Abstract
Insulin/IGF-1 signaling controls metabolism, stress resistance and aging in Caenorhabditis elegans by regulating the activity of the DAF-16/FoxO transcription factor (TF). However, the function of DAF-16 and the topology of the transcriptional network that it crowns remain unclear. Using chromatin profiling by DNA adenine methyltransferase identification (DamID), we identified 907 genes that are bound by DAF-16. These were enriched for genes showing DAF-16-dependent upregulation in long-lived daf-2 insulin/IGF-1 receptor mutants (P=1.4e−11). Cross-referencing DAF-16 targets with these upregulated genes (daf-2 versus daf-16; daf-2) identified 65 genes that were DAF-16 regulatory targets. These 65 were enriched for signaling genes, including known determinants of longevity, but not for genes specifying somatic maintenance functions (e.g. detoxification, repair). This suggests that DAF-16 acts within a relatively small transcriptional subnetwork activating (but not suppressing) other regulators of stress resistance and aging, rather than directly regulating terminal effectors of longevity. For most genes bound by DAF-16∷DAM, transcriptional regulation by DAF-16 was not detected, perhaps reflecting transcriptionally non-functional TF ‘parking sites'. This study demonstrates the efficacy of DamID for chromatin profiling in C. elegans.
Keywords: aging, C. elegans, DAF-16/FoxO, DamID chromatin profiling, transcriptional networks
Introduction
A major challenge in modern biology is to identify the genes and processes that control aging and longevity, and susceptibility to aging-related illness. Here, a breakthrough was the discovery that mutations in single genes, for example daf-2, can increase lifespan in C. elegans (Kenyon, 2010). daf-2 encodes the cell surface receptor in an insulin/insulin-like growth factor 1 signaling (IIS) pathway that inactivates the FoxO class transcription factor (TF) DAF-16 (Lin et al, 1997; Ogg et al, 1997). Loss of daf-16 suppresses daf-2 mutant traits, including longevity (Kenyon et al, 1993). The role of IIS (including FoxO TFs) as a regulator of aging shows evolutionary conservation, with effects seen in fruit flies, mice and possibly humans (Kenyon, 2010).
Understanding DAF-16 function is key to discovering how IIS controls aging. Microarray studies have shown that IIS regulates many genes (Murphy et al, 2003; McElwee et al, 2007). It is unknown what proportion are directly regulated by DAF-16. Previously, 103 potential DAF-16 direct targets were identified using chromatin immunoprecipitation (ChIP) (Oh et al, 2006). Of these, some influenced aging and/or were IIS regulated (e.g. 9 upregulated and 1 downregulated in daf-2 versus daf-16; daf-2 (McElwee et al, 2007)). We have now used a genome-wide, multi-platform approach to identify DAF-16 regulatory targets.
For chromatin profiling, we have used DNA adenine methyltransferase identification (DamID) (van Steensel and Henikoff, 2000), to our knowledge not used previously in C. elegans. This involves expression of DAF-16 tagged with a bacterial DNA adenine methyltransferase (DAM), which methylates A in the sequence GATC. When the DAM fusion protein binds to DNA, GATC sequences within ∼2 kb get methylated. This creates restriction sites for the enzyme DpnI (GAmTC), allowing methylated regions to be specifically amplified and hybridized to tiling arrays (Figure 1A). To identify genuine direct regulatory targets of DAF-16, we cross-referenced DAF-16-regulated genes with genes with high levels of DAF-16∷DAM methylation relative to a DAM-only control.
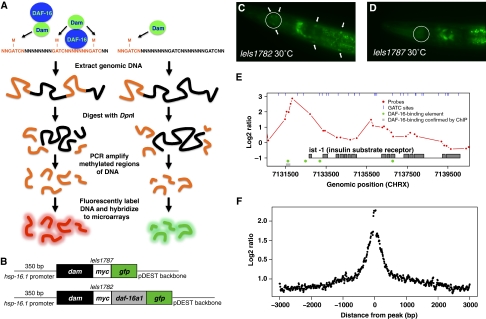
Figure 1.
The DAM identification (DamID) procedure in C. elegans. (A) How DamID works. A fusion protein consisting of DNA adenine methyltransferase (DAM) and the protein of interest methylates GATC sites near binding sites. Genomic DNA is digested with DpnI, which cuts only methylated GATC sites. Adaptors are added, and DNA is digested with DpnII (which cuts at unmethylated GATC sites) to assure selective amplification of methylated DNA. A parallel DAM-only experiment is also performed to control for non-specific methylation. Samples are then labeled and hybridized to arrays. (B) Schematic of plasmid constructs used for preparation of transgenic strains. (C, D) Transgene expression. Nuclear localization of GFP was detected in UL1782 animals (expressing DAM∷DAF-16∷GFP) (marked with arrows in C) in body wall muscle and anterior bulb of pharynx (circled) following heat shock. UL1787 animals (expressing DAM∷GFP) (D) do not show nuclear localization. (E) DAF-16∷DAM methylation profile for ist-1, one of several evolutionarily conserved FoxO targets identified. (F) Average distribution of methylation (DAF-16∷DAM versus DAM) from peak center for 1135 peaks identified.
Results and discussion
Creating C. elegans DamID strains
To identify DAF-16 direct targets using DamID, we created C. elegans lines expressing either DAM alone [UL1787 (leIs1787 [phsp-16.1∷dam∷gfp unc-119] unc-119(ed3) III)], or the DAF-16a1 isoform fused to DAM [UL1782 (leIs1782 [phsp-16.1∷dam∷daf-16a1∷GFP unc-119] unc-119(ed3) III) (Figure 1B; see Supplementary information and Supplementary Figure S7 for strain characterization). Next, we verified that transgenes showed the expected expression patterns. Ubiquitous expression of GFP was visible in both lines only after heat shock, as expected from prior descriptions of hsp-16.1 (Stringham et al, 1992). GFP showed nuclear localization in UL1782 but not UL1787 (Figure 1C and D), consistent with known effects of heat shock on DAF-16 (Henderson and Johnson, 2001). The DamID procedure was attempted both with and without heat shock, with far better results in the latter case (Supplementary information and Supplementary Figure S8).
Identifying DAF-16-binding sites
DAM methylation profiles derived from three biological replicates fed on daf-2 RNAi (no heat shock) showed a high degree of correlation between replicates (Supplementary Figure S1). We identified sequences with methylation peaks in the top 1% of smoothed log2 ratios (log2 ratio>1.5) and within the gene boundary or 2 kb upstream of the translational start. This defined 1135 methylation peaks and 907 associated genes (Supplementary Data Set S1), with a false discovery rate of <5% (see Materials and methods). As an example of DamID data for an individual gene showing strong DAF-16 binding, Figure 1E shows ist-1, whose ortholog (IRS-2) is a FoxO target in mammals (Ide et al, 2004; Puig and Tjian, 2005). On average, the resolution of binding site localization (i.e. log2 ratio>1.5) was <500 bp (Figure 1F).
DAF-16 binding was enriched in intergenic regions immediately upstream of gene coding regions and in introns but underrepresented in exons (Figure 2A). There was also an unexpected enrichment of peaks on the X chromosome. To try to minimize their dominance in subsequent analysis, X-linked peaks were ranked separately from autosomal peaks (Supplementary Figure S2), but this had little effect on the final results. In addition, ChIP-PCR tests confirmed binding of DAF-16 to X (see below), arguing against artefactual binding of DAM to X.
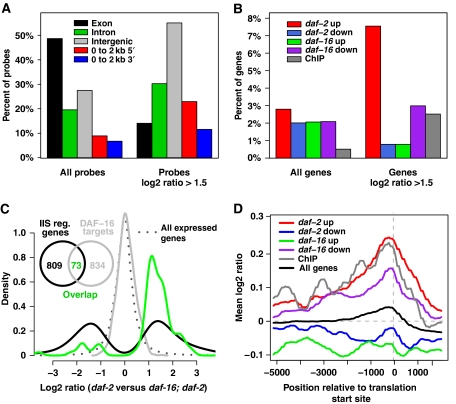
Figure 2.
Features of DAF-16∷DAM methylation. (A) Percent of probes that can be mapped to intergenic, exonic and intronic regions that show DAF-16 binding (i.e. log2 ratio DAF-16∷DAM versus DAM methylation >1.5). Probes were also mapped to regions that were 0–2 kb upstream (5′) or 0–2 kb downstream (3′) of a gene. (B) Percent of genes with DAF-16 binding. Genes were identified by ChIP (Oh et al, 2006), or by microarray analysis as upregulated or downregulated in a daf-2 mutant relative to a daf-16; daf-2 mutant (McElwee et al, 2007) or a daf-16 mutant relative to wild type (Budovskaya et al, 2008). Note: daf-16 down refers to genes that are putative regulatory targets of DAF-16 (i.e. genes that are downregulated when daf-16 is not expressed). (C) Direct targets of DAF-16 are upregulated. Density of log2 ratio for mRNA (daf-2 versus daf-16; daf-2) for genes significantly upregulated or downregulated in reference data set (IIS reg. genes) (McElwee et al, 2007), DAF-16 targets identified by DamID, and the overlap between the two gene lists. (D) Mean log2 ratio (DAF-16∷DAM versus DAM methylation) for the above groups of genes at positions relative to the translational start site.
Genes previously identified as potential DAF-16 targets that were within 5 kb of ChIP-derived DNA fragments (Oh et al, 2006) were enriched among the 907 genes defined by DamID (P=7.6e−07, Fisher's exact test). Similarly, targets showed enrichment of genes upregulated but not downregulated in daf-2 versus daf-16; daf-2 (McElwee et al, 2007) (P=1.4e−11) or similar comparisons (Murphy et al, 2003) (P=2.8e−06). Downregulated genes were underrepresented (P=0.007) and in general there was much greater binding of DAF-16 to the promoters of upregulated genes than downregulated genes (Figure 2B–D). Also enriched were genes encoding proteins with increased abundance in daf-2 versus daf-16; daf-2 mutants (Dong et al, 2007) (P=8.5e−07) but not proteins with decreased abundance (Supplementary Data Set S1). Analysis of the mRNA and protein studies demonstrate an enrichment of DAF-16-binding elements (DBEs) (Furuyama et al, 2000) in upregulated genes and an underrepresentation in downregulated genes (Supplementary Table SI). Thus, the association of DAF-16 binding only with transcriptional activation appears to be real and not a DamID artefact.
Cross-referencing chromatin and transcript profiles
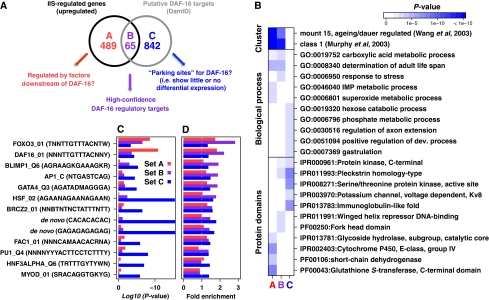
Given the relationship between DAF-16 binding and upregulation in IIS studies, we cross-referenced our DAF-16 targets to 554 genes upregulated in daf-2 versus daf-16; daf-2 (McElwee et al, 2007). This defined three sets of genes (Figure 3A). Set A contains 554−65=489 genes identified by microarray analysis as IIS regulated but likely not directly regulated by DAF-16. Set B contains the 65 genes identified by DamID as DAF-16 targets and which show IIS regulation. Set C contains 907−65=842 genes identified by DamID as bound by DAF-16, but with no IIS regulation detected using the reference microarray data set. Set C likely includes genes where regulation by DAF-16 is subtle or operational only under other conditions, plus genes where DAF-16 binding does not influence expression (Figure 2C).
Figure 3.
Cross-referencing of chromatin and transcriptome profiling. (A) Venn diagram showing relationship between genes identified by microarray analysis and DamID analysis as DAF-16 regulated. This figure also includes an interpretation of the relationship between DAF-16 and the genes within Sets A, B and C. This hypothesis will require further validation. (B) Overrepresentation of expression clusters (Cluster), protein domains and GO biological processes among genes in Sets A, B, and C. (C) Selected regulatory motifs that are overrepresented (P-value<0.0001) in Sets A, B, and C. (D) Fold enrichment of matches to motifs for genes in Sets A, B, and C.
We compared the pattern of gene category overrepresentation in each gene set using the expression analysis systematic explorer (EASE) application (Hosack et al, 2003) (full data in Supplementary Data Set S1). As expected, Sets A and B showed enrichment of class 1, a set of genes independently defined as upregulated in long-lived IIS mutants (Murphy et al, 2003), but Set C did not (Figure 3B), confirming that few of the genes in Set C are IIS/DAF-16 regulated. Previous studies have shown upregulation by DAF-16 of many drug metabolizing enzymes (DMEs), for example cytochrome P450 oxidases and glutathione S-transferases (McElwee et al, 2007). These categories are not enriched among Sets B or C but are enriched in Set A (Figure 3B), suggesting that DAF-16 upregulation of DMEs is indirect.
Given that they show both IIS/DAF-16 regulation and DAF-16 binding, the 65 genes in Set B represent high-confidence DAF-16 regulatory targets (Supplementary Table SII; Supplementary Figure S3). We used ChIP-PCR to verify endogenous DAF-16 binding to selected genes from this set (Supplementary information). DAF-16 binding was verified in peaks associated with akt-1, akt-2, fkh-7, hil-1, and ist-1 (Supplementary Figure S4). We also confirmed IIS regulation of 23/65 by mining additional microarray data sets (Supplementary Data Set S1), and a further 10/65 using real-time PCR (Supplementary Table SII). In addition, we confirmed IIS regulation by examining expression of promoter∷GFP reporter genes for three genes in Set B (Supplementary Figure S5).
The 65 DAF-16 direct targets are enriched for genes already known to influence lifespan, including akt-1, akt-2, coq-1, dao-3, hsp-12.6, mdl-1, and skn-1. Also overrepresented are TFs and genes in the IIS pathway (Figure 3B; Supplementary Table SII; Supplementary Figure S6A). The latter suggests that when IIS is low, DAF-16 increases insulin sensitivity by upregulating expression of IIS pathway proteins, as does FoxO in fruit flies and mammals (Puig and Tjian, 2005). DAF-16 targets include other stress response genes, including SEK-1 that activates DAF-16 (Kondo et al, 2005), and subunits of AMP-activated protein kinase (Supplementary Figure S6B), which is required for daf-2 mutant longevity (Apfeld et al, 2004).
Promoter analysis showed DBE (Furuyama et al, 2000) and other forkhead motifs were highly enriched, especially in the 65 genes (Set B) that show DAF-16 binding and IIS regulation. Also enriched are motifs known to be associated with DAF-16, including heat shock factor response element and GATA-like motif elements (Murphy et al, 2003; McElwee et al, 2004; Budovskaya et al, 2008), plus factors that may create nucleosome-free regions of DNA that allow TFs to bind (Figure 3C and D; Supplementary Table SIII).
Conclusions
Our findings demonstrate that DamID is an effective alternative to ChIP for chromatin profiling in C. elegans. Unlike ChIP, DamID requires no antibodies and fewer sample manipulations (e.g. no DNA–protein cross-linking) and its sensitivity could allow identification of TF targets via tissue-specific expression of DAM-tagged TFs. However, not all ChIP DAF-16 targets were identified by DamID and vice versa (e.g. pie-1 (Curran et al, 2009) and sod-3 (Oh et al, 2006) were not identified as DAF-16 targets by DamID), and a possible disadvantage of DamID is that transgene expression may differ from native gene expression. Moreover, the resolution of binding-site identification is likely to be less than ChIP-Seq (Johnson et al, 2007), although DamID resolution could in principle be improved by replacing tiling arrays with massive-parallel sequencing (‘Dam-seq').
Cross-referencing of DAF-16-related DamID and transcript profile data reveals that most genes to which DAF-16 binds are not detectably DAF-16 regulated in daf-2 mutants, and that most of the genes regulated downstream of DAF-16 are not directly regulated by it. The former observation raises the possibility that the majority of DAF-16-binding sites do not mediate effects on gene expression and are ‘parking sites' for proteins not engaged in regulatory activity.
Direct regulatory targets of DAF-16 include genes whose orthologs are FoxO regulated in mammals (e.g. ist-1, mdl-1, and R11A5.4/PEPCK). This raises the possibility that evolutionarily ancient FoxO targets control traits such as longevity. The targets of DAF-16 identified here provide a new picture of DAF-16 acting as a transcriptional activator (but not repressor) within a relatively small subnetwork of target genes to increase lifespan. Future analysis of this subnetwork should help to reveal the downstream, terminal effector genes that control diapause, metabolism and aging.
Materials and methods
Construction and analysis of DamID strains
Strains used for DamID trials: Control, UL1787 (leIs1787 [phsp-16. 1∷dam∷myc∷gfp pDEST unc-119] unc-119(ed3) III); Test, UL1782 (leIs1782 [phsp-16.1∷dam∷myc∷daf-16a1∷gfp pDEST unc-119] unc-119(ed3) III). Transgenic lines were generated using bombardment (see Supplementary information).
DamID analysis
In the main experiment described here, the two DamID strains were subjected to daf-2 RNAi from hatching at 20°C, much as previously described (Dillin et al, 2002), and using the same daf-2 RNAi feeding plasmid. Three biological replicates were conducted per trial, with one hybridization per biological replicate. Nematodes were raised from eggs prepared by hypochlorite treatment of gravid adults, and maintained at 20°C for ∼96 h before harvesting.
Methylated DNA was purified and amplified from C. elegans gDNA much as previously described (Greil et al, 2006). Briefly, genomic DNA was isolated using a DNAeasy kit (Qiagen) and then digested overnight with DpnI (New England Biolabs (NEB)) to cut at GAmTC sites. DpnI was inactivated (80°C, 20 min) and genomic DNA ligated to double-stranded adaptors with T4-ligase (Roche). DNA fragments were then digested with DpnII (NEB) for 1 h at 37°C to cut unmethylated GATC sites thereby preventing PCR amplification of unmethylated DNA. Methylated DNA was amplified using adaptor-specific primers, and PCR products were hybridized at the NimbleGen facility (NimbleGen Systems, Inc.) to a C. elegans whole genome tiling array (Design ID: 2155; Design Name: 2005-05-25_CE2_WG_CGH_T), for which the array probes were later aligned to WormBase 190 (Array Express E-MEXP-2414).
Identifying methylation peaks
For each hybridization, the Cy3 channel and Cy5 channel were quantile normalized in R using packages available in Bioconductor (Gentleman et al, 2004). Log2 ratio of DNA methylation of DAF-16∷DAM versus DAM was scaled between the biological conditions and replicates. A running average within a 500-bp window was used to smooth the log2 ratios.
Peaks of methylation were identified with the Ringo package (Toedling et al, 2007). Peaks were defined as having three consecutive probes in the top 5% of smoothed log2 ratios, and the maximum log2 ratio within the peak had to be in the top 1% of probes (Supplementary Data Set S1). To obtain an approximate estimate of the number of false positives in the data, observed log2 ratios (after normalization) were randomly re-distributed, smoothed and peaks detected. In 100 random trials, the maximum number of peaks identified was 33, suggesting a false discovery rate <5%.
Gene category analysis
Annotations for the genome of C. elegans were taken from Wormbase and (Wang and Kim, 2003). Overrepresentation or underrepresentation of a functional category for a set of genes as compared to all coding genes was calculated using Fisher's exact test executed in R and, when possible, EASE (Hosack et al, 2003).
Sequence analysis
To identify sequence motifs that were overrepresented in the data, we used Possum (http://biowulf.bu.edu/MotifViz/) to generate log-likelihood scores for position weight matrices (PWMs) (TRANSFAC 12.1). Within 0–2 kb upstream of genes, we defined scores above 8 as matches to a PWM. PWMs were defined as overrepresented in a set of genes relative to all coding genes if the number of genes with matches to a particular PWM was much greater than expected (P-value<0.0001, Fisher's exact test). Fold enrichment was calculated by dividing the number of genes within a gene set that had matches to a PWM to the number of genes expected to match the PWM based on the total number of genes in the genome that had matches to the PWM.
Data deposition: Array Express
ArrayExpress accession: E-MEXP-2414.
Supplementary Material
Supplementary Results, Materials and methods, Supplementary figures S1–8, Supplementary tables SI–III
Excel file containing the following Worksheets:A. GFF3 file showing DAF-16 binding sitesB. 907 genes identified as bound by DAF-16 by DamIDC. Table showing overlap with other datasetsD. EASE of Set A genesE. EASE of Set B genesF. EASE of Set C genes
Acknowledgments
We thank N Alic and L Partridge for useful discussion, and M Riesen for technical assistance. This study was supported by grants from the Wellcome Trust (066750, 081394) (RD, DG, JJM, ES, JMAT, JMT), the European Commission (FP6-518230, LifeSpan 036894) (DG) and the Medical Research Council (G0800339) (ES); Ghent University (GOA 12050101), the Fund for Scientific Research Flanders (G.0025.06) and the European Commission (LSHM-CT-2004-512020) (JRV, FM); and the National Cancer Institute (NCI 4 R33 CA097516-02) (IAH, JRH).
Footnotes
The authors declare that they have no conflict of interest.
References
- Apfeld J, O'Connor G, McDonagh T, DiStefano PS, Curtis R (2004) The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev 18: 3004–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK (2008) An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Wu X, Riedel CG, Ruvkun G (2009) A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature 459: 1079–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Crawford D, Kenyon C (2002) Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298: 830–834 [DOI] [PubMed] [Google Scholar]
- Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR III (2007) Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317: 660–663 [DOI] [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, Mori N (2000) Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J 349: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil F, Moorman C, van Steensel B (2006) DamID: mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase. Methods Enzymol 410: 342–359 [DOI] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE (2001) daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol 11: 1975–1980 [DOI] [PubMed] [Google Scholar]
- Hosack D, Dennis GJ, Sherman B, Lane H, Lempicki R (2003) Identifying biological themes within lists of genes with EASE. Genome Biol 4: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide T, Shimano H, Yahagi N, Matsuzaka T, Nakakuki M, Yamamoto T, Nakagawa Y, Takahashi A, Suzuki H, Sone H, Toyoshima H, Fukamizu A, Yamada N (2004) SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat Cell Biol 6: 351–357 [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B (2007) Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudener A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464 [DOI] [PubMed] [Google Scholar]
- Kenyon CJ (2010) The genetics of ageing. Nature 464: 504–512 [DOI] [PubMed] [Google Scholar]
- Kondo M, Yanase S, Ishii T, Hartman PS, Matsumoto K, Ishii N (2005) The p38 signal transduction pathway participates in the oxidative stress-mediated translocation of DAF-16 to Caenorhabditis elegans nuclei. Mech Ageing Dev 126: 642–647 [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C (1997) daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278: 1319–1322 [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Piper MD, Thomas JH, Patel DS, Selman C, Withers DJ, Thornton JM, Partridge L, Gems D (2007) Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol 8: R132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D (2004) Shared transcriptional signature in C. elegans dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem 279: 44533–44543 [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon CJ (2003) Genes that act downstream of DAF-16 to influence the lifespan of C. elegans. Nature 424: 277–284 [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G (1997) The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999 [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA (2006) Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 38: 251–257 [DOI] [PubMed] [Google Scholar]
- Puig O, Tjian R (2005) Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev 19: 2435–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringham E, Dixon D, Jones D, Candido E (1992) Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol Biol Cell 3: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W (2007) Ringo—an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics 8: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S (2000) Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol 18: 424–428 [DOI] [PubMed] [Google Scholar]
- Wang J, Kim SK (2003) Global analysis of dauer gene expression in Caenorhabditis elegans. Development 130: 1621–1634 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Results, Materials and methods, Supplementary figures S1–8, Supplementary tables SI–III
Excel file containing the following Worksheets:A. GFF3 file showing DAF-16 binding sitesB. 907 genes identified as bound by DAF-16 by DamIDC. Table showing overlap with other datasetsD. EASE of Set A genesE. EASE of Set B genesF. EASE of Set C genes