Preface
Staphylococcus aureus is notorious for its ability to become resistant to antibiotics. Infections caused by antibiotic-resistant strains often occur in epidemic waves initiated by one or a few successful clones. Methicillin-resistant S. aureus (MRSA) is prominently featured during these epidemics. Historically associated with hospitals and other healthcare settings, MRSA now has emerged as a widespread cause of community infections. So-called community or community-associated MRSA spreads rapidly among healthy individuals. Outbreaks of community MRSA infections have been reported worldwide and community MRSA strains are now epidemic in the United States. There is reason for concern because MRSA often are or can readily become resistant to multiple antibiotics, thus limiting treatment options.
Introduction
Staphylococcus aureus is naturally susceptible to virtually every antibiotic that has ever been developed. Resistance is often acquired by horizontal transfer to genes from outside sources, although chromosomal mutation and antibiotic selection are also important. This exquisite susceptibility of S. aureus led to Alexander Fleming’s discovery of penicillin, ushering in the “antibiotic era.” Penicillin was truly a miracle drug: uniformly fatal infections could be cured. Yet, by the mid-1940s, only a few years after its introduction into clinical practice, penicillin resistance was encountered in hospitals and within a decade it had become a significant problem in the community. S. aureus is remarkable in its ability to acquire resistance to any antibiotic.
A fundamental biological property of S. aureus is the ability to asymptomatically colonize normal people. Approximately 30% of humans are asymptomatic nasal carriers of S. aureus1, 2; i.e., S. aureus is normal flora. S. aureus carriers are at higher risk of infection and they are presumed to be an important source of spread of S. aureus strains among individuals. The primary mode of transmission of S. aureus is by direct contact, usually skin-to-skin contact with a colonized or infected individual, although contact with contaminated objects and surfaces or might also play a role3–6. Various host factors, including loss of the normal skin barrier, presence of underlying diseases such as diabetes and acquired immunodeficiency syndrome, or defects in neutrophils function predispose to infection.
Infections caused by antibiotic-resistant strains of S. aureus have reached epidemic proportions globally7. The overall burden of staphylococcal disease, particularly that caused by methicillin resistant S. aureus strains (MRSA), is increasing in many countries in both healthcare and community settings8–13. In the United States the emergence of community-associated MRSA (CA-MRSA) strains as a major cause of skin and soft-tissue infections14, 15 accounts for much of this increase. The rapidity and extent to which CA-MRSA strains have spread has been remarkable. In addition to the United States CA-MRSA strains have been reported from Canada, Asia, South America, Australia, and throughout Europe, including Norway, the Netherlands, Denmark, and Finland, countries with historically low prevalence of MRSA.12, 16–29 Globally, CA-MRSA strains have shown a remarkable diversity in the number of different clones that have been identified.
In addition to increasing prevalence and incidence CA-MRSA strains appear to be especially virulent. Overwhelming and tissue-destructive infections, such as necrotizing fasciitis and fulminant, necrotizing pneumonia30–32, which have been associated with CA-MRSA strains, were rarely seen prior to their emergence. The factor or factors responsible for this hypervirulent behavior of CA-MRSA are not known, but PVL, which has been epidemiologically associated with severe skin infections and pneumonia caused by methicillin-susceptible S. aureus (MSSA) strains33, has been proposed as a potential leading candidate.
Antibiotics arguably constitute the most concentrated selective pressure ever brought to bear on S. aureus in its long co-evolutionary history with mankind. The consequences of this selective pressure in conjunction with horizontal and vertical gene transfer are the subject of this review. Given their critical importance as therapeutic agents, the story will focus on resistance to penicillins and the structurally related beta-lactam antibiotics.
Epidemic Waves of Antibiotic Resistant Staphylococcus aureus
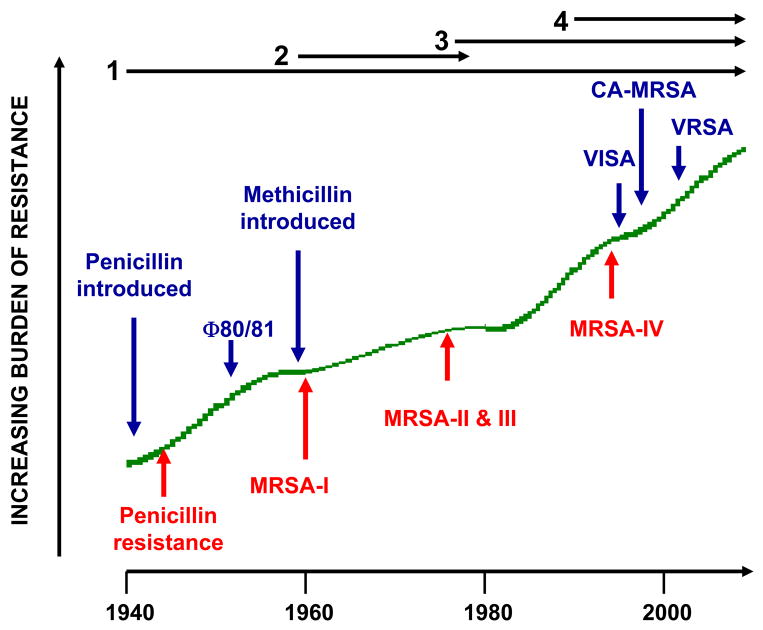
Emergence of antibiotic resistance by S. aureus can be visualized as a series of waves (Figure 1). The first wave began in the mid-1940s as the proportion of infections caused by penicillin-resistant S. aureus rose in hospitals 34, 35. These strains produced a plasmid-encoded penicillinase that hydrolyzes the beta-lactam ring of penicillin essential for its antimicrobial activity. Penicillin-resistant strains then were observed to cause community infections; by the early 1950s and 1960s they had become pandemic 36. These infections, both in hospitals and the community, were caused primarily by a S. aureus clone known as phage-type 80/81 36–39. Pandemic phage-type 80/81 S. aureus infections largely disappeared after the introduction of methicillin 40, but the prevalence of penicillinase-producing strains of other S. aureus lineages has remained high ever since.
Figure 1.
A timeline of the four waves of antibiotic resistance in Staphylococcus aureus. Wave 1, which continues today, began shortly after the introduction of penicillin into clinical practice in the 1940s. The first pandemic antibiotic resistant strains, from lineage named phage type 80/81 (Φ80/81), were penicillin resistant and produced PVL (Panton-Valentine leucocidin). Wave 2 began almost immediately upon the introduction of methicillin into clinical practice with isolation of the first MRSA (Archaic clone), which contained type I SCCmec (MRSA-I) and extended into the 1970s in the form of the Iberian clone. Wave 3 began in the mid-to-late 1970s with emergence of new MRSA strains, which contained novel SCCmec, types, II and III (MRSA-II and III), marking the on-going worldwide pandemic of MRSA in hospitals and healthcare facilities. The upsurge in vancomycin usage for treatment of MRSA infections eventually led to emergence of vancomycin intermediate S. aureus (VISA) strains. Wave 4, which began in the mid-to-late 1990s, marks the emergence of MRSA strains in the community. Community MRSA strains where susceptible to most antibiotics other than beeta-lacams, were unrelated to hospital strains, contained a novel, smaller, more mobile type IV SCCmec (MRSA-IV), and a variety of virulence factors, including PVL. Vancomycin-resistant S. aureus (VRSA) strains, of which 10 or so have been isolated exclusively in healthcare settings, were first identified 2002.
Introduction of methicillin marks the onset of the second wave of resistance. The first reports of a S. aureus strain that was resistant to methicillin were published in 1961 41, 42. Although the specific gene, mecA, the methicillin resistance determinant which encodes the low affinity penicillin binding protein, PBP 2a (also referred to as PBP 2′), was not identified until more than 20 years later, it was appreciated early on that the resistance mechanism was different from penicillinase-mediated resistance because there was no drug inactivation. Unlike penicillinase-mediated resistance, which is narrow in its spectrum, methicillin resistance is broad beta-lactam antibiotic class resistance to penicillins, cephalosporins, and carbapenems. Among the very earliest of MRSA clinical isolates is the archetypal strain COL, a member of the “archaic” clone of MRSA and perhaps the most studied MRSA strain, which was isolated from a patient in Colindale, United Kingdom in 1960 42. COL is a member of the most successful of all MRSA lineages, which includes both hospital and community-associated strains.
These archaic clone of MRSA strains circulated in hospitals throughout Europe until the 1970s 43. There were isolated reports of MRSA from hospitals in the United States44, 45, but the rest of the world was largely spared and these early MRSA never gained a foothold in the community. By the 1980s for unclear reasons archaic MRSA strains had largely disappeared from European hospitals, marking the end of the second and the beginning of the third wave of antibiotic. Descendants of the archaic MRSA clone (e.g., the Iberian and Rome clones46) and other highly successful MRSA lineages emerged (Table 1) 47–49, constituting the third wave of antibiotic resistance. Outbreaks of infections caused by MRSA strains were reported in hospitals in the United States in late 1970s and by the mid-1980s were endemic50, 51. These strains swept the globe leading to the worldwide pandemic of MRSA in hospitals that continues to the present time. Although global in distribution and impact, MRSA was still confined mainly to hospitals and other institutional healthcare settings, such as long-term care facilities. The ever increasing burden of MRSA infections in hospitals led to more usage of vancomycin, the last remaining antibiotic to which MRSA strains were reliably susceptible, and under this intensive selective pressure vancomycin intermediate S. aureus (VISA, which are not inhibited in vitro at vancomycin concentrations below 4 to 8 μg/ml)52 and vancomcyin-resistant S. aureus (VRSA, inhibited only at concentrations of 16 μg/ml or more)53 strains of MRSA emerged.
Table 1.
Lineages of common nosocomial MRSA.
| Clonal Complex | Multilocus sequence type | Common names for specific MRSA clones | Comment |
|---|---|---|---|
| CC5 | ST5 | USA100 and NewYork/Japan clone | Most common US healthcare-associated MRSA, SCCmecII |
| ST5 | EMRSA-3 | SCCmecI | |
| ST5 | USA800/Pediatric clone | Prevalent in Argentina, Colombia, United States; SCCmecIV | |
| ST5 | HDE288/Pediatric clone (Portugal) | SCCmecVI | |
| CC8 | ST250 | Archiac | First MRSA clone identified, COL strain as an example; SCCmecI |
| ST247 | Iberian clone and EMRSA-5 | Descendant of COL-type strains, SCCmecI | |
| ST239 | Brazilian/Hungarian clone, | SCCmecIII | |
| ST239 | EMRSA-1 | Eastern Australian epidemic clone of 1980s, SCCmecIII | |
| ST8 | AUS-2 and AUS-3 | SCCmecII | |
| ST8 | Irish-1 | Common nosocomial isolate in the 1990s in Europe and the US, | |
| ST8 | USA500 and EMRSA-2,-6 | SCCmecIV | |
| CC22 | ST22 | EMRSA-15 | International clone, prominent in Europe and Australia, SCCmecIV |
| CC30 | ST36 | USA200 and EMRSA-16 | Single most abundant cause of MRSA infections in United Kingdom; second most common cause of MRSA infections in US hospitals in 2003, SCCmecII |
| CC45 | ST45 | USA600 and Berlin | SCCmecII |
The MRSA invasion of the community constitutes the fourth and latest wave of antibiotic resistance. Some of the earliest cases of community-associated MRSA (CA-MRSA) infections occurred in indigenous populations in Western Australia in the early 1990s54–56. These MRSA strains were distinguishable from contemporary clones (i.e., genotypes) circulating in Australian hospitals by their pulsed field gel electrophoresis patterns and susceptibility to most antibiotics other than beta-lactams, suggesting that they were either remote, feral descendants of hospital strains or community strains that had acquired mecA by horizontal gene transfer. In the US, the first well-documented cases of MRSA infection that were truly community associated occurred in otherwise healthy children in 1997–99 57. These children had no risk factors for MRSA and all died with overwhelming infection, suggesting that these community MRSA strains were especially virulent. Like their Australian counterparts, these CA-MRSA isolates were unrelated to hospital clones and were susceptible to most antibiotics The CA-MRSA epidemic in the US can be traced to the early 1990s, based on retrospective data from 1993–1995 showing a dramatic increase in MRSA infections in Chicago among children lacking risk factors for hospital-associated MRSA exposure58. CA-MRSA has since been reported in numerous populations including American Indians and Alaska natives 59; Pacific Islanders 60; athletes 4; jail and prison inmates 61; men who have sex with men 62; contacts of patients with CA-MRSA infection 63; military personnel 61; adult emergency room patients 14; and children in day care centers 64. CA-MRSA clones have also gained a foothold in hospitals and are increasingly identified as a cause of hospital-onset and heathcare-associated infections 10, 12, 25, 65, 66.
The epidemic wave of CA-MRSA in the United States, and Canada as well67, 68, is actually two overlapping epidemics. The USA400 clone, which was isolated from the pediatric cases described above, was most prevalent prior to 2001 3, 57, 69. USA400 remains a common cause of community-onset disease in among indigenous populations in Alaska and the Pacific Northwest 70. A second epidemic clone, USA300, which is unrelated to USA400 and has largely displaced it in most other locations, emerged between 1999 and 2001, and now causes the vast majority of CA-MRSA infections in the United States3, 4, 71–74.
Outbreaks and epidemics of CA-MRSA now occur worldwide and with a similar epidemiology, although the specific clones that have emerged vary with geographical location. CA-MRSA strains are not merely escapees from healthcare facilities; their genotypes indicate that they are not closely related to endemic hospital clones and these community strains are susceptible to numerous antibiotics to which hospital strains are routinely resistant. Two molecular markers not found in typical hospital MRSA are strongly associated with emergence of CA-MRSA regardless of geographical origin: a specific cassette element encoding mecA and genes encoding Panton-Valentine leukocidin (PVL). These markers are discussed in detail below.
Skin and soft-tissue infections are the most common type of CA-MRSA infection, accounting for approximately 90% of cases, of which 90% are abscesses and/or cellulitis with purulent drainage 14, 15. CA-MRSA strains also appear to be especially virulent with the capacity to cause fulminant, overwhelming infections, such as necrotizing fasciitis, necrotizing pneumonia, bone and joint infections accompanied by septic thromboembolic disease 31, 75–77, purpura fulminans with or without Waterhouse-Friderichsen syndrome 78, orbital cellulitis and endophthalmitis 79, infections of the central nervous system 80, 81, and bacteremia and endocarditis 66, 82
Molecular epidemiology of Staphylococcus aureus in the antibiotic era
S. aureus Clonal Complexes
Robust sequence-based molecular methods for genotyping strains of S. aureus, and multilocus sequence typing (MLST) 83 in particular, have made the study of the evolutionary history of S. aureus possible (Box 1). MLST is performed by sequence analysis of approximately 450 bp internal fragments of seven housekeeping genes. Isolates that have identical sequences at all seven genetic loci are considered a clone, and assigned a unique sequence type (ST). Sequence types that differ by single nucleotide polymorphisms at fewer than three loci are considered closely related, and are grouped into clonal complexes (CC) (Figure 2). This is accomplished by application of the eBURST algorithm (http://eburst.mlst.net), which uses multilocus sequence typing data to group closely related strains into a clonal complex. It also predicts the probable founding clone (i.e., sequence type) of each group and recent evolutionary descent of all other strains within the clonal complex from the founder84, 85. The analysis can be further refined to identify specific subclones by the addition of other methods, such as spa typing86 pulsed field gel electrophoresis of genomic DNA, or by the presence of other genetic markers (e.g., toxin genes or specific plasmids).
Box 1. Genotyping is used to identify S. aureus strains and predict phylogeny
Multilocus sequence typing (MLST) is sequence based genotyping method. The method is based on single nucleotide variations (each variant is termed an allele) of 7 housekeeping genes in S. aureus, which provides a discriminatory allelic profile, known as sequence type (ST)83, for each bacterial isolate. MLST, because it indexes variations that accumulate slowly over time can be used to measure long periods of evolution among S. aureus lineages and is highly reproducible. S. aureus isolates having identity at 5 or more of the 7 housekeeping genes/loci based upon MLST are known as a clonal complex (CC)84, 87.
Pulsed-field gel electrophoresis (PFGE)has a somewhat more rapid clock speed than MLST and is suitable for evaluation of more recent evolution among groups of strains. The method relies on separation of SmaI-digested S. aureusgenomic DNA fragments according to size in an agarose gel by by pulsed-field electrophoresis. Related strains are clustered according to an 80% similarity coefficient 99. The CDC has developed a national PFGE database for S. aureus, which uses the “USA” designation (e.g., USA300 for the ST8, PVL-positive community associated MRSA).99
spa typing86 is based upon sequence analysis of variable number tandem repeats in the gene encoding protein A (Spa). Spa typing takes into account point mutations in the the repeat region as well as the number of repeat variations. The method is suitable for investigation of local or global S. aureusoutbreaks. This sequence-based analysis of a single target locus is a relatively inexpensive way of acquiring robust data that can be used to determine both epidemiological and phylogenetic relationships.
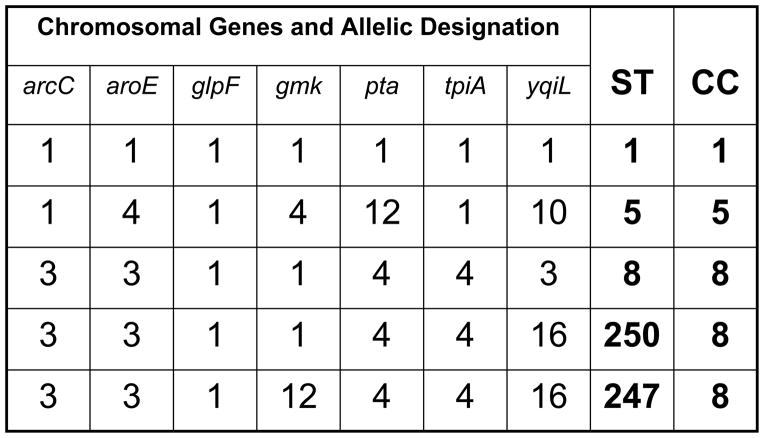
Figure 2. An example of multilocus sequence typing scheme and designation of clonal complexes.
Approximately 450 nucleotides of seven chromosomal “housekeeping” genes (arcC, carbamate kinase; aroE, shikimate dehydrogenase; glpF, glycerol kinase; gmk, guanylate kinase; pta, phosphate acetyltransferase; tpiA, triose phosphate isomerase yqiL, acetyl-CoA acetyltransferase), selected for their presumed absence of selective pressure and therefore relatively stable in nucleotide sequence, are sequenced. Each unique sequence within a gene locus is assigned a unique number. The numbers are concatenated in left-to-right in the order shown to provide a seven interger series of numbers, which is assigned a number designating this sequence type (ST). Strains which are identical at all seven loci are classified as the same ST. Strains differing at one or two loci, are related, but as they are not identical, are assigned different STs. Closely related STs are grouped into a clonal complex. In the example shown, ST1, ST5, and ST8 differ at most loci and thus are not closely related. ST250 and ST247 differ from each other at one locus and from ST8 at one or two loci, respectively. Thus, ST8, ST250, and ST247 are closely related and from a clonal complex, CC8, so designated because analysis of sequence identities and differences in a large collection of strains indicates ST8 is the founder of this clonal complex, the ancestor of both ST247 and ST250, and that ST247 is a descendant of ST250.
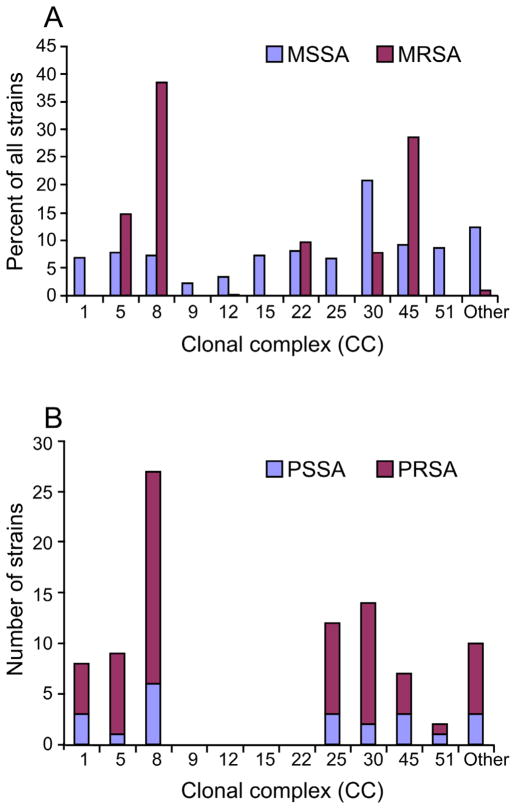
Studies 47, 83, 87–90 of MSSA strains, carriage isolates and hospital and community isolates causing disease, collected worldwide between 1961 through 2004, show that 88% of the strains can be assigned to one of one of 11 clonal complexes (CC1, CC5, CC8, CC9, CC12, CC15, CC22, CC25, CC30, CC45, and CC51/121 47, 84, 89–93), (Figure 3A). Percentages of isolates range between 2% and 9% for ten complexes; CC30 is an outlier, accounting for 21% of isolates.
Figure 3. Distribution of antibiotic-susceptible and -resistant S. aureus among clonal complexes.
a) MSSA (blue) versus MRSA (red) clonal complexes. b) Penicillin-susceptible S. aureus (PSSA, blue) versus penicillin-resistant S. aureus (PRSA, red) clonal complexes. Data in (a) were collected from 6 continents (1961–2004) and those in (b) are from a single study of 99 isolates collected in Copenhagen from 1957–73. See text for details.
Clonal complexes for contemporary isolates are almost certainly the same as those of strains circulating prior to 1940. For example, the ST5 lineage, the founder of CC5, is estimated to have existed for over 2000 years 94. Furthermore, when Gomes and colleagues 95 genotyped 22 penicillin-susceptible and 77 penicillin-resistant MSSA blood culture isolates dating from 1957 to 1973 by the Statens Serum Institute of Copenhagen, which has collected and maintained every blood culture isolate from patients in Denmark from 1957 to the present, they found that 86% of the isolates fell into 7 clonal complexes, the two most common being CC8 and CC30, which together accounted for 46% of the isolates (Figure 3B). The distributions of penicillin-sensitive and penicillin-resistant isolates were similar. Relatively few isolates were tested and all originated from a single country, which probably accounts for the absence of isolates from CC9, CC12, CC15, or CC22.
CC8 and CC30 have given rise to epidemics during each of the four waves of antibiotic resistance. The first well-characterized pandemic of antibiotic resistant S. aureus attributable to a single clone was caused by phage type 80/81 strains, which belong to CC30 96. Originally isolated in Australia in 1953 39, phage type 80/81 strains were penicillin-resistant and caused both hospital and community outbreaks on a global scale96. Phage type 80/81 strains are prevalent in strain collections dating back to 1927; these strains were considered to be highly transmissible and particularly virulent, and were also among the first to be identified as penicillin resistant 37. Phage type 80/81 isolates in a collection dating to the 1950s and 1960s have been shown almost uniformly to possess genes for PVL96, which is reminiscent of the association of PVL and resistance to methicillin in the contemporary epidemic CA-MRSA strains. For unknown reasons, phage type 80/81 strains virtually disappeared in the early 1960s, coincident with the first use of semi-synthetic penicillins, which are resistant to penicillinase. Modern descendents of the ST30/CC30 lineage include the PVL-positive southwest Pacific (SWP) clone of CA-MRSA in Australia and hospital associated ST36 EMRSA16 clone, a major cause of nosocomial infections and bacteremia in the both Australia and the United Kingdom 96–98.
MRSA Clonal Complexes
The very first MRSA clinical isolates, of which COL is an example, were ST250 and members of CC8. ST250 MRSA strains circulated in the UK and Europe prior to the 1970s, but never established a presence in the United States, and had largely disappeared by the 1980s. However, other highly successful clones emerged, including the ST247 Iberian/EMRSA5 clone, which is closely related to ST250. No fewer than nine other endemic nosocomial clones are descendants of the ST8 founder of this lineage. The CA-MRSA strain USA300 (which is PVL-positive) that is prevalent in the US is also ST8 99.
MSRA strains generally been concentrated into a subset of the S. aureus clonal complexes, including CC1, CC5, CC8, CC22, CC30, and CC45, although as discussed below CA-MRSA have exhibited some diversity. These clonal complexes were widespread prior to emergence of methicillin resistance 43, 95, indicating that superior epidemicity preceded acquisition of drug resistance and that the adaptations and innovations that make clones successful also may favor their adaptation to antibiotic selective pressures.
Staphylococcal Chromosome Cassette mec, SCCmec
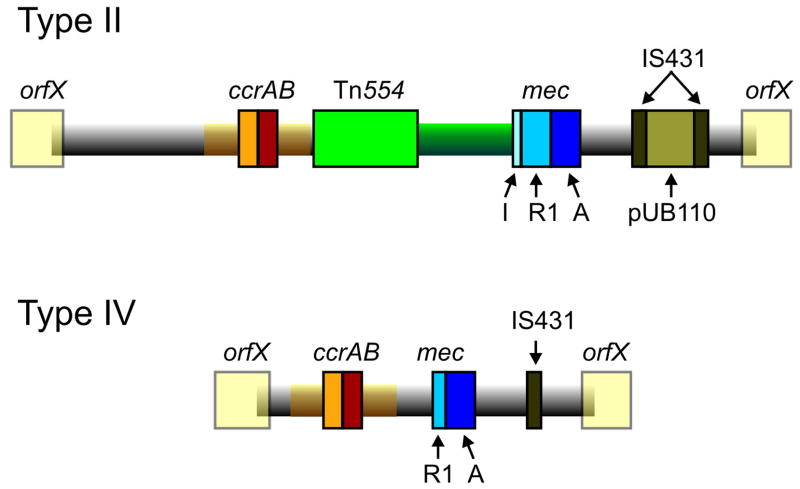
The discovery by Hiramatsu and colleagues that mecA is always found within a mobile cassette element was a great advance for understanding the biology of methicillin resistance and provided an additional tool for determining evolutionary relationships among MRSA 100. This element, SCCmec (staphylococcal chromosome cassette mec) is integrated into orfX, a S. aureus gene of unknown function (Figure 4). To date eight SCCmec allotypes, designated I through VIII 49, 100–104, have been described (Table 2) along with numerous subtypes and more are likely to be identified as sequence data become available for more MRSA strains (see http://www.staphylococcus.net/for additional descriptions and information). Similar elements are present in coagulase-negative staphylococci, which are commensual organisms that are normal skin flora of humans and other mammals 105. gene complexes, mec and ccr (the recombination/excision locus that encodes the gene or genes that mediates integration and excision of the whole cassette into orfX), are used to classify SCCmec types (Table 2). There are also other differences among the various SCCmecs, particularly in insertion sequences and antimicrobial resistance genes, but as these are themselves mobile elements, they have not proven useful in classification of major types, although they are useful in defining subtypes. class A mec gene complex (class A mec) is the prototype complex. It contains PBP 2a-encoding mecA, the complete mecR1 and mecI regulatory genes upstream of mecA, and the hypervariable region (HVR) and insertion sequence IS431 downstream of mecA. The class B mec gene complex is composed of mecA, a truncated mecR1 resulting from the insertion of IS1272 upstream of mecA, HVR, and IS431 downstream of mecA. The class C mec gene complex contains mecA and truncated mecR1 by the insertion of IS431 upstream of mecA, HVR, and IS431 downstream of mecA. There are two distinct class C mec gene complexes. In the class C1 mec gene complex, the IS431 elements upstream and downstream of mecA both have the same orientation. In the class C2 mec gene complex, the orientation of IS431 upstream of mecA is reversed. C1 and C2 are regarded as different mec gene complexes since they have likely evolved independently. The mecA, mecR1, and mecI sequences are highly conserved with >99% nucleotide sequence identity. The ccr complex consists of two adjacent genes ccrA and ccrB in SCCmec I–IV, VI, and VIII, and ccrC in V and VII. MRSA strains isolated prior to 1990, all nosocomial isolates, contained predominantly SCCmecI–III. Community MRSA isolates overwhelming contain SCCmecIV or SCCmecIV subtypes or, less commonly, SCCmecV 28, 106. SCCmec IV is increasingly identified in contemporary hospital MRSA strains as well.
Figure 4. Comparison of methicillin-resistance cassettes typical of hospital- or community MRSA.
Type II mec (SCCmecII) is most abundant in hospitals whereas Type IV mec (SCCmecIV) is present in the most abundant CA-MRSA strains. Transposon Tn554 encodes resistance to macrolide-lincosomide-streptogramin B antibiotics and spectinomycin. SCCmecII encodes resistance to multiple antibiotics whereas SCCmecIV encodes resistance to methicillin alone. IS431, insertion sequence 431. See text for additional details.
Table 2.
Comparison of SCCmec allotypes.
|
SCCmec Allotypes |
||||||||
|---|---|---|---|---|---|---|---|---|
| Feature* | I | II | III | IV | V | VI | VII | VIII |
| Size (kb) | 34 | 53 | 67 | 21–24 | 28 | 24 | 49 | 32 |
| mec complex | B | A | A | B | C2 | B | C1 or C2 | A |
| ccr complex | 1 | A2B2 | A3B3 | A2B2 | C | A4B4 | C2, C8 | A4B4 |
| IS431 (n) | 1 | 2 | 4 | 1 | 2 | 1 | 1 | 1 |
| Tn554 (n) | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 1 |
| pUB110 | − | + | − | − | − | − | − | − |
| pT181 | − | − | + | − | − | − | − | − |
| pI258 | − | − | + | − | − | − | − | − |
| Other resistance genes | None | Erm, tobra | Erm, tet, Hg++ | None | None | None | None | erm and spc |
mec complex A has intact regulatory genes, mecR1-mecI upstream of mecA; mec complex B has regulatory gene deletions from IS1272 insertion; mec complex C1 and C2 have regulatory gene deletions from IS431insertion ccr complex is the recombinase locus; IS431 = insertion sequence; pUB110, pT181, and pI258 are plasmids integrated at insertion sequences; erm = erythromycin, tobra = tobramycin, tet = tetracycline.
The three epidemic waves of MRSA correspond to evolutionary changes in SCCmec. The early MRSA strains, COL and other CC8 strains that circulated in the UK and Denmark in the early 1960s, all carried type I SCCmec. These clones were replaced in the 1980s by new, and arguably more successful, lineages that eventually became established in hospitals throughout the world. These clones, predominantly CC5 and CC8, carried type II or III SCCmecs (e.g., New York/Japan EMRSA, EMRSA-16 in Australia and the United Kingdom, Brazilian clone, Hungarian clone), or the type IA variant of the archaic SCCmec type I (Iberian clone). Why types II and III were more successful than type I SCCmec is not known, but it could be that the recombinase genes, which are defective in type I SCCmec but functional in types II and III100, limited the potential for horizontal gene transfer of type I SCCmec into new genomes.
What gave rise to the most recent worldwide epidemic wave of community MRSA, is the “invention” of SCCmecIV, which appears to have evolved from type I SCCmec, although it has type 2 ccrAB 107. Originally identified in the community-associated MW2/USA400 strain, the first occurrence of type IV SCCmec in S. aureus may have been in the ST5 “Pediatric” clone that was circulating in hospitals in the late 1980s and 1990s 108. The ultimate origins of mecA and SCCmec elements may never be known, but there is good evidence suggesting that coagulase-negative staphylococci are the sources 109–111.
The success of SCCmecIV is borne out by two observations. First, it is the most widely distributed among S. aureus isolates. It has been found in 9 distinct MRSA clonal complexes or sequence types, compared to only 2 such lineages for type I, 3 for type II, and 2 for type III 107. Second, CA-MRSA strains containing SCCmecIV have growth rates faster than hospital MRSA strains carrying other SCCmec types and these growth rates are no different from MSSA isolates 106. In a rabbit bacteremia model fitness and virulence of USA300, which carries SCCmec type IVa, was indistinguishable from its isogenic methicillin-susceptible variant 112. Thus, the type IV methicillin-resistance cassette appears to exact little or no cost in fitness for the organism.
Epidemiology of Community-Associated MRSA
As mentioned above, the earliest reported cases of CA-MRSA infection in the US were caused by a USA400 strain, MW2 57. MW2 is closely related to the PVL-negative clone, WA-1, which is an important CA-MRSA in Australia and to the MSSA strain 476 in the United Kingdom.55 USA400 has been supplanted by USA300, which is by far the most frequent cause of CA-MRSA infections in the US 113. The USA300 clone seems to be particularly well adapted to the community with reports of CA-MRSA infections caused by USA300 or its close relatives in Australia and Denmark and outbreaks of CA-MRSA in Columbia114–116. USA300 strains can also cause healthcare-associated infections65, 66, 117, 118
While there is evidence of international spread of these USA300 and USA400,18, 23, 119, 120 CA-MRSA strains unrelated to either have been responsible for infections outside of the United States. ST80 in is the predominant clone circulating in Europe, ST59 in Taiwan, and ST30 in Eastern Australia, demonstrating that CA-MRSA strains have evolved in separate geographical regions 21–23. There also can be considerable strain diversity in CA-MRSA from country to country. For example in Australia 45 distinct clones of CA-MRSA have been identified23; many of these are related to well-known MRSA lineages, but others appear to be novel. Diversity of CA-MRSA isolates has been noted by others as well18, 27, 114, 119, 120. In the United Kingdom the vast majority of CA-MRSA infections are caused by EMRSA-15 (ST22) and EMRSA-16 (ST36), which are also important hospital clones121; ST80 is also present, but it accounts only for a small proportion of isolates122. A CA-MRSA strain, ST 398, of swine origin and transmissible to humans has also been described123, 124.
The epidemiology of CA-MRSA is quite similar regardless of country of origin. Isolates tend not to be multiple drug-resistant, SCCmec types IV and V are typically present, and infections of skin and soft tissue are the most common. The presence of PVL among CA-MRSA isolates is more variable. For example in Australia and the United Kingdom most CA-MRSA clones do not produce PVL23, 121 and prevalence of PVL among the more common CA-MRSA isolates from Denmark ranged from 17% to 100%120. On the other hand isolates of clones that typically do not carry PVL genes, e.g., EMRSA-15 and EMRSA-16, have been found on occasion to be PVL-positive121.
Nasal carriage of MRSA has increased in parallel with the emergence of MRSA as a community pathogen, which is not unexpected given that approximately 30% of people have asymptomatic nasal colonization with S. aureus. Between 2001 and 2004 carriage of MRSA strains in a US population based study approximately doubled from 0.8% to 1.5% 2 and the percentage of community-associated MRSA genotypes increased from7% to 24.2% 88. Although the sites of carriage (e.g., nasal versus groin versus other) and the relationship between carriage of CA-MRSA strains and disease is not entirely clear, CA-MRSA strains, especially USA300, appear to be more easily transmitted than other strains,125 which could account for increasing carriage rates in the community. Thus, no individual or group can be considered not at risk for CA-MRSA infection.
Virulence of Community-Associated MRSA
Compared to infections caused by healthcare-associated MRSA strains and community MSSA, CA-MRSA infections have been associated with fulminant and lethal infections and worse clinical outcomes30, 77, 126, giving rise to the clinical impression that CA-MRSA strains, especially USA300, are more virulent than other strains. Much of what the information about the unique virulence properties of CA-MRSA is based on studies of USA300 strains, the most extensively investigated clone. The USA300 core genome (chromosome excluding mobile genetic elements) is quite similar to that of the early MRSA strain, COL 127. Yet, studies in animal models indicate that USA300 is more virulent than COL128, 129. Expression of virulence factors by USA300 is high and USA300 130, 131 and closely related strains are more lethal than more distant relatives and cause more extensive disease in animal models of infection129, 130. The major difference between COL and USA300 genomes resides in mobile genetic elements, which include prophages, plasmids, pathogenicity islands, and transposons, acquired through horizontal gene transfer. These elements encode factors that may impact transmission, antibiotic resistance, and virulence. Prophages ΦSa2 and ΦSa3, which are present in USA300 strains and not in COL, could contribute to the noted differences in virulence between these two lineages. Prophage ΦSa2 contains lukS-PV and lukF-PV, which encode PVL. Prophage ΦSa3 encodes staphylokinase, staphylococcal complement inhibitor (SCIN), and S. aureus chemotaxis inhibitory protein (CHIPS), all of which are modulators of the innate immune system 132, 133. ΦSa3 is present in strains other than CA-MRSA. A pathogenicity island, SaPI5, similar to the one that is in COL, is present in USA300. SaPI5 encodes two additional superantigens not present in COL, SEQ and SEK, which also are found in other MRSA and MSSA lineages. S. aureus produces many other molecules that promote host colonization, facilitate evasion of the innate immune system, and/or alter immune responses (Tables 3–6) 131, 134, 135. Most of these molecules are not unique to CA-MRSA. The virulence factors more commonly found in CA-MRSA compared to other strains, those that are linked by epidemiology to CA-MRSA infections, or those that have been studied in animal models of CA-MRSA infection are discussed below.
Table 3.
Virulence factors of Staphylococcus aureus that interfere with bacterial killing.
| Target cell, host factor or response | Gene(s) | Protein or molecule | Putative function/effect on immune system |
|---|---|---|---|
| Antimicrobial peptides | aur | Zinc metalloproteinase aureolysin, Aur | Degrades LL-37 |
| dlt operon | Dlt operon, DltABCD | Promotes resistance to cationic antimicrobial peptides and group IIA phospholipase A2 | |
| icaA, icaD, icaB, icaC, icaR | Polysaccharide intercellular adhesin, PIA | Resistance to cationic antimicrobial peptides | |
| isdA, isdB | Iron-regulated surface determinants of S. aureus, IsdA and IsdB | Resistance to antimicrobial peptides, skin fatty acids, and neutrophil reactive oxygen species | |
| mprF | Multiple peptide resistance factor, MprF | Promotes resistance to cationic antimicrobial peptides | |
| sak | Staphylokinase | Inhibits host α-defensins | |
| Oxygen-mediating bacterial killing | ahpC, ahpF | Alkyl hydroperoxide reductase subunits C and F, AhpC and AhpF | Promotes resistance to ROS |
| crtM, crtN | Carotenoid pigment, staphyloxanthin (S. aureus golden pigment) | Promotes resistance to reactive oxygen species | |
| isdA, isdB | Iron-regulated surface determinants of S. aureus, IsdA and IsdB | Resistance to neutrophil reactive oxygen species | |
| sodA, sodM | Superoxide dismutase, SodA, SodM | Promotes resistance to reactive oxygen species |
Table 6.
Superantigens produced by Staphylococcus aureus.
| Target cell, host factor or response | Gene(s) | Protein or molecule | Putative function/effect on immune system |
|---|---|---|---|
| T-cells | sea, seb, secn, sed, see, seg, seh, sei, sej, sek, sel, sep | Staphylococcal enterotoxins; SEA, SEB, SECn, SED, SEE, SEG, SEH, SEI, SEJ, SEK, SEL, and SEP | Activate T-cells (superantigen) |
| tst | Toxic shock syndrome toxin-1, TSST-1 | Activates T-cells (superantigen) |
Panton-Valentine leukocidin (PVL)
PVL has been studied extensively since its discovery by Panton and Valentine 70 years ago136. The role of PVL in the marked epidemicity and enhanced virulence of CA-MRSA is subject of debate. PVL is comprised of two subunits, LukS-PV and LukF-PV 137 that are encoded by prophage ΦSa2 138 that is acquired by horizontal gene transfer. LukS-PV and LukF-PV are secreted by the bacterium. These subunits bind to specific membrane receptors, yet to be identified, and associate to form pores in the membrane of host leukocytes139, 140. At high concentrations (200 nM) PVL causes lytic cell death, but at sublytic concentrations (5 nM), PVL appears to partially activate neutrophils in a phenomenon often called priming, as they release potent mediators of inflammation, such as leukotriene B4, interleukin-8, and neutophil granule contents through exocytosis 141–143. In addition, PVL primes neutrophils for enhanced release of reactive oxygen species upon stimulation with the widely used neutrophil agonist N-formylpeptide (fMLP) 144. Thus, PVL could contribute to pathogenesis by causing an exaggerated inflammatory response and injury to the host. Several lines of evidence, largely circumstantial, indicate that PVL is associated with severe skin infections and severe necrotic hemorrhagic pneumonia 33, 145, 146. Both the phage type 80/81 penicillin-resistant strains that were associated with numerous outbreaks and severe disease in the 1950s and USA300, now the leading cause of skin and soft tissue infections in the United States and a cause of extremely severe infections, produce PVL. The epidemiologic association between PVL and emergence of genetically unrelated (i.e., different and unrelated sequence types) CA-MRSA strains that are geographically dispersed is striking.
There are other observations, however, that call into question the presumption that PVL is driving the CA-MRSA epidemic. It is found infrequently in other common and quite successful community strains. For example, PVL genes are present in only ~1–10% of MSSA clinical isolates 89, 147, 148. And although USA300 and USA400 are both PVL-positive, it is USA300 that has become the predominant CA-MRSA clone in the US. This suggests that factors other than PVL are important for the recent emergence of CA-MRSA.
Experimental evidence does not provide a clear picture. Voyich et al. found that USA300 and USA400 wild-type and isogenic PVL-deletion strains (Δpvl) strains caused virtually identical courses of infection in mouse abscess and sepsis models and that there was no difference in neutrophil phagocytosis or lysis after uptake, although because these experiments were conducted with culture supernates the results could reflect the action of multiple lytic factors149. Similar results in a rat pneumonia model were reported by Montgomery and Daum150. Bubeck Wardenburg et al. showed that USA300 and USA400 wild-type and isogenic PVL-deletion strains were equally virulent in these mouse abscess and pneumonia models 151, 152. Diep et al. used two rabbit bacteremia models to compare hematogenous dissemination of wild-type and Δpvl CA-MRSA strains to major organs 153. Although PVL did not promote seeding of lungs, spleen or blood by USA300, there was a modest, transient contribution of PVL to colonization of the kidneys. However, in series of experiments using the same USA300 wild-type and mutant (Δpvl) strain pair as Voyich et al 149, Brown et al found that the parent was more virulent than the Δpvl mutant in murine pneumonia and abscess models and that disease caused by the wild-type strain was attenuated by immunization with recombinant LukF-PV or LukS-PV 154. In addition, Labandeira-Rey et al found evidence suggesting that PVL may play a role in murine model of staphylococcal pneumonia 155. Direct instillation of high doses of purified toxin provoked an inflammatory response in the lung and reduced survival. These investigators, using a laboratory strain transduced with PVL-encoding bacteriophage to establish infection, reported worse outcome for the PVL-producing variant. However, in addition to presence or absence of PVL, this laboratory construct has major alterations in global gene expression that confounded interpretation of the data. As PVL has no impact on protein or gene expression in USA300 or USA400 153, it is possible that factors other than PVL accounted for the experimental results. The data in aggregate suggest that the contribution of PVL to CA-MRSA pathogenesis may be relatively minor or perhaps dependent on an as yet unidentified bacterial factor or host-susceptibility component.
Alpha-hemolysin (Hla or alpha-toxin)
This pore-forming toxin causes destruction of a wide-range of host cells, including epithelial cells, erythrocytes, fibroblasts, and monocytes and is lethal in animal models when injected in purified form 156. Alpha-hemolysin is ubiquitous among clinical isolates, although some strains lack an active alpha-toxin. Recent studies by Bubeck Wardenburg et al 151 demonstrated that alpha-hemolysin is essential for USA300 and USA400 to cause lethal pneumonia in a murine model. Toxin levels produced by these strains in vitro correlate with severity of lung disease 130, 151, 157.
Alpha-type phenol-soluble modulins (PSMa)
Alpha-type phenol-soluble modulins (PSMα) from a novel group of peptides in S. aureus that have some similarity to phenol-soluble modulins (PSMs) of S. epidermidis 131. High expression of PSMα could contribute to the enhanced virulence of CA-MRSA; PSMs are produced at much higher levels in vitro by prominent CA-MRSA strains, including USA300 and USA400, compared to hospital MRSA strains 131. PSMα peptides recruit, activate, and ultimately lyse human neutrophils, thus promoting S. aureus pathogenesis and contribute significantly to USA300 and USA400 virulence in mouse abscess and sepsis models. The studies by Wang et al131 are the first to identify molecules of CA-MRSA that account at least in part for the enhanced virulence of USA300 and USA400.
Arginine catabolic mobile element (ACME)
ACME is a 30.9 kilobase segment of DNA that appears to be unique to USA300 112. This element is adjacent to SCCmecIV and is mobilized by the recombinases encoded on SCCmec. This DNA element contains two potential virulence factors including a cluster of arc genes that encode an arginine deiminase pathway and Opp-3, which encodes an oligopeptide permease operon158, 159. Deletion of ACME but not SCCmec has been shown to decrease fitness of USA300 in a rabbit bacteremia model 112. Thus, ACME could contribute to the fitness and epidemic spread of USA300.
Although mobile genetic elements such as ACME are likely to play a role in transmission of CA-MRSA, there are differences in virulence potential and human disease manifestation even among similar USA300 isolates. For example, Kennedy et al used comparative whole genome sequencing to determine whether USA300 arose by convergent evolution toward a hypervirulent phenotype or from a recent common ancestor of high virulence potential 113. Eleven USA300, which included those from a wide range of clinical syndromes and from different geographic locations in the US, were examined. The strains differed by only a few single nucleotide polymorphisms (SNPs), ranging from 11 to 408 in number compared to the USA300 reference strain FPR3757 genome. Phylogenetic analysis indicated that eight of the strains, differing on average by 32 SNPs from the reference strain and 50 SNPs from each other, clustered together with the reference strain and had descended from a recent common ancestor. These 9 closely related isolates comprise the epidemic USA300 clone; 8 of the 9 were ACME positive and all contained the same SCCmec type IVa subtype. The two other strains, both of which lacked ACME and carried a different SCCmec subtype, type IVb, were outliers. Unexpectedly, the virulence of the more closely related isolates was variable in animal infection models. Some of these isolates had caused dramatically different disease syndromes in humans (e.g., necrotizing pneumonia versus abscess in isolates that differ by only 23 SNPs), which serves to underscore the importance of host factors in disease presentation and severity.
Treatment in the Era of Community-Associated MRSA
CA-MRSA has had a profound impact on empirical therapy of suspected staphylococcal infection. Most beta-lactam antibiotics, including all orally available agents, no longer can be assumed to be effective for a variety of common staphylococcal infections and skin and soft-tissue infections in particular. In regions where CA-MRSA is prevalent antimicrobial therapy, if it is indicated for treatment of staphylococcal infection, should be active against MRSA strains. Yet, there are few clinical data to support the use of agents other than vancomycin, daptomycin, or linezolid. The oral agents that are recommended for treatment of CA-MRSA skin and soft tissue infections, despite lack of rigorous clinical studies, include clindamycin, long-acting tetracyclines (doxycycline and minocycline), TMP-SMX, and, as adjunctive agents to be used in combination, rifampin and fusidic160–162.
Surgical incision and drainage is the treatment of choice for cutaneous abscesses; adjunctive antimicrobial therapy is of little or no benefit in most cases 14, 15, 163, 164. Antibiotic therapy after drainage of CA-MRSA abscesses is not routinely recommended unless the patient has severe or extensive disease, or has rapid progression in the presence of associated cellulitis; has signs and symptoms of systemic illness; is very old or very young or has medical comorbidities or immune suppression (e.g., diabetes mellitus, HIV infections, neoplastic disease); or has an abscess in area that is difficult to drain or an abscess that is associated with septic phlebitis 160.
Vancomycin is still is the preferred drug for treatment of serious MRSA infections. However, prolonged, persistent, or recurrent bacteremia during therapy 165, 166, high rates of microbiological and clinical failures 167, nephrotoxicity 168, and increasing prevalence of non-susceptible strains 169, 170 limit its effectiveness. Randomized clinical trials of alterative agents such as linezolid and daptomycin show that they are comparable, or more precisely, non-inferior, but not superior, to standard therapy 171–176. and drug toxicity remain concerns regardless the choice of agent.
One or more compounds under development are likely to become available for treatment of MRSA infections in the near future 177, 178. Telavancin, dalbavancin, and oritavancin are vancomycin derivatives that rapidly kill S. aureus in a concentration-dependent manner in vitro. Whether more rapid killing will translate into improved efficacy over vancomycin for more serious infections, such as endocarditis or bacteremia, remains to be determined. Carbapenems and cephalosporins that bind PBP 2a, the penicillin-binding protein that mediates methicillin resistance, with much higher affinity than the currently available beta-lactams, have been developed 179. Two cephalosporins, ceftobiprole and ceftaroline 180, 181, have been shown to be clinically effective for treatment of MRSA skin and soft infections. An issue with these and the other anti-MRSA beta-lactams under development is that they are very broad spectrum for the targeted treatment of MRSA infection. Further studies are needed to define their eventual role in therapy of MRSA infections.
The vancomycin-derivatives and anti-MRSA beta-lactams, which can only be administered intravenously, do not address the need for orally active agents. Orally bioavailable oxazolidinones active against MRSA are in early stages of development 182.
Several non-traditional approaches to treatment and prevention of MRSA infections have been or are under investigation. These include lysostaphin 183, antimicrobial peptides 184 and other natural products (e.g., tea tree oil) 185, and anti-staphylococcal vaccines 186. There are major challenges in the development of these agents, including prohibitively expensive cost, potential for hypersensitivity with repeated administration of protein products, short half lives with systemic administration, and short-lived or partially protective immunity with vaccines, as was the case with an anticapsular vaccine that proved to be ineffective 187. These are years away from the clinic, if they make it at all. Prudent use of agents that are now available is essential to avoid further erosion of the antimicrobial armamentarium.
Concluding Remarks
S. aureus is an extraordinarily adaptable pathogen with a proven ability to develop resistance. Especially concerning is the steadily erosion in the effectiveness of beta-lactam antibiotics during a relatively brief 60-year time period. Although details vary the basic themes of each successive wave of antibiotic resistance are similar. Resistance, often as a consequence of horizontal gene transfer, is initially encountered in hospitals and healthcare institutions, where the selective pressures for resistance are greatest. Resistant strains are contained within hospitals temporarily, but eventually though a series of modifications and adjustments, invariably find their way into or arise from within the community to emerge as fully fit and virulent pathogens. Understanding of the forces that direct the evolution of virulent and drug-resistant organisms is imperfect, but overuse and misuse of antibiotics is clearly a contributing factor. Discovery and development of new antimicrobials, while necessary, is unlikely to solve the problem of drug resistance for very long. New technologies leading to improved and more rapid diagnostics, a better understanding of pathogenesis of staphylococcal disease, and non-antimicrobial approaches to prevention and treatment of infection will also be needed to forestall the coming of the post-antibiotic era.
Table 4.
Hemolysins and anti-platelet factors produced by Staphylococcus aureus.
| Target cell, host factor or response | Gene(s) | Protein or molecule | Putative function/effect on immune system |
|---|---|---|---|
| Erythrocytes | hla, hly | Alpha-hemolysin (α-hemolysin), Hla | Causes cell lysis (also affect epithelial cells, fibroblasts, and monocytes) |
| hld | Delta-hemolysin, Hld | Causes cell lysis | |
| hlgA, hlgB, hlgC | Gamma-hemolysin subunits A, B, and C; HlgA, HlgB, HlgC; two-component leukocidin | Causes cell lysis | |
| Platelets | clfA | Clumping factor A, ClfA | Causes platelet activation |
| fnbA, fnbB | Fibronectin-binding proteins A and B, FnbA and FnbB | Causes platelet activation | |
| katA | Catalase, KatA | Detoxifies hydrogen peroxide | |
| sodA, sodM | Superoxide dismutase, SodA, SodM | Promotes resistance to reactive oxygen species |
Table 5.
Leucocidins and anti-phagocytic factors produced by Staphylococcus aureus.
| Target cell, host factor or response | Gene(s) | Protein or molecule | Putative function/effect on immune system |
|---|---|---|---|
| Polymorphonuclear leukocytes | cap5 or cap8 genes | Capsular polysaccharide | Inhibits phagocytosis |
| clfA | Clumping factor A, ClfA | Inhibits phagocytosis | |
| eap | Extracellular adherence protein, Eap | Inhibits leukocyte adhesion | |
| hlgA, hlgB, hlgC | Gamma-hemolysin subunits A, B, and C; HlgA, HlgB, HlgC; two-component leukocidin | Causes cell lysis | |
| lukD, lukE | Leukocidin D and E; LukD and LukE; two-component leukocidins | Causes leukocyte lysis | |
| lukS-PV, lukF-PV | Leukocidin S-PV and F-PV subunits; two-component leukocidin, PVL | Causes phagocyte lysis | |
| psm | Phenol-soluble modulin-like peptides, PSMs | Cause leukocyte lysis | |
| sbi | IgG-binding protein, Sbi | Sequesters host IgG | |
| scn | Staphylococcal inhibitor of complement, SCIN | Inhibits complement | |
| ssl5 | Staphylococcal superantigen- like 5, SSL5 | Binds P-selectin glycoprotein ligand-1 and inhibits neutrophil rolling | |
| Chemotaxis | chp | Chemotaxis inhibitory protein of S. aureus, CHIPS | Inhibits chemotaxis |
| ecb | Extracellular complement- binding protein, Ecb | Inhibits C5a generation | |
| efb | Extracellular fibrinogen- binding protein, Efb | Inhibits C5a generation | |
| sbi | IgG-binding protein, Sbi | Sequesters host IgG | |
| scn | Staphylococcal inhibitor of complement, SCIN | Inhibits complement | |
| ssl7 | Staphylococcal superantigen- like 7, SSL7 | Binds to C5a and IgA |
Acknowledgments
This article was supported in part by the Intramural Research Program of the NIAID, NIH (FRD) and by NIH, NIAID grant number R01 AI070289 (HFC).
References
- 1.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10:505–20. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorwitz RJ, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis. 2008;197:1226–34. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- 3.Miller LG, Diep BA. Clinical practice: colonization, fomites, and virulence: rethinking the pathogenesis of community-associated methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46:752–60. doi: 10.1086/526773. [DOI] [PubMed] [Google Scholar]
- 4.Kazakova SV, et al. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N Engl J Med. 2005;352:468–75. doi: 10.1056/NEJMoa042859. [DOI] [PubMed] [Google Scholar]
- 5.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–32. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 6.Muto CA, et al. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus. Infect Control Hosp Epidemiol. 2003;24:362–86. doi: 10.1086/502213. [DOI] [PubMed] [Google Scholar]
- 7.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368:874–85. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan SL, et al. Three-year surveillance of community-acquired Staphylococcus aureus infections in children. Clin Infect Dis. 2005;40:1785–91. doi: 10.1086/430312. [DOI] [PubMed] [Google Scholar]
- 9.Hersh AL, Chambers HF, Maselli JH, Gonzales R. National trends in ambulatory visits and antibiotic prescribing for skin and soft-tissue infections. Arch Intern Med. 2008;168:1585–91. doi: 10.1001/archinte.168.14.1585. [DOI] [PubMed] [Google Scholar]
- 10.Klevens RM, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama. 2007;298:1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 11.Hope R, Livermore DM, Brick G, Lillie M, Reynolds R. Non-susceptibility trends among staphylococci from bacteraemias in the UK and Ireland, 2001–06. J Antimicrob Chemother. 2008;62(Suppl 2):ii65–74. doi: 10.1093/jac/dkn353. [DOI] [PubMed] [Google Scholar]
- 12.Laupland KB, Ross T, Gregson DB. Staphylococcus aureus bloodstream infections: risk factors, outcomes, and the influence of methicillin resistance in Calgary, Canada, 2000–2006. J Infect Dis. 2008;198:336–43. doi: 10.1086/589717. [DOI] [PubMed] [Google Scholar]
- 13.EARSS Annual Report. 2007 http://www.rivm.nl/earss/news/index.jsp.
- 14.Moran GJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–74. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 15.Fridkin SK, et al. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005;352:1436–44. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
- 16.Larsen A, Stegger M, Goering R, Sorum M, Skov R. Emergence and dissemination of the methicillin resistant Staphylococcus aureus USA300 clone in Denmark (2000–2005) Euro Surveill. 2007;12 [Google Scholar]
- 17.Larsen AR, et al. Epidemiology of European community-associated methicillin-resistant Staphylococcus aureus clonal complex 80 type IV strains isolated in Denmark from 1993 to 2004. J Clin Microbiol. 2008;46:62–8. doi: 10.1128/JCM.01381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wannet WJ, et al. Emergence of virulent methicillin-resistant Staphylococcus aureus strains carrying Panton-Valentine leucocidin genes in The Netherlands. J Clin Microbiol. 2005;43:3341–5. doi: 10.1128/JCM.43.7.3341-3345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deurenberg RH, et al. Cross-border dissemination of methicillin-resistant Staphylococcus aureus, Euregio Meuse-Rhin region. Emerg Infect Dis. 2009;15:727–34. doi: 10.3201/eid1505.071618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandenesch F, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–84. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stam-Bolink EM, Mithoe D, Baas WH, Arends JP, Moller AV. Spread of a methicillin-resistant Staphylococcus aureus ST80 strain in the community of the northern Netherlands. Eur J Clin Microbiol Infect Dis. 2007;26:723–7. doi: 10.1007/s10096-007-0352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang YC, Hwang KP, Chen PY, Chen CJ, Lin TY. Prevalence of methicillin-resistant Staphylococcus aureus nasal colonization among Taiwanese children in 2005 and 2006. J Clin Microbiol. 2007;45:3992–5. doi: 10.1128/JCM.01202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nimmo GR, Coombs GW. Community-associated methicillin-resistant Staphylococcus aureus (MRSA) in Australia. Int J Antimicrob Agents. 2008;31:401–10. doi: 10.1016/j.ijantimicag.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Kanerva M, et al. Community-associated methicillin-resistant Staphylococcus aureus, 2004–2006, Finland. J Clin Microbiol. 2009 doi: 10.1128/JCM.00771-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SH, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated bloodstream infections in Korea. Infect Control Hosp Epidemiol. 2009;30:146–55. doi: 10.1086/593953. [DOI] [PubMed] [Google Scholar]
- 26.Gardella N, et al. Community-associated methicillin-resistant Staphylococcus aureus, eastern Argentina. Diagn Microbiol Infect Dis. 2008;62:343–7. doi: 10.1016/j.diagmicrobio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Francois P, et al. Methicillin-resistant Staphylococcus aureus, Geneva, Switzerland, 1993–2005. Emerg Infect Dis. 2008;14:304–7. doi: 10.3201/eid1402.070229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang H, Hedin G, Li G, Nord CE. Genetic diversity of community-associated methicillin-resistant Staphylococcus aureus in southern Stockholm, 2000–2005. Clin Microbiol Infect. 2008;14:370–6. doi: 10.1111/j.1469-0691.2007.01941.x. [DOI] [PubMed] [Google Scholar]
- 29.Conly JM, Johnston BL. The emergence of methicillin-resistant Staphylococcus aureus as a community-acquired pathogen in Canada. Can J Infect Dis. 2003;14:249–51. doi: 10.1155/2003/197126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francis JS, et al. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin Infect Dis. 2005;40:100–7. doi: 10.1086/427148. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez BE, et al. Pulmonary manifestations in children with invasive community-acquired Staphylococcus aureus infection. Clin Infect Dis. 2005;41:583–90. doi: 10.1086/432475. [DOI] [PubMed] [Google Scholar]
- 32.Kallen AJ, et al. Staphylococcus aureus community-acquired pneumonia during the 2006 to 2007 influenza season. Ann Emerg Med. 2009;53:358–65. doi: 10.1016/j.annemergmed.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 33.Lina G, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–32. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 34.Kirby W. Extraction of a highly potene penicillin inactivator from penicillin resistant Staphylococci. Science. 1944;99:452–53. doi: 10.1126/science.99.2579.452. [DOI] [PubMed] [Google Scholar]
- 35.Barber M, Rozwadowska-Dowzenko M. Infection by penicillin-resistant staphylococci. Lancet. 1948;1:641–44. doi: 10.1016/s0140-6736(48)92166-7. [DOI] [PubMed] [Google Scholar]
- 36.Roundtree P, Freeman V. Infections caused by a particular phage type of Staphylococcus aureus. Med J Aust. 1956;42:157–61. [PubMed] [Google Scholar]
- 37.Blair JE, Carr M. Distribution of Phage Groups of Staphylococcus aureus in the Years 1927 through 1947. Science. 1960;132:1247–1248. doi: 10.1126/science.132.3435.1247. [DOI] [PubMed] [Google Scholar]
- 38.Bynoe ET, Elder RH, Comtois RD. Phage-typing and antibiotic-resistance of staphylococci isolated in a general hospital. Can J Microbiol. 1956;2:346–58. doi: 10.1139/m56-041. [DOI] [PubMed] [Google Scholar]
- 39.Roundtree P, Beard M. Further observations on infections with phage type 80 staphylococci in Australia. Med J Aust. 1958;2:789–95. [PubMed] [Google Scholar]
- 40.Jevons MP, Parker MT. The evolution of new hospital strains of Staphylococcus aureus. J Clin Pathol. 1964;17:243–50. doi: 10.1136/jcp.17.3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barber M. Methicillin-resistant staphylococci. J Clin Pathol. 1961;14:385–93. doi: 10.1136/jcp.14.4.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jevons M. “Celbenin”-resistant staphylococci. Br Med J. 1961;1:124–5. [Google Scholar]
- 43.Crisostomo MI, et al. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc Natl Acad Sci U S A. 2001;98:9865–70. doi: 10.1073/pnas.161272898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrett FF, McGehee RF, Jr, Finland M. Methicillin-resistant Staphylococcus aureus at Boston City Hospital. Bacteriologic and epidemiologic observations. N Engl J Med. 1968;279:441–8. doi: 10.1056/NEJM196808292790901. [DOI] [PubMed] [Google Scholar]
- 45.Bran JL, Levison ME, Kaye D. Survey for methicillin-resistant staphylococci. Antimicrob Agents Chemother. 1972;1:235–6. doi: 10.1128/aac.1.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mato R, et al. Clonal types and multidrug resistance patterns of methicillin-resistant Staphylococcus aureus (MRSA) recovered in Italy during the 1990s. Microb Drug Resist. 2004;10:106–13. doi: 10.1089/1076629041310109. [DOI] [PubMed] [Google Scholar]
- 47.Enright MC, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci U S A. 2002;99:7687–92. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson DA, Enright MC. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:3926–34. doi: 10.1128/AAC.47.12.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infect Genet Evol. 2008;8:747–63. doi: 10.1016/j.meegid.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Crossley K, Landesman B, Zaske D. An outbreak of infections caused by strains of Staphylococcus aureus resistant to methicillin and aminoglycosides. II. Epidemiologic studies. J Infect Dis. 1979;139:280–7. doi: 10.1093/infdis/139.3.280. [DOI] [PubMed] [Google Scholar]
- 51.Peacock JE, Jr, Marsik FJ, Wenzel RP. Methicillin-resistant Staphylococcus aureus: introduction and spread within a hospital. Ann Intern Med. 1980;93:526–32. doi: 10.7326/0003-4819-93-4-526. [DOI] [PubMed] [Google Scholar]
- 52.Hiramatsu K, et al. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–3. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 53.Weigel LM, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–71. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 54.O’Brien FG, et al. Diversity among community isolates of methicillin-resistant Staphylococcus aureus in Australia. J Clin Microbiol. 2004;42:3185–90. doi: 10.1128/JCM.42.7.3185-3190.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coombs GW, et al. Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J Clin Microbiol. 2004;42:4735–43. doi: 10.1128/JCM.42.10.4735-4743.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25:97–108. doi: 10.1016/0195-6701(93)90100-e. [DOI] [PubMed] [Google Scholar]
- 57.From the Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus--Minnesota and North Dakota, 1997–1999. Jama. 1999;282:1123–5. [PubMed] [Google Scholar]
- 58.Herold BC, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. Jama. 1998;279:593–8. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 59.Baggett HC, et al. Community-onset methicillin-resistant Staphylococcus aureus associated with antibiotic use and the cytotoxin Panton-Valentine leukocidin during a furunculosis outbreak in rural Alaska. J Infect Dis. 2004;189:1565–73. doi: 10.1086/383247. [DOI] [PubMed] [Google Scholar]
- 60.Community-associated methicillin-resistant Staphylococcus aureus infections in Pacific Islanders--Hawaii, 2001–2003. MMWR Morb Mortal Wkly Rep. 2004;53:767–70. [PubMed] [Google Scholar]
- 61.Aiello AE, Lowy FD, Wright LN, Larson EL. Meticillin-resistant Staphylococcus aureus among US prisoners and military personnel: review and recommendations for future studies. Lancet Infect Dis. 2006;6:335–41. doi: 10.1016/S1473-3099(06)70491-1. [DOI] [PubMed] [Google Scholar]
- 62.Diep BA, et al. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann Intern Med. 2008;148:249–57. doi: 10.7326/0003-4819-148-4-200802190-00204. [DOI] [PubMed] [Google Scholar]
- 63.Johansson PJ, Gustafsson EB, Ringberg H. High prevalence of MRSA in household contacts. Scand J Infect Dis. 2007;39:764–8. doi: 10.1080/00365540701302501. [DOI] [PubMed] [Google Scholar]
- 64.Adcock PM, Pastor P, Medley F, Patterson JE, Murphy TV. Methicillin-resistant Staphylococcus aureus in two child care centers. J Infect Dis. 1998;178:577–80. doi: 10.1086/517478. [DOI] [PubMed] [Google Scholar]
- 65.Liu C, et al. A population-based study of the incidence and molecular epidemiology of methicillin-resistant Staphylococcus aureus disease in San Francisco, 2004–2005. Clin Infect Dis. 2008;46:1637–46. doi: 10.1086/587893. [DOI] [PubMed] [Google Scholar]
- 66.Seybold U, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis. 2006;42:647–56. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 67.Gilbert M, et al. Outbreak in Alberta of community-acquired (USA300) methicillin-resistant Staphylococcus aureus in people with a history of drug use, homelessness or incarceration. Cmaj. 2006;175:149–54. doi: 10.1503/cmaj.051565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mulvey MR, et al. Community-associated methicillin-resistant Staphylococcus aureus, Canada. Emerg Infect Dis. 2005;11:844–50. doi: 10.3201/eid1106.041146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stemper ME, Shukla SK, Reed KD. Emergence and spread of community-associated methicillin-resistant Staphylococcus aureus in rural Wisconsin, 1989 to 1999. J Clin Microbiol. 2004;42:5673–80. doi: 10.1128/JCM.42.12.5673-5680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.David MZ, Rudolph KM, Hennessy TW, Boyle-Vavra S, Daum RS. Molecular epidemiology of methicillin-resistant Staphylococcus aureus, rural southwestern Alaska. Emerg Infect Dis. 2008;14:1693–9. doi: 10.3201/eid1411.080381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pan ES, et al. Increasing prevalence of methicillin-resistant Staphylococcus aureus infection in California jails. Clin Infect Dis. 2003;37:1384–8. doi: 10.1086/379019. [DOI] [PubMed] [Google Scholar]
- 72.Pannaraj PS, Hulten KG, Gonzalez BE, Mason EO, Jr, Kaplan SL. Infective pyomyositis and myositis in children in the era of community-acquired, methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2006;43:953–60. doi: 10.1086/507637. [DOI] [PubMed] [Google Scholar]
- 73.Diep BA, Sensabaugh GF, Somboona NS, Carleton HA, Perdreau-Remington F. Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leucocidin. J Clin Microbiol. 2004;42:2080–4. doi: 10.1128/JCM.42.5.2080-2084.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chavez-Bueno S, et al. Inducible clindamycin resistance and molecular epidemiologic trends of pediatric community-acquired methicillin-resistant Staphylococcus aureus in Dallas, Texas. Antimicrob Agents Chemother. 2005;49:2283–8. doi: 10.1128/AAC.49.6.2283-2288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gonzalez BE, et al. Severe Staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. Pediatrics. 2005;115:642–8. doi: 10.1542/peds.2004-2300. [DOI] [PubMed] [Google Scholar]
- 76.Nourse C, Starr M, Munckhof W. Community-acquired methicillin-resistant Staphylococcus aureus causes severe disseminated infection and deep venous thrombosis in children: literature review and recommendations for management. J Paediatr Child Health. 2007;43:656–61. doi: 10.1111/j.1440-1754.2007.01153.x. [DOI] [PubMed] [Google Scholar]
- 77.Miller LG, et al. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med. 2005;352:1445–53. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- 78.Adem PV, et al. Staphylococcus aureus sepsis and the Waterhouse-Friderichsen syndrome in children. N Engl J Med. 2005;353:1245–51. doi: 10.1056/NEJMoa044194. [DOI] [PubMed] [Google Scholar]
- 79.Rutar T, et al. Ophthalmic manifestations of infections caused by the USA300 clone of community-associated methicillin-resistant Staphylococcus aureus. Ophthalmology. 2006;113:1455–62. doi: 10.1016/j.ophtha.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 80.Munckhof WJ, Krishnan A, Kruger P, Looke D. Cavernous sinus thrombosis and meningitis from community-acquired methicillin-resistant Staphylococcus aureus infection. Intern Med J. 2008;38:283–7. doi: 10.1111/j.1445-5994.2008.01650.x. [DOI] [PubMed] [Google Scholar]
- 81.Sifri CD, Park J, Helm GA, Stemper ME, Shukla SK. Fatal brain abscess due to community-associated methicillin-resistant Staphylococcus aureus strain USA300. Clin Infect Dis. 2007;45:e113–7. doi: 10.1086/522171. [DOI] [PubMed] [Google Scholar]
- 82.Chua T, et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus bloodstream isolates in urban Detroit. J Clin Microbiol. 2008;46:2345–52. doi: 10.1128/JCM.00154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–30. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Turner KM, Hanage WP, Fraser C, Connor TR, Spratt BG. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 2007;7:30. doi: 10.1186/1471-2180-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shopsin B, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–63. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feil EJ, et al. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–16. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tenover FC, et al. Characterization of Staphylococcus aureus isolates from nasal cultures collected from individuals in the United States in 2001 to 2004. J Clin Microbiol. 2008;46:2837–41. doi: 10.1128/JCM.00480-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goering RV, et al. Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from global clinical trials. J Clin Microbiol. 2008;46:2842–7. doi: 10.1128/JCM.00521-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hallin M, et al. Genetic relatedness between methicillin-susceptible and methicillin-resistant Staphylococcus aureus: results of a national survey. J Antimicrob Chemother. 2007;59:465–72. doi: 10.1093/jac/dkl535. [DOI] [PubMed] [Google Scholar]
- 91.Feng Y, et al. Evolution and pathogenesis of Staphylococcus aureus: lessons learned from genotyping and comparative genomics. FEMS Microbiol Rev. 2008;32:23–37. doi: 10.1111/j.1574-6976.2007.00086.x. [DOI] [PubMed] [Google Scholar]
- 92.Feil EJ, Enright MC. Analyses of clonality and the evolution of bacterial pathogens. Curr Opin Microbiol. 2004;7:308–13. doi: 10.1016/j.mib.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 93.Lindsay JA, et al. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J Bacteriol. 2006;188:669–76. doi: 10.1128/JB.188.2.669-676.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nubel U, et al. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2008;105:14130–5. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gomes AR, Westh H, de Lencastre H. Origins and evolution of methicillin-resistant Staphylococcus aureus clonal lineages. Antimicrob Agents Chemother. 2006;50:3237–44. doi: 10.1128/AAC.00521-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robinson DA, et al. Re-emergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet. 2005;365:1256–8. doi: 10.1016/S0140-6736(05)74814-5. [DOI] [PubMed] [Google Scholar]
- 97.Cox RA, Conquest C, Mallaghan C, Marples RR. A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16) J Hosp Infect. 1995;29:87–106. doi: 10.1016/0195-6701(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 98.Johnson AP, et al. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS) J Antimicrob Chemother. 2001;48:143–4. doi: 10.1093/jac/48.1.143. [DOI] [PubMed] [Google Scholar]
- 99.McDougal LK, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–20. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ito T, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323–36. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Deurenberg RH, et al. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2007;13:222–35. doi: 10.1111/j.1469-0691.2006.01573.x. [DOI] [PubMed] [Google Scholar]
- 102.Ito T, et al. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother. 2004;48:2637–51. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ma XX, et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2002;46:1147–52. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oliveira DC, Milheirico C, de Lencastre H. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob Agents Chemother. 2006;50:3457–9. doi: 10.1128/AAC.00629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ruppe E, et al. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob Agents Chemother. 2009;53:442–9. doi: 10.1128/AAC.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Okuma K, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol. 2002;40:4289–94. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lina G, et al. Staphylococcal chromosome cassette evolution in Staphylococcus aureus inferred from ccr gene complex sequence typing analysis. Clin Microbiol Infect. 2006;12:1175–84. doi: 10.1111/j.1469-0691.2006.01548.x. [DOI] [PubMed] [Google Scholar]
- 108.Oliveira DC, Tomasz A, de Lencastre H. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb Drug Resist. 2001;7:349–61. doi: 10.1089/10766290152773365. [DOI] [PubMed] [Google Scholar]
- 109.Hanssen AM, Kjeldsen G, Sollid JU. Local variants of Staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative Staphylococci: evidence of horizontal gene transfer? Antimicrob Agents Chemother. 2004;48:285–96. doi: 10.1128/AAC.48.1.285-296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanssen AM, Ericson Sollid JU. SCCmec in staphylococci: genes on the move. FEMS Immunol Med Microbiol. 2006;46:8–20. doi: 10.1111/j.1574-695X.2005.00009.x. [DOI] [PubMed] [Google Scholar]
- 111.Wu S, Piscitelli C, de Lencastre H, Tomasz A. Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb Drug Resist. 1996;2:435–41. doi: 10.1089/mdr.1996.2.435. [DOI] [PubMed] [Google Scholar]
- 112.Diep BA, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–30. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 113.Kennedy AD, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A. 2008;105:1327–32. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bartels MD, Boye K, Rhod Larsen A, Skov R, Westh H. Rapid increase of genetically diverse methicillin-resistant Staphylococcus aureus, Copenhagen, Denmark. Emerg Infect Dis. 2007;13:1533–40. doi: 10.3201/eid1310.070503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gottlieb T, Su WY, Merlino J, Cheong EY. Recognition of USA300 isolates of community-acquired methicillin-resistant Staphylococcus aureus in Australia. Med J Aust. 2008;189:179–80. doi: 10.5694/j.1326-5377.2008.tb01962.x. [DOI] [PubMed] [Google Scholar]
- 116.Arias CA, et al. MRSA USA300 clone and VREF--a U.S.-Colombian connection? N Engl J Med. 2008;359:2177–9. doi: 10.1056/NEJMc0804021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maree CL, Daum RS, Boyle-Vavra S, Matayoshi K, Miller LG. Community-associated methicillin-resistant Staphylococcus aureus isolates causing healthcare-associated infections. Emerg Infect Dis. 2007;13:236–42. doi: 10.3201/eid1302.060781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gonzalez BE, et al. Community-associated strains of methicillin-resistant Staphylococccus aureus as the cause of healthcare-associated infection. Infect Control Hosp Epidemiol. 2006;27:1051–6. doi: 10.1086/507923. [DOI] [PubMed] [Google Scholar]
- 119.Tristan A, et al. Global distribution of Panton-Valentine leukocidin--positive methicillin-resistant Staphylococcus aureus, 2006. Emerg Infect Dis. 2007;13:594–600. doi: 10.3201/eid1304.061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Larsen AR, et al. Emergence and characterization of community-associated methicillin-resistant Staphyloccocus aureus infections in Denmark, 1999 to 2006. J Clin Microbiol. 2009;47:73–8. doi: 10.1128/JCM.01557-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rollason J, et al. Epidemiology of community-acquired meticillin-resistant Staphylococcus aureus obtained from the UK West Midlands region. J Hosp Infect. 2008;70:314–20. doi: 10.1016/j.jhin.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 122.Holmes A, et al. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J Clin Microbiol. 2005;43:2384–90. doi: 10.1128/JCM.43.5.2384-2390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huijsdens XW, et al. Community-acquired MRSA and pig-farming. Ann Clin Microbiol Antimicrob. 2006;5:26. doi: 10.1186/1476-0711-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Loeffler A, et al. First isolation of MRSA ST398 from UK animals: a new challenge for infection control teams? J Hosp Infect. 2009;72:269–71. doi: 10.1016/j.jhin.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 125.Crum NF, et al. Fifteen-year study of the changing epidemiology of methicillin-resistant Staphylococcus aureus. Am J Med. 2006;119:943–51. doi: 10.1016/j.amjmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 126.Davis SL, et al. Epidemiology and outcomes of community-associated methicillin-resistant Staphylococcus aureus infection. J Clin Microbiol. 2007;45:1705–11. doi: 10.1128/JCM.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–9. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 128.Voyich JM, et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–19. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 129.Li M, et al. Evolution of virulence in epidemic community-associated MRSA. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0900743106. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Montgomery CP, et al. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis. 2008;198:561–70. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- 131.Wang R, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–4. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 132.Rooijakkers SH, et al. Early expression of SCIN and CHIPS drives instant immune evasion by Staphylococcus aureus. Cell Microbiol. 2006;8:1282–93. doi: 10.1111/j.1462-5822.2006.00709.x. [DOI] [PubMed] [Google Scholar]
- 133.van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Strijp JA. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J Bacteriol. 2006;188:1310–5. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Deleo FR, Diep BA, Otto M. Host Defense and Pathogenesis in Staphylococcus aureus Infections. Infect Dis Clin North Am. 2009;23:17–34. doi: 10.1016/j.idc.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li M, et al. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol. 2007;66:1136–47. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- 136.Wright J. Staphylococcal leucocidin (Neisser-Wechsberg type) and antileucocidin. Lancet. 1936;227:1002–5. [Google Scholar]
- 137.Woodin AM. Purification of the two components of leucocidin from Staphylococcus aureus. Biochem J. 1960;75:158–65. doi: 10.1042/bj0750158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaneko J, Kimura T, Narita S, Tomita T, Kamio Y. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage phiPVL carrying Panton-Valentine leukocidin genes. Gene. 1998;215:57–67. doi: 10.1016/s0378-1119(98)00278-9. [DOI] [PubMed] [Google Scholar]
- 139.Meyer F, Girardot R, Piemont Y, Prevost G, Colin DA. Analysis of the specificity of Panton-Valentine leucocidin and gamma-hemolysin F component binding. Infect Immun. 2009;77:266–73. doi: 10.1128/IAI.00402-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Colin DA, Mazurier I, Sire S, Finck-Barbancon V. Interaction of the two components of leukocidin from Staphylococcus aureus with human polymorphonuclear leukocyte membranes: sequential binding and subsequent activation. Infect Immun. 1994;62:3184–8. doi: 10.1128/iai.62.8.3184-3188.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Konig B, Prevost G, Piemont Y, Konig W. Effects of Staphylococcus aureus leukocidins on inflammatory mediator release from human granulocytes. J Infect Dis. 1995;171:607–13. doi: 10.1093/infdis/171.3.607. [DOI] [PubMed] [Google Scholar]
- 142.Woodin AM, Wieneke AA. The participation of calcium, adenosine triphosphate and adenosine triphosphatase in the extrusion of the granule proteins from the polymorphonuclear leucocyte. Biochem J. 1964;90:498–509. doi: 10.1042/bj0900498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Genestier AL, et al. Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. J Clin Invest. 2005;115:3117–27. doi: 10.1172/JCI22684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Colin DA, Monteil H. Control of the oxidative burst of human neutrophils by staphylococcal leukotoxins. Infect Immun. 2003;71:3724–9. doi: 10.1128/IAI.71.7.3724-3729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gillet Y, et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359:753–9. doi: 10.1016/S0140-6736(02)07877-7. [DOI] [PubMed] [Google Scholar]
- 146.Gillet Y, et al. Factors predicting mortality in necrotizing community-acquired pneumonia caused by Staphylococcus aureus containing Panton-Valentine leukocidin. Clin Infect Dis. 2007;45:315–21. doi: 10.1086/519263. [DOI] [PubMed] [Google Scholar]
- 147.Kuehnert MJ, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. J Infect Dis. 2006;193:172–9. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- 148.Ellington MJ, et al. Is Panton-Valentine leucocidin associated with the pathogenesis of Staphylococcus aureus bacteraemia in the UK? J Antimicrob Chemother. 2007;60:402–5. doi: 10.1093/jac/dkm206. [DOI] [PubMed] [Google Scholar]
- 149.Voyich JM, et al. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis. 2006;194:1761–70. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 150.Montgomery CP, Daum RS. Transcription of inflammatory genes in the lung after infection with community-associated methicillin-resistant Staphylococcus aureus: A role for Panton-Valentine Leukocidin? Infect Immun. 2009 doi: 10.1128/IAI.00021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med. 2007;13:1405–6. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- 152.Bubeck Wardenburg J, Palazzolo-Ballance AM, Otto M, Schneewind O, DeLeo FR. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J Infect Dis. 2008;198:1166–70. doi: 10.1086/592053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Diep BA, et al. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS ONE. 2008;3:e3198. doi: 10.1371/journal.pone.0003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Brown EL, et al. The Panton-Valentine leukocidin vaccine protects mice against lung and skin infections caused by Staphylococcus aureus USA300. Clin Microbiol Infect. 2008 doi: 10.1111/j.1469-0691.2008.02648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Labandeira-Rey M, et al. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–3. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- 156.Bhakdi S, Tranum-Jensen J. Alpha-toxin of Staphylococcus aureus. Microbiol Rev. 1991;55:733–51. doi: 10.1128/mr.55.4.733-751.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Burlak C, et al. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol. 2007;9:1172–90. doi: 10.1111/j.1462-5822.2006.00858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Coulter SN, et al. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 159.Degnan BA, et al. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. Infect Immun. 1998;66:3050–8. doi: 10.1128/iai.66.7.3050-3058.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gorwitz RJ, et al. Strategies for clinical management of MRSA in the community: summary of an expert’s meeting convened by the Centers for Disease Control and Prevention. 2006 March; Available at: http://www.cdc.gov/ncidod/dhqp/ar_mrsa_ca.html.
- 161.Barton M, et al. Guidelines for the prevention and management of community-acquired methicillin-resistant Staphylococcus aureus: A perspective for Canadian health care practitioners. Can J Infect Dis Med Microbiol. 2006;17(Suppl C):4C–24C. [PMC free article] [PubMed] [Google Scholar]
- 162.Nathwani D, et al. Guidelines for UK practice for the diagnosis and management of methicillin-resistant Staphylococcus aureus (MRSA) infections presenting in the community. J Antimicrob Chemother. 2008;61:976–94. doi: 10.1093/jac/dkn096. [DOI] [PubMed] [Google Scholar]
- 163.Llera JL, Levy RC. Treatment of cutaneous abscess: a double-blind clinical study. Ann Emerg Med. 1985;14:15–9. doi: 10.1016/s0196-0644(85)80727-7. [DOI] [PubMed] [Google Scholar]
- 164.Lee MC, et al. Management and outcome of children with skin and soft tissue abscesses caused by community-acquired methicillin-resistant Staphylococcus aureus. Pediatr Infect Dis J. 2004;23:123–7. doi: 10.1097/01.inf.0000109288.06912.21. [DOI] [PubMed] [Google Scholar]
- 165.Khatib R, et al. Persistent Staphylococcus aureus bacteremia: Incidence and outcome trends over time. Scand J Infect Dis. 2009;41:4–9. doi: 10.1080/00365540802441711. [DOI] [PubMed] [Google Scholar]
- 166.Hawkins C, et al. Persistent Staphylococcus aureus bacteremia: an analysis of risk factors and outcomes. Arch Intern Med. 2007;167:1861–7. doi: 10.1001/archinte.167.17.1861. [DOI] [PubMed] [Google Scholar]
- 167.Dombrowski JC, Winston LG. Clinical failures of appropriately-treated methicillin-resistant Staphylococcus aureus infections. J Infect. 2008;57:110–5. doi: 10.1016/j.jinf.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lodise TP, Lomaestro B, Graves J, Drusano GL. Larger vancomycin doses (at least four grams per day) are associated with an increased incidence of nephrotoxicity. Antimicrob Agents Chemother. 2008;52:1330–6. doi: 10.1128/AAC.01602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Steinkraus G, White R, Friedrich L. Vancomycin MIC creep in non-vancomycin-intermediate Staphylococcus aureus (VISA), vancomycin-susceptible clinical methicillin-resistant S. aureus (MRSA) blood isolates from 2001–05. J Antimicrob Chemother. 2007;60:788–94. doi: 10.1093/jac/dkm258. [DOI] [PubMed] [Google Scholar]
- 170.Wang G, Hindler JF, Ward KW, Bruckner DA. Increased vancomycin MICs for Staphylococcus aureus clinical isolates from a university hospital during a 5-year period. J Clin Microbiol. 2006;44:3883–6. doi: 10.1128/JCM.01388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Arbeit RD, Maki D, Tally FP, Campanaro E, Eisenstein BI. The safety and efficacy of daptomycin for the treatment of complicated skin and skin-structure infections. Clin Infect Dis. 2004;38:1673–81. doi: 10.1086/420818. [DOI] [PubMed] [Google Scholar]
- 172.Shorr AF, Kunkel MJ, Kollef M. Linezolid versus vancomycin for Staphylococcus aureus bacteraemia: pooled analysis of randomized studies. J Antimicrob Chemother. 2005;56:923–9. doi: 10.1093/jac/dki355. [DOI] [PubMed] [Google Scholar]
- 173.Wunderink RG, Cammarata SK, Oliphant TH, Kollef MH. Continuation of a randomized, double-blind, multicenter study of linezolid versus vancomycin in the treatment of patients with nosocomial pneumonia. Clin Ther. 2003;25:980–92. doi: 10.1016/s0149-2918(03)80118-2. [DOI] [PubMed] [Google Scholar]
- 174.Weigelt J, et al. Linezolid versus vancomycin in treatment of complicated skin and soft tissue infections. Antimicrob Agents Chemother. 2005;49:2260–6. doi: 10.1128/AAC.49.6.2260-2266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kaplan SL, et al. Linezolid versus vancomycin for treatment of resistant Gram-positive infections in children. Pediatr Infect Dis J. 2003;22:677–86. doi: 10.1097/01.inf.0000078160.29072.42. [DOI] [PubMed] [Google Scholar]
- 176.Fowler VG, Jr, et al. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med. 2006;355:653–65. doi: 10.1056/NEJMoa053783. [DOI] [PubMed] [Google Scholar]
- 177.Lentino JR, Narita M, Yu VL. New antimicrobial agents as therapy for resistant gram-positive cocci. Eur J Clin Microbiol Infect Dis. 2008;27:3–15. doi: 10.1007/s10096-007-0389-y. [DOI] [PubMed] [Google Scholar]
- 178.Pan A, Lorenzotti S, Zoncada A. Registered and investigational drugs for the treatment of methicillin-resistant Staphylococcus aureus infection. Recent Pat Antiinfect Drug Discov. 2008;3:10–33. doi: 10.2174/157489108783413173. [DOI] [PubMed] [Google Scholar]
- 179.Koga T, et al. In vitro and in vivo antibacterial activities of CS-023 (RO4908463), a novel parenteral carbapenem. Antimicrob Agents Chemother. 2005;49:3239–50. doi: 10.1128/AAC.49.8.3239-3250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Parish D, Scheinfeld N. Ceftaroline fosamil, a cephalosporin derivative for the potential treatment of MRSA infection. Curr Opin Investig Drugs. 2008;9:201–9. [PubMed] [Google Scholar]
- 181.Anderson SD, Gums JG. Ceftobiprole: an extended-spectrum anti-methicillin-resistant Staphylococcus aureus cephalosporin. Ann Pharmacother. 2008;42:806–16. doi: 10.1345/aph.1L016. [DOI] [PubMed] [Google Scholar]
- 182.Shaw KJ, et al. In vitro activity of TR-700, the antibacterial moiety of the prodrug TR-701, against linezolid-resistant strains. Antimicrob Agents Chemother. 2008;52:4442–7. doi: 10.1128/AAC.00859-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dajcs JJ, et al. Lysostaphin is effective in treating methicillin-resistant Staphylococcus aureus endophthalmitis in the rabbit. Curr Eye Res. 2001;22:451–7. doi: 10.1076/ceyr.22.6.451.5486. [DOI] [PubMed] [Google Scholar]
- 184.Lawton EM, Ross RP, Hill C, Cotter PD. Two-peptide lantibiotics: a medical perspective. Mini Rev Med Chem. 2007;7:1236–47. doi: 10.2174/138955707782795638. [DOI] [PubMed] [Google Scholar]
- 185.Stapleton PD, Shah S, Ehlert K, Hara Y, Taylor PW. The beta-lactam-resistance modifier (−)-epicatechin gallate alters the architecture of the cell wall of Staphylococcus aureus. Microbiology. 2007;153:2093–103. doi: 10.1099/mic.0.2007/007807-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bubeck Wardenburg J, Schneewind O. Vaccine protection against Staphylococcus aureus pneumonia. J Exp Med. 2008;205:287–94. doi: 10.1084/jem.20072208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shinefield H, et al. Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis. N Engl J Med. 2002;346:491–6. doi: 10.1056/NEJMoa011297. [DOI] [PubMed] [Google Scholar]