed]
- Ramakers GJ. Rho proteins, mental retardation and the cellular basis of cognition. Trends Neurosci. 2002;25:191–199. doi: 10.1016/S0166-2236(00)02118-4. [DOI] [PubMed] [Google Scholar]
- Rauch A, Hoyer J, Guth S, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A. 2006;140:2063–2074. doi: 10.1002/ajmg.a.31416. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif R, Sales S, Hettwer S, et al. Specific cleavage of agrin by neurotrypsin, a synaptic protease linked to mental retardation. FASEB J. 2007;21:3468–3478. doi: 10.1096/fj.07-8800com. [DOI] [PubMed] [Google Scholar]
- Rejeb I, Saillour Y, Castelnau L, Julien C, Bienvenu T, Taga P, Chaabouni H, Chelly J, Ben Jemaa L, Bahi-Buisson N. A novel splice mutation in PAK3 gene underlying mental retardation with neuropsychiatric features. Eur J Hum Genet. 2008;16:1358–1363. doi: 10.1038/ejhg.2008.103. [DOI] [PubMed] [Google Scholar]
- Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeleveld N, Zielhuis GA, Gabreels F. The prevalence of mental retardation: a critical review of recent literature. Dev Med Child Neurol. 1997;39:125–132. doi: 10.1111/j.1469-8749.1997.tb07395.x. [DOI] [PubMed] [Google Scholar]
- Rogaeva A, Galaraga K, Albert PR. The Freud-1/CC2D1A family: transcriptional regulators implicated in mental retardation. J Neurosci Res. 2007;85:2833–2838. doi: 10.1002/jnr.21277. [DOI] [PubMed] [Google Scholar]
- Ropers HH. X-linked mental retardation: many genes for a complex disorder. Curr Opin Genet Dev. 2006;16:260–269. doi: 10.1016/j.gde.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Ropers HH, Hamel BC. X-linked mental retardation. Nat Rev Genet. 2005;6:46–57. doi: 10.1038/nrg1501. [DOI] [PubMed] [Google Scholar]
- Rousseau-Merck MF, Koczan D, Legrand I, Moller S, Autran S, Thiesen HJ. The KOX zinc finger genes: genome wide mapping of 368 ZNF PAC clones with zinc finger gene clusters predominantly in 23 chromosomal loci are confirmed by human sequences annotated in EnsEMBL. Cytogenet Genome Res. 2002;98:147–153. doi: 10.1159/000069802. [DOI] [PubMed] [Google Scholar]
- Ruano D, Abecasis GR, Glaser B, et al. Functional gene group analysis reveals a role of synaptic heterotrimeric G proteins in cognitive ability. Am J Hum Genet. 2010. [DOI] [PMC free article] [PubMed]
- Rumbaugh G, Adams JP, Kim JH, Huganir RL. SynGAP regulates synaptic strength and mitogen-activated protein kinases in cultured neurons. Proc Natl Acad Sci USA. 2006;103:4344–4351. doi: 10.1073/pnas.0600084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone S, De Sario A, Della Valle G, Bernardi G. The highest gene concentrations in the human genome are in telomeric bands of metaphase chromosomes. Proc Natl Acad Sci USA. 1992;89:4913–4917. doi: 10.1073/pnas.89.11.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomons GS, van Dooren SJ, Verhoeven NM, Cecil KM, Ball WS, Degrauw TJ, Jakobs C. X-linked creatine-transporter gene (SLC6A8) defect: a new creatine-deficiency syndrome. Am J Hum Genet. 2001;68:1497–1500. doi: 10.1086/320595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Dev. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- Sans N, Petralia RS, Wang YX, Blahos J, 2nd, Hell JW, Wenthold RJ. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci. 2000;20:1260–1271. doi: 10.1523/JNEUROSCI.20-03-01260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer IE, Wallace RH, Phillips FL, Hewson P, Reardon K, Parasivam G, Stromme P, Berkovic SF, Gecz J, Mulley JC. X-linked myoclonic epilepsy with spasticity and intellectual disability: mutation in the homeobox gene ARX. Neurology. 2002;59:348–356. doi: 10.1212/wnl.59.3.348. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/S0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Schmitz D, Mellor J, Breustedt J, Nicoll RA. Presynaptic kainate receptors impart an associative property to hippocampal mossy fiber long-term potentiation. Nat Neurosci. 2003;6:1058–1063. doi: 10.1038/nn1116. [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senderek J, Bergmann C, Stendel C, et al. Mutations in a gene encoding a novel SH3/TPR domain protein cause autosomal recessive Charcot-Marie-Tooth type 4C neuropathy. Am J Hum Genet. 2003;73:1106–1119. doi: 10.1086/379525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr EH. The ARX story (epilepsy, mental retardation, autism, and cerebral malformations): one gene leads to many phenotypes. Curr Opin Pediatr. 2003;15:567–571. doi: 10.1097/00008480-200312000-00004. [DOI] [PubMed] [Google Scholar]
- Shoichet SA, Hoffmann K, Menzel C, et al. Mutations in the ZNF41 gene are associated with cognitive deficits: identification of a new candidate for X-linked mental retardation. Am J Hum Genet. 2003;73:1341–1354. doi: 10.1086/380309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuang M, Liu J, Jia MX, Yang JZ, Wu SP, Gong XH, Ling YS, Ruan Y, Yang XL, Zhang D. Family-based association study between autism and glutamate receptor 6 gene in Chinese Han Trios. Am J Med Genet B Neuropsychiatr Genet. 2004;131B:48–50. doi: 10.1002/ajmg.b.30025. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Beaudet AL. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr Opin Genet Dev. 2007;17:182–192. doi: 10.1016/j.gde.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan A, Mateos JM, Kozlov SV, Cinelli P, Kistler AD, Hettwer S, Rulicke T, Streit P, Kunz B, Sonderegger P. Neurotrypsin cleaves agrin locally at the synapse. FASEB J. 2008;22:1861–1873. doi: 10.1096/fj.07-100008. [DOI] [PubMed] [Google Scholar]
- Stocco dos Santos RC, Castro NH, Lillia Holmes A, Becak W, Tackels-Horne D, Lindsey CJ, Lubs HA, Stevenson RE, Schwartz CE. Stocco Dos Santos X-linked mental retardation syndrome: clinical elucidation and localization to Xp11.3-Xq21.3. Am J Med Genet A. 2003;118A:255–259. doi: 10.1002/ajmg.a.20021. [DOI] [PubMed] [Google Scholar]
- Stromme P, Diseth TH. Prevalence of psychiatric diagnoses in children with mental retardation: data from a population-based study. Dev Med Child Neurol. 2000;42:266–270. doi: 10.1017/S0012162200000451. [DOI] [PubMed] [Google Scholar]
- Stromme P, Mangelsdorf ME, Shaw MA, et al. Mutations in the human ortholog of aristaless cause X-linked mental retardation and epilepsy. Nat Genet. 2002a;30:441–445. doi: 10.1038/ng862. [DOI] [PubMed] [Google Scholar]
- Stromme P, Mangelsdorf ME, Scheffer IE, Gecz J. Infantile spasms, dystonia, and other X-linked phenotypes caused by mutations in aristaless related homeobox gene, ARX. Brain Dev. 2002b;24:266–268. doi: 10.1016/S0387-7604(02)00079-7. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons FJ, Sperry LA, Dropik PL, Bodfish JW. The early development of stereotypy and self-injury: a review of research methods. J Intellect Disabil Res. 2005;49:144–158. doi: 10.1111/j.1365-2788.2004.00632.x. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Tarpey P, Parnau J, Blow M, et al. Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am J Hum Genet. 2004;75:318–324. doi: 10.1086/422703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Stevens C, Teague J, et al. Mutations in the gene encoding the sigma 2 subunit of the adaptor protein 1 complex, AP1S2, cause X-linked mental retardation. Am J Hum Genet. 2006;79:1119–1124. doi: 10.1086/510137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Raymond FL, Nguyen LS, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- Troester MM, Trachtenberg T, Narayanan V. A novel mutation of the ARX gene in a male with nonsyndromic mental retardation. J Child Neurol. 2007;22:744–748. doi: 10.1177/0883073807304000. [DOI] [PubMed] [Google Scholar]
- Tzschach A, Lenzner S, Moser B, et al. Novel JARID1C/SMCX mutations in patients with X-linked mental retardation. Hum Mutat. 2006;27:389. doi: 10.1002/humu.9420. [DOI] [PubMed] [Google Scholar]
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyguner O, Kayserili H, Li Y, et al. A new locus for autosomal recessive non-syndromic mental retardation maps to 1p21.1-p13.3. Clin Genet. 2007;71:212–219. doi: 10.1111/j.1399-0004.2007.00762.x. [DOI] [PubMed] [Google Scholar]
- Verpy E, Masmoudi S, Zwaenepoel I, et al. Mutations in a new gene encoding a protein of the hair bundle cause non-syndromic deafness at the DFNB16 locus. Nat Genet. 2001;29:345–349. doi: 10.1038/ng726. [DOI] [PubMed] [Google Scholar]
- Vervoort VS, Beachem MA, Edwards PS, et al. AGTR2 mutations in X-linked mental retardation. Science. 2002;296:2401–2403. doi: 10.1126/science.1072191. [DOI] [PubMed] [Google Scholar]
- Wagenstaller J, Spranger S, Lorenz-Depiereux B, et al. Copy-number variations measured by single-nucleotide-polymorphism oligonucleotide arrays in patients with mental retardation. Am J Hum Genet. 2007;81:768–779. doi: 10.1086/521274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/S0092-8674(00)81404-X. [DOI] [PubMed] [Google Scholar]
- Wilschanski M, Yahav Y, Yaacov Y, et al. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N Engl J Med. 2003;349:1433–1441. doi: 10.1056/NEJMoa022170. [DOI] [PubMed] [Google Scholar]
- Wing L, Gould J. Severe impairments of social interaction and associated abnormalities in children: epidemiology and classification. J Autism Dev Disord. 1979;9:11–29. doi: 10.1007/BF01531288. [DOI] [PubMed] [Google Scholar]
- Witzgall R, O’Leary E, Leaf A, Onaldi D, Bonventre JV. The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc Natl Acad Sci USA. 1994;91:4514–4518. doi: 10.1073/pnas.91.10.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M, Jakobsson J, Szulc J, Liao S, Quazzola A, Beermann F, Aebischer P, Trono D. The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J Biol Chem. 2007;282:34535–34541. doi: 10.1074/jbc.M705898200. [DOI] [PubMed] [Google Scholar]
- Wolfer DP, Lang R, Cinelli P, Madani R, Sonderegger P. Multiple roles of neurotrypsin in tissue morphogenesis and nervous system development suggested by the mRNA expression pattern. Mol Cell Neurosci. 2001;18:407–433. doi: 10.1006/mcne.2001.1029. [DOI] [PubMed] [Google Scholar]
- Wu Y, Arai AC, Rumbaugh G, et al. Mutations in ionotropic AMPA receptor 3 alter channel properties and are associated with moderate cognitive impairment in humans. Proc Natl Acad Sci USA. 2007;104:18163–18168. doi: 10.1073/pnas.0708699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yntema HG, Poppelaars FA, Derksen E, Oudakker AR, van Roosmalen T, Jacobs A, Obbema H, Brunner HG, Hamel BC, van Bokhoven H. Expanding phenotype of XNP mutations: mild to moderate mental retardation. Am J Med Genet. 2002;110:243–247. doi: 10.1002/ajmg.10446. [DOI] [PubMed] [Google Scholar]
- Zahir F, Friedman JM. The impact of array genomic hybridization on mental retardation research: a review of current technologies and their clinical utility. Clin Genet. 2007;72:271–287. doi: 10.1111/j.1399-0004.2007.00847.x. [DOI] [PubMed] [Google Scholar]
- Zemni R, Bienvenu T, Vinet MC, et al. A new gene involved in X-linked mental retardation identified by analysis of an X;2 balanced translocation. Nat Genet. 2000;24:167–170. doi: 10.1038/72829. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Luan Z, Liu A, Hu G. The scaffolding protein CASK mediates the interaction between rabphilin3a and beta-neurexins. FEBS Lett. 2001;497:99–102. doi: 10.1016/S0014-5793(01)02450-4. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Fischer DJ, Santos MF, Tigyi G, Pasteris NG, Gorski JL, Xu Y. The faciogenital dysplasia gene product FGD1 functions as a Cdc42Hs-specific guanine-nucleotide exchange factor. J Biol Chem. 1996;271:33169–33172. doi: 10.1074/jbc.271.52.33169. [DOI] [PubMed] [Google Scholar]
- Zhou H, Clapham DE. Mammalian MagT1 and TUSC3 are required for cellular magnesium uptake and vertebrate embryonic development. Proc Natl Acad Sci USA. 2009;106:15750–15755. doi: 10.1073/pnas.0908332106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JJ, Qin Y, Zhao M, Van Aelst L, Malinow R. Ras and rap control AMPA receptor trafficking during synaptic plasticity. Cell. 2002;110:443–455. doi: 10.1016/S0092-8674(02)00897-8. [DOI] [PubMed] [Google Scholar]
- Zweier C, de Jong EK, Zweier M, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in drosophila. Am J Hum Genet. 2009;85:655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]