Summary
Transcriptional regulation of the Nos2 gene encoding inducible nitric oxide synthase (iNOS) requires type I interferon (IFN-I) signaling and additional signals emanating from pattern recognition receptors. Here we showed sequential and cooperative contributions of the transcription factors ISGF3 (a complex containing STAT1, STAT2, and IRF9 subunits) and NF-κB to the transcriptional induction of the Nos2 gene in macrophages infected with the intracellular bacterial pathogen Listeria monocytogenes. NF-κB preceded ISGF3 at the Nos2 promoter and generated a transcriptional memory effect by depositing basal transcription factor TFIIH with the associated CDK7 kinase for serine 5 phosphorylation of the RNA polymerase II (pol II) carboxyterminal domain (CTD). Subsequent to TFIIH deposition by NF-κB, ISGF3 attracted the pol II enzyme and phosphorylation at CTD S5 occurred. Thus, STATs and NF-κB cooperate through pol II promoter recruitment and the phosphorylation of its CTD, respectively, as a prerequisite for productive elongation of iNOS mRNA.
Keywords: MOLIMMUNO, SIGNALING, RNA
Highlights
► The ISGF3 and NF-κB complexes are essential for iNOS induction by L. monocytogenes ► NF-κB attracts TFIIH-CDK7; the IFN-I-ISGF3 signal recruits RNA pol II ► CDK7 deposition by NF-κB generates a transcriptional memory effect for ISGF3 activity ► NF-κB and ISGF3 effectuate an unconventional mode of transcriptional initiation
Introduction
The production of nitric oxide (NO) occurs during innate immune responses to all classes of pathogens (Bogdan, 2001). The molecule has direct antimicrobial activity, contributes to cell signaling, and regulates cell survival (Bogdan, 2001; Zwaferink et al., 2008). Inducible nitric oxide synthase (iNOS), the enzyme encoded by the Nos2 gene and responsible for NO production during infection, is synthesized de novo as a response to the recognition of microbial molecular patterns. Studies with bacterial lipopolysacharide (LPS) or with pathogen-infected murine cells showed that full transcriptional induction of Nos2 and of NO production occurs only after synthesis of type I interferons (IFN-I) and signaling through the Janus kinase (JAK)-STAT pathway (Bogdan, 2001; Gao et al., 1998). Type II IFN (IFN-γ), produced by natural killer (NK) and T cells, also enhances mouse Nos2 induction by LPS in a manner requiring STAT1 activation by the IFN-γ receptor complex (IFNGR [Meraz et al., 1996]). Together the published work suggests that IFN receptor-activated STATs cooperate with non-IFN signals in the transcriptional regulation of Nos2.
Previous analyses of the murine Nos2 promoter revealed an IFN response region and binding sites for NF-κB (Kleinert et al., 2003). The IFN response region contains binding sites for STAT1 dimer (gamma IFN-activated site, GAS [Xie et al., 1993]) and interferon regulatory factors (IRF [Kamijo et al., 1994; Spink and Evans, 1997]). IFN-γ signaling leads to the formation of STAT1 homodimers and IRF1, both of which were shown to be essential for Nos2 induction by IFN-γ/LPS (Kamijo et al., 1994; Meraz et al., 1996). IFN-I causes formation of both STAT1 dimers and the ISGF3 complex, which comprise a STAT1/STAT2/IRF9 heterotrimer (Darnell, 1997; Schindler et al., 2007). It is unclear which of these complexes contributes to iNOS regulation by IFN-I and whether IFN-I, like IFN-γ, stimulate Nos2 transcription with strong dependence on IRF1 or other IRF family members.
The analysis of signals received by the Nos2 promoter directly from pattern recognition receptors emphasizes the role of NF-κB. Two sites for the transcription factor were identified (Kleinert et al., 2003; Lowenstein et al., 1993; Xie et al., 1994). Particularly the binding element proximal to the transcription start proved essential for the activity of the transfected promoter.
Listeria monocytogenes is a Gram-positive bacterial pathogen replicating in the cytoplasm of mammalian host cells. It is recognized by a variety of different pattern recognition receptors including toll-like receptors and NOD-like receptors (TLR and NLR, respectively) (Edelson and Unanue, 2002; Herskovits et al., 2007). In murine bone marrow-derived macrophages, a hitherto unknown cytoplasmic receptor initiates signaling to the IFN-I genes and subsequent release of IFN-I from the infected cells (Stetson and Medzhitov, 2006; Stockinger et al., 2004). Exclusion of L. monocytogenes from the cytoplasm, e.g., by mutation of its major virulence factor Listeriolysin O, completely abrogates the ability to stimulate IFN-I production (Stockinger et al., 2002). As with LPS, transcriptional induction of the Nos2 promoter was strongly diminished when either IFN-I production or signaling were disrupted (Stockinger et al., 2004). To continue this work, we now asked the question why the Nos2 gene, unlike classical IFN-I-stimulated genes (ISGs) or NF-κB target genes, requires input from both STATs and signals derived directly from pattern recognition receptors for maximal transcriptional induction. Combining an examination of transcription factor and signaling requirements for transcriptional induction with an analysis of transcription factor binding to the Nos2 promoter in situ, we conclude that NF-κB enhances carboxy-terminal domain (CTD) phosphorylation of RNA pol II, after recruitment of the enzyme by STATs.
Results
Cytoplasmic and Precytoplasmic Signals Synergize in Nos2 Induction
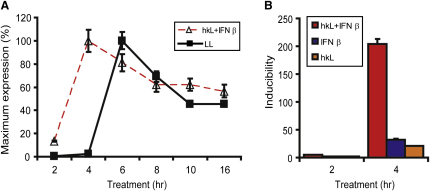
As discussed above, the innate immune response to L. monocytogenes results initially from plasma membrane and endosomal pattern recognition during entry and from cytoplasmic sensing after cytoplasmic escape. The Nos2 gene is paradigmatic for a large group of genes coregulated by pattern recognition receptors and IFN-I (Doyle et al., 2002; Toshchakov et al., 2002). To test whether IFN-I synthesis was the only essential signal for Nos2 induction derived from the cytoplasmic signaling, the two recognition phases were separated by treating macrophages with heat-killed L. monocytogenes (hkL) and with IFN-β either separately or together. Heat-killed Listeria are confined to phagosomes and cannot stimulate the cytoplasmic signal required for IFN-I production. hkL and IFN-β alone were poor inducers of iNOS mRNA synthesis (Figure 1). By contrast, both signals together synergized to produce the full-blown iNOS synthesis seen with viable L. monocytogenes. This result suggests that cytoplasmic signaling can indeed be recapitulated by providing IFN-I. In addition, it provides a valuable experimental tool to separate effects of non-IFN-I and IFN-I signals on the Nos2 promoter and to study each independently from the other. In agreement with IFN-I synthesis preceding Nos2 transcription, the kinetics of mRNA synthesis after infection with viable L. monocytogenes were delayed compared to the simultaneous treatment with hkL and IFN-β.
Figure 1.
Kinetics of iNOS Induction Determined by q-PCR
(A) Exposure of bone marrow-derived macrophages to living L. monocytogenes (LL) or to cotreatment with heat-killed Listeria (hkL) and IFN-β.
(B) Bone marrow-derived macrophages were treated with hkL, IFN-β, or a combination of both.
Error bars represent standard deviations from triplicate samples. The experiments were repeated at least three times.
Many genes expressed in macrophages infected with L. monocytogenes were found in a microarray experiment to display a pattern of regulation resembling that of the Nos2 gene. 38 genes showing the strongest synergy effect between IFN-β alone and the additional presence of L. monocytogenes-derived signals are shown in Figure S1 available online.
Signals and Transcription Factors Required for iNOS Regulation by L. monocytogenes
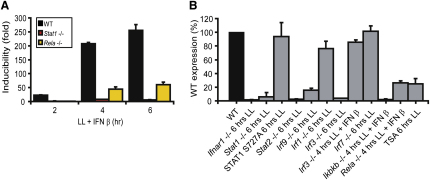
To examine transcription factor requirements for transcriptional induction of the Nos2 gene, we used bone marrow-derived macrophages from either wild-type or gene-targeted mice and infected them with L. monocytogenes (Figure 2). As expected, Nos2 expression required signaling through both the IFN and NF-κB pathways as deletion of either the Stat1 or Rela (NF-κB p65) genes strongly suppressed iNOS mRNA induction in infected macrophages (Figure 2A). More refined analyses confirmed the importance of the IFN-I receptor (Ifnar1−/− mice) and the NF-κB pathway (Rela−/− and Ikbkb −/− mice, deficient for NF-κB p65 and the IKKβ kinase, respectively) and established the importance of the ISGF3 subunits STAT1, STAT2, and IRF9 (Figure 2B). The diminished Nos2 expression observed upon interference with NF-kB signaling was not due to reduced IFN-I production as shown by the fact that addition of exogenous IFN-β did not rescue this effect. Use of macrophages derived from mice expressing STAT1 mutated at its S727 phosphorylation site (STAT1S727A) showed that phosphorylation of STAT1 at S727, important for full transcriptional induction of some IFN-γ-induced genes (Varinou et al., 2003), was not required for Nos2 expression. This contrasts with the reduced induction of Nos2 by IFN-γ early after treatment in STAT1S727A-expressing macrophages (Varinou et al., 2003). In further distinction from the IFN-γ response, the decrease resulting from IRF1 deficiency was marginal. Two additional members of the IRF family, IRF3 and IRF7, are active in L. monocytogenes-infected macrophages (Stockinger et al., 2009). IRF7 deficiency did not affect Nos2 expression. IRF3 deficiency reduced Nos2 induction, but the defect could be rescued by the addition of IFN-β, suggesting that it resulted from reduced IFN-β synthesis, but not from a direct effect on the Nos2 gene. The data suggest that IFN-I participate in Nos2 regulation during L. monocytogenes infection by deploying the ISGF3 complex, but not the ancillary activity of IRFs. The low levels of iNOS expression seen after treatment up to 6 hr with IFN-β alone (Figure 1) were strongly reduced in mice unable to form ISGF3 (data not shown). Interestingly, this differs from the regulation of Nos2 mRNA during the late stage of the IFN-I response, which has been shown to be independent of STAT1 (Plumlee et al., 2009).
Figure 2.
iNOS mRNA Induction by L. monocytogenes Requires Stat1, Stat2, IRF9, and NF-κB Signaling
(A) Bone marrow-derived macrophages of WT, Stat1−/−, and Rela−/− mice were infected with living L. monocytogenes (LL) for the times indicated. IFN-β was additionally present to compensate for potential defects in IFN-I production. iNOS mRNA expression was determined by q-PCR.
(B) Bone marrow-derived macrophages with the indicated genotypes were infected with living L. monocytogenes (LL) for 6 hr or a combination of LL and IFN-β (Ikbkb−/− + IFN-β; Rela−/− + IFNβ; Irf3−/− + IFN-β) for 4 hr. iNOS mRNA expression was determined by q-PCR.
To be able to compare data between individual experiments, genotype-specific expression is shown as percent induction found in wild-type macrophages. Error bars represent standard deviations from triplicate samples. The mentioned experiments were repeated at least three times.
A distinguishing feature of typical IFN-I-induced genes is that a deacetylation step is required for transcriptional induction, which can be inhibited with the histone deacetylase (HDAC) inhibitor trichostatin A (TSA [Nusinzon and Horvath, 2005]). Induced synthesis of Nos2 mRNA was TSA sensitive, suggesting that the activity of STATs on the Nos2 promoter abides by the same rules in this regard as the transcriptional activation of classical ISGs. MAP kinase pathways downstream of pattern recognition receptors (targeting ERK, JNK, and p38MAPK) were probed by pharmacological inhibition. None of the inhibitory drugs produced a significant reduction of L. monocytogenes-induced Nos2 expression (data not shown). In summary, the data from Figures 1 and 2 suggest that ISGF3 is the main signal derived from cytoplasmic signaling, recapitulated by the addition of exogeneous IFN-β, and that NF-κB is the major signal stimulated by hkL, provided by plasma membrane and/or endosomal pattern recognition receptors for Nos2 induction. Our further investigations therefore concentrated on the interaction between these two pathways.
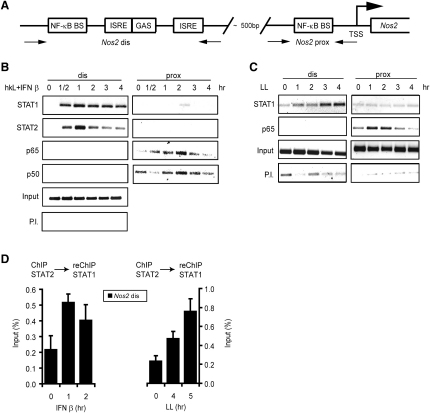
Binding of STATs and NF-κB to Nos2 Promoter Chromatin in Macrophages Infected with L. monocytogenes
Association of the ISGF3 complex with Nos2 chromatin was examined with antibodies against STAT1 and STAT2 for ChIP. Likewise, NF-κB binding was determined with antibodies to its p50 and p65 subunits. Amplification by polymerase chain reaction (PCR) was performed to reveal binding to the promoter-proximal region containing the essential NF-κB site as well as the more distal promoter containing the IFN response region and a second potential binding site for NF-κB (Figure 3A). Treating macrophages simultaneously with hkL and IFN-β stimulated binding of the ISGF3 subunits STAT1 and STAT2 with indistinguishable kinetics (Figure 3B). The same observation was made for the NF-κB subunits p50 and p65 with the notable exception that a reduction of constitutive p50 binding at the earliest time point after stimulation and preceding the phase of increased promoter binding was reproducibly observed. This finding is consistent with the reported negative regulation of NF-κB target genes by p50 homodimers in resting cells (Zhong et al., 2002). NF-κB association was found exclusively with the promoter-proximal, essential site, whereas no evidence for binding to the distal site was obtained. As expected, STAT binding was caused by treatment with IFN-I alone, whereas NF-κB binding occurred after exposure to hkL (data not shown). No evidence for interdependent binding of the two transcription factors was obtained. Consistently, infection with viable L. monocytogenes resulted in similar kinetics of NF-κB p65 binding, but STAT1 association now required prior IFN-I synthesis and was therefore delayed by about 2 hr compared to direct stimulation with IFN-I (Figure 3C). Thus, during infection, binding of NF-κB precedes that of STAT1 and STAT2. The simultaneous presence of these proteins was further examined via a ChIP-re-ChIP procedure (Figure 3D). It confirmed that after both IFN-β treatment and infection with L. monocytogenes, STAT1 could be reprecipitated from a STAT2 ChIP with the expected difference in binding kinetics (see Figure 1).
Figure 3.
Binding of STATs and NF-κB to the Nos2 Promoter
(A) Schematic drawing of the IFN response region and the NF-κB sites (NF-κB BS) in the Nos2 promoter (Kleinert et al., 2003). Binding of STATs and NF-κB to the Nos2 promoter in response to signals stimulated by exposure to L. monocytogenes.
(B) Bone marrow-derived macrophages were stimulated with hkL and IFN-β and the cells were processed for ChIP at the indicated time points. Antibodies used for ChIP are shown on the left, P.I. indicates controls performed with preimmune sera. The precipitates were amplified with primers flanking the proximal (NF-κB) or distal (STAT1, IRF) promoter regions as depicted in (A) and analyzed by gel electrophoresis.
(C) Bone marrow-derived macrophages were infected with viable L. monocytogenes and processed as described in (B).
(D) Bone marrow-derived macrophages were either treated with IFN-β or infected with living L. monocytogenes (LL) for the times indicated and processed for ChIP-Re-ChIP.
Antibodies used for ChIP and Re-ChIP are shown on top of the panels. The precipitates were amplified with primers flanking the distal Nos2 promoter region and analyzed by q-PCR. Error bars represent standard deviations from triplicate samples. The experiments were repeated at least three times.
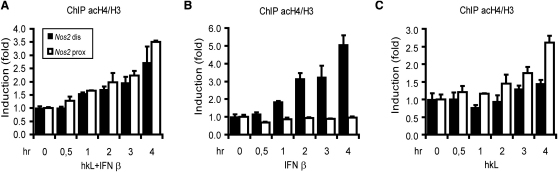
Acetylation of Histones in Proximity to the IFN Response Region and to the Promoter-Proximal NF-κB Site
Synergistic enhancement of transcriptional activation by ISGF3 and NF-κB might result from an interaction in the recruitment of histone acetylases to the Nos2 promoter. Acetylation of histone H4 at the proximal and distal promoter elements was assessed. To correct for histone eviction, data were normalized to the levels of total H3. Increases of histone acetylation are often rather subtle, so we verified significance and quantified our data by using a q-PCR protocol. All experiments were repeated at least five times. Black bars represent amplification of the distal promoter and white bars amplification of the proximal promoter. This convention is maintained through all subsequent figures.
Combined treatment of macrophages with IFN-I and hkL produced an increase of histone acetylation at both the proximal and distal promoter locations (Figure 4A). Treatment with IFN-I alone led to an increase of H4 acetylation almost exclusively at the distal IFN response region (Figure 4B). Conversely, hkL treatment alone caused an increase in H4 acetylation predominantly at the proximal NF-κB element (Figure 4C). Our findings suggest that ISGF3 and NF-κB indeed cooperate in producing hyperacetylated Nos2 promoter chromatin, but that their histone acetyl transferase (HAT)-recruiting activities show no signs of functional interdependence.
Figure 4.
Histone 4 Acetylation at the Nos2 Promoter
Bone marrow-derived macrophages were treated with hkL and IFN-β (A), IFN-β alone (B), or hkL alone (C) as indicated. ChIP was performed with antibodies to acetyl-histone 4 (acH4) and with antibodies to histone 3 (H3). The presence of distal (black) or proximal (white) Nos2 promoter fragments was determined by q-PCR. Data are expressed as increase of acH4 signals normalized to H3 signals to correct for histone eviction. The histograms thus denote the ratio of acetyl-histone 4 binding as a function of total histone 3 (acH4/H3). Error bars represent standard deviations from triplicate samples. All experiments were repeated at least five times.
Recruitment of RNA Polymerase II to the Nos2 Transcription Start Site
Pol II can be bound to transcription start sites in a poised state (Adelman et al., 2009; Koch et al., 2008; Margaritis and Holstege, 2008). Alternatively, the enzyme is recruited in response to the stimulus of gene activation (Adelman et al., 2009). To determine which situation applies to the macrophage Nos2 gene, we analyzed pol II association by ChIP. As shown in Figure 5A, infection with L. monocytogenes strongly increased pol II binding, suggesting that it occurs by regulated recruitment. Surprisingly, treatment with IFN-I alone also stimulated binding of pol II (Figure 5B). Association was somewhat, but not much, weaker than after the additional presence of hkL. In contrast to IFN-I, hkL alone did not stimulate pol II binding (Figure 5C). This result indicates (1) that the histone acetylation caused by NF-κB is not an absolute requirement for pol II binding and (2) that there is a mechanistic difference between ISGF3 and NF-κB in their mode of activating the Nos2 promoter. IFN and STAT-dependent recruitment of pol II predicts that binding of TFIID and its TBP subunit displays the same requirement. Figures 5D and 5E indeed show that both pol II and TBP binding was completely abrogated when Stat1−/−, Stat2−/−, or Irf9−/− macrophages were infected with L. monocytogenes.
Figure 5.
Recruitment of RNA Polymerase II to the Nos2 Promoter by L. monocytogenes-Derived Signals
Bone marrow-derived macrophages from wild-type mice (A–E) or Stat1−/−, Stat2−/−, and Irf9−/− mice (D, E) were infected with living L. monocytogenes (LL [A, D, E]), with IFN-β alone (B), heat-killed Listeria alone (hkL [C]), or with a combination of IFN-β and hkL (B, C) for the times indicated. The cells were processed for ChIP with antibodies against pol II (A–D) or TBP (E). The precipitated DNA was analyzed by q-PCR with primers amplifying the distal (black) and proximal (white) promoter regions. Panels (D) and (E) show a comparison of proximal promoter fragments in ChIP from WT (black), Stat1−/− (red), Stat2−/− (yellow), and Irf9−/− (green) macrophages. Error bars represent standard deviations from triplicate samples. The experiments were repeated at least three times.
Recruitment of TFIIH-CDK7 and Phosphorylation of the Pol II CTD
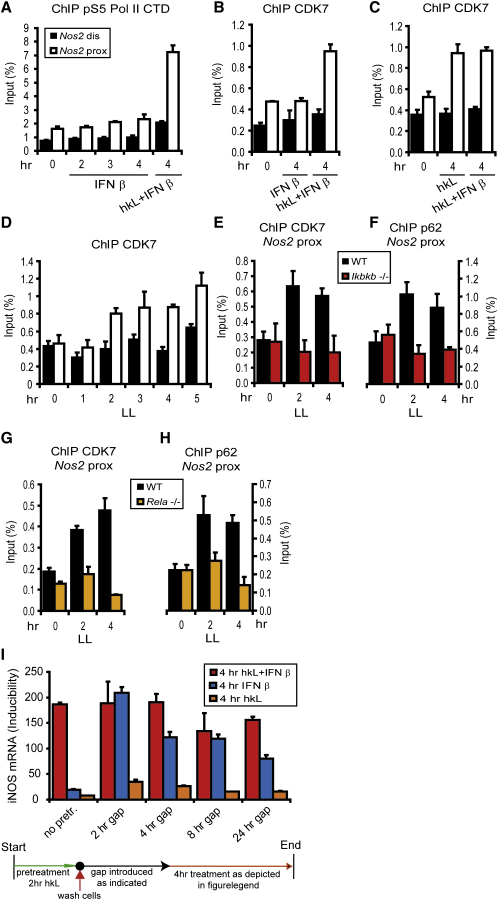
Pol II, once stably bound to the initiation site, must be phosphorylated at its CTD to associate with proteins required for promoter clearance, capping of the mRNA, and elongation (Chapman et al., 2008; Hirose and Ohkuma, 2007). Serine 5 (S5) of the CTD amino acid heptarepeat becomes phosphorylated first, followed by S2, to proceed to productive elongation. With NF-κB playing only a minor role in pol II recruitment, we wondered whether it might play a role in distinct steps of transcriptional initiation. We investigated CTD phosphorylation at S5 by using phosphospecific antibodies for ChIP. S5-phosphorylated pol II was precipitated from the Nos2 initiation site only after treatment with both hkL and IFN-I, but not after treatment with IFN-I alone (Figure 6A). This confirms our notion that NF-κB might be involved in regulating CTD phosphorylation. CTD S5 kinase activity is associated with the general transcription factor TFIIH. TFIIH usually joins the initiation complex only after pol II binding. It is a multiprotein transcription factor containing the CTD S5 kinase CDK7 and a number of additional subunits including p62 (Egly, 2001). As in the case of S5-phosphorylated pol II, CDK7 was associated with the Nos2 initiation site after stimulation with hkL and IFN-I, but not after treatment with IFN-I alone (Figure 6B). In contrast to IFN-I, hkL treatment alone produced as much CDK7 binding as the combined IFN-I-hkL treatment (Figure 6C). The kinetics of CDK7 binding as induced by L. monocytogenes demonstrated association with the Nos2 promoter at 2 hr postinfection (Figure 6D). At this time, NF-κB is associated with Nos2 chromatin, but no or very little ISGF3 is present (Figure 3C). Binding of CDK7 as well as that of TFIIH p62 was abrogated by both NF-κB p65 and IKKβ deficiency (Figures 6E–6H). Together, these data confirm the hypothesis that a TFIIH complex is recruited by NF-κB, providing kinase activity for the pol II CTD at S5. Comparing the kinetics of NF-κB and TFIIH binding in the course of infection suggested that TFIIH remains bound at the promoter even after dissociation of NF-κB (Figures 3, 6G, and 6H; Figure S2). We tested the possibility that NF-κB, by depositing TFIIH, primes the Nos2 promoter for subsequent ISGF3 activity, thus providing a “transcriptional memory” effect. To this end, macrophages were given a 2 hr pulse of hkL treatment, a period sufficient for CDK7 recruitment (Figure 6D). The pulsed cells were left without further stimulation for various intervals, followed by a 4 hr treatment with either IFN-β alone, hkL alone, or a combination of IFN-β and hkL. The data show that for at least 24 hr, the level achieved by IFN-β treatment of pulsed cells exceeded the level achieved by IFN-β treatment of unpulsed cells (Figure 6I). This result is in agreement with the notion of a transcriptional memory or priming effect of NF-κB-recruited CDK7.
Figure 6.
TFIIH-CDK7 Recruitment to the Nos2 Promoter and S5 Phosphorylation of the RNA Polymerase II CTD by L. monocytogenes-Derived Signals; Analysis of Nos2 Promoter Priming by hkL
(A–H) Bone marrow-derived macrophages from WT mice (A–H), Ikbkb−/− mice (E, F), or Rela−/− mice (G, H) were infected with living L. monocytogenes (LL [D–H]), with IFN-β alone (A, B), with hkL alone (C), or with a combination of IFN-β and hkL (A–C) for the times indicated. The cells were processed for ChIP with antibodies against S5-phosphorylated pol II (A), CDK7 (B–E, G), or the TFIIH subunit p62 (F, H). The precipitated DNA was analyzed by q-PCR with primers amplifying the distal (black) and proximal (white) promoter regions. Panels (E)–(H) show a comparison of proximal promoter fragments in ChIP from WT (black), Ikbkb−/− (red, E, F), and Rela−/− (orange, G, H) macrophages.
(I) Bone marrow-derived macrophages were pretreated with hkL for 2 hr or left without pretreatment followed by extensive washing of the cells. The cells were then left without treatment for different periods of time (indicated as hours gap). Thereafter cells were stimulated with hkL + IFN-β, IFN-β alone, or hkL alone for 4 hr. iNOS mRNA expression was determined by q-PCR.
Error bars represent standard deviations from triplicate samples. The experiments were repeated at least three times.
Pol II and CDK7 Recruitment by Interferon-Stimulated Genes or Classical NF-κB Target Genes
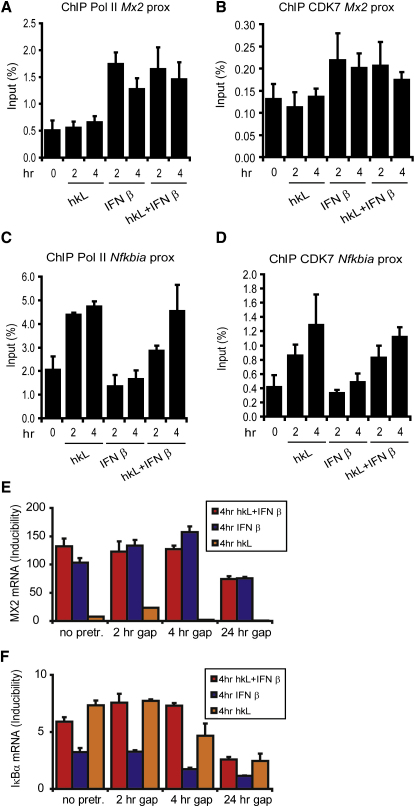
The results obtained by studying Nos2 regulation raise the question why the Nos2 gene requires both ISGF3 and NF-κB to achieve elongation competence. They also predict that genes induced by either IFN-I or the NF-κB pathway alone should demonstrate promoter binding of both CDK7 and pol II after single treatment with IFN-I or hkL. This assumption was tested by analyzing pol II and CDK7 binding to the promoters of the IFN-I-induced Mx2 gene or of the Nfkbia gene, which is activated by NF-κB for the production of IκBα to establish a feedback inhibition loop. The data summarized in Figures 7A–7D show that IFN-β treatment for 2 or 4 hr caused association of both pol II and CDK7 with the Mx2 but not the Nfkbia promoter. Conversely, treatment with hkL to activate the NF-κB pathway increased association of pol II and CDK7 with the Nfkbia but not the Mx2 promoter. Both stimuli provided together did not increase promoter binding beyond the level observed with single treatments. Consistent with this, transcriptional priming by the NF-κB pathway was not observed with either the Mx2 or the Nfkbia gene (Figures 7E and 7F). The data are consistent with our notion that the establishment of elongation competence by cooperative signals is a gene-specific attribute and a major contribution to the regulation of the Nos2 promoter by the transcription factors NF-κB and ISGF3.
Figure 7.
RNA Pol II and CDK7 Recruitment to the Proximal Promoter Regions of the IFN-Inducible Mx2 Gene and the Gene Encoding IκB; Analysis of Mx2 and Nfkbia Promoter Priming by hkL
(A–D) Bone marrow-derived macrophages from wild-type mice were treated with hkL + IFN-β, hkL alone, or IFN-β alone for the times indicated. The cells were processed for ChIP with antibodies against pol II (A, C) or CDK7 (B, D). The precipitated DNA was analyzed by q-PCR with primers amplifying the proximal promoter regions of the Mx2 gene (A, B) and the Nfkbia gene (the gene encoding IκBα) (C, D).
(E and F) Bone marrow-derived macrophages were pretreated with hkL for 2 hr or left without pretreatment followed by extensive washing of the cells. The cells were then left without treatment for different periods of time (indicated as hours gap). Thereafter cells were stimulated with hkL + IFN-β, IFN-β alone, or hkL alone for 4 hr. Mx2 (E) and IκBα (F) mRNA expression was determined by q-PCR. Error bars represent standard deviations from triplicate samples. The experiments were repeated at least three times.
Discussion
NO production is a hallmark of innate immune responses, but its influence on infected cells or organisms varies. For some pathogens, NO is an important clearance mechanism (Bogdan, 2001). By contrast, L. monocytogenes stimulates macrophages to synthesize large quantities of NO, but appears to be relatively insensitive to its toxic effects under our experimental conditions (Zwaferink et al., 2008). The main effect of NO is to promote the death of Listeria-infected macrophages. Our studies of Nos2 regulation were prompted by the findings of several labs that the gene expression signature of cells infected with pathogens, or exposed to their pathogen-associated molecular patterns (PAMPs), results to a significant extent from cooperative signaling by pattern recognition and IFN-I receptors (Doyle et al., 2002; Toshchakov et al., 2002). By using the Nos2 gene as a well-studied example, we show that the need for cooperation between the ISGF3 complex and NF-κB arises from the inability of the former to provide CTD kinase activity and the lack of pol II recruitment by the latter. The prevalent mode of rendering a gene competent for transcription is to assemble a TFIID-TFIIB-pol II complex prior to the association with TFIIH (Roeder, 1996). The combined ISGF3 and NF-κB activity at the Nos2 promoter results in an unconventional transcription initiation complex assembly where TFIIH binds the promoter first to provide kinase activity for the subsequent recruitment of pol II. Three lines of evidence led us to this conclusion: (1) the kinetics of transcription factor and CDK7 binding during L. monocytogenes infection show that TFIIH-CDK7 recruitment occurs before pol II binding, (2) IFN-I alone is able to bring about the recruitment of pol II and hkL alone are able to stimulate CDK7 binding, and (3) CDK7 binding is abrogated in absence of the NF-κB pathway and TBP-pol II binding is abrogated in absence of ISGF3. CDK7 binding trails that of NF-κB by about 1 hr, suggesting that a complex forms at the promoter that is not preassembled and may require intermediate steps and partner proteins. Similarly, pol II binding occurs roughly 1 hr after the observed increase in STAT1 association. CDK7 remains associated with Nos2 chromatin once NF-κB p65 leaves the promoter (best seen in Figure S2), suggesting that the function of NF-κB is to load the promoter with CDK7-TFIIH, but not to maintain this association once it has been established.
To our knowledge this is the first time this mode of initiation complex assembly is shown for a gene in the context of the cellular genome and as a result of ISGF3-NF-κB interaction, although several recent studies are in line with our findings (Spilianakis et al., 2003). The most compelling evidence that TFIIH-CDK7 recruitment by NF-κB may be more widely used was provided in studies on the activation of the HIV LTR in response to TNF (Kim et al., 2006). Contrasting the situation with Nos2, an initiation complex including a hyperphosphorylated RNA pol II was preassembled at the LTR, but, similar to our findings with Nos2, elongation competence required TNF and NF-κB to attract TFIIH-CDK7. The authors propose that NF-κB both associates with TFIIH and stimulates release of the inhibitory CDK8 from the mediator complex. Genes induced by LPS differ concerning the rate-limiting regulatory step for the onset of transcription, consisting either in the release of an elongation block to a paused polymerase or the pol II recruitment step (Adelman et al., 2009). Whether and how NF-κB-mediated TFIIH recruitment contributes in both situations is not known. Therefore it will be of interest to determine to what extent this mechanism contributes to the large impact of the NF-κB pathway on infection-related gene expression and in how far Nos2 represents a paradigm valid for the many genes synergistically induced by STATs and NF-κB. The mechanism of pol II recruitment to the many genes regulated by NF-κB in absence of ISGF3 requires further investigation and, conversely, the mode of TFIIH recruitment to ISGF3 target genes in the absence of NF-κB remains to be clarified. This may generally be determined by cooperative transcription factors bound to their target promoters and/or by differences in the preexisting chromatin structure and composition.
Our studies allow some conclusions about the mechanism of ISGF3 action beyond the functional division of labor with NF-κB. At the Nos2 promoter, ISGF3 stimulated binding of RNA pol II without requiring STAT1 phosphorylation at S727 or the helper function of IRF1. This differs from the STAT1 dimer, which is transcriptionally more active with its transactivating domain phosphorylated and requires IRF1 to induce the expression of Nos2 and other genes in response to IFN-γ (Kamijo et al., 1994; Ramsauer et al., 2007; Varinou et al., 2003). Data from our lab addressing IFN-γ induction of the Gbp2 promoter showed that the STAT1 dimer alone cannot recruit RNA pol II (Ramsauer et al., 2007). This allows speculation that the STAT2 transactivating domain may generally supersede the requirement for STAT1 S727 phosphorylation and the ancillary activity of IRF1. In accordance with our findings about acetylation of the Nos2 promoter, STAT2-dependent transcriptional initiation via mediator and TFIID subunits correlates with the ability of the STAT2 TAD to contact the HATs GCN5 and PCAF (Lau et al., 2003; Paulson et al., 2002). Histone acetylation is an important regulatory step for both NF-κB and STAT target genes (Chen and Greene, 2004; Ramsauer et al., 2007). NF-κB as well as ISGF3-dependent acetylation of Nos2 promoter chromatin was restricted to the nucleosomes adjacent to their binding sites. This resembles virus-induced histone acetylation at the IFN-β promoter or the promoter of the IFN-I-induced Ifi-56K gene that was similarly restricted to a region around the transcription factor binding sites and the transcription start (Parekh and Maniatis, 1999).
Reviewing our findings and corroborating studies in the perspective of L. monocytogenes infection or pathogen infection in general raises the question why some, but not all, ISGs are coupled to the NF-κB pathway. IFN-I synthesis during infection occurs in response to nucleic acid PAMPs in the cytoplasm, when endosomal TLRs are stimulated, or when TLRs resident at the plasma membrane travel to late endosomes in the process of pathogen uptake (Kagan et al., 2008). With the notable exception of the cytoplasmic DNA receptor (Stetson and Medzhitov, 2006), all PRRs stimulating IFN-I synthesis will also stimulate the NF-κB pathway, thus providing both signals necessary for Nos2 induction. Vice versa, some PRRs capable of activating NF-κB are not normally coupled to IFN-I synthesis. Examples of these are TLR2, TLR5, and the NLR family receptors NOD1 and NOD2, which have been associated with IFN-I synthesis only in a limited number of cell types or under specific circumstances (Barbalat et al., 2009; Pandey et al., 2009). Furthermore, a large number of stress or inflammatory signals, most notably those emanating from the TNF receptor family, provide NF-κB activity without concomitant IFN-I production and signaling (Dempsey et al., 2003). We hypothesize that such receptors and their signals provide a TFIIH-dependent transcriptional memory effect for Nos2 expression, independently of pathogen uptake. Vigorous iNOS expression and NO production are limited, however, to situations where a pathogen is engulfed and processed by host cells, and when PAMPs appear in the late endosome and cytoplasm. This mechanism is consistent with our results in Figure 6 showing that the hkL-stimulated NF-κB pathway can provide the Nos2 promoter with transcriptional memory for a subsequent treatment with IFN-I. It ensures that large amounts of NO are made only when its antipathogen activity is needed inside cells. Continuing along these lines, the reason why classical ISGs do not require this prime-and-trigger mechanism may be that their products are less harmful and cells can afford to prepare for pathogen entry without running the risk of inflicting damage upon themselves (Zwaferink et al., 2008). Although our study provides a mechanism for signal integration and a potential paradigm for cooperativity between the STAT and NF-κB pathways during infection, further experiments must reveal the biological impact of STAT-NF-κB convergence.
Experimental Procedures
Reagents
Recombinant IFN-β was purchased from Biomedica (Nova Scotia, Canada) and added to culture medium to a final concentration of 250 U/ml. The inhibitors Trichostatin A (TSA) (WAKO Biochemicals, Osaka, Japan), SP600125 for c-JUN kinase inhibition (Sigma-Aldrich, St Louis, MO), SB203580 for p38MAPK inhibition (Sigma-Aldrich), and U0126 for MEK inhibition (Calbiochem, Nottingham, UK) were used in a final concentration of 150 nM, 25 nM, 4 nM, and 10 nM, respectively.
Bacteria and Infection
The Listeria monocytogenes strain LO28 was cultured in brain heart infusion broth overnight at 37°C. Infection of cells at MOI 10 was performed as described (Stockinger et al., 2002). Heat-killed Listeria (hkL) were generated by incubation of an overnight culture of LO28 in a waterbath at 70°C for 20 min.
Mice and Cells
Animal experiments were discussed and approved by the University of Veterinary Medicine, Vienna, institutional ethics committee and carried out in accordance with protocols approved by the Austrian law (GZ 680 205/67-BrGt/2003). Mice (WT C57BL/6, Ifnar1−/− [Muller et al., 1994], Stat1−/− [Durbin et al., 1996], STAT1S727A [Varinou et al., 2003], Stat2−/− [Park et al., 2000], Irf1−/− [Reis et al., 1994], Irf3−/− [Sato et al., 2000], Irf7−/− [Honda et al., 2005], Irf9−/− [Harada et al., 1996], and IkbkbΔ and RelaΔ [Greten et al., 2007]) were sacrificed for bone marrow between 7 and 10 weeks of age. All animals were in a C57BL/6 genetic background. The mice were housed under specific-pathogen-free conditions. Poly I:C-mediated deletion of IKKβ and NF-κB p65 in bone marrow cells was performed as described (Greten et al., 2007). Bone marrow-derived macrophages (BMDM) were obtained by culture of bone marrow in L-cell-derived colony-stimulating factor 1 as described previously (Baccarini et al., 1985).
RNA Preperation and qRT-PCR
RNA preparation was performed with NucleoSpin RNA II Kit purchased from Macherey-Nagel (Düren, Germany) according to the manufacturer's protocol. Quantitative real-time PCR was performed on Mastercycler ep realplex S, purchased from Eppendorf (Vienna, Austria). Primer for iNOS mRNA expression and qRT-PCR were described previously (Stockinger et al., 2004). Primer for MX2 and IκBα mRNA expression were as follows: MX2 fwd 5′-CCAGTTCCTCTCAGTCCCAAGATT-3′; MX2 rev 5′-TACTGGATGATCAAGGGAACGTGG-3′; IκBα fwd 5′-GCAATTTCTGGCTGGTGGG-3′; IκBα rev 5′-GATCCGCCAGGTGAAGGG-3′.
Chromatin Immunoprecipitation and Re-ChIP
Chromatin immunoprecipitation (ChIPs) were performed according to the protocol described in Nissen and Yamamoto (2000). Antibodies used were described recently (anti-STAT1C [Kovarik et al., 1998], anti-STAT2 [Park et al., 2000]), purchased from Santa Cruz (Santa Cruz, CA) and used at a 1:20 dilution (anti-NF-κB p65, anti-NF-κB p50, anti-RNA Pol II, anti-CDK7, and anti-p62-TFIIH) purchased from Bethyl (Montogomery, TX) and used in a dilution of 1:100 (anti-pS5 CTD Pol II), purchased from Abcam (Cambridgeshire, UK) and used in a dilution of 1:100 (anti-histone 3 and anti-TBP), or purchased from Upstate and used in a dilution of 1:100 (anti-acetyl histone 4). ChIP data were normalized to input and, in case of histone acetylation, further normalized to total H3 and to the untreated sample to correct for histone eviction. In the re-ChIP experiments, the immunecomplexes were eluted by adding 10 mM DTT and incubation for 30 min at 37°C. The samples were diluted 40-fold in RIPA-buffer and reimmunoprecipitated.
Primers used for PCR and q-PCR of the Nos2 promoter were as follows: iNOS dis fwd 5′-CCAACTATTGAGGCCACACAC-3′; iNOS dis rev 5′-GCTTCCAATAAAGCATTCACA-3′; iNOS prox fwd 5′-GTCCCAGTTTTGAAGTGACTACG-3′; iNOS prox rev 5′-GTTGTGACCCTGGCAGCAG-3′; Mx2 prox fwd 5′-ACCCAGCCAAGGCCCCCTTA-3′; Mx2 prox rev 5′-GCAGCTGCCAGGGCTCAGAC; IκBα prox fwd 3′-GGACCCCAAACCAAAATCG-5′; IκBα prox rev 3′-TCAGGCGCGGGGAATTTCC-5′.
Microarray Analysis
Macrophages were infected with an overnight culture of L. monocytogenes for 8 hr (MOI10) or treated 4 hr with IFN-β. RNA was extracted with Trizol and QIAGEN RNeasy Kit according to the manufacturers' protocol. 1 μg of RNA per sample was used for cDNA synthesis. cDNA syntheses and array-hybridizations were performed according to the manufacturers' protocol (Amersham-BioSciences; GE Healthcare).
Acknowledgments
We are grateful to M. Nakasato and T. Taniguchi for providing bone marrow from Irf9-deficient mice. We thank A. Jamieson and P. Kovarik for critical reading of our manuscript. This work was funded by the Austrian Research Foundation (FWF) through grants SFB-28 (to M.M. and T.D.) and P20522-B05 (to T.D.).
Published online: July 15, 2010
Footnotes
Supplemental Information includes two figures and can be found with this article online at doi:10.1016/j.immuni.2010.07.001.
Supplemental Information
References
- Adelman K., Kennedy M.A., Nechaev S., Gilchrist D.A., Muse G.W., Chinenov Y., Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc. Natl. Acad. Sci. USA. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarini M., Bistoni F., Lohmann Matthes M.L. In vitro natural cell-mediated cytotoxicity against Candida albicans: Macrophage precursors as effector cells. J. Immunol. 1985;134:2658–2665. [PubMed] [Google Scholar]
- Barbalat R., Lau L., Locksley R.M., Barton G.M. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat. Immunol. 2009;10:1200–1207. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan C. Nitric oxide and the immune response. Nat. Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- Chapman R.D., Heidemann M., Hintermair C., Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24:289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Chen L.F., Greene W.C. Shaping the nuclear action of NF-kappaB. Nat. Rev. Mol. Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- Darnell J.E., Jr. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Dempsey P.W., Doyle S.E., He J.Q., Cheng G. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. 2003;14:193–209. doi: 10.1016/s1359-6101(03)00021-2. [DOI] [PubMed] [Google Scholar]
- Doyle S., Vaidya S., O'Connell R., Dadgostar H., Dempsey P., Wu T., Rao G., Sun R., Haberland M., Modlin R., Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Durbin J.E., Hackenmiller R., Simon M.C., Levy D.E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- Edelson B.T., Unanue E.R. MyD88-dependent but Toll-like Receptor 2-independent innate immunity to Listeria: No role for either in macrophage listericidal activity. J. Immunol. 2002;169:3869–3875. doi: 10.4049/jimmunol.169.7.3869. [DOI] [PubMed] [Google Scholar]
- Egly J.M. The 14th Datta Lecture. TFIIH: From transcription to clinic. FEBS Lett. 2001;498:124–128. doi: 10.1016/s0014-5793(01)02458-9. [DOI] [PubMed] [Google Scholar]
- Gao J.J., Filla M.B., Fultz M.J., Vogel S.N., Russell S.W., Murphy W.J. Autocrine/paracrine IFN-alphabeta mediates the lipopolysaccharide-induced activation of transcription factor Stat1alpha in mouse macrophages: pivotal role of Stat1alpha in induction of the inducible nitric oxide synthase gene. J. Immunol. 1998;161:4803–4810. [PubMed] [Google Scholar]
- Greten F.R., Arkan M.C., Bollrath J., Hsu L.C., Goode J., Miething C., Goktuna S.I., Neuenhahn M., Fierer J., Paxian S. NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell. 2007;130:918–931. doi: 10.1016/j.cell.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H., Matsumoto M., Sato M., Kashiwazaki Y., Kimura T., Kitagawa M., Yokochi T., Tan R.S., Takasugi T., Kadokawa Y. Regulation of IFN-alpha/beta genes: evidence for a dual function of the transcription factor complex ISGF3 in the production and action of IFN-alpha/beta. Genes Cells. 1996;1:995–1005. doi: 10.1046/j.1365-2443.1996.870287.x. [DOI] [PubMed] [Google Scholar]
- Herskovits A.A., Auerbuch V., Portnoy D.A. Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog. 2007;3:e51. doi: 10.1371/journal.ppat.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J. Biochem. 2007;141:601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Kagan J.C., Su T., Horng T., Chow A., Akira S., Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo R., Harada H., Matsuyama T., Bosland M., Gerecitano J., Shapiro D., Le J., Koh S.I., Kimura T., Green S.J. Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science. 1994;263:1612–1615. doi: 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Bourgeois C.F., Pearson R., Tyagi M., West M.J., Wong J., Wu S.Y., Chiang C.M., Karn J. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 2006;25:3596–3604. doi: 10.1038/sj.emboj.7601248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinert H., Schwarz P.M., Forstermann U. Regulation of the expression of inducible nitric oxide synthase. Biol. Chem. 2003;384:1343–1364. doi: 10.1515/BC.2003.152. [DOI] [PubMed] [Google Scholar]
- Koch F., Jourquin F., Ferrier P., Andrau J.C. Genome-wide RNA polymerase II: Not genes only! Trends Biochem. Sci. 2008;33:265–273. doi: 10.1016/j.tibs.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Kovarik P., Stoiber D., Novy M., Decker T. Stat1 combines signals derived from IFN-gamma and LPS receptors during macrophage activation. EMBO J. 1998;17:3660–3668. doi: 10.1093/emboj/17.13.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J.F., Nusinzon I., Burakov D., Freedman L.P., Horvath C.M. Role of metazoan mediator proteins in interferon-responsive transcription. Mol. Cell. Biol. 2003;23:620–628. doi: 10.1128/MCB.23.2.620-628.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstein C.J., Alley E.W., Raval P., Snowman A.M., Snyder S.H., Russell S.W., Murphy W.J. Macrophage nitric oxide synthase gene: two upstream regions mediate induction by interferon gamma and lipopolysaccharide. Proc. Natl. Acad. Sci. USA. 1993;90:9730–9734. doi: 10.1073/pnas.90.20.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritis T., Holstege F.C. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- Meraz M.A., White J.M., Sheehan K.C., Bach E.A., Rodig S.J., Dighe A.S., Kaplan D.H., Riley J.K., Greenlund A.C., Campbell D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- Muller U., Steinhoff U., Reis L.F., Hemmi S., Pavlovic J., Zinkernagel R.M., Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Nissen R.M., Yamamoto K.R. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinzon I., Horvath C.M. Histone deacetylases as transcriptional activators? Role reversal in inducible gene regulation. Sci. STKE. 2005;2005:re11. doi: 10.1126/stke.2962005re11. [DOI] [PubMed] [Google Scholar]
- Pandey A.K., Yang Y., Jiang Z., Fortune S.M., Coulombe F., Behr M.A., Fitzgerald K.A., Sassetti C.M., Kelliher M.A. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh B.S., Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol. Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- Park C., Li S., Cha E., Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- Paulson M., Press C., Smith E., Tanese N., Levy D.E. IFN-Stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 2002;4:140–147. doi: 10.1038/ncb747. [DOI] [PubMed] [Google Scholar]
- Plumlee C.R., Lee C., Beg A.A., Decker T., Shuman H.A., Schindler C. Interferons direct an effective innate response to Legionella pneumophila infection. J. Biol. Chem. 2009;284:30058–30066. doi: 10.1074/jbc.M109.018283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsauer K., Farlik M., Zupkovitz G., Seiser C., Kroger A., Hauser H., Decker T. Distinct modes of action applied by transcription factors STAT1 and IRF1 to initiate transcription of the IFN-gamma-inducible gbp2 gene. Proc. Natl. Acad. Sci. USA. 2007;104:2849–2854. doi: 10.1073/pnas.0610944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis L.F., Ruffner H., Stark G., Aguet M., Weissmann C. Mice devoid of interferon regulatory factor 1 (IRF-1) show normal expression of type I interferon genes. EMBO J. 1994;13:4798–4806. doi: 10.1002/j.1460-2075.1994.tb06805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder R.G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- Schindler C., Levy D.E., Decker T. JAK-STAT signaling: From interferons to cytokines. J. Biol. Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- Spilianakis C., Kretsovali A., Agalioti T., Makatounakis T., Thanos D., Papamatheakis J. CIITA regulates transcription onset via Ser5-phosphorylation of RNA Pol II. EMBO J. 2003;22:5125–5136. doi: 10.1093/emboj/cdg496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink J., Evans T. Binding of the transcription factor interferon regulatory factor 1 to the inducible nitric oxide synthase promoter. J. Biol. Chem. 1997;272:24417–24425. doi: 10.1074/jbc.272.39.24417. [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Stockinger S., Materna T., Stoiber D., Bayr L., Steinborn R., Kolbe T., Unger H., Chakraborty T., Levy D.E., Muller M., Decker T. Production of type I IFN sensitizes macrophages to cell death induced by Listeria monocytogenes. J. Immunol. 2002;169:6522–6529. doi: 10.4049/jimmunol.169.11.6522. [DOI] [PubMed] [Google Scholar]
- Stockinger S., Reutterer B., Schaljo B., Schellack C., Brunner S., Materna T., Yamamoto M., Akira S., Taniguchi T., Murray P.J. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J. Immunol. 2004;173:7416–7425. doi: 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- Stockinger S., Kastner R., Kernbauer E., Pilz A., Westermayer S., Reutterer B., Soulat D., Stengl G., Vogl C., Frenz T. Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS Pathog. 2009;5:e1000355. doi: 10.1371/journal.ppat.1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshchakov V., Jones B.W., Perera P.Y., Thomas K., Cody M.J., Zhang S., Williams B.R., Major J., Hamilton T.A., Fenton M.J., Vogel S.N. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat. Immunol. 2002;3:392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- Varinou L., Ramsauer K., Karaghiosoff M., Kolbe T., Pfeffer K., Muller M., Decker T. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-gamma-dependent innate immunity. Immunity. 2003;19:793–802. doi: 10.1016/s1074-7613(03)00322-4. [DOI] [PubMed] [Google Scholar]
- Xie Q.W., Whisnant R., Nathan C. Promoter of the mouse gene encoding calcium-independent nitric oxide synthase confers inducibility by interferon gamma and bacterial lipopolysaccharide. J. Exp. Med. 1993;177:1779–1784. doi: 10.1084/jem.177.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q.W., Kashiwabara Y., Nathan C. Role of transcription factor NF-kappa B/Rel in induction of nitric oxide synthase. J. Biol. Chem. 1994;269:4705–4708. [PubMed] [Google Scholar]
- Zhong H., May M.J., Jimi E., Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol. Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- Zwaferink H., Stockinger S., Reipert S., Decker T. Stimulation of inducible nitric oxide synthase expression by beta interferon increases necrotic death of macrophages upon Listeria monocytogenes infection. Infect. Immun. 2008;76:1649–1656. doi: 10.1128/IAI.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.