Abstract
Layered on top of information conveyed by DNA sequence and chromatin are higher order structures that encompass portions of chromosomes, entire chromosomes, and even whole genomes1-3. Interphase chromosomes are not positioned randomly within the nucleus but instead adopt preferred conformations4-7. Disparate DNA elements co-localize into functionally defined aggregates or “factories” for transcription8 and DNA replication9. In budding yeast, Drosophila and many other eukaryotes, chromosomes adopt a Rabl configuration, with arms extending from centromeres adjacent to the spindle pole body to telomeres that abut the nuclear envelope10-12. Nonetheless, the topologies and spatial relationships of chromosomes remain poorly understood. Here we developed a method to globally capture intra- and inter-chromosomal interactions, and applied it to generate a map at kilobase resolution of the haploid genome of Saccharomyces cerevisiae. The map recapitulates known features of genome organization, thereby validating the method, and identifies new features. Extensive regional and higher order folding of individual chromosomes is observed. Chromosome XII exhibits a striking conformation that implicates the nucleolus as a formidable barrier to interaction between DNA sequences at either end. Inter-chromosomal contacts are anchored by centromeres and include interactions among tRNA genes, among origins of early DNA replication and among sites where chromosomal breakpoints occur. Finally, we constructed a three-dimensional model of the yeast genome. Our findings provide a glimpse of the interface between the form and function of a eukaryotic genome.
Chromosome conformation capture (3C) and its derivatives have been used to detect long-range interactions within and between chromosomes13-20. We developed a method for identifying chromosomal interactions genome-wide by coupling 4C14 and massively parallel sequencing (Figure 1 and Supplementary Methods). Because all 3C-based technologies are encumbered by low signal-to-noise ratios18,21, we established the method’s reliability by assessing: i) random inter-molecular ligations from each of five control libraries (Figure 2a, Supplementary Table 1 and 2, Supplementary Methods); ii) restriction site-based biases (Figure 2b, Supplementary Figure 1 and 2, Supplementary Table 3); iii) reproducibility between independent sets of experimental libraries that differed in DNA concentration at the 3C step, which critically influences signal-to-noise ratios (Supplementary Table 1, Figure 2b and 2c, Supplementary Figure 2, ); iv) consistency between the HindIII and EcoRI libraries (Supplementary Figure 3 to 5, Supplementary Table 4-8), v) a set of 24 chromosomal interactions using conventional 3C (Figure 2D, Supplementary Figure 6). These results argue that our method is reliable and robust (detailed in Supplementary Methods). We established yeast genome architecture features using interactions from the HindIII libraries at an FDR of 1%, and confirmed them with interactions from the EcoRI libraries at the same threshold.
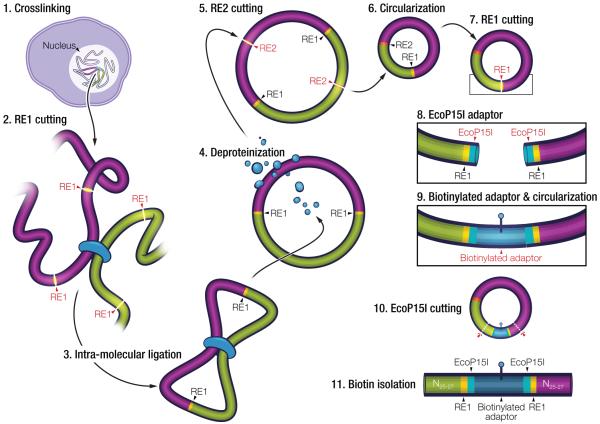
Figure 1.
Schematic depiction of the method. Our method relies on the 4C procedure by using cross-linking, two rounds of alternating restriction enzyme (RE) digestion (6 bp-cutter RE1 for the 3C-step digestion and 4 bp-cutter RE2 for the 4C-step digestion) and intra-molecular ligation. At step 7, each circle contains the 6 bp restriction enzyme recognition site originally used to link the two interacting partner sequences (RE1). Diverging from 4C, we re-linearize the circles using RE1, then sequentially insert two sets of adaptors, one of which permits digestion with a type IIS or type III restriction enzyme (such as Ecop15I). Following Ecop15I digestion, fragments are produced that incorporate interacting partner sequence at either end, which can be rendered suitable for deep sequencing (see Supplementary Methods).
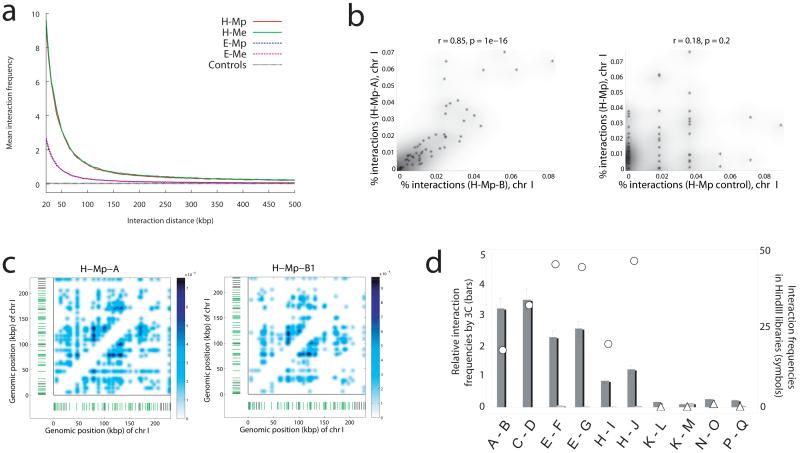
Figure 2.
Validation of the assay. a, Graph showing an inverse relationship between interaction frequency and genomic distance (20kb or larger, excluding self-ligations and adjacent ligations) separating interacting restriction fragments (either HindIII or EcoRI) in each of four experimental but none of five control libraries. Note, the five lines representing the five control libraries are very close to each other. b, The fraction of instances that each HindIII site along chromosome I was engaged in an intra-chromosomal interaction was highly correlated between two independently derived experimental H-Mp (HindIII-MspI) libraries (designated A and B, left panel) but was not correlated between experimental and non-cross linked control H-Mp libraries (right panel). c, Two-dimensional heat maps demonstrating broad reproducibility of interaction patterns within chromosome I for two independently derived H-Mp libraries. The chromosomal positions of mappable (green hatches) and un-mappable (black hatches) HindIII fragments are indicated. The binary interaction matrix of all interactions with an FDR threshold of 1% has been smoothed with a Gaussian of width 3 kb. d. High degree of correlation between absolute interaction frequencies as determined by our method (symbols) versus relative interaction frequencies as determined by conventional 3C using cross-linked (dark bars) and uncross-linked (light bars) libraries. Results for 10 potential long-range intra-chromosomal interactions are depicted, of which 6 passed (circles) and 4 did not pass (triangles) an FDR threshold of 1%. Error bars denote standard deviations over three experiments. Interaction sites are as follows. A: Chr III position 11811; B: Chr III position 290056; C: Chr III position 15939; D: Chr III position 314440; E: Chr I position 26147; F: Chr I position 191604; G: Chr I position 204567; H: Chr VI position 12007; I: Chr VI position 243206; J: Chr VI position 249743; K: Chr II position 238203; L: Chr II position 502988; M: Chr II position 512024; N: Chr IV position 236977; O: Chr IV position 447899; P, Chr IV position 239805; Q, Chr IV position 461284.
From our HindIII libraries, we identified 2,179,977 total interactions at an FDR of 1%, corresponding to 65,683 interactions between distinct pairs of HindIII fragments. We used these data to generate conformational maps of all 16 yeast chromosomes. The overall propensity of HindIII fragments to engage in intra-chromosomal interactions varied little between chromosomes, ranging from 436 interactions/HindIII fragment on chromosome XI to 620 interactions/HindIII fragment on chromosome IV (Supplementary Table 9). These results suggest broadly similar densities of self-interaction (intra-chromosomal interaction) between chromosomes and indicate that the density of self-interaction does not vary with chromosome size (Supplementary Figure 7).
Some large segments of chromosomes showed a striking propensity to interact with similarly sized regions of the same chromosome. For example, two regions on chromosome III (positions 30 kb-90 kb, and 105-185 kb) showed an excess of interactions (Figure 3 a,b). Such regions may represent a “zippering” of chromosomal segments, in which a large segment of DNA lies juxtaposed to a similar length segment (see also chromosome II: 20–200 kb and 250–430 kb; Supplementary Figure 8). In other cases, large segments of chromosomes were enriched for local interactions, such that a series of consecutive HindIII fragments spanning tens of kilobases interacted frequently with other HindIII fragments within the same segment (for example, regions in chromosomes IV, XIII, and XVI, Supplementary Figure 8). Conversely, many combinations of fragments showed few or no interactions, indicating highly improbable chromosome conformations. For example, centromeric regions tended to engage in relatively few long-range intra-chromosomal interactions (Supplementary Figure 8). Overall, the number of interactions involving any given HindIII fragment was strongly influenced by the interaction frequencies of neighboring HindIII fragments, demonstrating regional differences in the tendency for interaction within chromosomes.
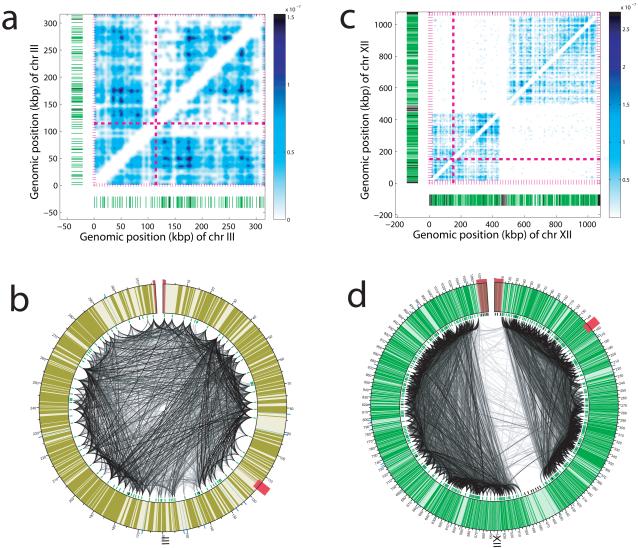
Figure 3.
Folding patterns of chromosomes III (a, b) and XII (c, d). The heat maps (a, c) and Circos diagrams (b, d) were generated using the intra-chromosomal interactions identified from the HindIII libraries at an FDR threshold of 1%. In the heat maps (a, c), the chromosomal positions of centromeres (dashed pink lines), telomeres (pink hatches), mappable (green hatches) and un-mappable (black hatches) HindIII fragments are indicated. Circos diagrams (b, d) depict each chromosome as a circle. Each arc connects two HindIII fragments and represents a distinct interaction. The shade of each arc, from very light grey to black, is proportional to the negative log of the p-value of the interaction. The chromosomal positions of centromeres (red hexagon), telomeres (red colored area), tRNA genes (blue outer thatches), mappable (green inner hatches) and un-mappable (black inner hatches) HindIII fragments are indicated. Black outer hatches and numbers mark genomic positions. Note that the two ends of chromosome XII (c, d) exhibit extensive local interactions but very little interaction with each other. Separating the ends of chromosome XII are 100–200 rDNA repeats, of which only two copies are depicted here (from coordinates 450 to 470 kb). Additional heat maps and Circos diagrams for all chromosomes are shown in Supplementary Figure 8.
Intra-chromosomal interactions between telomeric ends varied dramatically from one chromosome to another. Consistent with previous observations10,11, chromosomes III and VI exhibited high levels of enrichment of intra-chromosomal interactions between their telomeres (Supplementary Table 10). In contrast, the ends of chromosomes IV and XII showed no intra-chromosomal telomeric interactions (Supplementary Table 10). Also as previously observed10, the ratios of observed/possible intra-chromosomal telomeric interactions between the two ends of chromosomes V and XIV were less than 1/25 that of chromosome III (0.4 and 0.5 versus 13, respectively, Supplementary Table 10).
The conformation of chromosome XII differed strikingly from its counterparts. In contrast to the typical pattern of intra-chromosomal interactions enveloping the lengths of entire chromosomes (Figure 3 a, b, Supplementary Figure 8), chromosome XII segregated into three distinct segments (Figure 3 c, d). Regions of 430 kb at one end and 550 kb at the other end engaged in extensive local interactions; however, these two regions did not interact with each other. Extensive local interactions at either end of chromosome XII terminated abruptly at the boundaries of nucleolus-associated rDNA, where 100-200 rDNA repeats comprise 1-2 Mb of DNA.22 This finding indicates that rDNA, and by inference the nucleolus, acts as a near absolute barrier, blocking interactions between the chromosome ends.
For HindIII, a total of 639,607 intra-chromosomal and 8,119,614 inter-chromosomal interactions are possible. Thus, any given HindIII fragment end has a much larger universe of candidate fragments on other chromosomes with which to partner than fragments within the same chromosome. Nonetheless, a strong tendency for intra-chromosomal ligation resulted in 53.2% of observed interactions occurring between HindIII fragments within the same chromosome. The frequency of inter-chromosomal interactions was significantly enriched in the experimental versus control libraries, especially among inter-centromeric and inter-telomeric interactions (Supplementary Figure 10). In budding yeast, clustering of centromeres adjacent to the spindle pole body persists throughout the cell cycle.23 A clustering of centromeres marked the primary point of engagement between different chromosomes and was the most striking feature of the inter-chromosomal contacts (Figure 4, Supplementary Figures 9, 10). Of interactions of chromosome I with other chromosomes (at FDR of 1%) in both the HindIII and EcoRI libraries, the overwhelming majority lay within narrow 20 kb windows centered around their centromeres (Figure 4a, b). The centromeres of the other fifteen chromosomes demonstrated similar clustering (Supplementary Figure 9).
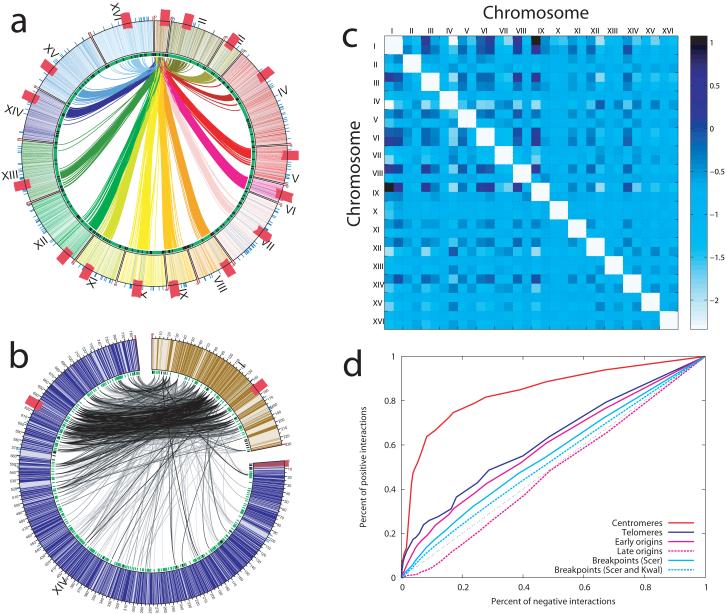
Figure 4.
Inter-chromosomal interactions. a, Circos diagram showing interactions between chromosome I and the remaining chromosomes. All 16 yeast chromosomes are aligned circumferentially, and arcs depict distinct inter-chromosomal interactions. Bold red hatch marks correspond to centromeres. To aid visualization of centromere clustering, these representations were created using the overlap set of intra-chromosomal interactions identified from both HindIII and EcoRI libraries at an FDR threshold of 1%. Additional heat maps and Circos diagrams are provided in Supplementary Figure 9. b, Circos diagram, generated using the intra-chromosomal interactions identified from the HindIII libraries at an FDR threshold of 1%, depicting the distinct interactions between a small and a large chromosome (I and XIV, respectively). Most of the interactions between these two chromosomes primarily involve the entirety of chromosome I, and a distinct region of corresponding size on chromosome XIV. c, Inter-chromosomal interactions between all pairs of the 32 yeast chromosomal arms (the 10 kb region starting from the midpoint of the centromere in each arm is excluded). For each chromosome, the shorter arm is always placed before the longer arm. Note that the arms of small chromosomes tend to interact with one another. The color scale corresponds to the natural log of the ratio of the observed versus expected number of interactions (see Supplementary Materials). d, Enrichment of interactions between centromeres, telomeres, tRNA genes, early origins of replication, and chromosomal breakpoints. To measure enrichment of strong interactions with respect to a given class of genomic loci, we use receiver operating curve (ROC) analysis.
Another chromosomal landmark that mediates inter-chromosomal contacts are telomeres,11,24 which congregate and form 5-8 foci within the interphase nucleus. Our data show widespread associations between pairs of telomeres on different chromosomes, with 88 of 450 possible telomere pairs associating (p=<0.02; another 30 pairs were not analyzed due to lack of mappable HindIII sites, Figure 4d, Supplementary Figure 11, Supplementary Table 11). The average size differences between the two corresponding chromosome arms of each of the 88 associated telomere pairs was much smaller than that of the remaining 450 pairs (199.0 kb versus 373.9 kb, p<10−6, unpaired two-tailed t-test). These results indicate that two telomeres positioned at similar distances from their corresponding centromeres are more likely to interact, as described.11 Nevertheless, there were exceptions. For example, the left arm of chromosome V and the right arm of chromosome XIV are of similar size (152 and 155 kb, respectively), but their telomeres did not associate (p=0.142, Supplementary Table 11), consistent with previous observations.11
We assessed whether specific categories of genes or other chromosomal features were enriched in interactions among their members. The 274 tRNA genes are dispersed throughout the yeast genome, yet clustering of tRNA genes has been observed in the nucleolus.25,26 Consistent with this finding, HindIII sites adjacent to tRNA genes were significantly enriched for interaction with sites neighboring other tRNA genes (Supplementary Figure 11). Using a hierarchical clustering algorithm, we identified two clusters of co-localized tRNA genes (Supplementary Figure 12), one that appears to be co-localized with the rDNA region on chromosome XII, consistent with nucleolar localization, and another that appears to be clustered with centromeres. There was an enrichment of interactions among early (but not late) origins of DNA replication (Figure 4d, Supplementary Figure 11). These early replication origins clustered into at least two discrete regions, consistent with their co-localization in replication factories (Supplementary Figure 13). Both tRNA genes and origins of early DNA replication associate with chromosomal breakpoints27, and we detected a significant co-localization of breakpoint sites (Figure 4d, Supplementary Figure 11). Finally, we asked whether other groups of genes were significantly enriched in interactions and found that they were not (Supplementary Figure 11).
The ratio of non-self to self interactions correlated inversely with chromosome size (Supplementary Figure 14). Smaller chromosomes (I, III and VI) had the strongest propensity to interact with other chromosomes, whereas the large chromosomes XII and IV were the most isolated. Considering each chromosome pair for the ratio of observed versus expected interactions, we found that interactions were much more prevalent between smaller chromosomes (I, III, VI, IX, and VIII) (Supplementary Figure 15). Only three pairs of larger chromosomes (IV and VII, IV and XII, and IV and XV) displayed relatively high enrichment ratios. Analyzing inter-chromosomal interactions among the 32 chromosome arms, we found that chromosome arms <250 kb in size were much more likely to interact with one another (Figure 4C). Notably, among the larger chromosome arms, the right arms of chromosomes IV and XII showed the highest interaction enrichments (Figure 4C). Similarly, the interaction pattern between any given chromosome pair was strongly influenced by the relative sizes of the partners. Chromosomes of similar size interacted along their entire lengths (Supplementary Figure 9); however, a smaller chromosome tended to interact along its length with a region of corresponding size within its larger partner. For example, chromosome I (230 kb in length) interacted preferentially within a region of chromosome XIV approximately 270 kb in length (between 510-780 kb) (Figure 4B and Supplementary Figure 9).
These observations can be explained by the Rabl configuration of yeast chromosomes. Tethered by their centromeres to one pole of the nucleus, the chromosome telomeres extend outward toward the nuclear membrane. Small chromosome arms are crowded within the thicket of the entire set of 32 arms, thereby making frequent contacts with other chromosomes. In contrast, the distal regions of the larger chromosome arms occupy relatively uncrowded terrain, making fewer contacts with other chromosomes.
To address the question of chromosome territories28,29, we compared the observed/expected ratios for intra-chromosomal versus inter-chromosomal interactions for all 32 chromosome arms (Supplementary Figure 16). Examining the entirety of each arm, we found a higher enrichment for the 16 intra-chromosomal pairings than all inter-chromosomal pairings, except for pairing between the two smallest arms (1R and 9R) (Supplementary Figure 16a). However, the preference for intra-chromosomal arm pairing versus inter-chromosomal arm pairing decreased with increasing distance from centromeres (Supplementary Figure 16 b-d). These observations indicate that yeast chromosome arms are highly flexible.
Combining our set of 4,097,539 total and 306,312 distinct interactions with known spatial distances that separate sub-nuclear landmarks,12 we derived a three-dimensional map of the yeast genome. To depict intra-chromosomal folding, we incorporated a metric that converts interaction probabilities into nuclear distances (assigning 130 bp of packed chromatin a length of 1 nm30) (Supplementary Figures 17, 18, Supplementary Methods). Using this ruler, we calculated the spatial distances between all possible pairings of the 16 centromeres (Supplementary Tables 14 and 15) The results are consistent with previous observations12.
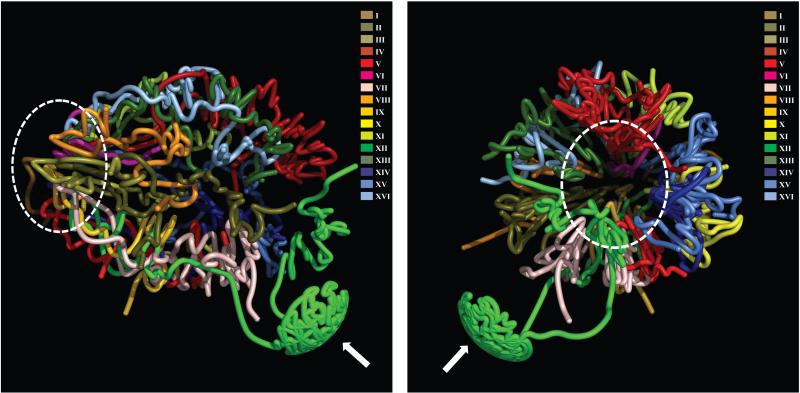
The resulting map resembles a water lily, with 32 chromosome arms jutting out from a base of clustered centromeres (Figure 5). Chromosome XII stretches its long arm across to the opposite nuclear pole, incorporating its rDNA repeats into the nucleolus, with the remainder of its long arm interacting with the long arm of chromosome IV. The map represents a coarse-grained image, a snapshot that ignores the dynamic nature of chromosomes. An additional feature constraining the resolution of the map is the population-based nature of the 3C technology, which cannot distinguish between interactions that occur at high probability in a small fraction of cells vs. those that occur at low probability in a majority of cells. Our results provide the first glimpse into the architecture of a eukaryotic genome at high resolution, highlighting the three-dimensional complexity of the genome of even this simple organism. While we do not understand how DNA sequence specifies this structure, further work should unveil its general organizing principles. With continuing developments in high throughput DNA sequence analysis, both the definition and comparative analysis of the high-resolution architectures of additional organisms will be increasingly feasible.
Figure 5.
Three-dimensional model of the yeast genome. Two views representing two different angles are provided. Chromosomes are colored as in Figure 4a (also indicated in the upper right). All chromosomes cluster via centromeres at one pole of the nucleus (the area within the dashed oval), while chromosome XII extends outward toward the nucleolus, which is occupied by rDNA repeats (indicated by the white arrow). After exiting the nucleolus, the remainder of chromosome XII interacts with the long arm of chromosome IV.
METHODS SUMMARY
A culture of a bar1 derivative of Saccharomyces cerevisiae BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 bar1::KanMX) was crosslinked with 1% formaldehyde for 10 min. Two sequential rounds of alternating restriction enzyme digestion, intra-molecular ligation, biotin-streptavidin-mediated purification, linear PCR amplification, and gel purification were carried out prior to construction of paired-end libraries. Libraries were paired-end sequenced using the Illumina Genome Analyzer 2, and sequence reads were mapped to the S. cerevisiae reference genome. To identify signal from background noise, we performed statistical confidence estimation. To estimate a false discovery rate (FDR), we eliminated self-ligations, ligations between adjacent restriction fragments, and ligations between restriction fragments separated by less than 20 kb at their midpoints. To account for the strong influence of genomic proximity on ligation frequency, we subdivided the remaining intra-chromosomal interactions into 5 kb bins as measured by the genomic distance between the midpoints of the two ligated fragments. Inter-chromosomal interactions were placed into a separate bin. In each bin, the observed interactions were ranked according to their sequence frequency and assigned a p value relative to all other possible interactions in the same bin. Lastly, the p value of each interaction was converted into a q value (defined as the minimal FDR threshold at which the interaction is deemed significant), and we used these values to rank interactions library-wide. After the true interaction sets were derived, further computational analyses were performed as described in detail in the online supplementary information.
Supplementary Material
Acknowledgements
We appreciate the advice and assistance of Michael Dorschner, the valuable comments of Sara Di Rienzi, Bonny Brewer and Breck Byers, and the assistance of Lu Zhang and Gary Schroth (Illumina Inc.) in performing sequencing. We thank Alex Brown for help with the 3D model. Supported by NIH grants P01GM081619, P41RR0011823, a post-doctoral fellowship (to MA) from the Natural Sciences and Engineering Research Council of Canada, and the Howard Hughes Medical Institute.
LITERATURE CITED
- 1.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature Rev. Genet. 8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 3.Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr. Opin. Genet. Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- 5.Gasser SM. Visualizing chromatin dynamics in interphase nuclei. Science. 2002;296:1412–1416. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- 6.Stone EM, Heun P, Laroche T, Pillus L, Gasser SM. MAP kinase signaling induces nuclear reorganization in budding yeast. Curr. Biol. 2000;10:373–382. doi: 10.1016/s0960-9822(00)00413-9. [DOI] [PubMed] [Google Scholar]
- 7.Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nature Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 9.Kitamura E, Blow JJ, Tanaka TU. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125:1297–1308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 2005;168:375–387. doi: 10.1083/jcb.200409091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schober H, et al. Controlled exchange of chromosomal arms reveals principles driving telomere interactions in yeast. Genome Res. 2008;18:261–271. doi: 10.1101/gr.6687808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger AB, et al. High-resolution statistical mapping reveals gene territories in live yeast. Nature Methods. 2008;5:1031–1037. doi: 10.1038/nmeth.1266. [DOI] [PubMed] [Google Scholar]
- 13.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 14.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nature Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 15.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nature Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 16.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 18.Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009;107:30–39. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nature Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 22.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 23.Jin Q, Trelles-Sticken E, Scherthan H, Loidl J. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol. 1998;141:21–29. doi: 10.1083/jcb.141.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gotta M, et al. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Rienzi SC, Collingwood D, Raghuraman MK, Brewer BJ. Fragile Genomic Sites Are Associated with Origins of Replication. Genome. Biol. Evol. 2009;2009:350–363. doi: 10.1093/gbe/evp034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haber JE, Leung WY. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc. Natl Acad. Sci. U S A. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz A, Fuchs J, Trelles-Sticken E, Scherthan H, Loidl J. Spatial organisation and behaviour of the parental chromosome sets in the nuclei of Saccharomyces cerevisiae x S. paradoxus hybrids. J. Cell Sci. 2002;115:3829–3835. doi: 10.1242/jcs.00066. [DOI] [PubMed] [Google Scholar]
- 30.Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl Acad. Sci. U S A. 2004;101:16495–16500. doi: 10.1073/pnas.0402766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.