Abstract
Brassinosteroid (BR) regulates gene expression and plant development through a receptor kinase-mediated signal transduction pathway1. Despite many components of the pathway identified, how the BR signal is transduced from the cell surface to the nucleus remains unclear2. Here we describe a complete BR signaling pathway by elucidating the key missing steps of the pathway. We show that phosphorylation of BSK1 by the BR receptor kinase BRI1 promotes BSK1 binding to the BSU1 phosphatase, and BSU1 inactivates the GSK3-like kinase BIN2 by dephosphorylating a conserved phospho-tyrosine residue (pTyr200). Mutations that affect phosphorylation/dephosphorylation of BIN2 pTyr200 (bin2-1, bin2-Y200F, and quadruple loss-of-function of BSU1-related phosphatases) support an essential role for BSU1-mediated BIN2 dephosphorylation in BR-dependent plant growth. These results demonstrate direct sequential BR activation of BRI1, BSK1, and BSU1, and inactivation of BIN2, leading to accumulation of unphosphorylated BZR transcription factors in the nucleus. This study establishes a fully connected BR signaling pathway and provides new insight into the mechanism of GSK3 regulation.
Steroid hormones are critical for development of all multicellular organisms and in plants brassinosteroids (BRs) play essential roles in a wide range of developmental and physiological processes3. Unlike animal steroid hormones, which act through nuclear receptors, BRs bind to a receptor kinase (BRI1) at the cell surface to activate the BR response transcription factors named BZR1 and BZR2 (also known as BES1) through a signal transduction pathway1, 4. Although many components have been identified and studied in detail, our understanding of the BR signaling pathway remains incomplete, with major gaps between the receptor kinases at the cell surface and downstream components in the cytoplasm and nucleus (Supplementary Information, Fig. S1a)1, 2.
The upstream BR-signaling components at the plasma membrane include BRI15, 6 and BAK17, 8 receptor kinases, a novel protein (BKI1) that inhibits BRI19, and the plasma membrane associated BR-signaling kinases (BSKs)10. BR binding to the extracellular domain of BRI1 causes disassociation of BKI1 from BRI19, 11 and induces association and trans-phosphorylation between BRI1 and its co-receptor BAK112, leading to activation of BRI1 kinase and phosphorylation of its substrates BSKs10. Proteomic and genetic studies demonstrated an essential role for BSKs in transducing the signal to the downstream components, but their direct target remains unknown10.
Downstream BR signaling involves the GSK3-like kinase BIN213, the Kelch-repeats-containing phosphatase BSU114, the 14-3-3 family of phosphopeptide-binding proteins15, and BZR1 and BZR2, which directly bind to DNA and regulate BR-responsive gene expression16–19. As a negative regulator of BR signaling, BIN2 phosphorylates BZR1 and BZR2 at numerous sites to inhibit their activities through multiple mechanisms2. These include accelerating proteasome-mediated degradation20, promoting nuclear export and cytoplasmic retention by the 14-3-3 proteins15, 21, and inhibiting DNA binding and transcriptional activity2, 15, 22. By contrast, the BSU1 phosphatase is a positive regulator of BR signaling14. Overexpression of BSU1 increases the dephosphorylated BZR2/BES1 and activates BR responses4, 14. However, BSU1 does not interact with or effectively dephosphorylate BZR2/BES1 in vitro and the biochemical function of BSU1 remains unknown4, 14. It is believed that BR induces rapid dephosphorylation of BZR1 and BZR2 by inhibiting BIN2 and/or activating BSU1. However, the mechanisms by which upstream BR signaling regulates BIN2 and BSU1 remain unclear (Supplementary Information, Fig. S1a)2, 23.
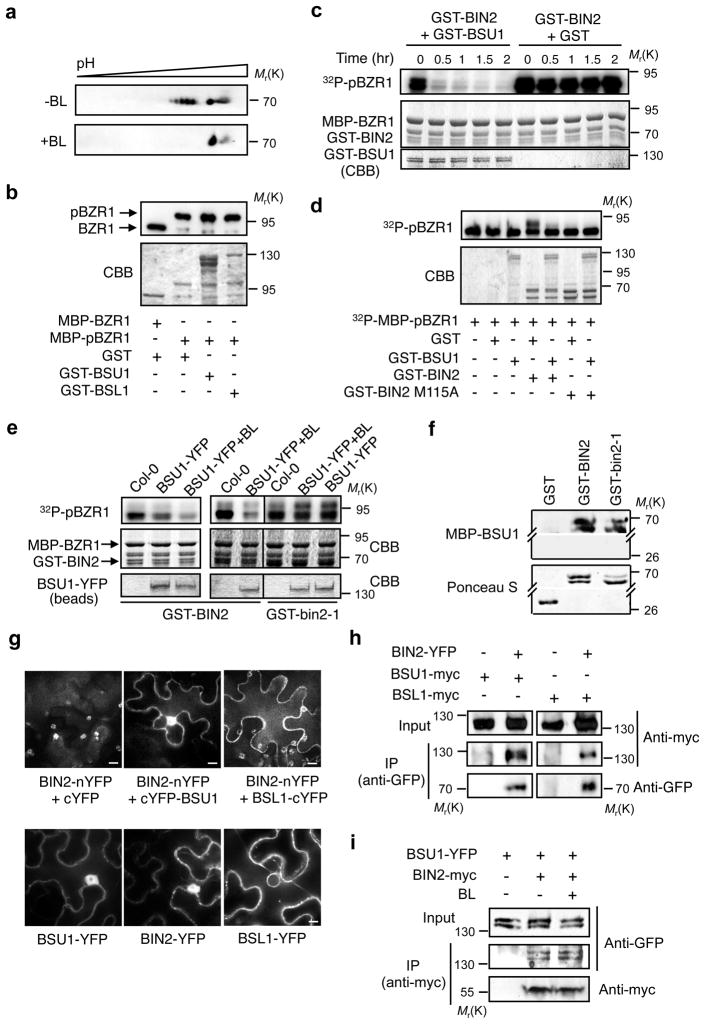
To understand how BR signaling regulates BIN2, we analyzed BR-induced changes of BIN2 using immunoblotting of 2-dimensional gel electrophoresis and found that treatment with brassinolide (BL, the most active BR) caused disappearance of the acidic forms and an increase of the basic forms of an epitope-tagged BIN2 protein (Fig. 1a), suggesting that BR induced dephosphorylation of BIN2. This result led us to test the possible role of BSU1 phosphatase in BR regulation of BIN2. Both BSU1 and its closest homolog BSL1 promote BR signaling in vivo (Supplementary Information, Fig. S2a)14 and show manganese-dependent phosphatase activities (Supplementary Information, Fig. S2b, c). BSU1 only partially reduced the phosphorylation of BZR1 when co-incubated with BIN2 and BZR1 (Supplementary Information, Fig. S3a, b)14, and failed to dephosphorylate BZR1 and BZR2 when added after BIN2 and ATP were removed from the kinase reaction (Fig. 1b; Supplementary Information, Fig. S3c, d). On the other hand, BSU1 most effectively reduced the BZR1 phosphorylation when pre-incubated with BIN2 before adding BZR1 (Fig. 1c). The BZR1 protein partially phosphorylated by BIN2 using radioactive 32P-γATP can be further phosphorylated by BIN2 using non-radioactive ATP, causing a mobility shift of the pre-labeled BZR1. Addition of BSU1 did not reduce the radioactivity of 32P-labeled BZR1 but abolished the mobility shift of the BZR1 band caused by BIN2 (Fig. 1d). These results indicate that BSU1 inhibits BIN2 kinase activity but does not dephosphorylate pre-phosphorylated BZR1 in vitro. The phosphatase domain of BSU1 reduced BIN2 phosphorylation of BZR1 whereas the Kelch repeat domain showed no effect (Supplementary Information, Fig. S3e)
Figure 1.
BSU1 directly inhibits BIN2 phosphorylation of BZR1. (a) BR induces dephosphorylation of BIN2. Total proteins of TAP-BIN2 transgenic plants treated with 0.25 μM brassinolide (BL) or mock solution for 2 hrs were analyzed by two-dimensional gel electrophoresis followed by immunoblotting using the peroxidase anti-peroxidase antibody that detects TAP-BIN2. (b) BSU1 does not dephosphorylate phospho-BZR1 in vitro. BIN2-phosphorylated MBP-BZR1 (BIN2 removed) was incubated with GST, GST-BSU1 or GST-BSL1 for 12 hrs and analyzed by immunoblotting using anti-MBP antibody. (c–e) BSU1 inhibits BIN2 but not bin2-1. (c) GST-BIN2 was pre-incubated with GST-BSU1 or GST for indicated time before MBP-BZR1 and 32P-γATP were added. (d) Partially phosphorylated 32P-MBP-pBZR1 was further incubated with GST-BIN2, GST-BSU1, or both, in the presence of non-radioactive ATP, and analyzed by autoradiography. GST-BIN2 M115A is a kinase-inactive mutant BIN2. (e) GST-BIN2 or GST-bin2-1 was first treated with BSU1-YFP immunoprecipitated from BR-treated (+BL, 0.25 μM BL for 30 min) or untreated 35S::BSU1-YFP plants, and then incubated with MBP-BZR1 and 32P-γATP. Col-0, immunoprecipitation from non-transgenic plant as control. CBB indicates Coomassie brilliant blue-stained gels. (f–i) BSU1 directly interacts with BIN2 and bin2-1. (f) Gel blot containing GST, GST-BIN2 and GST-bin2-1 was probed sequentially with MBP-BSU1 and anti-MBP antibody (upper) and then stained with Ponceau S (lower). (g) BiFC assay of interactions between BSU1 or BSL1 and BIN2. The indicated constructs were transformed into tobacco leaf cells. Bright spots in BIN2-nYFP+cYFP are chloroplast auto-fluorescence. Scale bar = 10 μm. (h) The proteins of tobacco leaves transiently transformed with the indicated constructs were immunoprecipitated with anti-GFP antibody, and the immunoblot was probed with anti-myc and anti-GFP antibody. (i) Arabidopsis plants (F1) expressing BSU1-YFP or co-expressing BSU1-YFP and BIN2-myc, grown on the medium containing BR biosynthetic inhibitor BRZ for 10 days, were treated with 10 μM MG-132 for 1 hr and then with 0.2 μM BL or mock solution for 15 min. Total protein extracts were immunoprecipitated with anti-myc antibodies, and the immunoblot was probed with anti-GFP and anti-myc antibodies. Full scan data of immunoblots and in vitro kinase/phosphatase assays are shown in Supplementary Information, Fig. S12.
We next examined whether BR and the bin2-1 mutation affect BSU1 inhibition of BIN2. A BSU1-YFP (yellow fluorescence protein) fusion protein was immunoprecipitated from transgenic Arabidopsis. Similar to recombinant GST-BSU1, BSU1-YFP from plants did not dephosphorylate the pre-phosphorylated BZR1 (Supplementary Information, Fig. S4a), but reduced BZR1 phosphorylation when co-incubated with BIN2 and BZR1 (Supplementary Information, Fig. S4b) or pre-incubated with BIN2 before adding to BZR1 (Fig. 1e). Moreover, BSU1-YFP from plants treated with BL more effectively inhibited BIN2 phosphorylation of BZR1 than that from untreated plants (Supplementary Information, Fig. S4b; Fig. 1e), suggesting that BR activates BSU1. The gain-of-function bin2-1 mutation that causes BR-insensitive phenotypes23 blocked the regulation by BSU1-YFP in vitro (Fig. 1e).
Several experiments demonstrated that BSU1 directly interacts with BIN2. First, GST-BIN2 was detected on a gel blot by MBP-BSU1 and anti-MBP antibody (Fig. 1f). Second, in vivo interaction was demonstrated by Bi-molecular Fluorescence Complementation (BiFC) assays24, in which tobacco cells co-transformed with BIN2 fused to the N-terminal half (nYFP) and BSU1 fused to C-terminal half (cYFP) of YFP showed a strong fluorescence signal (Fig. 1g). Furthermore, epitope-tagged BIN2 and BSU1 were co-immunoprecipitated and the amount of co-immunoprecipitation was increased by BR treatment (Fig. 1h, I; Supplementary Information, Fig. S5a), indicating that upstream BR signaling induces BSU1 binding to BIN2. Similarly, BSL1 also interacts with BIN2 in BiFC and co-immunoprecipitation assays (Fig. 1g, h). The bin2-1 mutant protein also interacted with BSU1 and BSL1 in these assays (Fig. 1f; Supplementary Information, Fig. S5a, b), suggesting that the bin2-1 mutation blocks BSU1 regulation of BIN2 without abolishing their physical interaction.
A BSU1-GFP protein was previously observed only in the nucleus14. In this study, the BSU1-YFP protein was detected predominantly in the nucleus but weakly in the cytoplasm. Interestingly, BSL1-YFP was excluded from the nucleus and localized exclusively in the cytoplasm and plasma membrane (Fig. 1g; Supplementary Information, Fig. S5c). In fact, BSL1 and its two other homologs have all been identified as plasma membrane proteins by recent proteomics studies25, suggesting that members of the BSU family can mediate upstream BR signaling at the plasma membrane as well as act in the cytoplasm and nucleus.
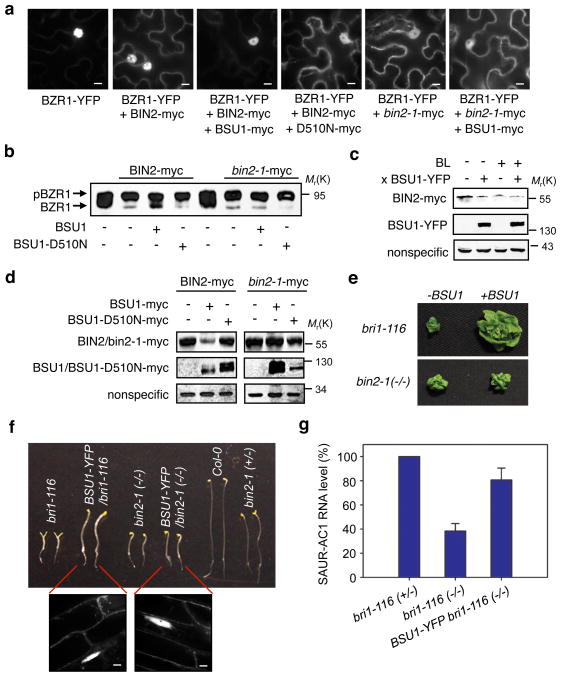
We further examined whether BSU1 inhibits BIN2 activity in vivo. We previously reported that BIN2 phosphorylation of BZR1 promotes BZR1 cytoplasmic retention by the 14-3-3 proteins while unphosphorylated BZR1 accumulates in the nucleus15. Co-expression of BIN2 with BZR1-YFP increased phosphorylation and cytoplasmic retention of BZR1-YFP. Such an effect of BIN2, but not of bin2-1, was canceled by the co-expressed BSU1 but not by a mutant BSU1 (BSU1-D510N), which has reduced phosphatase activity but normal localization (Fig. 2a, b; Supplementary Information, Fig. S6a, b).
Figure 2.
BSU1 regulates BIN2 but not bin2-1 in vivo. (a) Subcellular localization of BZR1-YFP in the cells co-transformed with the indicated constructs. The scale bar is 10 μm. (b) Immunoblots of BZR1-YFP proteins obtained from the tobacco leaves co-transformed with constructs indicated. The upper band is phosphorylated BZR1 and lower one unphosphorylated. (c) Overexpression of BSU1-YFP reduces the accumulation of BIN2-myc protein in a transgenic Arabidopsis line. Heterozygous 35S::BIN2-myc and 35S::BIN2-myc/35S-BSU1-YFP plants (F1) were treated with 0.25 μM BL or mock solution for 30 min. Immunoblot was probed with anti-myc or anti-GFP antibodies, and a non-specific band serves as loading control. (d) BSU1 reduces the accumulation of BIN2 but not that of bin2-1. BIN2- or bin2-1-myc levels were analyzed by anti-myc antibody in tobacco cells co-expressing myc-tagged BSU1 or BSU1-D510N mutant protein. A nonspecific band serves as loading control. (e) Overexpression of BSU1-YFP (+BSU1) partially rescues the bri1-116 mutant, but not the bin2-1 mutant. (f) Hypocotyl phenotypes of seedlings (genotype shown) grown in the dark on MS medium for 5 days. Bottom two panels show confocal images of BSU1-YFP in the plants indicated. The scale bar is 10 μm. (g) Quantitative RT-PCR analysis of SAUR-AC1 RNA expression in wild type (bri1-116 (+/−)), bri1-116 (−/−), and BSU1-YFP/bri1-116 plants. Error bars indicate standard error. Full scan data of immunoblots are shown in Supplementary Information, Fig. S12.
It was reported recently that BR treatment induces proteasome-mediated degradation of BIN223. We showed that BR treatment, or overexpression of BSU1-YFP but not the mutant BSU1-D510N, decreased the protein level of BIN2-myc but not of bin2-1 (Fig. 2c, d; Supplementary Information, Fig. S6c–f). Consistent with a BSU1 function upstream of BIN2 and downstream of BRI1, overexpression of BSU1 partly suppressed the dwarf phenotype of the bri1-116 null mutant but not that of the homozygous bin2-1 mutant (Fig. 2e, f; Supplementary Information, Fig. S7). Furthermore, expression of the BES1-target gene, SAUR-AC119, is increased in BSU1-YFP/bri1-116 plants (Fig. 2g). These results demonstrate that BSU1 acts between BRI1 and BIN2 in the BR signal transduction pathway.
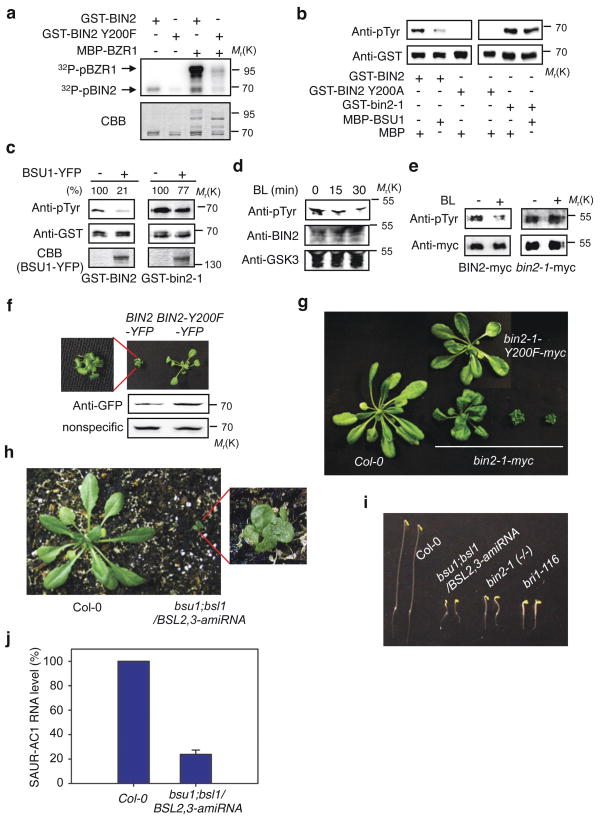
The direct interaction and the requirement of phosphatase activity of BSU1 suggest that BSU1 inhibits BIN2 by dephosphorylating BIN2. We analyzed the autophosphorylation sites of BIN2 in vitro using mass spectrometry, and identified Phospho-tyrosine 200 (pTyr200) of BIN2 as a major phosphorylation site (Supplementary Information, Fig. S8a). The same residue was recently detected as an in vivo phosphorylated site of BIN2 by a phosphoproteome analysis of Arabidopsis26. Mutation of Tyr200 to Phe (Y200F) in BIN2 greatly reduced its substrate phosphorylation (Fig. 3a), indicating an essential role of its phosphorylation for full BIN2 activity. The amino acid sequence flanking Tyr200 of BIN2 is highly conserved in mammalian GSK3s (Supplementary Information, Fig. S8b), and a monoclonal antibody for phospho-Tyr216 of human GSK3β27 specifically detected wild type GST-BIN2 but not the GST-BIN2 containing Y200A mutation or the kinase-inactivating M115A mutation (Fig. 4b; Supplementary Information, Fig. S8c), indicating specificity for the phospho-Tyr200 residue of BIN2. Immunoblot experiments using this antibody showed that BSU1 (Fig. 3b, c) greatly reduced Tyr200 phosphorylation of the wild type BIN2, but not that of the mutant bin2-1. BL treatment also reduced the in vivo phosphorylation of Tyr200 of BIN2 but not that of bin2-1 (Fig. 3d, e). These results demonstrate that BR signaling inhibits BIN2 through BSU1-mediated dephosphorylation of pTyr200, and the bin2-1 mutation causes BR insensitivity by blocking this dephosphorylation.
Figure 3.
BSU1 dephosphorylates pTyr200 of BIN2 but not bin2-1. (a) Tyr200 phosphorylation is required for BIN2 kinase activity. GST-BIN2 or GST-BIN2 Y200F was incubated with MBP-BZR1 and 32P-γATP. CBB, Coomassie brilliant blue-staining. (b, c) BSU1 dephosphorylates pTyr200 of BIN2 but not of bin2-1. Immunoblots of GST-BIN2, GST-BIN2 Y200A and GST-bin2-1 mutant proteins, incubated with MBP or MBP-BSU1 (b) or with BSU1-YFP immunoprecipitated from transgenic Arabidopsis (c), were probed with the anti-pTyr antibody and then with anti-GST antibody. (%) indicates relative signal level of pTyr200 normalized to total GST-BIN2 or GST-bin2-1 protein. (d, e) BR induces dephosphorylation of pTyr200 of BIN2 but not bin2-1. (d) The det2 mutant was treated with 10 μM MG132 for 1 hr prior to treatment with 0.2 μM BL for the indicated time. BIN2 protein was immunoprecipitated with a polyclonal anti-serum and immunoblotted with anti-pTyr, anti-BIN2 serum, and anti-GSK3 α/β antibody. (e) Transgenic plants expressing BIN2-myc or bin2-1-myc was pretreated with 10 μM MG132 and then treated with 0.25 μM BL (+BL) or mock solution (-BL). BIN2-myc and bin2-1-myc were immunoprecipitated by anti-myc antibody and gel blots were probed with antibodies indicated. (f, g) Phosphorylation of Tyr200 is required for BIN2 inhibition of plant growth. (f) Overexpression of BIN2-YFP but not BIN2-Y200F-YFP causes severe dwarf phenotypes in T1 generation. Upper left panel shows zoom-in view. Lower panel shows BIN2-YFP and BIN2-Y200F protein levels detected by anti-GFP antibodies. A nonspecific band serves as loading control. (g) Overexpression of bin2-1-myc but not bin2-1-Y200F-myc caused dwarf phenotypes. Seventy-six of a total 281 35S::bin2-1-myc transgenic T1 seedlings and none of a total 412 35S::bin2-1-Y200F-myc transgenic plants showed dwarf phenotype. (h–j) BSU1 family members play an essential role in BR signaling. (h) Eight of 27 bsu1;bsl1 double mutant plants transformed with an artificial microRNA construct targeting BSL2 and BSL3 (BSL2,3-amiRNA) showed dwarf phenotypes. Right panel shows zoom-in view of the quadruple mutant. (i) Phenotypes of 5-day old dark-grown seedlings of bsu1;bsl1/BSL2,3-amiRNA. (j) Quantitative RT-PCR analysis of SAUR-AC1 RNA expression. Bars indicate standard error. Full scan data of immunoblots and in vitro kinase/phosphatase assays are shown in Supplementary Information, Fig. S12.
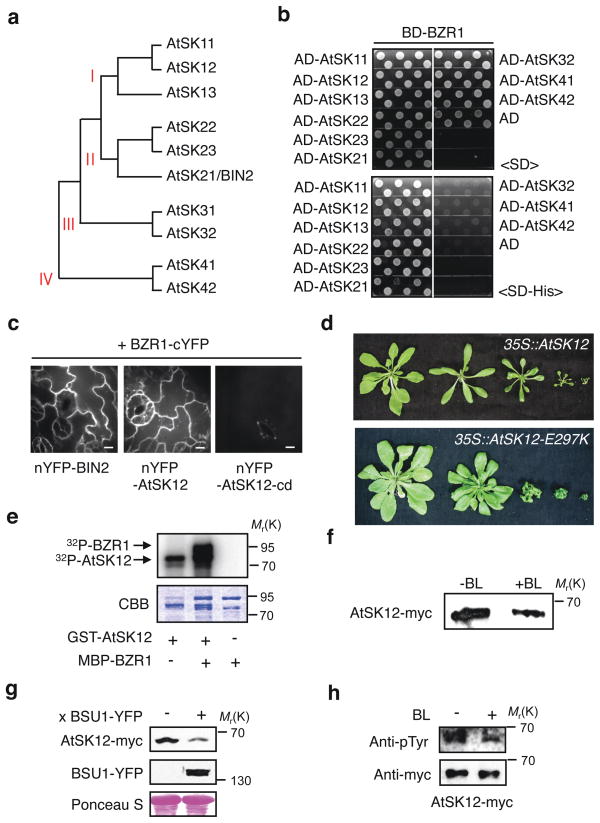
Figure 4.
Regulation of the BIN2 homolog, AtSK12 by BSU1-mediated tyrosine dephosphorylation. (a) Phylogenetic tree of the ten Arabidopsis GSK3/Shaggy-like kinases (AtSKs). (b) Six AtSKs specifically interact with BZR1 in yeast two-hybrid assays. Activation domain (AD) fused AtSKs were transformed into the cells containing DNA binding domain (BD) fused BZR1. Yeast clones were grown on Synthetic Dropout (SD) or SD-Histidine medium. (c) Both AtSK12 and BIN2 interact with BZR1 in BiFC assays. Transgenic Arabidopsis plants expressing nYFP-BIN2, nYFP-AtSK12 and nYFP-AtSK12-cd (C-terminal 29 amino acid deletion) were crossed to BZR1-cYFP plants, respectively. The seedlings of F1 generation were grown in white light for 7 days and YFP signals of epidermal cells were observed. The scale bar is 10 μm. (d) Various phenotypes of transgenic plants (T1) overexpressing WT AtSK12 or AtSK12-E297K. (e) AtSK12 phosphorylates BZR1 in vitro. GST-AtSK12 was incubated with MBP-BZR1 and 32P-γATP. CBB indicates Coomassie brilliant blue-stained gel. (f) BR induces degradation of AtSK12. Homozygous plants expressing AtSK12-myc were treated with 0.25 μM BL for 30 min. Proteins immunoprecipitated by anti-myc antibodies were blotted onto nitrocellulose membrane and probed by anti-myc antibody. (g) Overexpression of BSU1-YFP reduces the accumulation of AtSK12-myc protein in a transgenic Arabidopsis plant. (h) BR induces pTyr dephosphorylation of AtSK12. Homozygous AtSK12-myc plants were pretreated with 10 μM MG132 and then treated with 0.25 μM BL (+BL) or mock solution (-BL). AtSK12-myc was immunoprecipitated by anti-myc antibody and gel blots were probed with anti-pTyr and anti-myc antibodies. Full scan data of immunoblots and in vitro kinase/phosphatase assays are shown in Supplementary Information, Fig. S12.
To further confirm the role of Tyr200 phosphorylation for BIN2 regulation, a Y200F mutation was created. While overexpression of wild type BIN2 or mutant bin2-1 causes BR-insensitive dwarf phenotypes in transgenic Arabidopsis plants, overexpression of BIN2 or bin2-1 containing the Y200F mutation did not (Fig. 3f, g), indicating that Tyr200 phosphorylation is essential for BIN2 to inhibit BR-dependent plant growth and that dephosphorylation of pTyr200 is sufficient to inactivate BIN2. Consistent with an essential role of BSU1 and its homologs in inhibiting BIN2, suppressing BSL2 and BSL3 expression in the bsu1;bsl1 double mutant causes a severe dwarf phenotype and reduced expression of the BES1-target gene SAUR-AC1 (Fig. 3h–j). Taken together, these results demonstrate that dephosphorylation by the BSU1-related phosphatases is the primary mechanism of BIN2 inactivation and an essential step of BR signal transduction.
The Arabidopsis genome encodes 10 GSK3/Shaggy-like kinases (AtSKs), which are classified into four subgroups (Fig. 4a). A triple knockout mutant for group II AtSKs including BIN2 shows increased cell elongation but still accumulates phosphorylated BES1 and responds to BL, indicating that other GSK3-like kinases also act in BR signaling2, 22. We performed interaction study between BZR1 and nine AtSKs representing four subgroups, and found that all six AtSKs of subgroup I and II interact with BZR1 in yeast two-hybrid assays (Fig. 4b). We further examined the function of AtSK12 as a representative of subgroup I AtSKs in BR signaling. BiFC assays showed that AtSK12 interacts with BZR1 as does BIN2 in Arabidopsis, and deletion of the C-terminal 29 amino acids of AtSK12 abolished the interaction with BZR1 (Fig. 4c; Supplementary Information, Fig. S9a). Transgenic plants overexpressing AtSK12, or AtSK12-E297K corresponding to the bin2-1 gain-of-function mutation, displayed similar dwarf phenotypes as those overexpressing BIN2 or bin2-1 (Fig. 4d)13. Similar to BIN2, AtSK12 strongly phosphorylates BZR1 in vitro (Fig. 4e), is localized in both cytoplasm and nucleus independent of BR (Supplementary Information, Fig. S9b), is stabilized by the BR biosynthetic inhibitor brassinazole (BRZ) (Supplementary Information, Fig. S9c) and destabilized by BL or by overexpression of BSU1-YFP (Fig. 4f, g). Mass spectrometry analysis indicated that Tyr233 of AtSK12 (corresponding to Tyr200 of BIN2) was also phosphorylated (Supplementary Information, Fig. S9d). BR treatment greatly reduced phosphorylation of AtSK12 Tyr233, indicating similar mechanism of BR regulation as for BIN2 (Fig. 4h). These results indicate that BSU1-mediated tyrosine dephosphorylation is a common mechanism shared by at least two of six GSK3-like kinases that are likely involved in BR signaling.
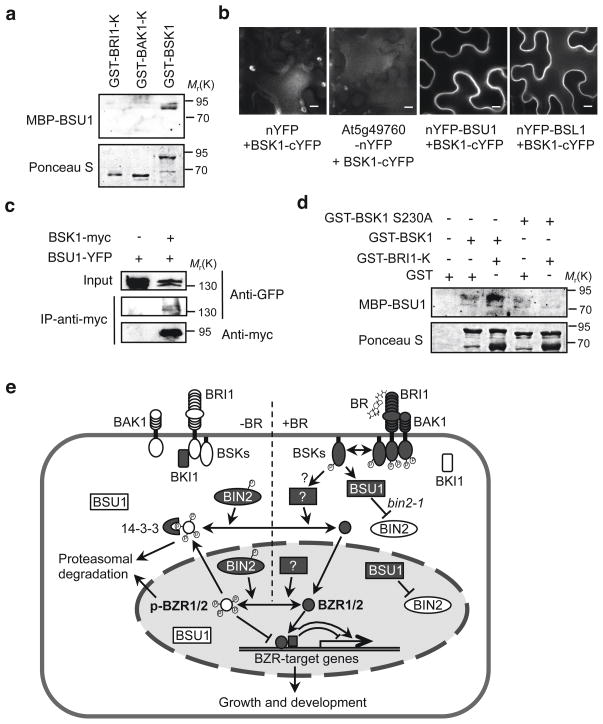
The function of BSU1 upstream of BIN2 suggests that it might be directly regulated by upstream components on the plasma membrane. We tested direct interaction of BSU1 with BRI1, BAK1 and BSK1 in vitro and found that the MBP-BSU1 protein interacted with BSK1 but not with BRI1 or BAK1 (Fig. 5a), which is consistent with BSK1 being downstream of BRI1 in the signaling pathway. BiFC assays showed that BSK1 interacts with both BSU1 and BSL1 in vivo (Fig. 5b), and the interaction was further confirmed by co-immunoprecipitation assays (Fig. 5c). We have previously shown that BRI1 phosphorylates BSK1 at Ser230. We found that phosphorylation of BSK1 by BRI1 increased while mutation of S230A abolished the binding of BSK1 to BSU1 (Fig. 5d), indicating that BRI1 phosphorylation of BSK1 at Ser230 increases its interaction with BSU1. These results demonstrate that BRI1 phosphorylation of BSK1 Ser230 promotes BSK1 binding to BSU1. Such interaction with BSK1 is likely to mediate BR activation of BSU1 in vivo, although we did not detect an effect of BSK1 on BSU1 activity in vitro (data not shown).
Figure 5.
BSK1 directly interacts with BSU1. (a) The GST fusion proteins of the kinase domains of BRI1 (GST-BRI1-K) and BAK1 (GST-BAK1-K) and full-length BSK1 (GST-BSK1) were separated by SDS-PAGE and blotted onto nitrocellulose membrane. The blot was probed sequentially with MBP-BSU1 and anti-MBP antibody (upper) and then stained with Ponceau S (lower). (b) BiFC assays show in vivo interaction between BSU1 or BSL1 and BSK1. Tobacco leaf epidermal cells were transformed with indicated constructs. At5g49760 is a receptor kinase unrelated to BR signaling used here as a negative control. Bright spots in nYFP+BSK1-cYFP and At5g49760-nYFP+BSK1-cYFP are chloroplast auto-fluorescence. The scale bar is 10 μm. (c) Total protein extracts obtained from Arabidopsis plants (F1) expressing BSU1-YFP or co-expressing BSU1-YFP and BSK1-myc were immunoprecipitated with anti-myc, and the immunoblot was probed with anti-GFP and anti-myc antibody. (d) Phosphorylation of BSK1 Ser230 by BRI1 enhances BSK1 binding to BSU1. GST-BSK1 or GST-BSK1 S230A was incubated with GST-BRI1-K or GST for 2 hrs. Overlay assay was performed as described in (a). Full scan data of immunoblots are shown in Supplementary Information, Fig. S12. (e) The BR signal transduction pathway. Filled objects indicate components in active states and open objects inactive states. In the absence of BR (-BR), BRI1 is kept in an inactive form with help of its inhibitor BKI1, and consequently BAK1, BSK1 and BSU1 are inactive, while BIN2 is active and phosphorylates BZR1 and BZR2 (BZR1/2), leading to their loss of DNA binding activity, exclusion from the nucleus by the 14-3-3 proteins, and degradation by the proteasome. In the presence of BR (+BR), BR binding to the extracellular domain of BRI1 induces dissociation of BKI1 and association and inter-activation between BRI1 and BAK1. Activated BRI1 then phosphorylates BSK1, which in turn dissociates from the receptor complex and interacts with and presumably activates BSU1. BSU1 inactivates BIN2 by dephosphorylating its pTyr200, allowing accumulation of unphosphorylated BZR1/2, likely with help of an unknown phosphatase. Unphosphorylated BZR1/2 accumulate in the nucleus and bind to promoters to regulate the expression of BR-target genes, leading to cellular and developmental responses.
Together our results bridge the last major gaps and elucidate a complete BR signaling pathway that involves multiple steps of sequential phosphorylation/dephosphorylation, transducing the signal from BRI1/BAK1 receptor kinase complex to BSK1, BSU1, BIN2, and BZR1 and BZR2 (Fig. 5e). In the absence of BR, BZR1 and BZR2 are inhibited by BIN2-catalyzed phosphorylation and consequent exclusion from the nucleus due to binding by the 14-3-3 proteins2, loss of DNA binding activity15, 22, and degradation by the proteasome20. BR binding to the extracellular domain of BRI1 activates BRI1 kinase through ligand-induced association and trans-phosphorylation with its co-receptor kinase BAK112. BRI1 then phosphorylates the BSK1 kinase at Ser23010, and this phosphorylation promotes BSK1 interaction with and activation of BSU1. Upon activation, BSU1 dephosphorylates BIN2 at the pTyr200 residue to inhibit its kinase activity, allowing accumulation of unphosphorylated BZR1 and BZR2 in the nucleus, where they regulate BR responsive gene expression and plant growth (Fig. 5e; Supplementary Information, Fig. S1b).
Signal transduction through cell surface receptor kinases is a fundamental mechanism for cellular regulation in living organisms. BRI1 is a member of the large family of leucine-rich-repeat receptor-like kinases (LRR-RLK), with over 220 members in Arabidopsis and 400 in rice28. Only a handful of these RLKs have been studied29. The BR signaling pathway described in this study represents the first complete RLK-mediated signaling pathway in plants and provides a paradigm for understanding other signal transduction pathways in plants.
Each BR signaling component in Arabidopsis is encoded by a small gene family with three to six members that have similar biochemical functions. BRI1 is the only essential gene for BR signaling that was identified by recessive mutations, yet two BRI1 homologs, BRL1 and BRL3, appear to mediate BR signaling in a tissue specific manner11, 30, 31. All the other components were identified either by gain-of-function mutations or by proteomic/biochemical approaches10. Single knockout of BIN2, BZR1, BES1, BSU1, and BSK1 caused no obvious phenotype or very subtle growth phenotypes, suggesting genetic redundancy among the members of each gene family. Our transgenic and biochemical studies provide strong evidence that six Arabidopsis GSK3s (Group I and II) are involved in BR signaling (Fig. 4), and loss-of-function data show that all four members of the BSU1 family contribute to BR signaling (Fig. 3h–j). As such, each step of BR signal transduction seems to be carried out by several members of the gene family, although only the founding member of each family is presented in the conceptual model of BR signal transduction (Fig. 5e). On the other hand, microarray data indicates very similar ubiquitous expression patterns for BRI1, BSK1, BSU1, BSL1, and BZR1 (Supplementary Information, Fig. S10)14, 31, supporting their major roles in BR responses.
Our study reveals BSU1-mediated pTyr200 dephosphorylation as the primary mechanism for regulating plant GSK3s in the BR signaling pathway. This tyrosine residue is absolutely conserved in all GSK3s identified so far, and its phosphorylation is required for kinase activity in Dictyostelium and mammals27, 32, 33. However, the phosphatase for this regulation has not been identified in these systems27, 33–35. BSU1 represents the first phosphatase that mediates dephosphorylation of this conserved tyrosine residue of GSK3s. BSU1 contains an N-terminal Kelch-repeat domain and a C-terminal phosphatase domain14, which dephosphorylates both phospho-Ser/Thr and phospho-Tyr residues (Supplementary Information, Fig. S2b, c, S11). The phosphatase domain of BSU1 shares about 45% sequence identity with mammalian protein phosphatase-1 (PP1). Interestingly, PP1 expressed in E. coli exhibits both Tyr and Ser/Thr phosphatase activity, although native PP1 expressed in mammalian cells is inactive on phospho-Tyr due to inhibition by inhibitor-236. It will be interesting to see if BSU1-related phosphatases mediate tyrosine dephosphorylation of GSK3s in mammals and other species.
METHODS
Materials
The bri1-5 mutant is in WS ecotype background, and all other Arabidopsis thaliana plants are in Columbia ecotype background. The det2, BIN2-myc, bin2-1-myc, AtSK12-myc and BSU1-YFP plants for Western blotting or in vitro kinase and phosphatase assays were sterilized with bleach and grown in agar plate containing half strength (x0.5) Murashige-Skoog (MS) medium under continuous light for 10 days. Tobacco (Nicotiana benthamiana) plants were grown in greenhouse under 16 h light/8 h dark cycles. All fusion proteins were expressed by the 35S promoter, unless indicated otherwise, in transient assays or in stable plant transformation experiments.
Phenotypic analysis of hypocotyls
Sterilized Arabidopsis seeds were planted on ×0.5 MS agar plate. Cold-treated agar plates were kept under white light for 6 hrs and vertically grown in the dark for 5 days. The seedlings were photocopied by digital camera.
In vitro kinase and phosphatase assays
MBP-BZR1 and GST-BIN2 proteins were expressed and purified from E. coli, and maltose or glutathione was removed from the proteins by ultrafiltration using Centricon 50 (Amicon Ultra, Millipore, Billerica, MA). To prepare fully phosphorylated BZR1 proteins, MBP-BZR1 protein was incubated with GST-BIN2 as 1 to 1 ratio in the kinase buffer containing 100 μM ATP at 30°C overnight. The protein mixture was incubated with glutathione Sepharose beads to remove GST-BIN2, then with amylose beads to purify MBP-pBZR1. Partially phosphorylated 32P-labeled pBZR1 and pBZR2 were prepared by the same method but MBP-BZR1 or MBP-BZR2 was incubated with GST-BIN2 at a 15 to 1 ratio for 3 hrs in the presence of 20 μCi 32P-γATP. For dephosphorylation, GST-BSU1 was incubated with fully phosphorylated MBP-pBZR1 and 32P-MBP-pBZR1 or 32P-MBP-pBZR2 for 12 or 16 hrs.
In vitro BIN2 inhibition assays were performed by 3 hrs co-incubation of MBP-BZR1, GST-BIN2, GST-BSU1 and 32P-γATP or pre-incubation of GST-BIN2 with GST-BSU1 for various time followed by adding MBP-BZR1 and 32P-γATP. To examine activities of partial BSU1, N-terminal Kelch (1–363th amino acid) and C-terminal phosphatase (364–793th amino acid) region were used. GST, GST-BSU1, GST-BSU1-Kelch and GST-BSU1-phosphatase were pre-incubated with GST-BIN2 for 1hr, and further incubated with MBP-BZR1 and 32P-γATP for 3hrs.
To test activities of BSU1-YFP, anti-GFP antibody-Protein A beads were used to immunoprecipitate BSU1-YFP from extracts of BSU1-YFP transgenic plants, and non-transgenic wild type plants were used as control. The beads were incubated with GST-BIN2 or GST-bin2-1 for 1 hr, and then the beads were removed. The BSU1-treated GST-BIN2 or GST-bin2-1 was further incubated with MBP-BZR1 and 32P-γATP for 3 hrs.
In vitro phosphatase assay using phospho-myelin basic protein was performed according to manufacturer’s protocol (New England Biolab, Beverly, MA). To examine tyrosine phosphatase activity of BSU1, 20 mM p-nitrophenyl phosphate was incubated with MBP-BSU1 in 50 uL of reaction buffer (50 mM Tris, pH 7.2, 20 mM NaCl, 5 mM DTT, 10 mM MgCl2). The reaction was quenched by the addition of 100 uL of 0.5 M NaOH after incubation at 30°C for 1 hr. p-Nitrophenol production was determined by measuring A405 (extinction coefficient, ε=1.78 × 104M−1cm−1).
Immunoprecipitation and co-immunoprecipitation
Plant materials were ground with liquid nitrogen and resuspended in IP buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5% Glycerol, 1% Triton X-100, 1 mM PMSF and 1× protease inhibitor cocktail (Sigma)). Filtered protein extracts were centrifuged at 20,000g for 10 min and resulting supernatant was incubated with anti-GFP antibody bound Protein A beads or anti-myc agarose beads for 1 hr. Beads were washed 5 times with washing buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.2% Triton X-100, 1 mM PMSF and 1× Protease inhibitor cocktail). The beads were resuspended with a small volume of kinase buffer (20 mM Tris, pH 7.5, 1 mM MgCl2, 100 mM NaCl and 1 mM DTT) and used for in vitro phosphatase assays, or immunoprecipitated proteins were eluted with buffer containing 2% SDS and analyzed by SDS-PAGE and immunoblotting.
Dephosphorylation of phospho-tyrosine 200 residue of BIN2
GST-BIN2 or GST-bin2-1 was incubated with MBP-BSU1 or BSU1-YFP beads for 3 hrs and subjected to immunoblotting. pTyr200 residue of BIN2 was detected by anti-phospho-GSK3α/β (Tyr279/216) monoclonal antibody, 5G-2F (Millipore, Temecula, CA) and re-probed with HRP conjugated anti-GST antibody (Santa Cruz Biotechnology, Santa Cruz, CA). The det2 plants were treated with 0.2 μM BL after 1 hr incubation with 10 μM MG132. Anti-BIN2 serum was developed in rabbits using GST-BIN2 as an immunogen. Monoclonal anti-GSK3α/β antibody was purchased from Invitrogen (Carlsbad, CA).
Site-directed mutagenesis
Point mutations were generated by site-directed mutagenesis PCR according to manufacturer’s protocol (Stratagene, La Jolla, CA). The primers used for different mutagenesis were: BIN2-Y200F-For, GAAGCCAACATTTCTTTCATCT GCTCACGATT; BIN2-Y200F-Rev, AAGCCAACATTTCTTTCATCTGCTCACGATT C; BIN2-Y200A-For, GAAGCCAACATTTCTGCCATCTGCTCACGATTC; BIN2-Y200A-Rev, GAATCGTGAGCAGATGGCAGAAATGTTGGCTTC; BIN2 M115A-For, CTTTTCTTGAACTTGGTTGCGGAGTATGTCCCTGAGA; BIN2 M115A-Rev, TCTC AGGGACATACTCCGCAACCAAGTTCAAGAAAAG; AtSK12 E297K-For, GAACA CCAACAAGGGAAAAAATCAAATGCATGAACCC; AtSK12 E297K-Rev, GGGTTC ATGCATTTGATTTTTTCCCTTGTTGGTGTTC, BSU1 D510N-For, CAATCAAAGT CTTCGGCAATATCCATGGACAATAC; BSU1 D510N-Rev, GTATTGTCCATGGAT ATTGCCGAAGACTTTGATTG.
Overexpression and knock-out/-down of BSU1-related phosphatases
Full-length cDNAs of BSU1 and BSL1 without stop codon were amplified by PCR using gene specific primers (BSU1-For, caccATGGCTCCTGATCAATCTTATCAATAT; BSU1-Rev, TTCACTTGACTCCCCTCGAGCTGGAGTAG; BSL1-For, caccATGGGCTCGA AGCCTTGGCTACATCCA; BSL1-Rev, GATGTATGCAAGCGAGCTTCTGTCAAAATC) from reverse transcription of Arabidopsis mRNA and cDNA clone (RIKEN, RAFL09-11-J01), respectively. The cDNAs were cloned into pENTR/SD/D-TOPO vectors (Invitrogen, Carlsbad, CA) and subcloned into gateway compatible pEarleyGate 101 or pGWB17 or pGWB20 or BiFC vectors15 by using LR reaction kit (Invitrogen). To test phenotypic suppression of bri1-116 and bin2-1 by BSU1, 35S::BSU1-YFP single plant was crossed into bri1-116 and bin2-1. The phenotype of F3 double homozygous plants was analyzed. To generate the quadruple loss-of-function mutant of bsu1, bsl1/BSL2,3-amiRNA, the double mutant of bsu1-1 (SALK_030721) and bsl1-1 (SALK_051383)37 was transformed with an artificial microRNA construct targeting both BSL2 and BSL3 genes (BSL2,3-amiRNA), which was designed by the Web MicroRNA Designer 238, using the oligo (TATTCATCAAAAAGGCGCGTG) and plasmid pRS30038. The DNA fragment of amiRNA was cloned into pEarleyGate 100 (pEG100) by using the Gateway cloning kit (Invitrogen), yielding BSL2,3-amiRNA/pEG100. The binary vector constructs were introduced into Agrobacterium strain GV3101 by electroporation and transformed into Arabidopsis by using the floral dipping method.
Quantitative RT-PCR
Quantitative real-time PCR analysis of SAUR-AC1 mRNA was performed as described by Gampala et al15 using gene specific primers (SAUR-AC1-for, AAGAGGATTCATGGCGGTCTATG; SAUR-AC1-rev, GTATTGTTAAGCCGCCCA TTGG). UBC (UBC-for, CAAATCCAAAACCCTAGAAACCGAA; UBC-rev, ATCTC CCGTAGGACCTGCACTG) was used to normalize the loading.
Yeast two-hybrid assays of AtSKs
The cDNA clones of AtSKs were obtained from ABRC (http://www.biosci.ohio-state.edu/pcmb/Facilities/abrc/abrchome.htm)39. All AtSKs cDNAs were subcloned into gateway compatible pGADT7 vector (Clontech). Nine AtSKs-pGADT7 constructs and empty pGADT7 vector were transformed into the cells containing BZR1-pGBKT7. Yeast clones were grown on Synthetic Dropout (SD) or SD-Histidine containing 2.5~10 mM 3-amino-1, 2, 4-triazole.
In vitro kinase assay of AtSK12
GST-AtSK12 (1 μg) was incubated with MBP-BZR1 (2 μg), 100 μM ATP and 32P-γATP (10 μCi) in the kinase assay buffer for 2 hrs. The reaction was terminated by addition of 2× SDS loading buffer and separated by 7.5% SDS-PAGE. Gel was stained with Coomassie brilliant blue followed by drying. The radioactivity was analyzed by Phospho-image screen using Typhoon 8600 Scanner (GE Healthcare).
Determination of in vitro phosphorylation sites of BIN2 and AtSK12
GST-BIN2 or GST-AtSK12 protein (25 μg) purified from E.coli was incubated with 100 μM ATP in the kinase buffer for 16 hrs at 30°C. Autophosphorylated GST-BIN2 or GST-AtSK12 was subjected to in-solution alkylation/tryptic digestion followed by LC-MS/MS analysis according to Gampala et al15.
Overlay Western blot
To test interaction of BSU1 with BIN2 or bin2-1 in vitro, a gel blot separating GST, GST-BIN2, GST-bin2-1 was incubated with 20 μg MBP-BSU1 in 5% non-fat dry milk/PBS buffer and washed four times. The blot was then probed with a polyclonal anti-MBP antibody. In the case of BSU1 overlay to BSK1, GST-BRI1-K, GST-BAK1-K and GST-BSK1 were separated by SDS-PAGE. To prepare phosphorylated BSK1, GST-BSK1 was incubated with GST-BRI1-K and 100 μM ATP in the kinase buffer for 2 hrs before SDS-PAGE. The blot was sequentially probed with MBP-BSU1 and a monoclonal anti-MBP antibody (New England Biolab, Beverly, MA).
Immunoblotting of 2-DE
Total proteins were extracted from BL-treated or untreated 35S::TAP-BIN2 plants for two-dimensional gel electrophoresis (2-DE) as described previously40. The amount of BL-treated and untreated TAP-BIN2 proteins was normalized with Western blot. Equal amount of TAP-BIN2 proteins was separated by 2-DE using an immobilized pH gradient gel strip (7 cm, pH 3–10 non-linear) and 7.5% SDS-PAGE gel. The blots were probed with anti-PAP antibody (Sigma, St. Louis, MO).
Transient transformation and confocal microscopy
Transformation by Agrobacterium infiltration, observations of subcellular localization and BiFC signal in tobacco or Arabidopsis were performed as described previously15. Fluorescence of YFP was visualized by using a spinning-disk confocal microscope (Leica Mirosystems, Heerbrugg, Germany).
Supplementary Material
Acknowledgments
We thank Dr. Joanne Chory for providing the bsu1-D seeds and BSU1 cDNA clone, and Drs. Devaki Bhaya and Kathryn Barton for comments on the manuscript. Research was supported by grants from NIH (R01GM066258), NSF (0724688), U.S. Department of Energy (DE-FG02-08ER15973), and the Herman Frasch Foundation. The UCSF Mass Spectrometry Facility (A.L. Burlingame, Director) is supported by the Biomedical Research Technology Program of the National Center for Research Resources, NIH NCRR RR01614, RR012961 and RR019934.
Footnotes
AUTHOR CONTRIBUTIONS
S.G. and A.L.B carried out LC-MS/MS analysis. Y.S1 and Z.D. were involved in 2-D PAGE immunoblot. J.X.S and Y.S3 developed BIN2 antiserum. W.T. made GST-BSK1 proteins. T.W.K. performed all other experiments. T.W.K and Z.W. designed the experiments, analyzed data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 2.Gendron JM, Wang ZY. Multiple mechanisms modulate brassinosteroid signaling. Curr Opin Plant Biol. 2007;10:436–441. doi: 10.1016/j.pbi.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clouse SD, Sasse JM. BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 4.Belkhadir Y, Wang X, Chory J. Arabidopsis brassinosteroid signaling pathway. Sci STKE. 2006;2006:cm5. doi: 10.1126/stke.3642006cm5. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 6.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 7.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 8.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 10.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinoshita T, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 14.Mora-Garcia S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gampala SS, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 17.Yin Y, et al. A crucial role for the putative Arabidopsis topoisomerase VI in plant growth and development. Proc Natl Acad Sci U S A. 2002;99:10191–10196. doi: 10.1073/pnas.152337599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 20.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu H, et al. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;19:2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 23.Peng P, Yan Z, Zhu Y, Li J. Regulation of the Arabidopsis GSK3-like Kinase BRASSINOSTEROID-INSENSITIVE 2 through Proteasome-Mediated Protein Degradation. Mol Plant. 2008;1:338–346. doi: 10.1093/mp/ssn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 25.Benschop JJ, et al. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics. 2007;6:1198–1214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Sugiyama N, et al. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol. 2008;4:193. doi: 10.1038/msb.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim L, Harwood A, Kimmel AR. Receptor-dependent and tyrosine phosphatase-mediated inhibition of GSK3 regulates cell fate choice. Dev Cell. 2002;3:523–532. doi: 10.1016/s1534-5807(02)00269-1. [DOI] [PubMed] [Google Scholar]
- 28.Shiu SH, et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson KL, Ingram GC. Sending the right signals: regulating receptor kinase activity. Curr Opin Plant Biol. 2005;8:648–656. doi: 10.1016/j.pbi.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Zhou A, Wang H, Walker JC, Li J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004;40:399–409. doi: 10.1111/j.1365-313X.2004.02214.x. [DOI] [PubMed] [Google Scholar]
- 31.Cano-Delgado A, et al. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131:5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- 32.Hughes K, Nikolakaki E, Plyte SE, Totty NF, Woodgett JR. Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. Embo J. 1993;12:803–808. doi: 10.1002/j.1460-2075.1993.tb05715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhat RV, et al. Regulation and localization of tyrosine216 phosphorylation of glycogen synthase kinase-3beta in cellular and animal models of neuronal degeneration. Proc Natl Acad Sci U S A. 2000;97:11074–11079. doi: 10.1073/pnas.190297597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim L, Kimmel AR. GSK3 at the edge: regulation of developmental specification and cell polarization. Curr Drug Targets. 2006;7:1411–1419. doi: 10.2174/1389450110607011411. [DOI] [PubMed] [Google Scholar]
- 35.Cole A, Frame S, Cohen P. Further evidence that the tyrosine phosphorylation of glycogen synthase kinase-3 (GSK3) in mammalian cells is an autophosphorylation event. Biochem J. 2004;377:249–255. doi: 10.1042/BJ20031259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacKintosh C, et al. Further evidence that inhibitor-2 acts like a chaperone to fold PP1 into its native conformation. FEBS Lett. 1996;397:235–238. doi: 10.1016/s0014-5793(96)01175-1. [DOI] [PubMed] [Google Scholar]
- 37.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 38.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada K, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 40.Deng Z, et al. A proteomics study of brassinosteroid response in Arabidopsis. Mol Cell Proteomics. 2007;6:2058–2071. doi: 10.1074/mcp.M700123-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.