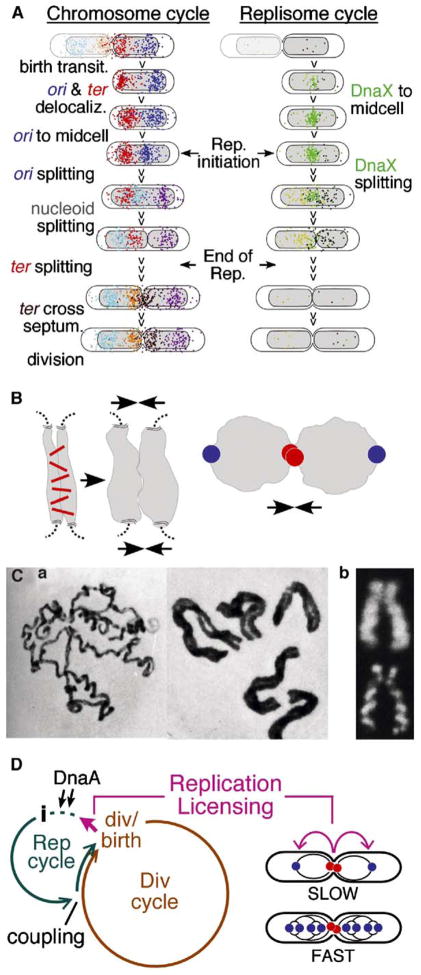
Summary
Chromosome and replisome dynamics were examined in synchronized E. coli cells undergoing a eukaryotic-like cell cycle. Sister chromosomes remain tightly colocalized for much of S phase and then separate, in a single coordinate transition. Origin and terminus regions behave differently, as functionally independent domains. During separation, sister loci move far apart and the nucleoid becomes bilobed. Origins and terminus regions also move. We infer that sisters are initially linked and that loss of cohesion triggers global chromosome reorganization. This reorganization creates the 2-fold symmetric, ter-in/ori-out conformation which, for E. coli, comprises sister segregation. Analogies with eukaryotic prometaphase suggest that this could be a primordial segregation mechanism to which microtubule-based processes were later added. We see no long-lived replication “factory”; replication initiation timing does not covary with cell mass, and we identify changes in nucleoid position and state that are tightly linked to cell division. We propose that cell division licenses the next round of replication initiation via these changes.
Introduction
All dividing cells face the same fundamental problems of chromosome management. Two of the most important are how to ensure that sister chromosomes always segregate regularly into the two daughter cells and how to ensure a one-to-one relationship between DNA replication and cell division. We have investigated these processes in E. coli with the idea that further understanding of events in this classical model organism might ultimately provide insights applicable to eukaryotic organisms.
E. coli has a single circular chromosome. Replication is initiated from a unique, genetically defined origin and proceeds bidirectionally. Converging forks are resolved approximately halfway around the chromosome in a broad “terminus region” that includes several specialized determinants. Eukaryotic replication is analogous, with each chromosome comprising multiple origins and specialized intervening “terminus regions” (e.g., for yeast, Cha and Kleckner, 2002).
At cell division, sister E. coli chromosomes occur in a 2-fold symmetric, mirror-image configuration: termini are located just to either side of midcell; and origins are located near the cell poles (e.g., Gordon et al., 2002; Lau et al., 2003; Viollier et al., 2004; Figure 1). Ensuing cell division creates sister cells having a highly asymmetric, polarized configuration (Figure 1, left). Thus, for E. coli, sister segregation comprises the program of chromosome dynamics that recreates a symmetrical sister chromosome configuration (Figure 1, right).
Figure 1. Chromosome Resymmetrization Mediates Chromosome Segregation.
Slowly growing wild-type cells just prior to division (left) and just after birth (right); images are overlays of phase contrast, DAPI (gray) and ori (blue) and ter (red) FISH exposures. Dashed box: ori and ter movements required to convert polarized newborn cell into a dividing cell with mirror-image symmetry.
Two models for E. coli sister segregation have been proposed. In one model, origin-proximal regions function analogously to eukaryotic centromeres: they lead sisters to opposite poles, with other regions following behind (Ben-Yehuda et al., 2003; Gordon et al., 2002). In the other model, replicating regions are drawn through a midcell-localized complex containing paired clockwise and counterclockwise replisomes, sometimes called a “replication factory.” As sister chromosomes emerge from this complex, they are extruded outward in opposite directions toward the poles; concomitantly, the terminus is drawn automatically into its final position at midcell (Lemon and Grossman, 1998). However, previous E. coli studies have suggested that sister regions outside of the origin may remain linked for a significant period of time after their replication (Sunako et al., 2001, and references therein).
A further important issue is that changes in chromosome disposition involve chromosome motion, which in turn requires mechanical force. “Origin-as-centromere” models invoke contractile filaments operating on the origins. The replisome extrusion model invokes forces generated by movement of replicating DNA through the replisome and compaction of replicated DNA in opposite halves of the cell (Lemon and Grossman, 2001).
Coordination between the DNA replication and cell growth/division cycles in E. coli is also a subject of active debate. For cells growing exponentially in steady state, each cell mass doubling must be accompanied by one round of cell division and one round of DNA replication. A long-standing model proposes that cell mass (or some closely correlated parameter) determines replication initiation, e.g., by mediating accumulation of a critical concentration of initiator protein DnaA, which in turn governs the timing of cell division (Donachie and Blakely, 2003, and references therein). However, cell mass/timing relationships predicted by this model are not always observed (Boye and Nordstrom, 2003; below; Wold et al., 1994).
Investigation of these questions in E. coli has been hindered by lack of a simple method for obtaining large populations of cells proceeding synchronously through the cell cycle under normal growth conditions. The current study uses a newly developed approach that overcomes previous limitations and permits high temporal resolution (Bates et al., 2005; below). Chromosomal events were examined in synchronous populations under relatively slow growth conditions, where E. coli undergoes a linear progression from G1 to S phase to G2 to chromosome segregation and division (M phase), thus facilitating interpretation of events and comparisons with the eukaryotic cell cycle. The results of this study provide new information regarding several basic aspects of E. coli chromosome and replisome dynamics and their coordination with cell division. Implications for analogous eukaryotic processes are discussed.
Results
Experimental Approach
Synchronous cell cultures were obtained using a baby cell column (Experimental Procedures; Bates et al., 2005). Exponentially growing cells are affixed to glass beads via their flagella. These beads are packed into a column through which fresh medium is flowing and flagellin synthesis is turned off. Thereafter, newly divided (“newborn”) cells that lack flagella flow off the column. Eluted cells collected over ~1% of a generation time comprise a uniform population of newborn cells which, upon further incubation in growth medium, proceed synchronously through at least two ensuing rounds of cell division. Baby cell cultures are in steady state, unperturbed by the synchronization process (Bates et al., 2005). To obtain sufficient cells for analysis of multiple events at each of a large number of time points after birth, multiple samples were collected in rapid succession from a single column and then incubated for appropriately varying amounts of time prior to analysis. Such populations exhibit highly reproducible cell cycles. In independent experiments, cell doubling times and the timing of assayed events within the cell cycle vary by <10 min (below; data not shown).
Timing of Landmark Events at Doubling Time (Td) = 125 min
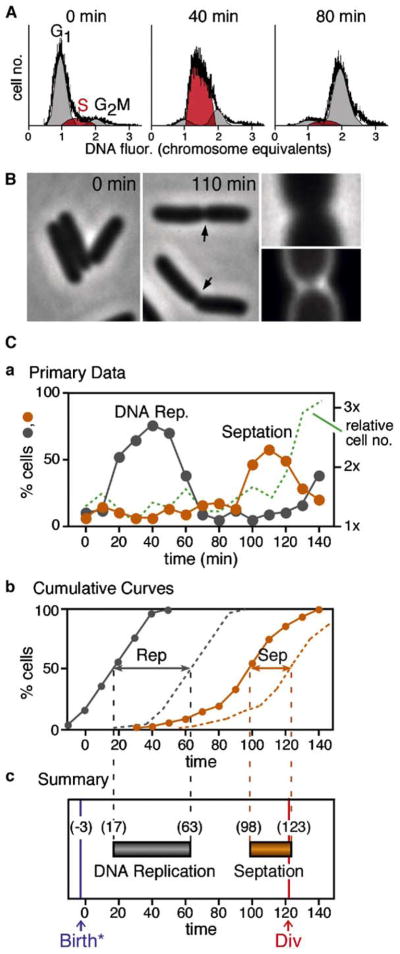
DNA replication was analyzed by flow cytometry. The percentage of cells undergoing bulk DNA replication at a particular time point is given by the percentage of cells with DNA contents intermediate between 1n and 2n (Figure 2A). The level of “S phase” cells increases to a maximum at t = 40 min and then decreases (Figure 2Ca). Septation is detected as appearance of indentations visible by phase contrast microscopy (Figure 2B). The percentage of septated cells increases to a maximum at t = 110 min and then decreases (Figure 2Ca). Cell division, manifested by increased cell concentration (Experimental Procedures), occurs at ~t = 120 min. (Figure 2Ca). Total interdivision time, given by the time between the first and second division cycles after elution, is 123–125 min (data not shown). Thus, actual cell “birth” precedes emergence of cells off the column by ~3–5 min.
Figure 2. Landmark Cell Cycle Events.
(A) Flow cytometry defines the distributions of DNA contents in a cell population, from which the percentage of cells in G1, S, and G2 phases can be calculated (Experimental Procedures).
(B) Visible cell septation is detected by phase contrast microscopy (arrows, middle panel); presence of a septal membrane ring can be verified by staining with fluorescent dye FM4-64 (right panel).
(C) Timing of landmark events at Td = 125min. (a) Percentages of cells undergoing DNA replication (gray) or cell septation (gold) and relative cell number (green) as a function of time. (b) Cumulative curves describing the kinetics of entry into (solid lines) and exit out of (dashed lines) the replication (gray) and septation (gold) stages. (c) Summary given by times at which 50% of cells have completed events in (b) plus timing of cell division from (a) and timing of cell birth, one division time earlier (text). Data from sample set I.
For a synchronous cell population, the progression of events can be described more precisely by an approach used previously for analysis of synchronous yeast meiosis (e.g., Hunter and Kleckner, 2001). (1) Any particular chromosomal or cellular stage has a particular duration or “life span,” which can be calculated as the area under the corresponding primary curve (in % cells × time) divided by the total percentage of cells that go through the stage. (2) Given such a life span, the area under a primary curve can be converted to a “cumulative curve” describing the percentage of cells that have entered the stage in question as a function of time. (3) The percentage of cells that has exited each stage is also given by a parallel curve displaced along the time axis by one life span. (4) “Time of occurrence” of an event is defined as the time at which 50% of cells have entered (or exited) the corresponding stage. In cell populations eluted from the baby-cell column, ~80% of cells are proceeding through S phase and other identified stages as a single synchronous wave while the remaining ~20% are a minority subpopulation of asynchronous cells (Bates et al., 2005). Since the rise and fall in primary data curves represent essentially only the synchronous cells, life spans are calculated by dividing the area under the primary curve by the percentage of such cells, 81% for the current experiment. Correspondingly, the resulting cumulative curves represent the behavior of this entire synchronous cell population and, as a result, rise to 100% (Bates et al., 2005).
A representative synchronous cell population growing under “standard conditions” behaved as follows (Figures 2Cb and 2Cc). Bulk DNA replication lasted 46 min and extended from t = 17 min to t = 63 min. These times are accurate to within 5 min (see the Supplemental Data available with this article online). The septation stage lasted 25 min and extended from t = 98 min through t = 123 min, when it is terminated by cell division. By independent measurements, division occurred at t = 121 min (above). This correspondence confirms the accuracy of cumulative curve analysis.
Sister Chromosome Relationships Evolve in Four Discrete Phases
Four chromosomal loci were probed simultaneously by fluorescence in situ hybridization (FISH): the replication origin (ori); the gln and lac loci, located respectively at ~133 and ~1084 kb clockwise from the origin; and a terminus region segment (ter) located directly opposite ori, near the dif locus involved in dimer resolution (Figure 3A). Each cell was visualized as a two-dimensional image in a single focal plane (e.g., Figure 3B). The number of FISH-staining foci for each locus present was scored in 50 cells at each time point. Detection was 75%–90% efficient at each locus as determined by the percentage of newborn cells exhibiting a signal (data not shown).
Figure 3. Sister Chromosome Cohesion and Separation.
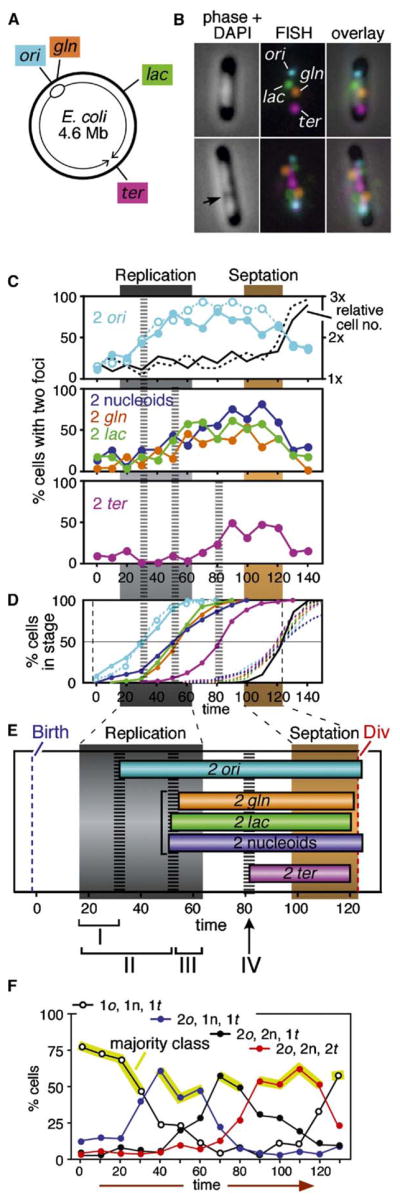
(A) Chromosomal positions of ori, gln, lac, and ter.
(B) Loci in (A) were visualized by four-color FISH; each probed locus may present either one or two staining foci. Nucleoids were visualized by DAPI staining and may be single- or bilobed (arrow).
(C) Percentage of cells containing two ori foci (top panel), two gln or two lac foci or two DAPI-staining bodies (middle panel), or two ter foci (bottom panel). Td = 125min. Closed circles—sample set II; open circles plus timing of replication and septation stages—sample set I, Figure 2C.
(D) Cumulative entry and exit curves (solid and dashed lines) for each two-focus stage and the two-nucleoid stage.
(E) Summary, with four stages in the evolution of sister chromosome relationships (text) indicated at bottom.
(F) Frequencies of cells containing different numbers of ori (o) and ter (t) foci and nucleoid(s) (n) (sample set I). Cell classes represented by <5% of total cells (e.g., 1o, 1n, 2t) were excluded from the analysis. Yellow lines identify majority class at each time point.
In newborn cells (t = 0), each of the four probed loci appears primarily as a single staining focus; two-focus cells then appear, first for ori (filled and open turquoise circles represent two independent experiments), then for gln and lac, essentially simultaneously (filled orange and green circles), and finally for ter (filled magenta circles) (Figure 3C). The frequency of two-focus cells then decreases coordinately at all loci at the time of (and due to) cell division (Figure 3C).
A locus may be represented as a single focus either because it has not yet been replicated or because it has been replicated but the two sister chromosomes remain tightly colocalized (e.g., Niki et al., 2000). When the times of appearance of two-focus cells as defined by FISH are compared with replication timing as defined by flow cytometry (Figure 2), three separation transitions emerge (Figures 3C–3E) which define four stages in the evolution of sister chromosome relationships (Figure 3E).
Stage I: Colocalization and Separation of Sister Origin Domains
DNA replication initiates at ~t = 17 min. Two-ori cells appear at ~t = 31 min. Thus, sister origins remain colocalized for ~14 min after initiation of replication, after which they separate. Since the nearby gln region does not exhibit sister separation until later, ori separation involves a small “origin domain.” Sister ori colocalization may result from direct linkage, with splitting resulting from loss of cohesion. Alternatively, since origin(s) are fixed at midcell from early in the cell cycle up to the time of sister ori separation (below), newly formed sister origins might be independently affixed to a common underlying entity and then released.
Stage II: Cohesion and Loss of Cohesion Partway through S Phase
At the time when separated sister gln and lac regions appear, replication has proceeded through ~75% of the chromosome (~t = ~50 min; Figure 3E). Since gln is immediately adjacent to ori and lac is located ~50% of the way around the chromosome, DNA replication has proceeded well beyond both loci before separation occurs. We infer that sister chromosomes remain tightly colocalized from the time of their formation until much of the chromosome is replicated, at which point they separate throughout most or all of the thus-far-replicated region (see also below). By this model, any locus between gln and some as-yet-undefined distance beyond lac would separate coordinately. We assume that this same pattern pertains symmetrically on both sides of the origin. Global colocalization of sister foci most likely reflects their direct linkage, i.e., “cohesion,” with separation of corresponding sister regions representing loss of cohesion plus actively promoted sister separation.
Stage III: Disposition of Late-Replicating Sister Regions
Loci in the terminus-proximal half of the chromosome have not yet been probed, but evidence below suggests that regions replicated after the major sister-splitting transition may separate immediately, without prolonged cohesion (below).
Stage IV: Appearance of Two ter Foci
Two ter foci appear well after the end of bulk DNA replication (t = 82 min versus t = 63 min) before onset of septation (t = 98 min) and long before cell division (t = 123 min; Figure 3D). We favor the idea that this event corresponds to separation of already-replicated sister ter’s, which may either be linked or independently colocalized (Discussion).
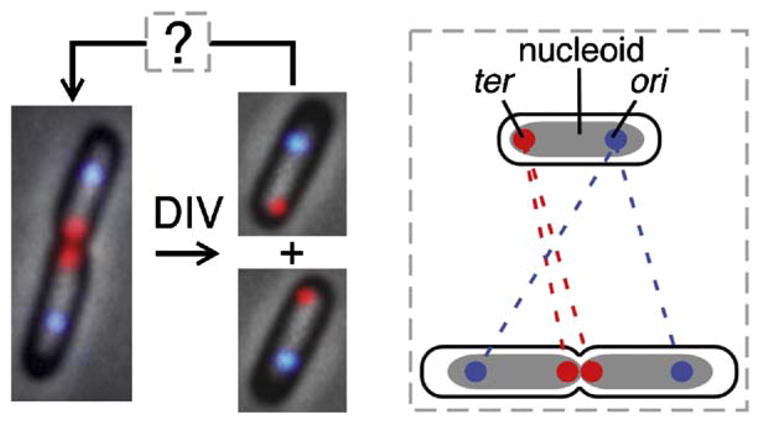
The Sister Separation Transition Involves Nucleoid Splitting and Wide Separation of Corresponding Sister Regions
Cells examined above by FISH were also examined for bulk chromosome status by staining with the fluorescent dye 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI). The nucleoid initially appears as a single cylindrical object (Figure 3B, top left) but, at later times, is bilobed or “split.” Nucleoid splitting is not an artifact of sample preparation: the same transition can be observed, with the same timing after birth, in living cells (data not shown).
Split-nucleoid cells appear at about the same time as cells containing two gln or two lac foci, as seen in both primary data and corresponding cumulative curves (Figures 3C and 3D). These data further support occurrence of a single underlying transition in which corresponding sister regions become widely separated into two different halves of the cell. Correspondingly, inspection of individual cells shows that, among cells containing two gln foci, two lac foci, and a bilobed nucleoid, all (>95%) exhibit one gln focus and one lac focus in each nucleoid lobe (Figure 4D; discussed further below). Importantly, this correlation holds even at the early time points, when such cells are first appearing, as expected if the three events are truly concomitant.
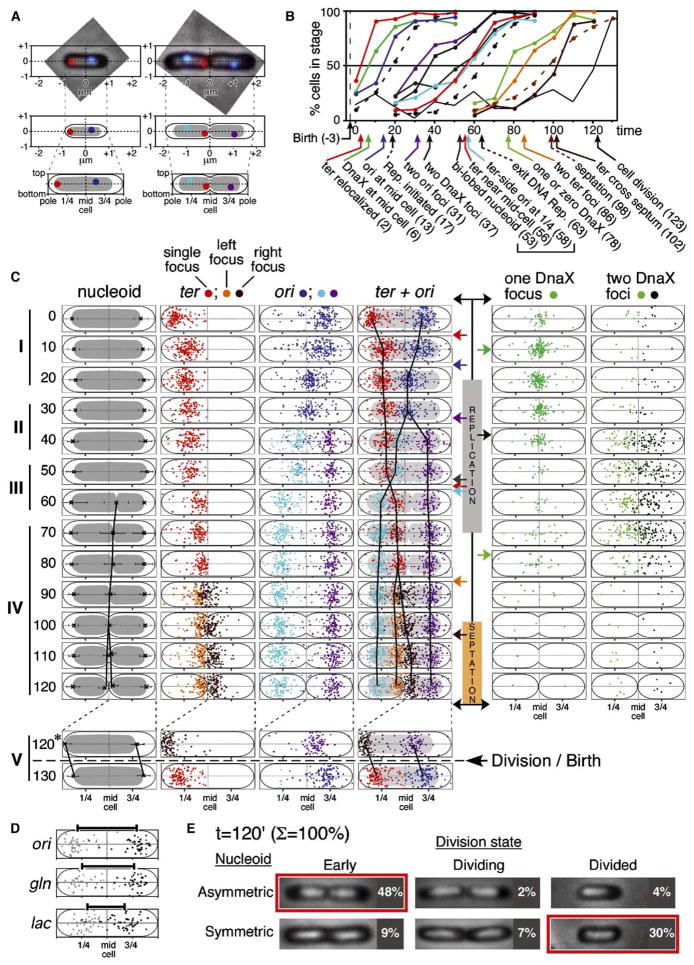
Figure 4. Spatial Dynamics of the Chromosome and Replisome.
Cellular positions of all ori and ter foci, all DnaX-GFP foci, and dimensional boundaries of all nucleoids and phase contrast cell outlines were recorded for each of 200 cells at 13 sequential time points (sample set I).
(A) Absolute positions of each entity of interest were defined with respect to x and y coordinates centered at the middle of the cell (e.g., top, middle). Absolute positions were then normalized to total cell length and width, giving corresponding relative positions (bottom).
(B) Cumulative curves describing occurrence of events of interest. Curves for DNA replication, septation, and division are from Figure 2. Data for numbers of foci and nucleoids from Figure 3F. For events involving focus position, an entity was defined as being “at” or “near” a particular location if it was ≤1/5 of the total cell length away from that location. The resolution of this approach is sufficient to resolve events separated in time by more than 5 min, as confirmed in several cases by individual cell analysis (e.g., Figures 5D and 5E).
(C) Left: normalized focus positions for cells in the majority class at each time point (defined by Figure 3F), drawn within a nucleoid of average size for that class (error bars = 1 SD). Vertical black lines are drawn through the mean positions of split nucleoids (inner boundaries only) and of ori and ter foci. Right: Positions of DnaX foci in all cells at each time point.
(D) Normalized positions of ori, gln, and lac foci in cells containing split nucleoids at t = 60 min in the four-color FISH experiment of Figure 3C. Bars represent the distance between the mean positions of the left (gray dots) and right (black dots) foci; “left” and “right” were defined arbitrarily.
(E) Cell division is accompanied by nucleoid repositioning (text).
Sequential ori splitting, nucleoid splitting, and ter splitting were confirmed in an independent experiment (Figure 3F).
Method of Positional Analysis
We next defined, as a function of time after cell birth, the spatial positions of origin and terminus foci and nucleoid boundaries as well as the numbers and position(s) of immnostaining foci of a replisome component, DnaX. In a single experiment (Figure 3F), 200 cells at each of 13 time points were analyzed. For each cell, the end nearest to a ter focus was designated as “left.” Relative to this orientation, the middle of the cell was determined and defined as (0,0). The position of each focus of interest was then marked and assigned x and y coordinates (Figure 4A, middle). Absolute focus positions were then normalized relative to total cell length and width, giving a corresponding set of “relative” positions (Figure 4A, bottom). Normalization corrects for significant variation in absolute cell lengths among cells of the same age/stage (Bates et al., 2005), thus giving much sharper positional distributions. It also facilitates monitoring of the 1/4 and 3/4 positions, which have special significance as the division sites in the next cell cycle. 31,000 data points were defined in this way.
Origin, Terminus, and Nucleoid Dynamics
At each time point, a majority of cells belongs to a particular class as defined by the “splitting transitions” described above (yellow line, Figure 3F). The overall pattern of ori, ter, and nucleoid dynamics is described by their positional distributions in these majority classes (Figure 4C). Cumulative curve analysis, which utilizes the data from all cells, gives precise timing of identified changes (Figure 4B).
I. Relaxation of ter and ori Positions and Migration of ori to Midcell Culminate in Initiation of DNA Replication
Newborn cells exhibit a single centrally positioned nucleoid with ter strongly localized at the end nearest the recent division site, and ori localized in the opposite half of the cell near the 3/4 position (Figure 4C, t = 0 min). Soon after birth, both loci become delocalized but remain within their respective halves of the cell (Figure 4C, t = 10 min). Then, ori’s become tightly colocalized at midcell while ter positions remain dispersed (Figure 4C, t = 20 min). DNA replication initiates, ~5 min after arrival of ori at midcell (Figure 4B). While 5 min is close to the resolution of our approach (above), initiation likely occurs with a midcell-localized initiation ensemble.
II. Sister Origins Remain at Midcell and Then Separate Widely and Asymmetrically
Sister ori’s remain together at midcell throughout their colocalization period. When colocalization is lost, sister origins become widely and rapidly separated, with no evidence of any long-lived intermediate state (Figure 4C, t = 40–50 min). Strikingly, the two origins behave asymmetrically. The origin in the “non-ter” side of the cell moves directly to the 3/4 position, where it remains until cell division (Figure 4C, purple dots). The “ter” side origin moves outward but does not reach the 3/4 position, instead becoming localized between midcell and ter (Figure 4C, turquoise dots). The distribution of ter positions does not change during these events (Figure 4C). About 20 min then elapses with little change in any determinant (Figure 4C).
III. Nucleoid Splitting Transition Is Accompanied by Global Reorganization throughout the Genome
Nucleoid splitting occurs at ~t = 60 min (Figures 4B and 4C). Strikingly, concomitant with this transition, ter and the ter-side origin switch positions (Figure 4C, compare t = 50 and 60 min): ter moves inward toward midcell while the ter-side origin becomes localized to the 1/4 position, symmetrical to its sister at the 3/4 position. These three changes appear simultaneously in the majority phenotype class and are contemporaneous by cumulative curve analysis, to the level of resolution of this approach (Figure 4B, t = 52–58 min). Correspondingly, >90% of cells containing split-nucleoids exhibit a fully symmetrical ori pattern and a ter near midcell. We infer that nucleoid splitting and ori and ter repositioning are all components of a single coordinate transition. Once this transition has occurred, the positions of both ter and the ter-side ori are essentially fixed, with no major changes until the time of division (Figure 4C).
Several important implications emerge. First, the nucleoid splitting transition involves a global reorganization that includes not only wide separation of the loci undergoing splitting (i.e., gln and lac, above) but also a dramatic reorganization of other regions (i.e., ori and ter). Moreover, it seems as though the failure of the ter-side ori to move directly to the 1/4 position at the time of origin splitting is due to the presence of impeding chromosomal regions, e.g., unreplicated regions including ter, which are then moved out of the way by the nucleoid splitting transition (see also below).
Second, this transition creates the basic 2-fold symmetric, mirror-image configuration that comprises sister chromosome segregation in E. coli. This effect is manifested not only by the emergence of the final ter-in ori-out configuration but also in the positions of interstitial regions. Positional analysis of cells stained for gln and lac in the multicolor FISH experiment described above reveals that, at the time point when bilobed nucleoids first appear as the majority class (t = 60 min), ori, gln, and lac are symmetrically disposed on either side of midcell in relation to their relative locations along the chromosome (Figure 4D).
When bilobed nucleoids first appear, they are asymmetric, with the ter-side lobe being larger than the non-ter lobe (Figure 3B, bottom-left panel; Figure 4C, t = 60 min); they also have a dumbbell shape due to a prominent central connection. Then, as replication proceeds, the two lobes become equal in size, symmetrically disposed within the cell and well separated with no central connection (Figure 4C, t = 60–90 min). Concomitantly, ter positions seem to creep a bit closer to midcell and to become more tightly clustered (Figure 4C, 60–80 min). These patterns could suggest that unreplicated regions tend to occur on the ter side of the cell and that, in the latter stages of replication, this region is depleted and newly created sister regions are incorporated directly into their corresponding domains as already established by the nucleoid splitting transition, without any (prolonged) period of cohesion.
IV. ter-Specific Events
ter becomes specifically localized immediately adjacent to midcell by the end of bulk DNA replication (above). Approximately twenty minutes later, the single ter focus becomes two ter foci that are usually, but not always, positioned on one side of midcell (Figure 4C, t = 90 min). Lastly, the ter closest to midcell moves across midcell into a symmetrical position with respect to its sibling, thus finalizing the 2-fold symmetrical chromosome configuration present at cell division (Figure 4C, t = 100–110 min), with sister ter’s specifically localized at the nucleoid/midcell junctions and sister origins located at the 1/4 and 3/4 positions. Interestingly, at this point, the inner edges of sister nucleoids are tightly juxtaposed to midcell while there is considerable space between their outer edges and the adjacent cell poles (Figure 4C, t = 110 min; Figure 4E, top left image).
V. The Birth Transition
Cell division is accompanied by outward translation of sister nucleoids, in opposite directions, away from mid-cell, to symmetrical positions within their respective emerging daughter cells (compare Figure 4C, t = 120 with t = 0; Figure 4E, top left panel with bottom right panel). Cell division and nucleoid translation are tightly coupled processes. In the t = 120 min sample (Figure 4E), most predivisional cells have asymmetrically positioned nucleoids; in cells undergoing division as seen by septum morphology, both nucleoid types occur; and most divided cells have symmetrically positioned nucleoids. Interestingly, during this transition, the origin and terminus regions retain their positions relative to the nucleoid rather than remaining at fixed positions with respect to the boundaries of the cell (Figure 4C, bottom). Thus, the nucleoid apparently translates within the cell as an internally static unit.
DnaX/Replisome Dynamics
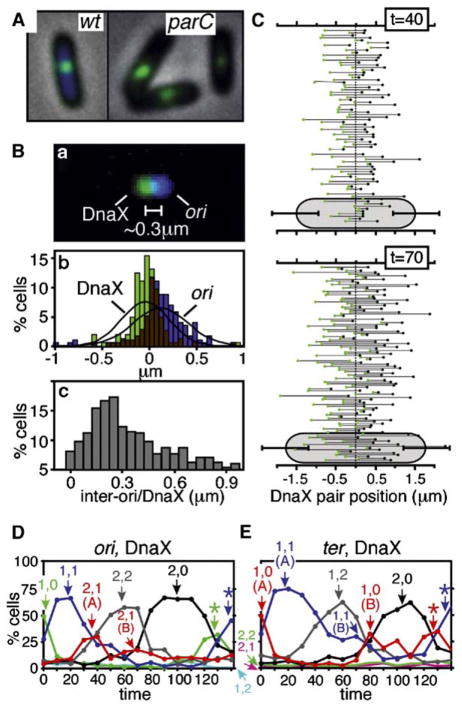
Wright and colleagues have shown that, in living cells, during DNA replication, foci of a DnaX-GFP fusion protein correspond to sites of DNA replication forks (personal communication). In the present study, foci of this same fusion protein were visualized using anti-GFP antibodies (Experimental Procedures). Since DnaX dynamics are not specifically correlated with sister-separation transitions, focus positions in all cells are presented separately (Figure 4C, right). These data, plus cumulative curve analysis (Figure 4B), reveal three new features of DnaX dynamics.
First, a prominent DnaX focus appears at midcell immediately following cell division (Figure 4C, t = 0–10 min). This midcell localization can occur even in the absence of any chromosome, as seen in anucleate cells formed in a partition-defective mutant (Figure 5A). DnaX appears at midcell prior to localization of ori. Interestingly, in DnaX/ori cofoci, DnaX has an average position exactly at midcell while ori occurs on average ~0.3 μm away, almost always on the “right” side of the cell, i.e., the side from which it would have approached (Figure 5B). Regular separation of ori and DnaX is not an artifactual result of image misalignment (Experimental Procedures). Given that replication initiates immediately after localization of ori to midcell (above), initiation occurs within the DnaX/ori midcell ensemble.
Figure 5. DnaX (Replisome) Dynamics.
(A) DnaX-GFP localizes to midcell not only in wild-type cells (left) but in anucleate parC1215(Ts) cells arising after growth at 42°C for 1 hr (right).
(B) Positions of DnaX-GFP and ori foci in cells containing a single focus of each type (from Figure 4C, t = 20 min; sample set I). (a) Overlay of DnaX and ori images from a typical cell. (b) All focus positions. (c) Inter-focus distances; mean = 0.3 μm.
(C) Positions of DnaX foci in two-focus cells at t = 40 and 70 min. Each line represents the pair of foci seen in a single cell. Mean cell lengths shown at bottom (error bars = 1 SD).
(D and E) Percentages of cells in different morphological categories plotted as a function of time; cells categorized by number of ori and DnaX foci (D) or ter and DnaX foci (E). Components of biphasic curves denoted by “(A)” and “(B).” *early classes reappearing after cell division.
Second, a single midcell DnaX focus remains through initiation of DNA replication and into S phase, implying that clockwise and counterclockwise replication fork ensembles are colocalized in an apparent macromolecular complex. After ~40% of “S phase,” cells exhibit two DnaX foci, which occur at highly variable, apparently independent positions throughout the nucleoid (Figure 4C, t = 40–70 min; Figure 5C). This transition implies splitting of clockwise and counterclockwise replisomes and their forks. It does not represent initiation of the next round of replication, which does not occur until after division (above), and does not represent precocious assembly of complexes in preparation for the next cycle, because two-focus cells are virtually absent during the last 40 min of the cell cycle. Replisome splitting occurs ~10 min after ori splitting (Figures 4B and 4C). The same sequence of events is seen by progressive appearance and disappearance of cells containing the appropriate numbers of ori and DnaX foci or ter and DnaX foci (Figures 5D and 5E).
Third, after completion of bulk DNA replication but prior to appearance of two ter foci, cells containing a single DnaX focus are transiently observed. Such cells are prominent at t = 70, 80 min (Figure 4C) and arise ~15 min after exit from DNA replication (Figure 4B). Correspondingly, cells containing one DnaX focus appear at late times in cells containing two ori foci (Figure 5D, 2,1 (B)) and in cells containing one ter focus (Figure 5E, 1,1 (B)), followed by cells containing one ter focus and no DnaX foci and, finally, two ter foci and no DnaX foci (Figure 5E, 1,0 (B) and 2,0). Late single foci still occur in dispersed positions. We infer that replisome ensembles tend to be lost sequentially from the chromosome. Since ter is flanked by sites that block progression of forks that progress beyond ter (Coskun-Ari and Hill, 1997), sequential encounters of the faster fork and then the slower fork with one such site could explain this pattern.
Comparison of Cellular and Chromosomal Events at Different Slow Growth Rates
To understand whether and how the observed events are linked to the time of cell birth, to the cell cycle as a whole, and/or to one another, we examined events of interest at two additional growth rates, one slower and one faster than the standard conditions analyzed above (Td = 90 and 300 min, versus Td = 125 min). We also examined origin, nucleoid, and terminus positions at Td = 90 min to assess temporal linkages amongst spatial changes prior to and during the nucleoid splitting transition.
Differences in Growth Rate Are Accommodated Primarily by Changes in the Length of “G2”
Analysis of landmark events (Figure 6A) shows that the period between cell birth and initiation of DNA replication, “G1,” is short and of similar length under all three conditions. S phase is also of similar length in all three conditions. Correspondingly, the major differences between the three situations is the length of the period between the end of bulk replication and cell division, i.e., G2. Septation occurs toward the end of this period and is also of similar length in the three conditions, though with some prolongation at slower growth rates. Notably, these comparisons all involve a single E. coli strain and thus can be attributed unambiguously to differences in growth conditions.
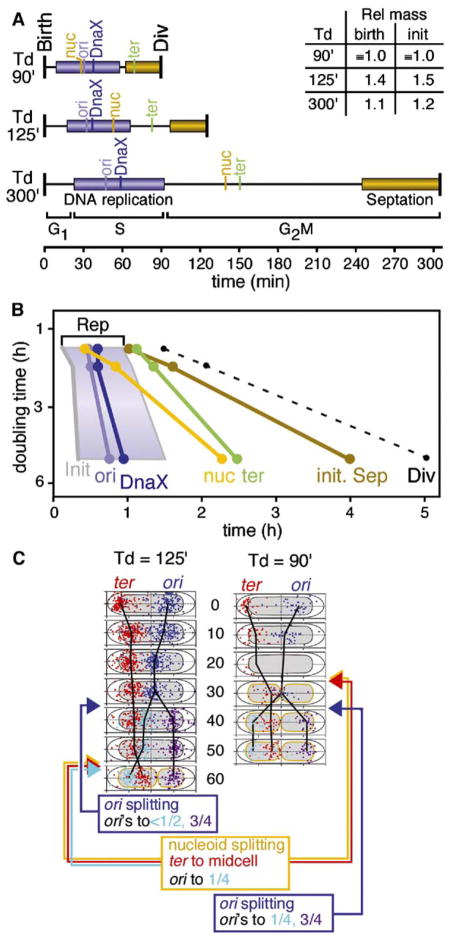
Figure 6. Event Timing Under Varied Growth Conditions.
(A) Timing of chromosome and replisome events in synchronized cells grown under three different conditions (text). Replication and septation are indicated by horizontal bars as above; splitting of ori, DnaX, ter, and the nucleoid and cell division, are indicated by vertical lines. Relative cell mass at birth was determined from average cell length at t = 0 (~birth) (L = 1.8, 2.5, and 2.0 μm for Td = 90, 125, and 300 min, respectively). Relative cell mass at the time of initiation was determined from average cell length at the time point corresponding to initiation of replication in each growth condition (L = 1.8, 2.7, and 2.1 μm, respectively).
(B) Alternate representation of data in (A); slope of line indicates extent to which timing of event after birth varies with doubling time. Period of DNA replication is shaded.
(C) Normalized positions of ori and ter FISH foci for Td = 90 min (A and B) as compared with those for Td = 125 min (above), described as in Figure 4C.
Replication Initiation Does Not Occur at a Specific Constant Cell Mass
Early models for replication control, developed for rapidly growing cells with multiple origins, proposed that initiation occurs when the ratio of cell mass to the number of origins reaches a specific fixed value which is independent of growth rate (Boye and Nordstrom, 2003; Wold et al., 1994). In the current study, DNA replication initiates at very similar times after birth under all three conditions (above) but relative cell masses do not vary in correlation with differences in growth rate; rather, cell mass at Td = 125 min is significantly greater than at the other two doubling times (e.g., at birth; Figure 6A, inset). As a result, among the three conditions, cell mass at the time of initiation varies by nearly a factor of two (Figure 6A, inset).
Origin Splitting and DnaX/Replisome Splitting Occur in Ordered Succession Just before Mid-S Phase
At all three growth rates, separation of sister origins away from midcell and separation of DnaX foci away from midcell always occur sequentially, about ten minutes apart, during the second quarter of S phase (Figures 6A and 6B). Thus, separation of sister origins may be linked directly to progression of DNA replication while replisome separation may be linked either to origin splitting and/or directly to replication progression as well.
The Timing of Nucleoid Splitting Varies Dramatically with Growth Rate, Independent of S Phase but in Parallel with Septation
In contrast to origin and replisome splitting, nucleoid splitting occurs at dramatically different times under the three growth conditions, in direct relation to growth rate (Figures 6A and 6B). At the fastest doubling time, this transition occurs early during DNA replication, prior to origin or replisome splitting; at the intermediate doubling time, it occurs in the latter part of S phase; and at the slowest doubling time, it occurs just after the end of bulk DNA replication. This pattern is strikingly similar to that exhibited by septation, whose onset also varies strongly with growth rate (above; Figures 6A and 6B). Thus, nucleoid splitting may be linked more closely to cell growth than any of the other assayed chromosomal events.
Terminus Splitting Always Occurs after S Phase and after Nucleoid Splitting
Terminus splitting exhibits a behavior intermediate between origin/DnaX splitting and nucleoid splitting, always occurring after completion of bulk DNA replication but with a somewhat longer delay as growth rate decreases (Figures 6A and 6B). At the slowest growth rate examined, terminus splitting immediately follows nucleoid splitting, raising the possibility that the latter transition (which places ter adjacent to midcell) is required for the former.
Early Nucleoid Splitting again Results in Movement of ter toward Midcell and Permits Subsequently Split Origins to Move Directly to the 1/4 and 3/4 Positions
Under standard conditions, Td = 125 min, origin splitting precedes nucleoid splitting, which then has two effects: (1) placing ter near midcell and (2) causing the ter-side origin to move to its final 1/4 position (above). At Td = 90 min, nucleoid splitting precedes origin splitting (above). Positional analysis (Figure 6C) reveals that nucleoid splitting is again accompanied by movement of ter toward midcell; indeed, this effect is, if anything, even more dramatic than at Td = 125 min. Moreover, nucleoid splitting has no effect on the position of sister origins, which remain colocalized at midcell. However, strikingly, when sister origins do separate, they now move symmetrically to the 1/4 and 3/4 positions, with no delay by the ter-side origin as seen under standard conditions. These findings strongly support the conclusion that nucleoid splitting is directly responsible for global changes in chromosome organization which place ter near midcell and which, under standard conditions, eliminate some type of spatial impediment to movement of the ter-side origin to the 1/4 position.
Discussion
High-resolution analysis of synchronous slowly growing E. coli populations establishes a more complete framework for understanding chromosome and replisome dynamics in this organism. The pattern of events described in detail for Td = 125 min is summarized in relation to progressive cell growth in Figure 7A.
Figure 7. Summary and Implications.
(A) Summary of events at Td = 125 min. ori and ter foci are depicted within nucleoids and cells of appropriate absolute sizes, prior to normalization (Figure 4).
(B) Interchromosome pushing forces could promote sister separation. (Left) Sister chromosomes linked by cohesion molecules accumulate a tendency to push one another apart. (Right) Two chromosomes that push one another apart will become symmetrically displayed on either side of any region of residual linkage.
(C) Features of late-stage eukaryotic chromosomes. (a) At pro-phase, sisters are tightly conjoined into a single unit; at prometaphase, sisters lie side-by-side (Sparrow et al., 1941, as cited in Kleckner et al., 2004). (b) Chromosome coils also arise at prometaphase and often exhibit opposite helical handedness (Boy de la Tour and Laemmli, 1988).
(D) We propose (text) that cell division per se licenses initiation of the next round of DNA replication. Coupling at late-cell cycle stage precludes initiation until and unless cell progresses through critical step of cell division. Such a mechanism can explain one-to-one linkage of cell division and replication initiation at both slow and fast growth rates.
I. Sister Colocalization and Separation with Accompanying Global Chromosome Movement
We show above that sister chromosomes remain together and then separate in a single coordinate transition involving most or all of the thus-far-replicated regions. This behavior does not include the origin or the terminus, which behave as spatially and functionally distinct domains with respect to sister disposition (below). We infer that sisters are specifically linked, that this linkage (i.e., “cohesion”) is lost at the time of the nucleoid splitting transition, and that sisters are then separated by some type of active mechanism. These observations confirm earlier suggestions of cohesion by Hiraga and colleagues (Sunako et al., 2001). Our results may also explain the failure of other studies to detect cohesion (e.g., Roos et al., 2001): those studies examined relatively fast growth rates, and we find that the duration of cohesion varies inversely with growth rate. The observed sister separation transition is especially remarkable in two respects. First, splitting appears to occur simultaneously throughout a very extensive region of the genome. Second, it is accompanied by a global change in chromosome disposition: the splitting sister chromosome regions become widely separated from one another; moreover, dramatic changes in position are also observed for regions not involved in the splitting process, e.g., the origin and terminus.
Source of the Force
What type of force could explain the coordinate global chromosome reorganization involved in the nucleoid splitting transition? We previously suggested that significant forces might be exerted by programmed chromatin expansion (Kleckner et al., 2004). In brief, as chromatin expands, it would tend to push against constraining features, which might be external (e.g., other chromatin) or internal (e.g., a meshwork of intersegment interconnections; Figure 7B, left). Such forces could explain the phenomena we observe here. Pushing of sister chromosomes against one another could promote both the release of connections and ensuing spatial separation. Moreover, as sisters separate, they might in turn push on other chromosomal regions, shoving them about the cell into the least “stressed” configuration. In fact, such effects are predicted specifically to produce the observed 2-fold symmetric mirror image configuration. If two linked chromatin masses push one another apart, the linked regions will remain at the interface and other regions will move outward, to greater or lesser extents according to their distance from the linked region(s) (Figure 7B, right).
MukB
The SMC family protein MukB, in interaction with partners MukE and MukF, has been implicated as a mediator of E. coli sister-chromatid cohesion (Sunako et al., 2001). On the other hand, mukB mutants also exhibit defective chromosome compaction (Weitao et al., 1999). It will be interesting to determine more precisely the direct roles of this complex in the formation of intersister connections.
Linkage to Cell Division Cycle
The timing of nucleoid splitting relative to the previous cell division varies strongly with growth rate, in parallel with the predivisional step of septation, and is independent of oriC and replisome splitting. We infer that nucleoid splitting is functionally linked to the overall cellular growth/division program and not to other chromosomal events, which appear to be related to the DNA replication program.
II. Loss of Sister Cohesion Mediates Sister Chromosome Segregation, Analogously to Eukaryotic Prometaphase
The nucleoid splitting transition places sister chromosomes in a 2-fold symmetric, ter-in and ori-out configuration, with loci in the splitting regions occupying positions that correspond to their positions along the chromosome. Thus, the events of this transition solve the fundamental problem of sister chromosome segregation (Figure 1). We infer that loss of sister chromosome cohesion triggers global chromosome movement which in turn mediates sister chromosome segregation
These findings suggest that chromosome segregation is mediated by a global internal change within the chromosomes. They specifically exclude the two prevalent models for bacterial sister segregation (Introduction). The replisome extrusion model implies that sisters are never linked and requires long-lived association and spatial constraining of clockwise and counterclockwise replisomes. We find that, in E. coli, sisters are initially linked and there is no long-lived association of the two replisomes; moreover, association can be lost either before or after (and thus independent of) sister splitting. The “origins as centromeres” model implies that localization of the origins to the 1/4 and 3/4 positions precedes and leads sister separation. We find that the opposite is true: sister segregation is mediated by the nucleoid splitting transition which can precede ori separation and which, in any event, is required for origin localization (to the 1/4 position).
Interestingly, sister separation during the E. coli nucleoid splitting transition is closely analogous to sister separation during prometaphase of the eukaryotic mitotic cell cycle. In both cases, sisters are initially colocalized along their lengths and then separate, in a single coordinate transition, into a side-by-side configuration where sisters are disposed in a 2-fold symmetric relationship (Introduction; Figure 7C). Thus, the E. coli process could represent a primordial sister segregation mechanism to which microtubule-based processes were later added. Interestingly, also, mitotic prometaphase is a period of pronounced chromosome expansion, in accord with such expansion as the force driving sister separation (Kleckner et al., 2004).
III. Three Other Phases of Sister Separation
Separation of sister origins and appearance of two ter foci are spatially and functionally independent of one another and of the major sister splitting transition. The timing of ter replication is not established, but we favor the view that appearance of two ter foci represents splitting of already-replicated ter’s. When sister foci appear, they are symmetrically disposed about the site of the prior single ter focus, as expected for splitting of tethered ter regions. Further, ter is likely replicated well before two ter foci appear: replisomes are lost prior to appearance of two ter foci, and such loss likely requires either progression of one fork past ter to a fork-blocking site or convergence of two forks, which probably does not usually occur exactly at ter. For ter and for ori, colocalization/separation could represent either cohesion/loss of cohesion or independent capture and release by a common underlying determinant.
Sister relationships in origin-distal regions remain to be determined, but we suggest that, after the nucleoid-splitting transition, newly synthesized sister regions move directly into the two already-established nucleoid domains (above).
IV. Midcell
The 1/4 and 3/4 positions are the sites of midcell in the subsequent cell cycle. Specific localization of ori to these positions confirms their early demarcation under linear cell cycle conditions, as known for fast growth conditions, and also suggests that ori might be specifically attached to some cellular determinant. How these sites are marked and the significance of ori localization to these positions remain important mysteries.
Appearance of a DnaX focus at midcell shortly after division is the earliest postdivision marking of midcell known thus far, preceding ori localization (above) and septum component FtsZ, which arises at midcell after replication is in progress (Td = 200 min; Lau et al., 2003). ori foci become located at a nearly fixed distance to a specific side of the midcell DnaX focus, suggestive of a direct relationship between the two determinants. Other replisome and chromosome components are likely also present in this ensemble. Prior localization of DnaX might recruit oriC and/or ensure spatial coordination between replication initiation and ensuing replication fork development. Localization of oriC to midcell might be required for replication initiation and/or for regular coupling of initiation to the cell division program. After replication initiates, the midcell ensemble is disassembled in an ordered sequence, in apparent correlation with progression of DNA replication. The mechanism and functional significance of disassembly also remain to be determined.
Later in the cell cycle, ter arrives immediately adjacent to midcell, as a result of the nucleoid-splitting transition. ter seems to be specifically captured at the time of this relocalization because (1) there is little movement thereafter and (2) late-disappearing DnaX foci, which should be approaching the ter region, appear to move specifically toward midcell, as if tracking along to an anchored ter. After two ter foci appear, the ter nearest to midcell moves across to a symmetrical position on the opposite side, as described previously (Lau et al., 2003; Li et al., 2003). Importantly, terminus-related events at midcell are spatially and functionally distinct from ori/DnaX-related events. DnaX occurs exactly at midcell, and ori usually occurs to the “right” and ter usually occurs to the “left”. Moreover, ter localization may precede or follow disassembly of the ori/ DnaX ensemble according to growth rate.
V. In E. coli, Clockwise and Counterclockwise Replisomes Are Colocalized Only Transiently
In Caulobacter, cells undergoing DNA replication during a linear cell cycle always contain a single focus of replisome components (Viollier et al., 2004); similarly, in B. subtilis, such components exhibit dynamic variation between a single focus and two nearby foci (Migocki et al., 2004). In contrast, in linearly cycling E. coli cells, single replisome foci are lost partway through “S phase.” Thereafter, the two replisomes are well separated and independently disposed throughout the nucleoid. This pattern reflects dynamic and independent movement of the two as shown by time-lapse studies (A. Wright, personal communication; D.B., unpublished data). The observed movements could represent motion of the replisome relative to a fixed nucleoid, motion of the nucleoid with little relative motion of the replisome, or a mixture of the two.
VI. The Division Transition
The current study reveals previously undescribed features of E. coli chromosomes as they traverse the cell division transition. Immediately prior to division, sister nucleoids closely juxtaposed to midcell and their origins and termini occur at specific positions relative to the cell boundaries, apparently due to capture by underlying cellular determinants (above). Concomitant with division, sister nucleoids translate away from mid-cell such that they occupy symmetrical positions within their two about-to-be daughter cells. During this transition, the origin and the terminus retain their same positions relative to the nucleoid but change positions relative to the cell boundaries. These features strongly suggest that, during this transition, (1) the nucleoid is effectively “frozen” and (2) origins and termini are released from whichever determinants previously linked them to fixed cellular positions. Moreover, after division, the origin and the terminus both lose their strong localization and occur at many positions within their respective halves of the cell, which suggests that the nucleoid has become “unfrozen” concomitant with cell division. The origin then becomes localized on the non-ter side of the midcell DnaX ensemble as if it eventually drifts to and is captured by the midcell ensemble.
VII. Coordination of DNA Replication and Cell Division
Replication Initiation Is More Closely Linked to Cell Division Than to Cell Mass
Early models proposed that initiation of DNA replication begins at a fixed ratio of cell mass to the origin number. The current study confirms and extends earlier findings showing that, in slowly growing cells, cell mass at the time of initiation and thus also the mass to origin ratio are far from constant and can vary over a range of at least 1.5-fold (above; Wold et al., 1994). Thus, direct or indirect sensing of cell mass does not appear to be crucial. We further find that the occurrence of replication-related events, e.g., DnaX dynamics, is much more tightly correlated with time after release from the baby cell column than with absolute cell size (Bates et al., 2005). This difference strongly suggests that initiation of DNA replication is more tightly linked to occurrence of the prior cell division (or some closely correlated event) than to cell mass, as pointed out to us by E. Boye (Bates et al., 2005).
A Licensing Model
How might occurrence of replication initiation be controlled so as to ensure a 1:1 relationship between cell division and initiation? We propose occurrence of cell division may per se license the chromosome(s) for the next round of replication initiation (Figure 7D). The exact timing of initiation after cell division would then be determined by other factors such as DnaA concentration. This model is simple and direct. Moreover, it easily accommodates the fact that in fast-growing cells, which contain multiple origins, all of those origins fire synchronously (Figure 7D). An interesting possibility is that “freezing” of the nucleoid effectively precludes steps needed for replication initiation and that licensing corresponds to “unfreezing” of the nucleoid. Inhibitory coupling of replication to division and release of that inhibition by cell division could be explained, e.g., by trans-acting signal transduction mechanisms or by cis effects of the division process on the state of the terminus region, which might then be transmitted throughout the chromosome.
Experimental Procedures
Bacterial Strains and Growth Conditions
All experiments were performed in NK9387, CM735 (Wold et al., 1994) carrying an isopropyl β-D-thiogalactopyranoside (IPTG)-inducible sticky flagellin tethering construct (Ptac::fliCst lacIQ) plus a dnaX allele encoding a C-terminal dnaX-GFP fusion construct (gift of A. Wright; Bates et al., 2005). The GFP fusion has no detectable effect on either doubling time or DNA replication (data not shown). Cells were grown at 30°C in AB minimal media with 0.2% alanine, 0.2% succinate, or 0.05% glucose as indicated, and supplemented with tryptophan, histidine, methionine, and thiamine at 20 μg/ml each.
Time-Course Analysis
Preparation and use of the baby cell column are described by (Bates et al., 2005). After column equilibration, a series of samples were collected in rapid succession and then incubated for different appropriate periods of time (text). Sample volumes were proportionally scaled with growth rate (AB succinate, 10 ml; AB alanine, 5 ml; AB Glucose, 2 ml), about 2–3 × 108 cells per sample. After the desired incubation time, one half of each sample was processed for FACS analysis (Bates et al., 2005) while the other half was processed for FISH/immunocytology (below).
Five Data Sets
Results presented for standard conditions (Td = 125 min) represent two independent sets of synchronized cell samples. Figures 2, 3F, 4, and 5 present results obtained from a single sample set I; Figures 3A–3E and 4D present results obtained from an independent sample set II. Timing of landmark events in set I are applicable to set II due to high reproducibility of kinetics among independent experiments performed under any given set of conditions (D.B. and N.K., unpublished data). Two additional sets of synchronized cells were obtained at different growth rates; Td = 90 and 300 min (Figure 6).
DNA Replication
The percentage of cells undergoing DNA replication was obtained by subtracting the percentage of cells containing full genome complements (either one or two fully replicated chromosomes) from the total. Since the DNA fluorescence histogram peaks corresponding to one and two chromosomes are partially obscured by cells in early and late stages of replication, these percentages were approximated by quantifying the “outer” half of the corresponding peaks (the sides of the peaks that do not overlap cells in the process of replication), and multiplying by 2 (details in the Supplemental Data available with this article online). This calculation was performed five independent times for each histogram and the results averaged.
Cell Concentration
Cell concentration was determined from the same FACS analyses used for analysis of replication (above) by comparing the rate of cell flow for each sample to that for a standard of known concentration.
FISH, DAPI, and Immunofluorescence
For FISH analysis, cells were fixed in 2.5% paraformaldehyde and hybridized with multiple fluorescently labeled probes (below; Supplemental Data). In sample set I and in samples representing other growth rates, DAPI was added at 150 μg/ml. In sample set II, one of the colors detected by multicolor FISH analysis overlaps significantly with DAPI fluorescence (below); thus, slides were first scored for FISH foci and then, after removal of coverslips, washed twice in 1× SSC, stained with DAPI, and scored for nucleoid status. For certain per-cell studies, these samples were also rescored for still-visible FISH foci. Note: late in the cell cycle, split sister nucleoids are well separated from one another (ca. 0.4 μm); in contrast, newly split nucleoids have a dumbbell appearance, with a prominent central connection, and thus are occasionally missed when the plane of focus runs through the connected region. This results in artificial flattening of the cumulative curve for nucleoid splitting.
FISH probes corresponded to 6 kb regions. Corresponding DNA fragments were obtained from plasmids by restriction digestion and gel purification (probe coordinates in the Supplemental Data), purified on a spin column (Qiagen), and chemically labeled with Alexa fluor (Ulysis DNA labeling system, Molecular Probes). Probe/ fluor combinations were ori/Alexa 594 and ter/Alexa 680 for all experiments except for the four-color FISH experiment (Figure 3), which used ori/Alexa 488, gln/Alexa 594, lac/Alexa 680, and ter/ Pacific Blue. The emission spectrum of the latter signal overlaps that of DAPI but, in favorable cases, was bright enough to be detected by a narrow band-pass filter that allowed five-color visualization (Figure 3B). When analyzed in combination with FISH, DnaX-GFP complexes were detected using anti-GFP antiserum immediately prior to in situ hybridization (Supplemental Data).
Fluorescence and phase contrast images were obtained using a Zeiss Axioplan2 microscope equipped with a Princeton CCD camera and analyzed using Metamorph software (Universal Imaging). Multiple fluorescent and phase contrast images taken for each field of cells were aligned to multiwavelength-emitting beads (Molecular Probes), placed in low concentration onto each slide as an internal control. At each time point in all experiments, 200 cells were analyzed for each feature of interest; the only exceptions were Figures 3A–3E and 6, Td = 90 min and 300 min, where 50 cells were analyzed per time point.
Supplementary Material
Acknowledgments
We thank Guido Guidotti for original insights leading to design of the baby cell column. We also thank Ken Marians for the parC strain, Pam Silver for GFP antibody, and, especially, Andrew Wright for the DnaX-GFP construct, sharing of unpublished data, and lively discussions, all essential to this work. We also thank Dave Sherratt and Anders Lobner-Olesen for sharing unpublished results. All research was supported by National Institutes of Health (NIH) grant GM25326 to N.K. and NIH postdoctoral grant GM20627 to D.B.
Footnotes
Supplemental Data include Supplemental Experimental Procedures and can be found online at http://www.cell.com/cgi/content/full/ 121/6/899/DC1/.
References
- Bates D, Epstein J, Fahrner KA, Berg HC, Kleckner N. The E. coli baby cell column: a novel cell synchronization method provides new insight into the bacterial cell cycle. Mol Microbiol. 2005;13:2097. doi: 10.1111/j.1365-2958.2005.04693.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- Boy de la Tour E, Laemmli UK. The metaphase scaffold is helically folded: sister chromatids have predominantly opposite helical handedness. Cell. 1988;55:937–944. doi: 10.1016/0092-8674(88)90239-5. [DOI] [PubMed] [Google Scholar]
- Boye E, Nordstrom K. Coupling the cell cycle to cell growth. EMBO Rep. 2003;4:757–760. doi: 10.1038/sj.embor.embor895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- Coskun-Ari FF, Hill TM. Sequence-specific interactions in the Tus-Ter complex and the effect of base pair substitutions on arrest of DNA replication in Escherichia coli. J Biol Chem. 1997;272:26448–26456. doi: 10.1074/jbc.272.42.26448. [DOI] [PubMed] [Google Scholar]
- Donachie WD, Blakely GW. Coupling the initiation of chromosome replication to cell size in Escherichia coli. Curr Opin Microbiol. 2003;6:146–150. doi: 10.1016/s1369-5274(03)00026-2. [DOI] [PubMed] [Google Scholar]
- Gordon GS, Shivers RP, Wright A. Polar localization of the Escherichia coli oriC region is independent of the site of replication initiation. Mol Microbiol. 2002;44:501–507. doi: 10.1046/j.1365-2958.2002.02901.x. [DOI] [PubMed] [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion: An asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J. A mechanical basis for chromosome function. Proc Natl Acad Sci USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD. The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev. 2001;15:2031–2041. doi: 10.1101/gad.913301. [DOI] [PubMed] [Google Scholar]
- Li Y, Youngren B, Sergueev K, Austin S. Segregation of the Escherichia coli chromosome terminus. Mol Microbiol. 2003;50:825–834. doi: 10.1046/j.1365-2958.2003.03746.x. [DOI] [PubMed] [Google Scholar]
- Migocki MD, Lewis PJ, Wake RG, Harry EJ. The midcell replication factory in Bacillus subtilis is highly mobile: implications for coordinating chromosome replication with other cell cycle events. Mol Microbiol. 2004;54:452–463. doi: 10.1111/j.1365-2958.2004.04267.x. [DOI] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- Roos M, van Geel AB, Aarsman ME, Veuskens JT, Woldringh CL, Nanninga N. The replicated ftsQAZ and minB chromosomal regions of Escherichia coli segregate on average in line with nucleoid movement. Mol Microbiol. 2001;39:633–640. doi: 10.1046/j.1365-2958.2001.02263.x. [DOI] [PubMed] [Google Scholar]
- Sunako Y, Onogi T, Hiraga S. Sister chromosome cohesion of Escherichia coli. Mol Microbiol. 2001;42:1233–1241. doi: 10.1046/j.1365-2958.2001.02680.x. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitao T, Nordstrom K, Dasgupta S. Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol Microbiol. 1999;34:157–168. doi: 10.1046/j.1365-2958.1999.01589.x. [DOI] [PubMed] [Google Scholar]
- Wold S, Skarstad K, Steen HB, Stokke T, Boye E. The initiation mass for DNA replication in Escherichia coli K-12 is dependent on growth rate. EMBO J. 1994;13:2097–2102. doi: 10.1002/j.1460-2075.1994.tb06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.