Abstract
The retromer complex is required for the efficient endosome-to-Golgi retrieval of the CIMPR, sortilin, SORL1, wntless and other physiologically important membrane proteins. Retromer comprises two protein complexes that act together in endosome-to-Golgi retrieval; the cargo-selective complex is a trimer of VPS35, VPS29 and VPS26 that sorts cargo into tubules for retrieval to the Golgi. Tubules are produced by the oligomerization of sorting nexin dimers. Here, we report the identification of five endosomally-localised proteins that modulate tubule formation and are recruited to the membrane via interactions with the cargo-selective retromer complex. One of the retromer-interacting proteins, strumpellin, is mutated in hereditary spastic paraplegia, a progressive length-dependent axonopathy. Here, we show that strumpellin regulates endosomal tubules as part of a protein complex with three other proteins that include WASH1, an actin-nucleating promoting factor. Therefore, in addition to a direct role in endosome-to-Golgi retrieval, the cargo-selective retromer complex also acts as a platform for recruiting physiologically important proteins to endosomal membranes that regulate membrane tubule dynamics.
Keywords: Retromer, Endosome, Tubule, Recruitment, Strumpellin
Introduction
Endosomal protein sorting has a vital role in a number of physiologically important processes including antigen presentation, macromolecular nutrient uptake, growth factor receptor signaling and downregulation, autophagy and lysosome biogenesis (for reviews, see Sadowski et al., 2009; Saksena and Emr, 2009; Sann et al., 2009; Seaman, 2008; Lee et al., 2008). Recent studies of inherited diseases have identified several examples of genes encoding proteins that function in endosomal protein sorting that, when mutated, result in a range of pathologies. A notable example is hereditary spastic paraplegias (HSP), the hallmark of which is a selective distal axonopathy. There is a striking localisation of many of the HSP-encoded proteins to the endosome, including the microtubule-severing protein spastin, the ubiquitin-ligase-interacting protein spartin, and NIPA1, a membrane protein that mediates bone morphogenic protein signaling at the endosome (Tsang et al., 2009; for a review, see Salinas et al., 2008). Despite this concentration of HSP proteins at endosomes, in most cases their function is unknown.
Much of the core machinery that carries out endosomal protein sorting is conserved in evolution, for example, the retromer complex (for reviews, see Attar and Cullen, 2009; Verges, 2008; Collins, 2008; Bonifacino and Hurley, 2008). Retromer mediates endosome-to-Golgi retrieval of lysosomal and vacuolar hydrolase receptors (e.g. the cation-independent mannose 6 phosphate receptor, CIMPR) along with other physiologically significant membrane proteins including wntless, which functions in WNT secretion, and SORL1, a protein that is genetically linked to late-onset Alzheimer's disease (Arighi et al., 2004; Seaman, 2004; Eaton, 2008; Nielsen et al., 2007; Rogaeva et al., 2007).
The retromer complex was first identified in yeast where it comprises five proteins encoded by vacuolar protein sorting (VPS) genes. The heteropentameric retromer complex can be functionally dissected into two subcomplexes: a cargo-selective complex formed from a conserved trimer of Vps35p, Vps29p and Vps26p and a ‘structural complex’ formed from a dimer of the sorting nexin (SNX) proteins Vps5p and Vps17p (Seaman et al., 1998). In mammals, SNX1, SNX2 with SNX5 and SNX6 provide the ‘structural’ role and can tubulate membranes through the C-terminal Bin, amphiphysin and Rvs (BAR) domains present in these proteins (Carlton et al., 2004; Wassmer et al., 2007). Additionally, SNX5 and SNX6 interact with the microtubule cytoskeleton via the p150glued protein that binds to dynein, thereby linking endosomal protein sorting to microtubules (Wassmer et al., 2009; Hong et al., 2009).
The interaction between the SNX component of retromer and p150glued is an example of how retromer-interacting proteins facilitate retromer in mediating endosome-to-Golgi retrieval. In yeast, the SNX3 homologue Grd19p binds to Ftr1p to sort Ftr1p into the retromer pathway (Strochlic et al., 2008). In mammalian cells, the EPS15 homology domain protein, EHD1, interacts with retromer and is required to stabilize SNX1-positive membrane tubules (Gokool et al., 2007a). Recruitment of the cargo-selective retromer complex to the endosomal membrane is mediated by the small GTPase RAB7 (Rojas et al., 2008; Seaman et al., 2009). Additionally, the cargo-selective retromer complex also interacts with a Rab GTPase-activating protein (GAP), TBC1D5, that can downregulate the VPS35-VPS29-VPS26 complex recruitment to the membrane (Seaman et al., 2009).
Identification of retromer-interacting proteins has been key to understanding the functioning of endosome-to-Golgi retrieval. In this study, we have identified five novel retromer-interacting proteins, characterized their interaction with the cargo-selective retromer complex and revealed a role for the retromer-interacting proteins in regulating endosomal tubule dynamics. One of the five retromer-interacting proteins is strumpellin (KIAA0196), a protein encoded by the Spg8 gene that is mutated in HSP (Valdmanis et al., 2007). Here, we show that strumpellin forms a complex with three other proteins – KIAA1033, FAM21 and WASH1, a member of the WASp, Wave family of actin-nucleating promoting factors (Linardopoulou et al., 2007; Liu et al., 2009) – and that the WASH1 complex requires the cargo-selective retromer complex for recruitment to the endosomal membrane. We also report the detailed analysis of the retromer-TBC1D5 interaction and find that a mutant of VPS29 that abolishes the interaction with TBC1D5 is the same mutation that, in yeast, abolishes the interaction between the cargo-selective retromer subcomplex and the structural SNX Vps5p/Vps17p complex. These data shed new light on the functioning of the retromer complex at endosomal membrane and reveal that the cargo-selective VPS35-VPS29-VPS26 complex acts not only to recruit cargo for endosome-to-Golgi retrieval but also as a platform onto which other large endosomal membrane regulating complexes are recruited.
Results
Identification of retromer-interacting proteins
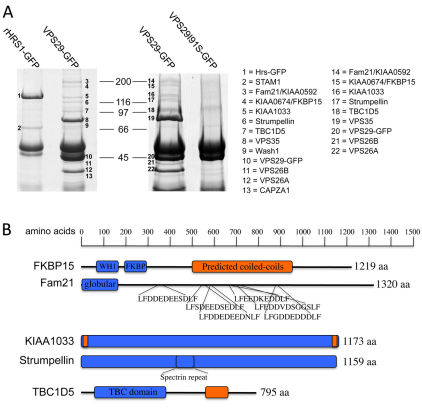
In order to identify proteins that interact with retromer we carried out a series of native immunoprecipitation (IP) experiments. In Fig. 1A, the results of two native IPs from VPS29-GFP-expressing cells and either HRS1-GFP or VPS29I91S-GFP-expressing cells are shown. HRS1, like retromer, is endosomally localised (Bache et al., 2003) and the VPS29I91S mutant cannot interact with VPS35 (Collins et al., 2005). These proteins therefore are negative controls in this experiment. The proteins that were identified by mass spectrometry are listed as 1–22. In addition to FAM21, KIAA1033 and strumpellin, a protein called KIAA0674 (FKBP15) was detected in the VPS29-GFP lanes along with TBC1D5, which we have previously reported as a retromer-interacting protein (Seaman et al., 2009). WASH1 (band 9) and CAPZa (an actin-capping protein, band 13) were also identified in one of the native IPs, but their presence was variable between experiments. We were also able to co-IP strumpellin and KIAA1033 with the retromer cargo-selective complex using anti-VPS26 antisera to IP from lysates of mouse embryonic fibroblasts (see supplementary material Fig. S1). In none of the native IPs did we observe a band(s) corresponding to any of the SNX proteins that function in the retromer pathway.
Fig. 1.
Identification and features of retromer-interacting proteins. (A) HeLa cells stably expressing GFP-tagged VPS29, HRS1 or a mutant of VPS29 (I91S) that cannot bind VPS35 were lysed and treated with anti-GFP. The IPs were analysed by SDS-PAGE. Several proteins associated with VPS29-GFP, but not with HRS1 or the VPS29I91S mutant, are shown. The bands were excised and subjected to MALDI-TOF mass spectrometry. The proteins identified are listed as 1–22. (B) The domain architecture of the retromer-interacting proteins. Globular domains are shown as rectangles and predicted coiled-coil domains are coloured orange; unstructured regions are depicted as straight lines.
Features and conservation of the retromer-interacting proteins
The retromer-interacting proteins were investigated initially using bioinformatic tools. The features (e.g. domains) present in the FKBP15, FAM21, KIAA1033, strumpellin and TBC1D5 proteins are shown in Fig. 1B. FKBP15 has a WASp homology domain 1 (WH1) and a proline-isomerase (FK506 binding) domain in its N-terminal region and a large central coiled-coils domain. FAM21 is predicted to be largely unstructured with the exception of a globular domain at its N-terminus. The unstructured region of FAM21 contains several repeated patches of negatively charged residues (usually Asp or Glu) flanked by a Leu-Phe dipeptide.
Both KIAA1033 and strumpellin are predicted to be globular proteins. Strumpellin has a central spectrin-repeat domain and KIAA1033 has two small regions of coiled-coils at its N- and C-termini but no other features or domains. Of the retromer-interacting proteins identified, only TBC1D5 has a domain that suggests a function – namely the TBC domain that is present in the family of Rab GTPase activating proteins (GAPs) (Fukuda, 2008). In addition to the TBC domain, TBC1D5 also has a region of predicted coiled-coils in the C-terminal half of the protein.
The VPS35-VPS29-VPS26 components of the retromer complex are conserved in all eukaryotes (Dacks and Field, 2007). The retromer-interacting proteins identified in Fig. 1A are also conserved although it is not possible to identify homologues in all eukaryotes. The degree of identity between the human retromer-interacting proteins and homologues present in other metazoan eukaryotes is shown in Table 1. There is no clear homologue of the FKBP15 protein in either Drosophila or C. elegans, and FAM21 does not appear to be present in C. elegans.
Table 1.
Conservation of the retromer-interacting proteins
Characterisation of the TBC1D5-retromer interaction
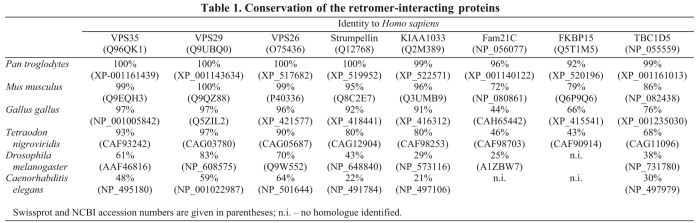
As TBC1D5 is the only retromer-interacting protein with a domain that suggests a function, we chose to investigate the interaction between TBC1D5 and retromer first. We have previously shown that overexpression of TBC1D5 results in the cargo-selective retromer complex (i.e. VPS35-VPS29-VPS26) being displaced from the endosomal membrane (Seaman et al., 2009). Structural studies have revealed that TBC domains catalyze the hydrolysis of GTP by Rab proteins through the concerted action of conserved Arg and Glu residues (Pan et al., 2006). TBC1D5 has these conserved Arg and Glu residues within its TBC domain (R169 and Q204). Therefore, we generated a double mutant of TBC1D5 using R169A and Q204A (hereafter termed RQ mutant) and transiently transfected GFP-tagged wild-type TBC1D5 or TBC1D5 RQ mutants into HeLa cells. In Fig. 2A, the expression of wild-type TBC1D5 caused VPS26 to dissociate from the membrane and its labelling became diffuse. By contrast, expression of the TBC1D5 RQ mutant did not cause VPS26 to become cytosolic and there was clear colocalisation between the GFP-TBC1D5 RQ mutant and VPS26. Cells transfected with either wild-type GFP-TBC1D5, GFP-TBC1D5 RQ or empty vector were blind-scored to assess the membrane association of VPS26 by comparing transfected cells with untransfected cells within the same field. In Fig. 2B, we found that ~75% of cells expressing the GFP-TBC1D5 were scored as having displaced (i.e. cytosolic) VPS26, whereas <10% of cells expressing the GFP-TBC1D5 RQ mutant were scored as having cytosolic VPS26. This demonstrates, therefore, that TBC1D5 requires a functional catalytic TBC domain to displace the cargo-selective retromer complex from the membrane.
Fig. 2.
The catalytic domain of TBC1D5 is required for its retromer-recruitment regulating activity. (A) HeLa cells were transiently transfected with wild-type GFP-TBC1D5 or a mutant (R169A and Q204A, termed TBC1D5 RQ) that lacks catalytic activity. The wild-type GFP-TBC1D5 causes VPS26 to become cytosolic, whereas the RQ mutant colocalises with VPS26 and does not redistribute the cargo-selective complex into the cytoplasm. Scale bar: 20 μm. (B) HeLa cells transiently transfected with GFP-TBC1D5, GFP-TBC1D5 RQ or empty vector were blind scored for ‘normal’ (i.e. membrane-associated) or ‘displaced’ (i.e. cytosolic) VPS26 (n, number of transfected cells scored). In ~75% of cells expressing the wild-type TBC1D5, VPS26 was scored as displaced, whereas <10% of cells expressing GFP-TBC1D5 RQ were scored as displaced.
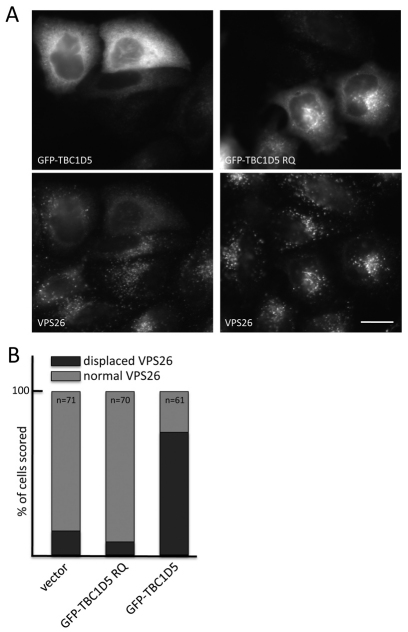
In order to further characterise the TBC1D5-retromer interaction, we employed the yeast two-hybrid (Y2H) system in which growth on –His plates is indicative of an interaction. In Fig. 3A, TBC1D5 was tested against each of the cargo-selective retromer complex components. We show that TBC1D5 interacted directly with VPS29 but not with VPS26 or VPS35. Two conserved areas of surface hydrophobicity, centered around the V90 and I91 residues and the L152 residue, respectively, regulate the interactions of VPS29 and the assembly of the retromer complex. Mutation of the V90 or I91 residues of VPS29 (or equivalent residues in yeast Vps29p) blocks the interaction with VPS35/Vps35p, but mutation of the L152 residue has no effect on the VPS29-VPS35 interaction but does abolish assembly of the heteropentameric complex in yeast (Collins et al., 2005). In Fig. 3B, the VPS29L152E mutant was no longer able to interact with TBC1D5 but retained the ability to interact with VPS35. The L152 residue of VPS29, therefore, is crucial for the interaction with TBC1D5.
Fig. 3.
TBC1D5 interacts with VPS29 through the L152-centered hydrophobic patch. (A) TBC1D5 cloned into the Y2H ‘prey’ vector, pGAD424, was tested against VPS35, VPS29 and VPS26 expressed from the Y2H ‘bait’ vector, pGBT9. Growth on –His plates was observed for the pGBT9-VPS29 and pGAD424-TBC1D5 combination. (B) Mutation of the L152 residue on VPS29 blocks the interaction with TBC1D5 but not with VPS35. (C) Truncations of TBC1D5 (tagged with GFP) were tranfected into HeLa cells. Twenty-four hours after transfection, the cells were lysed and the lysates IPed with anti-GFP. Only full-length TBC1D5 retains a robust interaction with retromer.
We next sought to define the region of TBC1D5 that binds to VPS29. A series of truncation mutants of TBC1D5 were generated and expressed in yeast via the Y2H ‘prey’ vector (pGAD424). We found that only the full-length TBC1D5 retains a robust interaction with VPS29 (supplementary material Fig. S2A). To confirm the data obtained using the Y2H system, full-length or truncated TBC1D5 (tagged with GFP) was transiently transfected into HeLa cells. Lysates were prepared and the GFP-TBC1D5 protein was immunoprecipitated using anti-GFP antisera. In Fig. 3C, full-length wild-type TBC1D5 co-IPed both VPS35 and VPS26, demonstrating that it is competent to interact with VPS29. Truncations of TBC1D5 abolished the interaction with the VPS35-VPS29-VPS26 complex, although a weak interaction with the GFP-TBC1D5 1–548 construct was observed. Point mutations of TBC1D5 to mutate the crucial residues required for the catalytic action of the TBC domain (R169A, Q204A, R169A and Q204A), or mutation of residues predicted to be phosphorylated (SST43AAA), did not affect the interaction of TBC1D5 with VPS35-VPS29-VPS26.
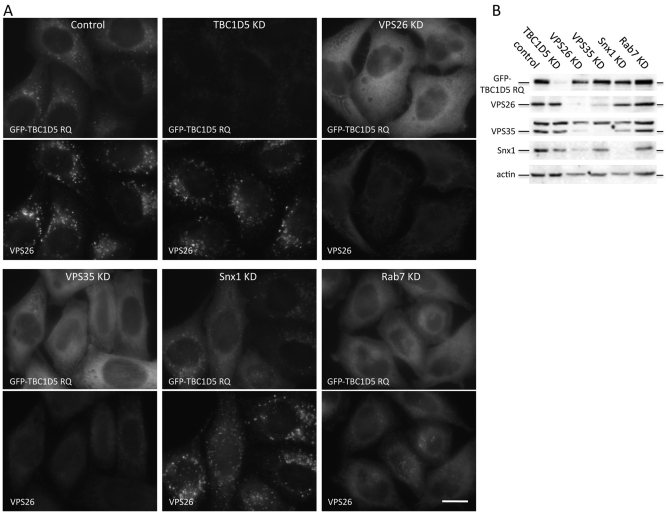
As shown in Fig. 2, the GFP-tagged TBC1D5 RQ mutant colocalised with retromer when transiently transfected into cells. We next investigated whether the interaction with retromer is required for the membrane association of TBC1D5. In Fig. 4, cells stably expressing GFP-TBC1D5 RQ were treated with siRNA to abolish expression of TBC1D5, VPS26, VPS35, SNX1 or RAB7 and the localisation of GFP-TBC1D5 RQ was assessed by microscopy. As shown in Fig. 4A, in control cells, GFP-TBC1D5 RQ colocalised with VPS26. When TBC1D5 expression was ablated by RNAi, the GFP-TBC1D5 RQ signal was lost and was no longer visible by fluorescence microscopy or western blotting (Fig. 4A,B). The siRNA knockdown (KD) of VPS26 or VPS35 both resulted in GFP-TBC1D5 RQ becoming cytosolic, but KD of SNX1 had no effect. We have previously shown that RAB7 is required to mediate recruitment of the VPS35-VPS29-VPS26 complex (Seaman et al., 2009) and, in Fig. 4A, loss of RAB7 also caused GFP-TB1D5 RQ to become cytosolic. These data show that the cargo-selective retromer complex is required for the localisation of TBC1D5 to the endosomal membrane.
Fig. 4.
TBC1D5 requires the cargo-selective retromer complex to target to endosomes. (A) HeLa cells stably expressing GFP-TBC1D5 RQ were treated with siRNA to knockdown (KD) TBC1D5, VPS26, VPS35, SNX1 or RAB7. Loss of VPS26, VPS35 or RAB7 results in GFP-TBC1D5 redistributing into the cytoplasm but SNX1 KD does not affect GFP-TBC1D5 localisation. Scale bar: 20 μm. (B) Western blot of lysates from cells treated as in A confirms the efficacy of the siRNA targeting TBC1D5, VPS26, VPS35 and SNX1.
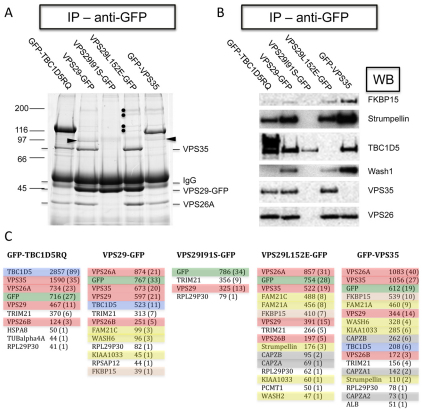
To further examine the interactions of the cargo-selective retromer complex in vivo, a series of native IP experiments were performed. In Fig. 5, HeLa cells stably expressing GFP-tagged versions of TBC1D5 RQ mutant VPS29, VPS29I91S, VPS29L152E or VPS35 were lysed and the GFP-tagged proteins were immunoprecipitated and analysed by SDS-PAGE (Fig. 5A). GFP-TBC1D5 RQ co-IPed VPS35 and VPS26, as did VPS29. The VPS29 IP lane also contained bands corresponding to TBC1D5, strumpellin, KIAA1033, FKBP15 and FAM21 proteins but these proteins were absent in the VPS29I91S lane. The IP from cells expressing VPS29L152E cells had an almost identical protein profile to the wild-type VPS29 lane except that the band corresponding to TBC1D5 is absent. The IP from cells expressing GFP-VPS35 was essentially the same as that from VPS29-GFP cells.
Fig. 5.
In vivo analysis of the interactions between the cargo-selective complex and the various retromer-interacting proteins identified in Fig. 1. (A) Cells stably expressing GFP-TBC1D5 RQ, VPS29-GFP, VPS29I91S-GFP, VPS29L152E-GFP or GFP-VPS35 were lysed and the lysates were treated with anti-GFP. After washes, the IPs were analysed by SDS-PAGE. GFP-TBC1D5 RQ co-IPed the retromer cargo-selective (VPS35-VPS29-VPS26) complex. Endogenous TBC1D5 (arrowhead) co-IPed with VPS29-GFP and GFP-VPS35 but was not observed in the VPS29I91S or the VPS29L152E lanes. Bands corresponding to the other retromer-interacting proteins (FAM21, FKBP15, KIAA1033 and strumpellin, highlighted with dots) were observed in the VPS29, VPS29L152E and VPS35 lanes. (B) Samples identical to those in A were analysed by western blotting, confirming that TBC1D5 does not co-IP with the VPS29L152E mutant. (C) Samples identical to those in A were analysed by LC-MSMS. The different proteins have been colour-coded as follows: red, retromer; green, GFP; blue, TBC1D5; pink, FKBP15; grey, CAPZa and CAPZb; and yellow, strumpellin, KIAA1033, FAM21 and WASH. Proteins considered to be contaminants were left unshaded, e.g. TRIM21 is a contaminant in these native IPs and is present owing to its IgG binding domain (Keeble et al., 2008). No peptides corresponding to TBC1D5 were detected in the VPS29L152E sample and no peptides from SNX proteins were detected in any samples.
When the samples shown in Fig. 5A were western blotted using antibodies against many of the retromer-interacting proteins identified in Fig. 1, it was observed that TBC1D5 is absent in the VPS29L152E lane but was weakly detected in the VPS29I91S lane, consistent with a direct interaction between VPS29 and TBC1D5 (see Fig. 3). To extend these observations, the native IP shown in Fig. 5A was repeated and the whole sample subjected to tryptic digestion and mass spectrometry so that all of the proteins present could be identified (Fig. 5C). As shown in Fig.5, the various proteins have been colour-coded so that a rapid visual comparison can be made. The numbers are the Mascot score and the number of peptides identified from the respective protein is shown in parentheses. Full details of the mass spectrometric data is shown in supplementary material Table S1. The data from the mass spectrometric analysis supports the gel and western blot shown in Fig. 5A,B. TBC1D5 is absent in the sample from the VPS29L152E native IP and none of the SNX proteins that mediate tubulation (i.e. SNX1, SNX2, SNX5 and SNX6) are present.
The retromer cargo-selective complex interacts with a large multimeric protein complex through FAM21 and WASH1
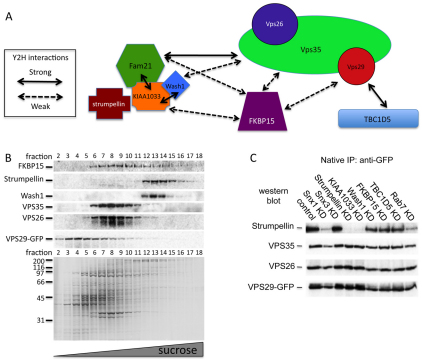
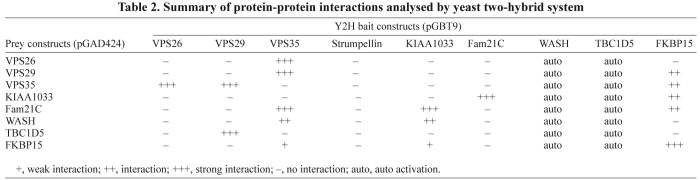
In Fig. 3, we used the Y2H system to show that TBC1D5 directly binds to VPS29. Therefore, we employed the Y2H system again to elucidate the interactions between the retromer VPS35-VPS29-VPS26 complex and the various retromer-interacting proteins identified in Figs 1 and 5. Shown in supplementary material Figs S2B and S2C are the results of several tests of possible binary interactions. We could not detect any interactions for strumpellin and we found that WASH1 and TBC1D5 would autoactivate when expressed from the pGBT9 ‘bait’ vector. These data are summarized in Fig. 6A and Table 2.
Fig. 6.
Yeast two-hybrid (Y2H) and biochemical analysis of interactions between the retromer cargo-selective complex and retromer interacting proteins. (A) A cartoon diagram of the Y2H interactions of the retromer cargo-selective complex and the various retromer-interacting proteins reported herein. (B) Cells expressing VPS29-GFP were lysed and subjected to centrifugation on a 5-30% sucrose velocity gradient. Fractions were collected and analysed by SDS-PAGE (lower panel) and western blotting (upper panel). VPS35 and VPS26 co-fractionated. WASH1 and strumpellin also co-fractionated in the later (larger complex) fractions. FKBP15 was detected in multiple fractions. (C) VPS29-GFP cells were treated with siRNA to KD various proteins and then lysed. Anti-GFP was used to IP the VPS29-GFP and associated proteins. The IPs were analysed by western blotting. The association of strumpellin with the retromer cargo-selective complex is most strongly affected by KD of KIAA1033, RAB7 or SNX1.
Table 2.
Summary of protein-protein interactions analysed by yeast two-hybrid system
Although strumpellin did not appear to interact with any of the retromer or retromer-interacting proteins by Y2H, we believe that strumpellin might constitute part of a large complex that also contains FAM21, KIAA1033 and WASH1 and that strumpellin directly binds KIAA1033. This hypothesis is supported first by the observation that strumpellin is repeatedly detected in our native IPs along with FAM21 and KIAA1033 (Fig. 1; supplementary material Fig. S1). Second, in Fig. 6B, we observed that strumpellin co-migrates with WASH1 on sucrose velocity gradients and strumpellin is no longer able to co-IP with VPS29-GFP after KD of KIAA1033 (Fig. 6C); however, KD of FKBP15 or TBC1D5 did not affect the interaction with VPS29-GFP.
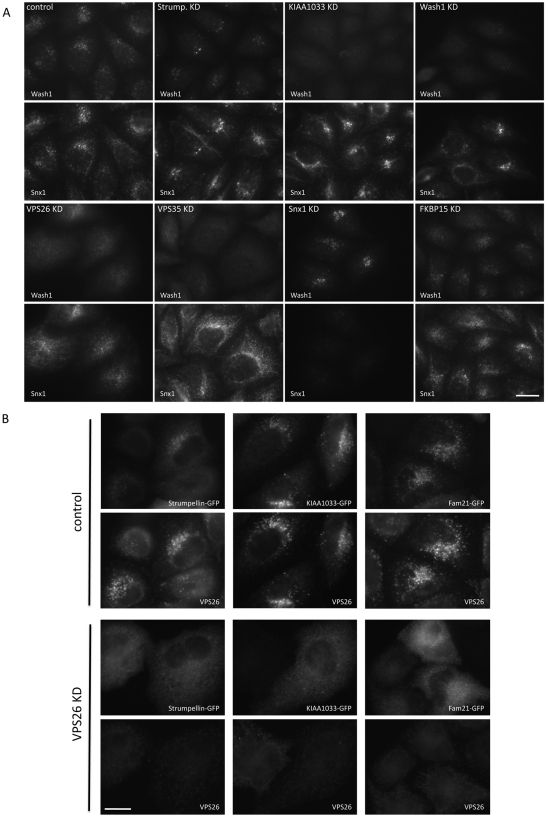
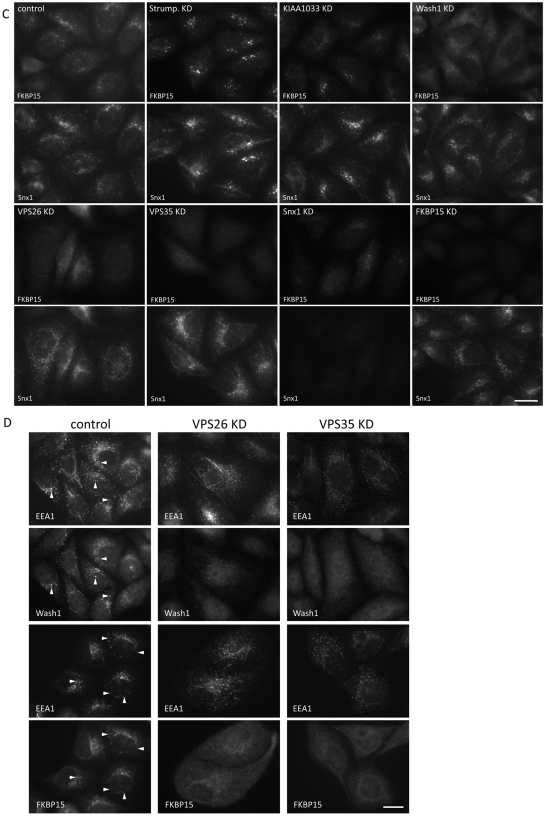
The localisation of the actin-nucleating promoting factor WASH1 was investigated by immunofluorescence. In Fig. 7A, we show that WASH1 colocalised with SNX1, consistent with WASH1 localising to the endosome. Loss of strumpellin by siRNA KD caused a marked reduction in WASH1 labelling, and KD of KIAA1033 appeared to abolish WASH1 localisation. We also noticed in these experiments that the SNX1 labelling was much more tubular after strumpellin or KIAA1033 KD. RNAi-mediated KD of VPS26 or VPS35 resulted in a loss of WASH1 localisation to the endosomal membrane but SNX1 KD did not redistribute WASH1 from endosomes. Additionally, we show that RNAi of either VPS26 or VPS35 resulted in the loss of FAM21 from the membrane (supplementary material Fig. S3C) and, using a biochemical assay to measure the amount of strumpellin or WASH1 that is membrane-associated, we found that KD of VPS35 causes a notable shift of both strumpellin and WASH1 from the pelletable (membrane-associated) to the soluble (cytosolic) fraction, consistent with VPS35 being required for the membrane association of the WASH1 complex (supplementary material Fig. S3D,E).
Fig. 7.
WASH1 and FKBP15 require the cargo-selective retromer complex for their membrane association. (A) HeLa cells treated with siRNA were fixed and labelled with antibodies against WASH1 and SNX1. WASH1 localisation is lost after KD of KIAA1033, VPS26 or VPS35 but SNX1 or FKBP15 KD did not affect WASH1 localisation. Scale bar: 20 μm. (B) HeLa cells were transiently transfected with GFP-tagged constructs of strumpellin, KIAA1033 and FAM21. In control cells, the GFP-tagged constructs colocalise with VPS26 but, after VPS26 KD, no membrane association is observed. Scale bar: 20 μm. (C) Cells were treated as in A but this time were labelled with antibodies against FKBP15 and SNX1. FKBP15 localisation was most strongly affected by KD of VPS35, VPS26 and WASH1. Scale bar: 20 μm. (D) Cells treated with siRNA to silence VPS26 or VPS35 were labelled with antibodies against EEA1 and WASH1 or FKBP15. In control cells, EEA1 partially colocalises with both WASH1 and FKBP15 (indicated with arrowheads), but in VPS26 KD or VPS35 KD cells, the membrane association of WASH1 and FKBP15 is lost. EEA1 remains associated with punctate endosomal structures. Scale bar: 20 μm.
The anti-strumpellin antisera did not work for microscopy; therefore, in order to examine the localisation of the strumpellin, KIAA1033 and FAM21 proteins, GFP-tagged constructs were generated. In Fig. 7B, HeLa cells were transfected with the strumpellin-, KIAA1033- and FAM21-GFP constructs. In control cells, the GFP-tagged proteins colocalised with VPS26 but, after siRNA KD of VPS26, there was no membrane localisation of the GFP-tagged constructs observed.
When the localisation of FKBP15 was investigated, it was observed that the FKBP15 protein behaved in a similar fashion to WASH1 in that its localisation to the membrane required the retromer cargo-selective complex but, unlike WASH1, FKBP15 localisation was not perturbed by KD of strumpellin, KIAA1033 or WASH1 (see Fig. 7C). Additionally, both WASH1 and FKBP15 partially colocalised with the endosomal marker protein EEA1 (Fig. 7D). Although EEA1 remained associated with punctate tubulovesicular structures after siRNA KD of VPS26 or VPS35, neither WASH1 nor FKBP15 were membrane-associated (Fig. 7D). These data demonstrate that the cargo-selective retromer complex is required for the membrane association of the WASH1 complex and FKBP15.
The WASH1 complex regulates endosomal tubules
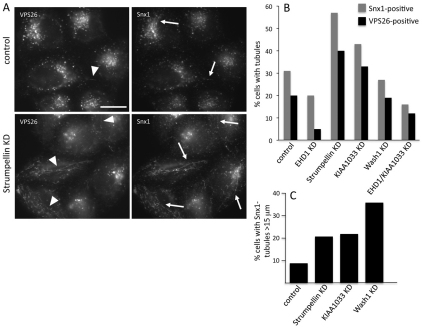
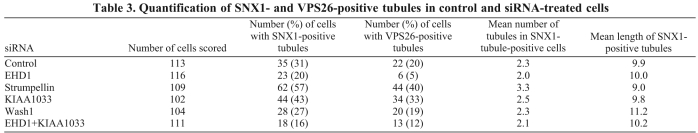
In Fig. 7, we noticed that siRNA KD of strumpellin or KIAA1033 results in an apparent increase in SNX1-labelled tubules and, in Fig. 8A, we confirmed this observation and found that these tubules were also positive for VPS26, which we rarely observe tubule-associated. Previously, we have reported that the EHD1 protein is required to stabilise SNX1 tubules and thereby facilitates endosome-to-Golgi retrieval (Gokool et al., 2007a). Therefore, we carried out a morphometric analysis of the SNX1 and VPS26 tubules in cells that had undergone KDs of strumpellin, KIAA1033, WASH1, EHD1 or a double KD of KIAA1033 and EHD1. At least 100 cells for each condition were scored blind and the data is shown in Fig. 8B (see also Table 3). Strumpellin or KIAA1033 KD led to a substantial increase in SNX1- and VPS26-positive tubules, whereas EHD1 KD had the opposite effect. The KIAA1033 and EHD1 double KD resulted in a phenotype more like the EHD1 KD. The WASH1 KD did not alter the number of either SNX1 or VPS26 tubules observed but did result in a marked increase in the number of tubules that were 15 μm or longer (Fig. 8C).
Fig. 8.
Increased retromer-positive tubules observed after loss of strumpellin or KIAA1033. (A) HeLa cells treated with siRNA to KD strumpellin were labelled with antibodies against VPS26 and SNX1. Fine tubules observed after strumpellin KD are readily observed but less frequently seen in control cells. Arrows indicate tubules labelled with SNX1; arrowheads indicate VPS26-labelled tubules. Scale bar: 20 μm. (B) HeLa cells treated with various siRNA were labelled with antibodies against VPS26 or SNX1 (as in A) and then were scored for SNX1- or VPS26-positive tubules in a blind experiment. More than 100 cells for each KD were observed. The strumpellin or KIAA1033 KDs increase the frequency of SNX1- and VPS26-positive tubules but the tubules still require EHD1 function. (C) Tubules were measured in 10 cells that had at least three SNX1-tubules. Although WASH1 KD did not increase the number of tubules observed, it did result in the observed tubules being much longer.
Table 3.
Quantification of SNX1- and VPS26-positive tubules in control and siRNA-treated cells
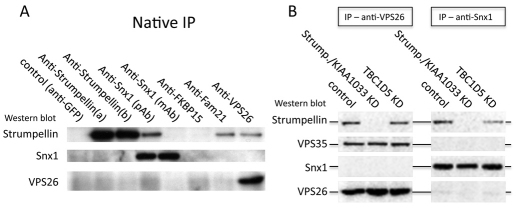
The increased number and length of SNX1-tubules observed after a strumpellin or KIAA1033 KD suggests that the strumpellin-containing WASH1 complex regulates SNX1 tubule formation and therefore could associate with SNX1. We tested this hypothesis by performing native IPs from HeLa cells. In Fig. 9A, we show that anti-SNX1 can co-IP strumpellin, although the interaction is only apparent when we used a polyclonal anti-SNX1 antisera. Strumpellin also co-IPed with anti-VPS26 or anti-FAM21. Interestingly, we did not observe a co-IP of SNX1 with anti-VPS26 or vice-versa – an observation that is consistent with the native IP data shown in Figs 1 and 5.
Fig. 9.
The VPS35-VPS29-VPS26 retromer complex and SNX1 interact separately with the strumpellin complex. (A) Native IPs were performed on lysates from HeLa cells. Both VPS26 and SNX1 co-IP strumpellin but they do not co-IP each other. (B) Native IPs of VPS26 and SNX1 were performed on control HeLa cells or cells in which strumpellin and KIAA1033 or TBC1D5 expression had been silenced with siRNA. Loss of TBC1D5 does not increase the interaction between the cargo-selective complex and SNX1. Therefore, it seems probable that the cargo-selective complex is not linked to the SNX1-complex through strumpellin and that TBC1D5 does not compete with SNX proteins for binding to VPS29.
One reason why there is no detectable interaction between the cargo-selective retromer complex and SNXs in mammalian lysates is that TBC1D5 could be competing for the binding site on VPS29 that in yeast mediates the Vps35p/Vps29p/Vps26p assembly with Vps5p/Vps17p. The yeast Saccharomyces cerevisiae has no TBC1D5 homologue and therefore no competition for binding of Vps29p could occur, resulting in yeast retromer functioning as a stable heteropentamer. We therefore tested whether siRNA KD of TBC1D5 enables an interaction between VPS35-VPS29-VPS26 and SNX1.
Also, as we had observed an increase in the amount of VPS26 associated with SNX1-tubules after loss of strumpellin or KIAA1033, we wondered whether we could detect an increased association between the VPS35-VPS29-VPS26 complex and SNX1 after KD of strumpellin or KIAA1033. Fig. 9B shows the results of native IPs using polyclonal anti-VPS26 or anti-SNX1 from control cells, or cells where a strumpellin and KIAA1033 KD or TBC1D5 KD has been performed. Even after KD of strumpellin and KIAA1033 or TBC1D5, no interaction between the VPS35-VPS29-VPS26 complex and SNX1 was detectable.
The role of the retromer-interacting proteins in endosome-to-Golgi retrieval
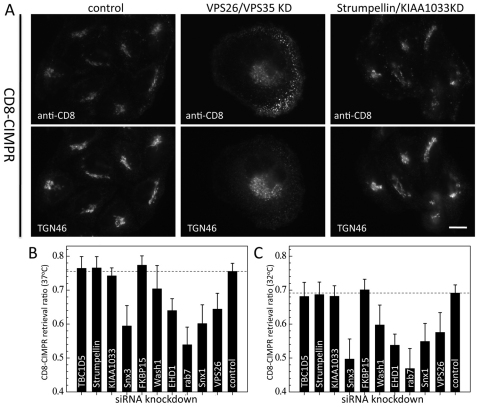
The question that we next sought to address is whether the retromer-interacting proteins function in endosome-to-Golgi retrieval. Initially, using a cell line expressing a CD8-CIMPR reporter and our antibody-uptake assay (Seaman, 2004; Seaman, 2007; Gokool et al., 2007a; Seaman et al., 2009), we examined endosome-to-Golgi retrieval after cells had been treated with siRNA to KD both VPS26 and VPS35 or strumpellin and KIAA1033. The VPS26 and VPS35 double KD resulted in the endocytosed antibody accumulating in peripheral structures but the strumpellin and KIAA1033 double KD did not appear to strongly inhibit endosome-to-Golgi retrieval as the endocytosed anti-CD8 colocalised with TGN46 (Fig. 10A).
Fig. 10.
Analysis of the role of the retromer-interacting proteins in endosome-to-Golgi retrieval. (A) CD8-CIMPR-expressing cells were treated with siRNA to double KD VPS26 and VPS35 or strumpellin and KIAA1033. The cells were incubated with anti-CD8 for 15 minutes and then chased for 30 minutes. In control cells, endocytosed anti-CD8 colocalises with TGN46. Double KD of VPS26 and VPS35 causes the anti-CD8 to accumulate in peripheral structures but double KD of strumpellin and KIAA1033 does not appear to block retrieval. Scale bar: 20 μm. (B) Quantitative analysis of endosome-to-Golgi retrieval. In control cells, 75% of cell-associated anti-CD8 is coincident with the GFP-GMX33, giving a retrieval ratio of 0.75. RNAi KD of VPS26, SNX1, RAB7 or SNX3 inhibits retrieval, lowering the ratio; however, KD of TBC1D5, strumpellin, KIAA1033 or FKBP15 also did not affect endosome-to-Golgi retrieval at 37°C. (C) As in B, but the assay was performed at 32°C.
In order to more quantitatively examine endosome-to-Golgi retrieval and allow comparison of the roles of several proteins in endosome-to-Golgi retrieval, we have adapted our antibody-uptake assay for 96-well plates and imaging by an automated microscope. The assay utilises a cell line stably expressing both a GFP-tagged protein that localises to the Golgi, namely, GMX33/GOLPH3/VPS74 (Wu et al., 2000; Snyder et al., 2006; Schmitz et al., 2008; Dippold et al., 2009; Wood et al., 2009) and the CD8-CIMPR reporter protein.
Cells are incubated for 15 minutes with a mouse monoclonal anti-CD8 that binds to the CD8-CIMPR at the cell surface. The cells are then chased for 30 minutes to allow the anti-CD8 to endocytose and be delivered to the Golgi. After fixation, the cells are labeled with rabbit anti-GFP to enhance the GFP-GMX33 signal, followed by Alexa Fluor 488 anti-rabbit, Alexa Fluor 555 anti-mouse and a blue whole-cell stain. The cells are then imaged in three colours and the amount of anti-CD8 coincident with the Golgi is quantified along with the total anti-CD8 present within the whole-cell mask (determined by the blue whole-cell stain; see supplementary material Fig. S4). Endosome-to-Golgi retrieval is expressed as a ratio of Golgi-localised anti-CD8 divided by the total cell-associated anti-CD8 fluorescence. From each well, 250 ‘objects’ are measured – an object being the blue whole-cell stain.
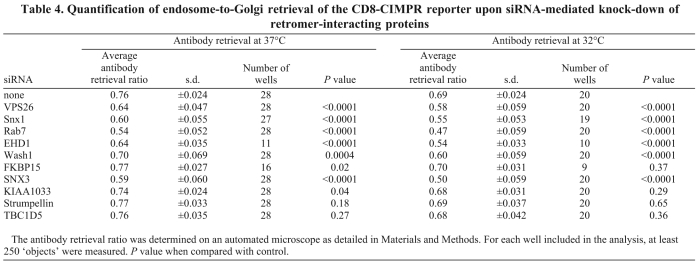
In Fig. 10B, the results from multiple endosome-to-Golgi retrieval assays conducted at 37°C are shown. Control cells treated only with oligofectamine consistently generated a retrieval ratio of ~0.75, which indicates that 75% of the endocytosed anti-CD8 is coincident with the Golgi. Silencing of VPS26, SNX1, RAB7, EHD1 and SNX3 generated clear significant defects in endosome-to-Golgi retrieval, but loss of expression of TBC1D5, strumpellin, KIAA1033 or FKBP15 did not. The WASH1 KD produced a modest endosome-to-Golgi retrieval defect when the assay was conducted at 37°C. In order to determine if the loss of TBC1D5, FKBP15, strumpellin or KIAA1033 was having only a kinetic effect on endosome-to-Golgi retrieval, we repeated the assay but at the lower temperature of 32°C to slow down endosome-to-Golgi retrieval. In Fig. 10C, the graph shows that at the lower temperature the control cells had a retrieval ratio of ~0.69, consistent with retrieval occurring more slowly at the lower temperature. Even at 32°C, the TBC1D5, strumpellin, KIAA1033 and FKBP15 KDs did not affect endosome-to-Golgi retrieval but the loss of WASH1 now generated a more significant retrieval defect. The data obtained from multiple experiments was analysed for statistical significance and is shown in Table 4.
Table 4.
Quantification of endosome-to-Golgi retrieval of the CD8-CIMPR reporter upon siRNA-mediated knock-down of retromer-interacting proteins
Discussion
Interactions of the cargo-selective retromer complex
In this report we have identified five novel interacting proteins for the retromer cargo-selective complex and have characterised the interaction of a sixth protein, TBC1D5, with the VPS29 protein.
Of the six retromer-interacting proteins studied here, four proteins appear to form a complex that we have called the WASH1 complex after one of its members, the WASp, Wave homologue, WASH1. The other members of this complex are FAM21, KIAA1033 and strumpellin. We have determined the protein-protein associations both within the WASH1 complex and also between the WASH1 complex and the retromer cargo-selective complex using the yeast two hybrid (Y2H) system. Interaction between the WASH1 complex and the VPS35-VPS29-VPS26 complex is mediated by the direct binding of VPS35 with FAM21. VPS35 and also VPS29 can interact with KIAA1033, although the Y2H assay indicated that these were much weaker interactions. FAM21 interacts strongly with KIAA1033, whereas WASH1 can bind to KIAA1033 strongly and VPS35 weakly.
Although we could not detect any interactions for strumpellin, we believe that strumpellin might bind directly to KIAA1033 as siRNA KD of KIAA1033 prevents the association of strumpellin with the VPS35-VPS29-VPS26 complex. Strumpellin is mutated in hereditary spastic paraplegia (HSP) (Valdmanis et al., 2007) and, interestingly, the KIAA1033 gene is located at a region of chromosome 12 that contains the locus for another gene mutated in HSP (Schüle et al., 2009). It remains to be determined, however, whether mutations in KIAA1033 also cause HSP.
While this manuscript was in preparation, two reports were published that also described complexes containing the WASH1 protein. The paper by Derivery et al. (Derivery et al., 2009) describes a complex of WASH1 with KIAA1033, strumpellin (KIAA0196) and FAM21 (KIAA0592) along with the actin-capping proteins CAPZa and CAPZb, but did not detect any components of retromer associated with the WASH1 complex. In this report, we also find CAPZa and CAPZb as relatively minor components in native IPs from VPS29-GFP cells. The paper from Gomez and Billadeau (Gomez and Billadeau, 2009) describes a complex of WASH1 and FAM21 that associates with SNX1 and SNX2. Here, we find that FAM21 and WASH1 can directly bind to VPS35 but we do not detect any SNXs in our native IPs of the retromer cargo-selective complex; however, we do find strumpellin associated with SNX1 by native IP. The data that we present here extends the recently published data by revealing a set of novel interactions between the cargo-selective retromer complex (VPS35-VPS29-VPS26) and the WASH1 complex and crucially demonstrates that these interactions are vital for the endosomal recruitment of the WASH1 complex.
The loss of the retromer cargo-selective complex results in the WASH1 complex being unable to associate with endosomal membranes. After siRNA KD of VPS26 or VPS35, WASH1 becomes cytosolic. Similarly, strumpellin and FAM21 redistribute into the cytoplasm after VPS35 KD (see supplementary material Fig. S3C-E). As SNX1 remains membrane-associated after VPS26 or VPS35 KD, the reported association between FAM21 and the SNXs (Gomez and Billadeau, 2009) cannot be required for the membrane recruitment of the WASH1 complex.
Membrane tubule disregulation as a possible cause of HSP
We show here that KD of strumpellin or KIAA1033 causes increased SNX1- and VPS26-positive tubules, whereas loss of WASH1 results in greater tubule length. The tubules observed after KD of KIAA1033 retain a requirement for EHD1 to stabilize them. The studies by Derivery et al. and Gomez and Billadeau (Derivery et al., 2009; Gomez and Billadeau, 2009) also reported increased endosomal tubules after loss of the WASH1 complex, and Derivery et al. identified dynamin 2 and the ARP2/3 complex as additional WASH1-complex-interacting proteins. We did not find dynamin 2 or ARP2/3 associated with retromer in any of our native IPs and therefore it seems probable that the binding of these proteins by the WASH1 complex occurs after the WASH1 complex is recruited to the endosomal membrane through its association with the retromer VPS35-VPS29-VPS26 complex.
Although substantially increased tubules were observed after loss of the WASH1 complex, the KD of strumpellin or KIAA1033 had no effect on endosome-to-Golgi retrieval (although we cannot rule out the possibility that cargo proteins other than CIMPR might be affected by loss of function of strumpellin or KIAA1033 function), whereas the KD of VPS26, SNX1, EHD1, RAB7 or SNX3 produced a significant endosome-to-Golgi deficit. We conclude, therefore, that increased endosomal tubulation is less detrimental to the functioning of the endosome-to-Golgi retrieval pathway than the loss of the tubule-stabilising function of EHD1. There might, however, be examples where tubule disregulation leads to more profound physiological consequences such as the HSP-causing mutations in strumpellin. HSP is a neurodegenerative disease in which there is a length-dependent progressive axonpathy affecting the longest axons of the main central motor pathway, the corticospinal tracts. Recently, three other HSP proteins have been implicated in membrane tubule formation. Thus overexpression of mutant spastin results in dramatic tubulation of the endoplasmic reticulum (ER), whereas atlastin and REEP1 act in concert with reticulons to regulate tubule formation in the reticular ER (Sanderson et al., 2006; Hu et al., 2009; Orso et al., 2009; Muriel et al., 2009).
The finding that another HSP protein, namely strumpellin, is involved in regulating tubulation, this time at endosomes, suggests that in very long axons there is a requirement for tight regulation of tubule dynamics. It remains to be seen whether the pathological defect in these HSPs is in the ER, or endosome tubulation per se, or whether there are other axon-specific tubulation processes that require these proteins. It is worth noting that although strumpellin and KIAA1033 are conserved in many simple and ancient eukaryotes, these proteins, unlike retromer, have no homologues in yeast and therefore are less likely to be essential components of the endosome-to-Golgi retrieval pathway.
Although it is not yet known why mutations in strumpellin cause HSP, it is interesting to note that there might be another connection between the retromer-interacting proteins and neurodegenerative disease. The ataxin 1 protein that, when mutated, is the most common cause of hereditary ataxia has been found to bind to both TBC1D5 and also FAM21 in a large scale Y2H analysis (Lim et al., 2006). We do not detect ataxin 1 in our native IPs, although that does not preclude an interaction. Ataxin 1 is believed to shuttle between the nucleus and cytoplasm to regulate transcription of certain genes required for the maintenance of Purkinje cells. Perhaps a transient endosomal localisation of ataxin 1 could enable an interaction with neurotropic receptors that are present in neuronal endosomes. Interestingly, strumpellin and KIAA1033 were both detected in native IPs of transcriptional complexes from Drosophila, suggesting a possible role in regulating transcription (Hochheimer et al., 2002).
The function of the FKBP15 protein and its relationship to retromer is somewhat enigmatic. We can demonstrate interactions between FKBP15 and the VPS35-VPS29-VPS26 complex and also the WASH1 complex, suggesting that FKBP15 might undergo multiple interactions with these complexes and, like WASH1, FKBP15 also requires the VPS35-VPS29-VPS26 complex for its membrane association. Unlike strumpellin or KIAA1033, FKBP15 does not appear to have a role in regulating tubule dynamics as we did not observe any obvious changes in the appearance of SNX1-tubules in FKBP15 KD cells. Similarly, loss of FKBP15 did not cause an endosome-to-Golgi retrieval defect. FKBP15 (also called WAFL/KIAA0674/FKBP133) has been shown to associate with endosomes and has a reported role in regulating endosomal membrane traffic (Viklund et al., 2009). In a separate study, FKBP15 was also found to be required for nerve growth cone collapse (Nakajima et al., 2006), suggesting that, like strumpellin, FKBP15 might exhibit stronger phenotypes in neuronal tissues.
The interaction of TBC1D5 with VPS35-VPS29-VPS26 and its implications for the mammalian retromer complex
The association of the TBC1D5 protein with the retromer cargo-selective complex is perhaps most intriguing. Like the WASH1 complex and also FKBP15, TBC1D5 requires the retromer cargo-selective complex to be localised to endosomes and also requires a catalytically active TBC domain to cause the cargo-selective complex to redistribute from the membrane to the cytoplasm. This is significant because it reveals that the ability of TBC1D5 to downregulate the recruitment of the cargo-selective complex is likely to be through TBC1D5 acting on a Rab protein – probably RAB7.
We now show that TBC1D5 binds directly to VPS29 and that the L152E mutation abolishes that interaction. What is most intriguing is that the equivalent of the L152E mutation in yeast Vps29p (L252E) prevents the assembly of the cargo-selective complex (Vps35p/Vps29p/Vps26p) with the SNX proteins (Vps5p/Vps17p), but this mutation in yeast results in only a ~40% carboxypeptidase Y (CPY) sorting defect (Collins et al., 2005). Similarly, we have previously shown that truncation of the N-terminal region of Vps5p that binds to the Vps35p/Vps29p/Vps26p complex resulting in disruption of the heteropentameric retromer complex also results in a ~40% CPY sorting defect (Seaman and Williams, 2002). In yeast, therefore, the heteropentameric retromer complex can also function reasonably efficiently as two separate complexes. Given that we do not detect SNXs in our native IPs of the human VPS35-VPS29-VPS26 complex, an observation that is consistent with our previous data (Gokool et al., 2007) and data from others (Rojas et al., 2007), we must ask ourselves whether retromer in higher eukaryotes is a heteropentamer (as it is most frequently depicted) (e.g. Attar and Cullen, 2009) or rather two separate subcomplexes that perform either cargo-selective or membrane-tubulation functions. The data presented here favours the latter working model and perhaps the heteropentameric configuration of yeast retromer should be regarded as the exception rather than the rule. Consistent with this view are the reports of SNX proteins functioning in the absence of the retromer cargo-selective complex (Nisar et al., 2010).
In higher eukaryotes, the strumpellin-containing complex might functionally link the cargo-selective VPS35-VPS29-VPS26 complex to SNXs but, as we cannot detect complexes of strumpellin associated with both VPS35-VPS29-VPS26 and SNX1, it seems probable that this link is sequential and, as loss of strumpellin (or KIAA1033) expression does not affect endosome-to-Golgi retrieval, is not required for the functioning of the endosome-to-Golgi retrieval pathway. Because we cannot identify a homologue for TBC1D5 in Saccharomyces cerevisiae, nor can we identify strumpellin, KIAA1033, FAM21 or FKBP15 homologues, the retromer complex in yeast might in fact be a very basic and simple form of retromer that can perform all the essential functions.
Although many of the retromer-interacting proteins do not appear to significantly affect retromer or endosome-to-Golgi retrieval, the retromer-cargo selective (VPS35-VPS29-VPS26) complex is clearly required for the membrane localization of the WASH1 complex, FKBP15 and TBC1D5. As the retromer VPS35-VPS29-VPS26 complex is ubiquitously expressed, conserved and reasonably abundant, it makes an effective recruiting platform that other proteins can employ to mediate their endosomal membrane localisation in order to perform their respective function(s).
Materials and Methods
Chemicals and general reagents
Biochemicals were purchased from Sigma Aldrich (Poole, Dorset, UK). Molecular biology reagents were purchased from New England Biolabs (Hitchin, Herts, UK). 125I-protein A used in western blotting was supplied by Perkin Elmer (Waltham, MA, USA). Effectene used for transfection of HeLa cells was obtained from Qiagen (Crawley, Sussex, UK).
Web-based bioinformatic analyses
Secondary structure prediction to identify regions of ‘globularity’ or unstructured regions was performed using the GlobPlot utility (http://globplot.embl.de/). Regions of predicted coiled-coils were based upon the output from the Coils program on the EMBnet server (http://www.ch.embnet.org/software/COILS_form.html) and potential motifs such as the spectrin repeat in strumpellin were identified using the MotifScan database (http://myhits.isb-sib.ch/cgi-bin/motif_scan). Other domains such as the FK506 domain in FKBP15 were identified using a basic blast search at the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Antibodies and cloning and generation of GFP-tagged constructs
Polyclonal antibodies against VPS26, VPS35 and SNX1 have been described previously (Seaman, 2004). Anti-strumpellin antisera used for western blotting was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-FKBP15 and anti-FAM21 antisera used for immunofluorescence or IPs, respectively, were purchased from Abcam (Cambridge, UK). Anti-WASH1 was purchased from Sigma. Monoclonal anti-SNX1 and anti-EEA1 antibodies were purchased from BD biosciences (Franklin Lakes, NJ, USA). The polyclonal anti-strumpellin, anti-TBC1D5 and anti-FKBP15 antisera used in native IPs or western blotting was made against GST-fusion proteins expressed in E. coli (see below).
The cDNA clones for FKBP15, strumpellin, KIAA1033 and FAM21 were obtained from the Kazusa DNA Research Institute (Japan). The FKBP15 cDNA was amplified by PCR and subsequently cloned into the pGEX 4T2 vector (Invitrogen, Paisley, UK) for production of a GST-FKBP15 fusion protein in bacteria. The fusion protein was purified using glutathione-sepharose and used as an antigen for antibody production. After the terminal bleed, the antiserum was affinity-purified using the GST-FKBP15 fusion protein coupled to CNBr-sepharose. The strumpellin, KIAA1033 and FAM21 clones were amplified by PCR and then subsequently cloned into the pEGFP-N1-3 vector (Clontech, Mountain View, CA, USA) for expression as GFP-tagged fusions in mammalian cells. The VPS29-GFP along with the I91S and L152E mutants are described in Collins et al. (Collins et al., 2005).
Site-directed mutagenesis of TBC1D5 was performed using the QuickChange Kit (Agilent Technologies, La Jolla, CA, USA) following the manufacturer's instructions. The GFP-TBC1D5 construct has been previously described (Seaman et al., 2009). The GFP-TBC1D5 RQ mutant construct was subcloned into pIRESneo2 so that a stable cell line expressing the construct could be produced. The GFP-VPS35 construct has been described previously (Gokool et al., 2007b) and this was cloned into pIRESneo2, also so that a stable cell line could be produced.
Fluorescence microscopy
Fluorescence microscopy was performed essentially as previously described (see Seaman et al., 2009), except that the images were captured using a Hamamatsu CCD camera (Sewickly, PA, USA) controlled by the SimplePCI software package.
To assess the effect of wild-type TBC1D5 or the RQ mutant on VPS26 localisation, cells were fixed 24 hours after transient transfection and labelled with rabbit anti-VPS26 and mouse anti-GFP. For each construct, ~30 micrographs were captured over a timed period (20 minutes). The micrographs were then scored blind and the localisation of VPS26 was assessed as being ‘normal’ (i.e. membrane associated) or ‘displaced from the membrane’ (i.e. cytosolic) in cells brightly fluorescent for GFP when compared with untransfected cells in the same field.
For the tubule quantification experiments, cells were treated with siRNA on day 1 and day 3, seeded onto coverslips on day 4 and fixed on day 5. Cells were permeabilised and incubated at room temperature with monoclonal anti-SNX1 antibody and rabbit anti-VPS26 for 1 hour followed by Alexa Fluor 488 anti-mouse IgG and Alexa Fluor 594 anti-rabbit IgG, also for 1 hour. Cells were imaged with an Orca-R CCD camera (Hamamatsu, Japan) mounted on a full-field Zeiss microscope (Carl Zeiss SMT, Cambridge, UK) using a 63× 1.3 NA oil-immersion objective. The imager did not know the cell treatment and imaged 25 fields on each coverslip. She then counted at least 100 cells for each treatment and scored: (1) the number of cells that had SNX1-positive tubules; (2) the number of cells that had VPS26-positive tubules; (3) the number of SNX1 tubules in the SNX1-tubule-positive cells; and (4) the length of these SNX1-positive tubules.
RNAi
Duplexed siRNA oligonucleotides for RNAi-mediated knockdowns were purchased from Dharmacon (Lafayette, CO, USA). Oligofectamine and Optimem were purchased from Invitrogen (Paisley, UK). HeLa cells were treated with siRNAs twice using a five-day knockdown protocol in which the cells were transfected with siRNA on days 1 and 3 and then assayed (e.g. fixed for microscopy or lysed for native IP) on day 5. Concentrations of siRNA and oligofectamine used were as described previously (Seaman, 2004).
Native IPs and yeast two-hybrid assay
The native IPs were performed essentially as previously described (Seaman, 2007; Seaman et al., 2009). Initial IPs were performed using a MES–digitonin lysis buffer (0.1 M MES, 1% w/v digitonin, 1 mM MgAcetate, 200 μM sodium orthovanadate, 0.5 mM EGTA; supplementary material Fig. S1), but in subsequent IPs the lysis buffer was PBS with 1% v/v Triton. Buffers were supplemented with protease inhibitors. For the larger-scale native IPs that were analysed by SDS-PAGE and/or mass spectrometry, 4–6 140 mm tissue culture dishes of cells for each cell line were typically used and the IPs were combined at the end.
Yeast two hybrid (Y2H) assays were carried out using the Clontech Matchmaker system that we have used previously (Gokool et al., 2007b), in which the two genes of interest are expressed in the pGBT9 (‘bait’) and pGAD424 (‘prey’) vectors to generate fusions with the GAL4 binding protein and GAL4 activation protein, respectively. The HF7c strain was used as the reporter strain with growth on plates lacking histidine (–His) as the readout, indicating that an interaction has occurred. All of the pGBT9 and pGAD424 constructs used in this study were sequenced to confirm that a correct, in-frame fusion had been made. Yeast transformations were carried out as described previously (Reddy and Seaman, 2001).
Velocity gradient centrifugation
Sucrose gradients of 5–30% (w/v) were prepared [using the MES lysis buffer (see above) but with no digitonin] as a series of 1 ml steps of 5%, 7.5%, 10%, 13%, 16%, 19%, 22%, 25% and 28%, followed by 2 ml of 30%. One millilitre of lysate from cells stably expressing VPS29-GFP (lysed in the MES–digitonin buffer and cleared by centrifugation at 10,000 g for 5 minutes) was loaded onto the top of the gradient and centrifuged at 170,000 g for 16 hours in a Beckman SW40 rotor. Twenty-four 0.5 ml fractions were collected and analysed for retromer and retromer-interacting proteins by SDS-PAGE and western blotting. Preliminary analysis of the fractions indicated that all of the proteins of interest were between fractions 2–18.
Mass spectrometry
The mass spectrometric identification of excised gel bands using MALDI-TOF was performed as described in Seaman et al. (Seaman et al., 2009). For the analysis of IPed proteins using LC-MSMS, the following protocol was used: following filter-assisted sample preparation (FASP) (Wisniewski et al., 2009), digestion and StageTip desalting, the tryptic peptides were analysed by LC-MSMS using a nanoAcquity (Waters, Milford, MA) coupled to an LTQ-Orbitrap XL (Thermo Fisher, Waltham, MA). Raw spectra were processed using MaxQuant version 1.0.12.31 (Cox and Mann, 2008) and .msn files were searched using Mascot Daemon 2.2.2 against IPI human database release 20090422 (Kersey et al., 2004). Carbamidomethyl was included as a fixed modification and oxidation; N-acetylation (protein) and deamidation were included as variable modifications. Data was searched with a peptide mass tolerance of 7 ppm and a fragment mass tolerance of 0.5 Da. Peptides with an ion score below 29 were excluded and proteins identified by a single, unique peptide were manually validated and required a minimum of six consecutive b- or y-ions.
Automated quantitative endosome-to-Golgi retrieval assay
HeLa cells stably transfected with a GFP-GMX33 construct and CD8-CIMPR construct were seeded into 96-well tissue culture plates containing siRNA that had been pre-incubated with oligofectamine. After 72 hours, the cells were incubated with media containing the monoclonal anti-CD8 antibody for 15 minutes at room temperature, washed with buffer and then incubated with pre-warmed media (either 37°C or 32°C) for a further 30 minutes at 37°C or 32°C. The cells were then fixed with 4% paraformaldehyde, permeabilised and then labelled with rabbit-polyclonal anti-GFP antisera. After washes, the cells were incubated with Alexa Fluor 488 anti-rabbit, Alexa Fluor 555 anti-mouse and the Dharmacon whole-cell stain before further washing and then imaging using a Cellomics ArrayScan V microscope with a 40× objective lens (Zeiss). The microscope was programmed to focus on blue whole-cell stain and each cell was designated as an ‘object’. Two hundred and fifty objects were imaged per well and the amount of red anti-CD8 fluorescence present in the green Golgi mask relative to the total anti-CD8 fluorescence within the blue whole-cell mask was calculated. This value is the endosome-to-Golgi retrieval ratio.
For the various KDs, cells from multiple wells were imaged and the retrieval ratio was calculated. The minimum number of wells imaged was for FKBP15 at 32°C where nine wells were imaged. The total number of objects in these nine wells was ~2250 (250 × 9). In other KD experiments, >20 wells were imaged, equating to ~5000 cells.
Supplementary Material
Acknowledgments
This work is supported by funding from the MRC and the Wellcome Trust. M.N.J.S. is supported by an MRC Senior Non-Clinical Research Fellowship, E.R. is a Wellcome Trust Senior Research Fellow in Clinical Science and C.F. is a recipient of an MRC PhD studentship. We are grateful to our colleagues Folma Buss and Scottie Robinson for critical reading of this manuscript. Deposited in PMC for release after 6 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/21/3703/DC1
References
- Arighi C. N., Hartnell L. M., Aguilar R. C., Haft C. R., Bonifacino J. S. (2004). Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165, 123-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attar N., Cullen P. J. (2009). The retromer complex. Adv Enzyme Regul. 50, 216-236 [DOI] [PubMed] [Google Scholar]
- Bache K. G., Raiborg C., Mehlum A., Stenmark H. (2003). STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278, 12513-12521 [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Hurley J. H. (2008). Retromer. Curr. Opin. Cell Biol. 20, 427-436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J., Bujny M., Peter B. J., Oorschot V. M., Rutherford A., Mellor H., Klumperman J., McMahon H. T., Cullen P. J. (2004). Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr. Biol. 14, 1791-1800 [DOI] [PubMed] [Google Scholar]
- Collins B. M. (2008). The structure and function of the retromer protein complex. Traffic 9, 1811-1822 [DOI] [PubMed] [Google Scholar]
- Collins B. M., Skinner C. F., Watson P. J., Seaman M. N., Owen D. J. (2005). Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nat. Struct. Mol. Biol. 12, 594-602 [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008). MaxQuant enables high peptide identification rates, individualised p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367-1372 [DOI] [PubMed] [Google Scholar]
- Dacks J. B., Field M. C. (2007). Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell Sci. 120, 2977-2985 [DOI] [PubMed] [Google Scholar]
- Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D., Gautreau A. (2009). The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 17, 712-723 [DOI] [PubMed] [Google Scholar]
- Dippold H. C., Ng M. M., Farber-Katz S. E., Lee S. K., Kerr M. L., Peterman M. C., Sim R., Wiharto P. A., Galbraith K. A., Madhavarapu S., et al. (2009). GOLPH3 bridges phosphatidylinositol-4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell 139, 337-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S. (2008). Retromer retrieves wntless. Dev. Cell 14, 4-6 [DOI] [PubMed] [Google Scholar]
- Fukuda M. (2008). Regulation of secretory vesicle traffic by Rab small GTPases. Cell. Mol. Life Sci. 65, 2801-2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokool S., Tattersall D., Seaman M. N. (2007a). EHD1 interacts with retromer to stabilize SNX1 tubules and facilitate endosome-to-Golgi retrieval. Traffic 8, 1873-1886 [DOI] [PubMed] [Google Scholar]
- Gokool S., Tattersall D., Reddy J. V., Seaman M. N. (2007b). Identification of a conserved motif required for Vps35p/Vps26p interaction and assembly of the retromer complex. Biochem. J. 408, 287-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T. S., Billadeau D. D. (2009). A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699-711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A., Zhou S., Zheng S., Holmes M. C., Tjian R. (2002). TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420, 439-445 [DOI] [PubMed] [Google Scholar]
- Hong Z., Yang Y., Zhang C., Niu Y., Li K., Zhao X., Liu J. J. (2009). The retromer component SNX6 interacts with dynactin p150(Glued) and mediates endosome-to-TGN transport. Cell Res. 19, 1334-1349 [DOI] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Zhu P. P., Voss C., Rismanchi N., Prinz W. A., Rapoport T. A., Blackstone C. (2009). A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138, 549-561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeble A. H., Khan Z., Forster A., James L. C. (2008). TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc. Natl. Acad. Sci. USA 105, 6045-6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004). The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985-1988 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Barral S., Reitz C. (2008). The neuronal sortilin-related receptor gene SORL1 and late-onset Alzheimer's disease. Curr. Neurol. Neurosci. Rep. 8, 384-391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Hao T., Shaw C., Patel A. J., Szabó G., Rual J. F., Fisk C. J., Li N., Smolyar A., Hill D. E., et al. (2006). A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 125, 801-814 [DOI] [PubMed] [Google Scholar]
- Linardopoulou E. V., Parghi S. S., Friedman C., Osborn G. E., Parkhurst S. M., Trask B. J. (2007). Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 3, e237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Abreu-Blanco M. T., Barry K. C., Linardopoulou E. V., Osborn G. E., Parkhurst S. M. (2009). Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development 136, 2849-2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriel M. P., Dauphin A., Namekawa M., Gervais A., Brice A., Ruberg M. (2009). Atlastin-1, the dynamin-like GTPase responsible for spastic paraplegia SPG3A, remodels lipid membranes and may form tubules and vesicles in the endoplasmic reticulum. J. Neurochem. 110, 1607-1616 [DOI] [PubMed] [Google Scholar]
- Nakajima O., Nakamura F., Yamashita N., Tomita Y., Suto F., Okada T., Iwamatsu A., Kondo E., Fujisawa H., Takei K., et al. (2006). FKBP133: a novel mouse FK506-binding protein homolog alters growth cone morphology. Biochem. Biophys. Res. Commun. 346, 140-149 [DOI] [PubMed] [Google Scholar]
- Nielsen M. S., Gustafsen C., Madsen P., Nyengaard J. R., Hermey G., Bakke O., Mari M., Schu P., Pohlmann R., Dennes A., et al. (2007). Sorting by the cytoplasmic domain of the amyloid precursor protein binding receptor SorLA. Mol. Cell. Biol. 27, 6842-6851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisar S., Kelly E., Cullen P. J., Mundell S. J. (2010). Regulation of P2Y receptor traffic by sorting nexin 1 is retromer independent. Traffic 11, 508-519 [DOI] [PubMed] [Google Scholar]
- Orso G., Pendin D., Liu S., Tosetto J., Moss T. J., Faust J. E., Micaroni M., Egorova A., Martinuzzi A., McNew J. A., et al. (2009). Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460, 978-983 [DOI] [PubMed] [Google Scholar]
- Pan X., Eathiraj S., Munson M., Lambright D. G. (2006). TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442, 303-306 [DOI] [PubMed] [Google Scholar]
- Reddy J. V., Seaman M. N. (2001). Vps26p, a component of retromer, directs the interactions of Vps35p in endosome-to-Golgi retrieval. Mol. Biol. Cell 12, 3242-3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaeva E., Meng Y., Lee J. H., Gu Y., Kawarai T., Zou F., Katayama T., Baldwin C. T., Cheng R., Hasegawa H., et al. (2007). The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 39, 168-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R., Kametaka S., Haft C. R., Bonifacino J. S. (2007). Interchangeable but essential functions of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of mannose 6-phosphate receptors. Mol. Cell. Biol. 27, 1112-1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R., van Vlijmen T., Mardones G. A., Prabhu Y., Rojas A. L., Mohammed S., Heck A. J., Raposo G., van der Sluijs P., Bonifacino J. S. (2008). Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J. Cell Biol. 183, 513-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski L., Pilecka I., Miaczynska M. (2009). Signaling from endosomes: location makes a difference. Exp. Cell Res. 315, 1601-1609 [DOI] [PubMed] [Google Scholar]
- Saksena S., Emr S. D. (2009). ESCRTs and human disease. Biochem. Soc. Trans. 37, 167-172 [DOI] [PubMed] [Google Scholar]
- Salinas S., Proukakis C., Crosby A., Warner T. T. (2008). Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet. Neurol. 7, 1127-1138 [DOI] [PubMed] [Google Scholar]
- Sanderson C. M., Connell J. W., Edwards T. L., Bright N. A., Duley S., Thompson A., Luzio J. P., Reid E. (2006). Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum. Mol. Genet. 15, 307-318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sann S., Wang Z., Brown H., Jin Y. (2009). Roles of endosomal trafficking in neurite outgrowth and guidance. Trends Cell Biol. 19, 317-324 [DOI] [PubMed] [Google Scholar]
- Schmitz K. R., Liu J., Li S., Setty T. G., Wood C. S., Burd C. G., Ferguson K. M. (2008). Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev. Cell. 14, 523-534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüle R., Bonin M., Dürr A., Forlani S., Sperfeld A. D., Klimpe S., Mueller J. C., Seibel A., van de Warrenburg B. P., Bauer P., Schöls L. (2009). Autosomal dominant spastic paraplegia with peripheral neuropathy maps to chr12q23-24. Neurology 72, 1893-1898 [DOI] [PubMed] [Google Scholar]
- Seaman M. N. (2004). Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165, 111-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M. N. (2007). Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J. Cell Sci. 120, 2378-2389 [DOI] [PubMed] [Google Scholar]
- Seaman M. N. (2008). Endosome protein sorting: motifs and machinery. Cell. Mol. Life Sci. 65, 2842-2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M. N., Williams H. P. (2002). Identification of the functional domains of yeast sorting nexins Vps5p and Vps17p. Mol. Biol. Cell 13, 2826-2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M. N., McCaffery J. M., Emr S. D. (1998). A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142, 665-681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M. N., Harbour M. E., Tattersall D., Read E., Bright N. (2009). Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J. Cell Sci. 122, 2371-2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder C. M., Mardones G. A., Ladinsky M. S., Howell K. E. (2006). GMx33 associates with the trans-Golgi matrix in a dynamic manner and sorts within tubules exiting the Golgi. Mol. Biol. Cell. 17, 511-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic T. I., Schmiedekamp B. C., Lee J., Katzmann D. J., Burd C. G. (2008). Opposing activities of the SNX3-retromer complex and ESCRT proteins mediate regulated cargo sorting at a common endosome. Mol. Biol. Cell. 19, 4694-4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang H. T., Edwards T. L., Wang X., Connell J. W., Davies R. J., Durrington H. J., O'Kane C. J., Luzio J. P., Reid E. (2009). The hereditary spastic paraplegia proteins NIPA1, spastin and spartin are inhibitors of mammalian BMP signalling. Hum. Mol. Genet. 18, 3805-3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdmanis P. N., Meijer I. A., Reynolds A., Lei A., MacLeod P., Schlesinger D., Zatz M., Reid E., Dion P. A., Drapeau P., et al. (2007). Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am. J. Hum. Genet. 80, 152-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verges M. (2008). Retromer: multipurpose sorting and specialization in polarized transport. Int. Rev. Cell Mol. Biol. 271, 153-198 [DOI] [PubMed] [Google Scholar]
- Viklund I. M., Aspenström P., Meas-Yedid V., Zhang B., Kopec J., Agren D., Schneider G., D'Amato M., Olivo-Marin J. C., Sansonetti P., et al. (2009). WAFL, a new protein involved in regulation of early endocytic transport at the intersection of actin and microtubule dynamics. Exp. Cell Res. 315, 1040-1052 [DOI] [PubMed] [Google Scholar]
- Wassmer T., Attar N., Bujny M. V., Oakley J., Traer C. J., Cullen P. J. (2007). A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer. J. Cell Sci. 120, 45-54 [DOI] [PubMed] [Google Scholar]
- Wassmer T., Attar N., Harterink M., van Weering J. R., Traer C. J., Oakley J., Goud B., Stephens D. J., Verkade P., Korswagen H. C., et al. (2009). The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev. Cell 17, 110-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359-363 [DOI] [PubMed] [Google Scholar]
- Wood C. S., Schmitz K. R., Bessman N. J., Setty T. G., Ferguson K. M., Burd C. G. (2009). PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J. Cell Biol. 187, 967-975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. C., Taylor R. S., Lane D. R., Ladinsky M. S., Weisz J. A., Howell K. E. (2000). GMx33: a novel family of trans-Golgi proteins identified by proteomics. Traffic 1, 963-975 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.