Abstract
Protein modification by SUMO conjugation has emerged to be an important regulatory event. Recently, the mechanisms through which SUMO elicits its effects on target proteins have been elucidated. One of these is the noncovalent association between SUMO and coregulatory proteins via SUMO interaction motifs (SIMs). We therefore searched for additional binding proteins to elucidate how SUMO acts as a signal to potentiate novel noncovalent interactions with SUMO-binding proteins. We identified an E3 ligase, Pc2, as a SUMO-binding protein with two functionally distinct SIMs. Here, we focus on the role of SIM2 and demonstrate that it is crucial for many of the documented Pc2 functions, which converge on determining its E3 ligase activity. One role of SUMO binding in this context is the subnuclear partitioning of the active form of Ubc9 (SUMO∼Ubc9) by Pc2. The significance of the SIM2-dependent functions of Pc2 is demonstrated in the control of the precise expression of lineage-specific genes during embryonic stem cell differentiation.
The modification of proteins by conjugation with small ubiquitin-like modification (SUMO) has emerged as an important regulatory posttranslational event. Sumoylation can modulate a diverse range of cellular processes including important roles in controlling genome stability and gene transcription (7, 9, 13). Four SUMO paralogues (designated SUMO-1, -2, -3, and -4) have been identified in mammals (40). SUMO conjugation is part of an enzymatic cascade involving a heterodimeric E1-activating enzyme (SAE1/2), an E2-conjugating enzyme (Ubc9), and a growing number of distinct E3 ligases (9, 13). The activated SUMO is transferred from SAE1/2 to Ubc9 via a thioester linkage between diglycine residues at the extreme C terminus of mature SUMO proteins and the active-site cysteine of Ubc9. The SUMO moiety is subsequently ligated onto an acceptor lysine residue of a substrate in a process that can be enhanced by the involvement of an E3 ligase, although at least in vitro, Ubc9 is sufficient to promote substrate sumoylation (9, 13). The specificity of conjugation is enhanced by embedding the target lysines within the consensus sequence core motif, ΨKXE (where Ψ is a bulky hydrophobic residue) (21, 31). A number of extended SUMO consensus motifs, including the synergy control (SC) motif (41), the phosphorylation-dependent sumoylation motif (PDSM) (11, 46), and the negatively charged amino acid-dependent sumoylation motif (NDSM) (45), have been identified, which serve to further increase the specificity of substrate modification beyond this core motif. These are characterized by surrounding proline residues (SC motif) or a downstream cluster of negatively charged amino acids (NDSM) or S/TP phosphorylation sites (PDSM). However, the sumoylation of several substrates has also been demonstrated to take place on sites that do not conform to these motifs (13).
Structural and mutational analyses have revealed the importance of the ΨKXE motif for the interaction of substrates with Ubc9 and their subsequent sumoylation (9). However, this interaction confers limited substrate specificity. Indeed, an important basic patch on the surface of Ubc9 has been identified, which is required for the efficient binding and sumoylation of NDSM-containing substrate proteins and provides an additional specificity determinant (45). Structural information revealed that phosphorylation in the context of the PDSM also promotes interactions with a basic surface on Ubc9 that is distinct from the catalytic site (25). In addition, a recent study also demonstrated that the sumoylation of Ubc9 can regulate target discrimination of protein sumoylation through a mechanism involving interactions between the substrate and SUMO interaction motifs (SIMs) in SUMO-modified Ubc9 (16). A further level of specificity determination within the SUMO pathway came from the discovery of E3 ligases (9, 13). These E3 ligases physically interact with Ubc9, SUMO, and substrates, which increase the rate of SUMO conjugation to substrates. In addition, many E3 ligases are themselves sumoylated and localized to distinct subnuclear structures. For example, RanBP2 associates with the nuclear pore complex, the PIAS family of proteins is found in subnuclear bodies, and polychrome 2 (Pc2) is located in nuclear polycomb group (PcG) bodies (9, 15, 18, 28, 33).
Protein sumoylation is a dynamic process, and regulation can occur at all levels of the SUMO pathway. Indeed, a recent study demonstrated that global sumoylation events can be regulated by reactive oxygen species (ROS) by the induced formation of a reversible disulfide bridge between Ubc9 and the E1-activating enzyme (2). In a different regulatory mechanism, an E3 ligase, Pc2, can be phosphorylated by HIPK2 upon DNA damage, which in turn controls Pc2 sumoylation, intranuclear localization, and E3 ligase activity toward its substrates (32). Moreover, changes in transcription factor activity induced by the SUMO pathway have been shown to be regulated by extracellular signals (8, 48, 51).
The physiological consequences of SUMO modification are typically mediated by effector proteins that noncovalently associate with SUMO via their SIMs. In this context, the SIM acts as a docking motif for specifying interactions with a sumoylated protein. SIMs are short hydrophobic peptide sequences that, in some cases, are preceded, or followed, by a stretch of acidic residues (1, 6, 10, 20, 23, 27, 30, 36, 38, 42, 44). SIMs have been found in enzymes of SUMO machinery (10, 28, 36), SUMO substrates (20, 24, 35, 36), and proteins involved in the SUMO-dependent repression of gene transcription (20). The SIMs in these proteins have been shown to function in the assembly of nuclear bodies (36), the dynamic distribution of a protein between subnuclear compartments, the regulation of substrate sumoylation levels, the modulation of transcriptional activity (20), and the targeting of polysumoylated substrates for ubiquitin-mediated proteolysis (42, 44).
To gain more insights into the functional significance of SUMO as a signal that mediates noncovalent interactions with SUMO-binding proteins, we performed a yeast two-hybrid screen and identified Pc2 (also known as CBX4) as a novel SUMO-binding protein. Pc2 is a member of the PcG class of proteins, which typically assemble into multiprotein complexes. At least two distinct polycomb-repressive complexes (PRCs) exist, PRC1 and PRC2 (19). Chromatin binding by PRC1/2 serves to initiate and maintain the gene repression of a large cohort of developmental regulators, such as Hox gene clusters, whose activation promotes the onset of cellular differentiation. In addition, the multiple enzymatic activities within PRCs have been shown to be essential for gene silencing during development (3, 26, 29). The components of PRCs have thus been shown to be involved in embryonic stem (ES) cell (ESC) self-renewal and pluripotency (29). Pc2 is an essential component of PRC1 and has been shown to function to promote protein sumoylation through its SUMO E3 ligase activity (14, 15).
Pc2 contains two SIMs. We focused mainly on one of these (SIM2). The SIM2 motif is required for the sumoylation of both Pc2 itself and one of its substrates, CtBP1. Mechanistically, SIM2 promotes the E3 ligase activity of Pc2 and allows Pc2 to relocalize the active form of Ubc9. The functional relevance of our findings is provided by the observation that SIM2 in Pc2 is essential for its ability to control lineage-specific gene expression during mouse ESC (mESC) differentiation.
MATERIALS AND METHODS
Plasmid construction, mutagenesis, and small interfering RNA (siRNA).
The following plasmids were used in yeast two-hybrid assays. pAS2926 [pGAD-Pc2(297-558)], pAS2924 [pGAD-Pc2(297-496)], pAS2921 [pGAD-Pc2(297-404)], pAS2925 [pGAD-Pc2(400-558)], pAS2930 [pGAD-Pc2(400-544)], pAS2931 (Pc11) [pGAD-Pc2(400-536)], pAS2932 [pGAD-Pc2(400-521)], pAS2922 [pGAD-Pc2(400-496)], pAS2927 [pGAD-Pc2(420-558)], pAS2928 [pGAD-Pc2(439-558)], pAS2929 [pGAD-Pc2(455-558)], pAS2923 [pGAD-Pc2(480-558)], pAS2938 [pGAD-Pc2(455-553)], and pAS2939 [pGAD-Pc2(455-549)] were constructed by the ligation of NcoI/XhoI-cleaved PCR fragments (created with primer pairs SY73/SY78, SY73/SY77, SY73/SY76, SY74/SY78, SY74/SY100, SY74/SY101, SY74/SY102, SY74/SY77, SY97/SY78, SY98/SY78, SY99/SY78, SY75/SY78, SY99/SY120, and SY121/SY99 and template pACT-11) into the same sites of pGAD. pAS2933 [pGAD-Pc2(455-558)ΔSIM2], pAS2935 [pGAD-Pc2(455-558)T544A], pAS2936 [pGAD-Pc2(455-558)C547S], pAS2940 [pGAD-Pc2(455-558)I3A], pAS2934 [pGAD-Pc2(455-558)ΔPIDLR], pAS2937 [pGAD-Pc2(455-558)E494A], and pAS2941 [pGAD-Pc2(455-558)K553R] were constructed by QuikChange mutagenesis (Stratagene) by using the following primer pairs on template pAS2929: SY107, SY110, SY111, SY123, SY108, SY122, and SY124. pAS2920 (ypSY4) (pGBKT7-SUMO-1Δgg) was constructed by the ligation of an NcoI/SalI-cleaved PCR fragment (created with primer pair SY42/SY46 and template pAS2985) into the same sites of pGBKT7.
For bacterial expression, pAS2942 [encoding glutathione S-transferase (GST)-Pc2(459-558)], pAS2943 [encoding GST-Pc2(459-498)], and pAS2944 [encoding GST-Pc2(530-558)] were constructed by the ligation of BamHI/XhoI-cleaved PCR products (created with primer pairs SY133/SY78, SY133/SY135, and SY134/SY78 and template pAS2929) into the same sites of pGEX6P-1. pAS2974 (encoding GST-SUMO-1) was constructed by the ligation of an NcoI/SalI-cleaved PCR fragment (created with primer pair SY42/SY46 and template pAS2985) into the same sites of pGEXKG. pAS2976 (encoding GST-SUMO-2) was constructed by the ligation of a BamHI/EcoRI-cleaved PCR fragment into the same sites of pGEXKG. pAS2975 [encoding GST-SUMO-1(I34A/F36A)] was constructed by QuikChange mutagenesis using primer SY154 on template pAS2974. pAS2069 [encoding GST-Elk-1(201-260)] was described previously (49). pAS2946 [encoding His-Pc2(459-558)] was constructed by the ligation of an NcoI/XhoI-cleaved PCR fragment (created with primer pair SY99/SY78 and template pACT-11) into the same sites of pET30b. pAS2947 [encoding His-Pc2(459-558)ΔSIM2] and pAS2948 [encoding His-Pc2(459-558)I3A] were constructed by QuikChange mutagenesis using primers SY107 and SY123, respectively, on the pAS2946 template. pAS2969 (encoding His-SUMO-1) (kindly provided by V. deLorenzo) was described previously (22).
The following plasmids were used in mammalian cell transfections. pAS2949 [pCMV5-T7-Pc2(1-558)], pAS2962 [pCMV5-Flag-Pc2(1-558)], pAS2990 [pCCS2-YFP-Ubc9(1-158)], pAS2956 [pCS2-CFP-CtBP1(1-440)], and pAS2954 [pCMV5-Flag-CtBP1(1-440)] (kindly provided by D. Wotton) were described previously (14, 15). pAS2964 [pCMV5-Flag-Pc2(1-558)ΔSIM1], pAS2963 [pCMV5-Flag-Pc2(1-558)ΔSIM2], and pAS2965 [pCMV5-Flag-Pc2(1-558)ΔSIM1/ΔSIM2] were kindly provided by D. Wotton. pAS2950 [pCMV5-T7-Pc2(1-558)ΔSIM2], pAS2952 [pCMV5-T7-Pc2(1-558)K492R], pAS2953 [pCMV5-T7-Pc2(1-558)E494A], and pAS2995 [pCMV5-T7-Pc2(1-558)AILA] were created by QuikChange mutagenesis using the following primer-template combinations: SY107 and pAS2949, SY109 and pAS2949 SY122 and pAS2949, and SY160 and pAS2949, respectively. pAS2957 [peGFP-Pc2(1-558)] and pAS2958 [peGFP-Pc2(1-558)ΔSIM2] were created by the ligation of EcoRI/XbaI-cleaved products from pAS2949 and pAS2950, respectively, into the same sites of peGFP-C2. pAS2977 (pCMV-GAL-SUMO-1), pAS2978 (pCMV-GAL-SUMO-1Δgg), pAS2979 (pCMV-GAL-SUMO-2), and pAS2980 (pCMV-GAL-SUMO-2Δgg) were created by the ligation of SalI/BamHI-cleaved PCR products into the same sites of pAS571 (pCMV-GAL). pAS2981 (pHcRed1-C1-SUMO-1), pAS2982 (pHcRed1-C1-SUMO-1Δgg), pAS2983 (pHcRed1-C1-SUMO-2), and pAS2984 (pHcRed1-C1-SUMO-2Δgg) were created by the ligation of SalI/BamHI-cleaved PCR products from pAS2977, pAS2978, pAS2979, and pAS2980, respectively, into XhoI/BamHI sites of pHcRed1-C2. pAS2985 (pCDNA3-HA-SUMO-1), pAS2986 (pCDNA3-HA-SUMO-2), pAS2987 [pCDNA3-HA-SUMO-2(K11R)], and pAS2989 (pCDNA3-HA-SUMO-3) (kindly provided by Ron Hay) were described previously (43). pAS2988 [pCDNA3-HA-SUMO-2(D62R)] was created by QuikChange mutagenesis using primer SY152 on template pAS2986. pAS2424 [pCDNA3-(Myc)6-Ubc9] was constructed by the ligation of an EcoRI/XbaI-cleaved PCR product into the same sites of pCDNA3-(Myc)6. pAS2991 [pCDNA3-(Myc)6-Ubc9(R13E/K14E)], pAS2992 [pCDNA3-(Myc)6-Ubc9(H20D)], and pAS2993 [pCDNA3-(Myc)6-Ubc9(C93S)] were created by QuikChange mutagenesis using the following primers on template pAS2424: SY159, SY150, and SY152, respectively. Details of PCR primers are available upon request.
Yeast two-hybrid screen and interaction assays.
The bait protein Gal4 DNA-binding domain [Gal4(DBD)]-SUMO-1Δgg was used in a yeast two-hybrid screen against a human fetal brain cDNA library, and subsequent verification and interaction-site mapping using yeast two-hybrid assays for the indicated bait and prey proteins were performed according to the manufacturer's protocol (Clontech).
Protein-protein interaction assays.
In vitro His pulldown assays were performed by using 5 μg of recombinant His6-SUMO-1 immobilized onto 20 μl 50% (vol/vol) MagneHis Ni particles (Promega) along with 1 μg of the indicated recombinant GST fusion proteins in 100 μl of 1/2 Mega binding buffer (50 mM HEPES [pH 7.9], 5 mM imidazole, 0.025% Tween 20, and 150 mM NaCl) for 2 h at room temperature. The beads were washed three times with 800 μl of 1/2 Mega wash buffer (50 mM HEPES [pH 7.9], 10 mM imidazole, 0.025% Tween 20, and 150 mM NaCl) before bound proteins were eluted by the addition of Laemmli sample buffer. The bound species were detected by Western blotting.
GST pulldown assays were performed essentially as described previously (37) by using GST-SUMO fusion proteins expressed and purified from Escherichia coli and whole-cell lysates from 293T cells transfected with Pc2 expression constructs.
Fluorescence spectroscopy measurements.
Fluorescence measurements were performed by using an FP-750 spectrofluorimeter. Data were analyzed by Spectra Manager software and subsequently plotted with Excel. For tyrosine emission spectra, the proteins were excited at 280 nm, and emission spectra were collected between 290 and 450 nm. The peak fluorescence (tyrosine; 305 nm) was selected for further analysis.
Cell culture and transfection.
293T cells were grown and transfection experiments were carried out as described previously (50). Feeder-free E14 mouse embryonic stem cells were cultured at 37°C with 5% CO2 and were maintained on gelatin (Millipore)-coated dishes in knockout Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS), 2.5 mM Glutamax-1 supplement, 1.2 mM nonessential amino acids, 0.06 mM 2-mercaptoethanol (Gibco), and 1,000 U/ml of leukemia inhibitory factor (LIF; Millipore). A cell line stably expressing an inducible RNA interference (RNAi) construct against Pc2 (pSuperior-based plasmid) under the control of the Tet regulatory element was generated by a sequential cloning strategy. Briefly, a plasmid encoding the Tet repressor (pCDNA6-TR) was transfected into E14 murine ESCs (mESCs), and a stable line was selected by using 4 μg/ml of blasticidin in ESC medium. This cell line was subsequently transfected with a pSuperior-siPc2 construct and further selected in ESC medium containing 8 μg/ml of blasticidin and 350 μg/ml of G418.
The transfection of siRNA and overexpression plasmids was performed by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions along with 50 nM siRNA and 1 ng of expression plasmids.
Confocal microscopy imaging.
Cells were transfected with constructs encoding fluorescence protein-tagged fusion proteins as indicated. After 24 h, cells were fixed with 4% formaldehyde for 15 min at room temperature. DNA was stained by 4′,6-diamidino-2-phenylindole (DAPI). Cells were analyzed by fluorescence confocal microscopy (API Delta Vision). Images represent a projection of multiple optical confocal sections.
SUMO assays, biochemical fractionation, and Western blotting.
In vivo sumoylation assays were carried out by using either total cell lysates made in SUMO lysis buffer as described previously (5) or nickel affinity purification under denaturing conditions as described previously (47). In vitro sumoylation assays were performed as described previously (45), by using 100 ng SAE1/2 (Biomol), 2 to 4 μg of GST-SUMO-1/SUMO-2, 100 ng of Ubc9, and 1 to 2 μg of Pc2.
For biochemical fractionation, experiments were carried out as described previously (32). Briefly, cellular proteins were extracted in NP-40 lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 15 mM MgCl2, 5 mM EDTA, 1 mM dithiothreitol [DTT]) containing protease inhibitors (Roche), 10 mM N-ethylmaleimide (NEM), and 10 μg/ml E64 (Sigma). Lysates were centrifuged, and the supernatant was retained (NP-40-soluble fraction). The remaining pellets were washed twice with NP-40 lysis buffer and extracted by boiling in SDS sample buffer (insoluble fraction).
Western blotting was carried out with Supersignal West Dura Extended Duration substrate (Pierce), as described previously (50), using the primary antibodies anti-GST, anti-extracellular signal-regulated kinase (anti-ERK), anti-Myc (Santa Cruz), anti-T7 (Novagen), antihemagglutinin (anti-HA) (12CA5; Roche), anti-Flag (Sigma), anti-SUMO-1, anti-SUMO-2 (Biomol), anti-Ubc9 (Santa Cruz), and anti-Pc2 (Abgent). Data were visualized with a Bio-Rad Fluor-S MultiImager and Quantity One software (Bio-Rad).
Alkaline phosphatase staining.
The detection of alkaline phosphatase, which is indicative of undifferentiated state of ES cells, was carried out by using a commercial alkaline phosphatase detection kit (Millipore).
Quantitative RT-PCR.
Total RNA was harvested with an RNeasy kit (Qiagen). Forty nanograms of RNA was used in a one-step reverse transcription (RT)-PCR with Quantitect SYBR green reagent (Qiagen) and the following primers: 18S forward primer 5′-TCAAGAACGAAAGTCGGAGGTT-3′ and reverse primer 5′-GGACATCTAAGGGCATCACAG-3′, Pc2 forward primer 5′-ATATAACACGTGGGAACCAGAG-3′ and reverse primer 5′-TACTGATGGTGCTTCTTGCT-3′, Oct4 forward primer 5′-GTGAAGTTGGAGAAGGTGGA-3′ and reverse primer 5′-GCTGAACACCTTTCCAAAGAG-3′, Msx1 forward primer 5′-AGAAGATGCTCTGGTGAAGG-3′ and reverse primer 5′-ATAGACAGGTACTGCTTCTGG-3′, and fgf5 forward primer 5′-GATGGCAAAGTCAATGGCTC-3′ and reverse primer 5′-GTAAATTTGGCACTTGCATGG-3′. The RT-PCR primers used for the amplification of Pc2 recognize both human and mouse sequences.
Results shown are representative of at least two independent experiments measured at least in triplicate. Error bars show standard deviations (n = 3). Statistical significance was determined by using paired t tests.
RESULTS
Identification of Pc2 as a SUMO-interacting protein.
To identify proteins that interact with SUMO and hence potentially functionally couple sumoylated proteins to downstream regulatory events, a yeast two-hybrid screen of a human fetal brain cDNA library was performed. To enrich for proteins interacting in a noncovalent manner, we used a SUMO-1Δgg protein fused to the Gal4 DNA-binding domain [Gal4(DBD)] as the bait. This form of SUMO cannot be covalently conjugated to substrates. More than 1 × 106 independent colonies were screened, from which we identified 56 positive colonies, and 1 of the resulting candidates was identified as a partial cDNA sequence encoding Pc2 (amino acids 297 to 558) (Fig. 1A). No previously known SUMO-interacting proteins were identified.
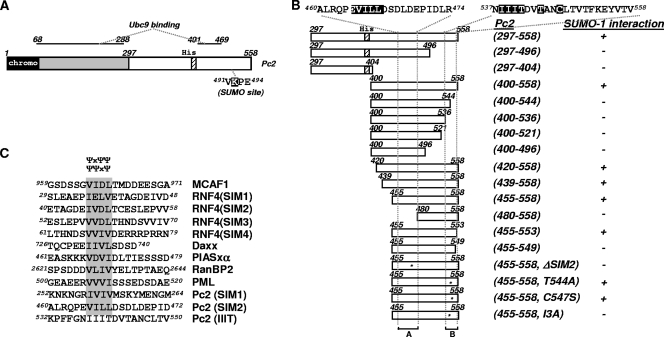
FIG. 1.
Identification of a SIM in Pc2 by yeast two-hybrid assays. (A) The domain structure of Pc2 is shown schematically: the chromodomain is indicated by a black box, the polyhistidine stretch is indicated by a striped box, and a white box indicates the region identified by a yeast two-hybrid screen. The binding regions of Ubc9 in Pc2 are indicated. (B) Schematic representation of a series of truncated Pc2 proteins fused to the Gal4(AD) in yeast expression vectors, which were used in yeast two-hybrid assays (left). The amino acids at each end of the deletion mutants are numbered. Amino acid sequences targeted for point mutation or deletion are highlighted in a black box, and their relative positions within the constructs are indicated by asterisks. The nomenclature of Pc2 expression constructs is indicated. A qualitative assessment of the relative binding of SUMO-1Δgg fused to the GAL4(DBD) is shown. “−” represents no growth, and “+” indicates growth and, hence, binding. (C) Amino acid sequence alignment of known SIMs with putative SIMs in Pc2. SIMs are shaded. The core consensus SIMs, ΨΨXΨ and ΨXΨΨ, are indicated.
To confirm this interaction, Saccharomyces cerevisiae strain AH109 was cotransformed with plasmids expressing SUMO-1Δgg and Pc2(297-558). Yeast cells that contained both Gal4(DBD)-SUMO-1Δgg (a bait) and a C-terminal fragment of Pc2 fused to the Gal4 transcription activation domain [Gal4(AD)] [Gal4(AD)-Pc2(297-558)] (an interacting prey) were able to grow. In contrast, yeast cells expressing Gal4(AD)-Pc2(297-558) and Gal4(DBD) alone in the absence of a SUMO-1 fusion did not grow (see Fig. S1A in the supplemental material). Thus, the SUMO moiety is required to observe interactions with Pc2.
Next, we sought to delineate the regions of Pc2 that bind to SUMO-1 by yeast two-hybrid assays. For this purpose, a series of truncated Pc2 proteins were constructed as Gal4(AD) fusions and tested for their abilities to interact with SUMO-1 (Fig. 1B and see Fig. S1B and C in the supplemental material). The results demonstrate that the region encompassing amino acids 455 to 558 is the minimum interacting region. Upon the deletion of either amino acids 455 to 480 or 544 to 558, the interaction is lost. The first of these regions contains the amino acid sequence EVILL, which resembles sequences that are characteristic of the recently identified SUMO interaction motifs (SIMs) (20, 30, 35, 36, 38, 44) (Fig. 1C), and we refer to this as SIM2. The second region contains the motif IIIT, which, although hydrophobic, does not match the consensus SIM sequence (Fig. 1C). To establish whether these regions act as SIMs, further mutational analyses were performed, and yeast two-hybrid analysis demonstrated that these motifs (SIM2 and the IIIT motif) are critical determinants for the binding of Pc2 to SUMO-1 (Fig. 1C and see Fig. S1D and E in the supplemental material). Importantly, both of these mutants were expressed to levels similar to those of the wild-type (WT) protein (Fig. S1F).
PML and Daxx can be sumoylated and also contain SIMs (20, 36). However, their ability to bind to SUMO was demonstrated to be independent from their ability to be modified with SUMO. As the E3 ligase Pc2 has been shown to be a SUMO substrate (15), we therefore examined whether the sumoylation status of Pc2 might influence its ability to interact with SUMO-1. The SUMO modification-defective mutant of Pc2 (E494A) displayed a positive interaction with SUMO-1 in a yeast two-hybrid assay (see Fig. S1E in the supplemental material). This indicates that, at least in the heterologous yeast system, the noncovalent interaction between Pc2 and SUMO-1 is uncoupled from the SUMO conjugation of Pc2.
Pc2 physically interacts with SUMO.
To determine whether Pc2 interacts with SUMO directly, we performed an in vitro pulldown analysis with components that were expressed in and purified from E. coli cells. Various GST-Pc2 fusion proteins were incubated with hexahistidine-tagged SUMO-1 (His-SUMO-1) immobilized on nickel-conjugated magnetic beads. Subsequently, the interactions were monitored by immunoblotting (IB). GST-Pc2(459-558) showed a strong interaction with SUMO-1 (Fig. 2A, lane 3), while the GST-Elk-1(R-motif) control was not pulled down by SUMO-1 (Fig. 2A, lane 2). To establish whether the SIM2 or IIIT motif represented a bona fide SIM and exhibited SUMO-binding properties in isolation, we divided the Pc2 fragment into two parts. SUMO binding was detectable by GST-Pc2(459-498) (containing SIM2) albeit at a reduced level compared to that of GST-Pc2(459-558). However, SUMO binding by GST-Pc2(530-558) (containing the IIIT motif) was undetectable (Fig. 2A, lanes 4 and 5). These results demonstrate that Pc2 and SUMO-1 physically associate with each other and that the region containing the SIM2 motif in Pc2 is required and sufficient for this interaction in vitro.
FIG. 2.
In vitro binding between Pc2 and SUMO. (A and B) Nickel affinity pulldown assays of the indicated His-tagged proteins. Schematic diagrams show series of bacterially expressed GST-Pc2 (A) and His6-Pc2 (B) fusion proteins used in the in vitro pulldown assays. The nomenclature of Pc2 constructs is indicated on the left. The relative positions within the constructs targeted for mutation or deletion are indicated by asterisks. A qualitative assessment of relative binding of GST-Pc2 or SUMO fusion proteins to the His6-SUMO-1 or His6-Pc2 fusion proteins is indicated on the right. “−” represents no binding, “+” illustrates weak binding, and “++” indicates strong binding. A representative immunoblot from the pulldown assays is shown underneath the schematics. (A) Interactions between Pc2 and SUMO-1 with the indicated GST-Pc2 fusion proteins and His6-tagged SUMO-1 (top) (anti-GST immunoblot [IB]). Inputs of His6-SUMO-1 (middle) (Ponceau stained) and GST-Pc2 fusion proteins (bottom) (Coomassie stained) are shown. (B) Interactions between Pc2 and SUMO-1 with the indicated GST-SUMO fusion proteins (top) (anti-GST IB). Inputs of His6-Pc2 fusion proteins (bottom) (Ponceau stained) and GST-SUMO fusion proteins (right) (Coomassie stained) are shown. (C) Concentration-dependent interactions between WT and ΔSIM2 His6-Pc2 fusion proteins (indicated on the right) and His6-SUMO-1 were analyzed by fluorescence spectrometry. The data are plotted as a change in fluorescence intensity due to SUMO binding, ΔI [I(Pc2 + SUMO-1) − (IPc2 + ISUMO-1)] normalized to ΔI at 1 μM Pc2, versus concentrations of Pc2 (x axis [Pc2]) and are presented as the averages of data from three independent experiments (standard errors of the means [SEMs] are shown; n = 3). (D) GST pulldown assays of the indicated Pc2 derivatives with GST-SUMO-1. The source of Pc2 derivatives was lysates from 293T cells transfected with the indicated full-length Pc2 proteins. Binding of Pc2 was detected by IB (top) (anti-Flag). Inputs of GST-SUMO-1 (middle) (Ponceau stained) and Pc2 (bottom) (anti-Flag immunoblot) proteins are shown. A schematic diagram showing the locations of SIM1 and SIM2 in Pc2 is shown at the top.
We next sought to establish whether SIM2 and the IIIT motif identified by yeast two-hybrid analyses are critical determinants for the direct interaction between Pc2 and SUMO-1 in the context of the intact C-terminal region of Pc2. Further GST pulldown experiments were performed. Whereas the ability of Pc2 to associate with SUMO-1 was reduced by mutating the IIIT motif (I3A mutant), no interaction was observed upon the deletion of SIM2 (ΔSIM2), which lacks the amino acid stretch EVILL (Fig. 2B, lanes 4 to 6), demonstrating a critical role for SIM2 in SUMO binding. These results were confirmed by monitoring the association kinetics of SUMO-1 with wild-type Pc2 and a ΔSIM2 mutant version of Pc2 by measuring changes in fluorescence. The binding of Pc2 was severely impaired in the ΔSIM2 mutant (Fig. 2C). In addition to SUMO-1, there are two additional functional SUMO forms in the cell, SUMO-2 and SUMO-3. All three SUMO proteins have identical structural folds, but although SUMO-2 and -3 are highly similar (88% identity) at the amino acid level, SUMO-1 is more divergent, sharing only 44% sequence identity with SUMO-2 (40). Because of this sequence deviation, we also tested the binding of Pc2 to SUMO-2 and found that Pc2 bound to SUMO-2 but with reduced levels compared to those of SUMO-1 (Fig. 2B, lanes 3 and 4, and see Fig. S2A in the supplemental material). SIM2 is also important for binding to SUMO-2 (see Fig. S2B).
We next investigated whether SIM2 is required for SUMO associations in the context of full-length Pc2. Interestingly, there is an IVIV motif (referred to as SIM1), which is located toward the N terminus of Pc2 and resembles the SIMs in other proteins (Fig. 1C) (23). A series of full-length Pc2 constructs lacking one or both of these SIMs were transfected into 293T cells, and the lysates were used in pulldown assays with GST-SUMO-1 or GST-SUMO-2 fusion proteins. As expected, the binding of full-length Pc2 to the SIM-binding-defective SUMO-1 derivative SUMO-1(I34A/F36A) was lost, validating the assay (see Fig. S2C in the supplemental material). The deletion of the either SIM2 or the IVIV motif (ΔSIM1) in Pc2 caused significant reductions in SUMO-1 (Fig. 2D, lanes 1 to 3) or SUMO-2 (see Fig. S2D) interactions. However, the residual SUMO-binding activity was abolished when both SIM1 and SIM2 in Pc2 were deleted (Fig. 2D and see Fig. S2D, lane 4, in the supplemental material).
Together, these results suggest that SIM1 and SIM2 in Pc2 are critical determinants for its association with SUMO-1 and SUMO-2 and that SIM2 is sufficient for this interaction in vitro, thus validating its role as a SIM. However, the hydrophobic patch (IIIT) does not appear to act as a classical SIM in SUMO binding but appears to work together with SIM2 to facilitate its interactions with SUMO. These observations are broadly consistent with our findings obtained by yeast two-hybrid analyses (Fig. 1), although in the latter case, in the absence of the IIIT motif, the SIM2 region was insufficient to score as a positive interaction, possibly due to falling below a critical threshold in this assay.
The SUMO-binding activity of Pc2 is required for its E3 ligase activities.
A number of reports have revealed close relationships between SUMO conjugation and SUMO-binding activity in substrates such as Ubc9, Daxx, and BLM (16, 20, 52). In addition to its function as an E3 ligase for other substrates, Pc2 itself has also been found to be a SUMO substrate; thus, SUMO binding might be important for its modification by SUMO (14, 15). In this context, Pc2 most likely acts as the relevant E3 ligase. Multiple slower-migrating forms of Pc2 representing sumoylated species were detected when Pc2 was coexpressed with three different SUMO paralogues (see Fig. S3A, lanes 6 to 8, in the supplemental material). Pc2 was more efficiently modified by SUMO-2 and -3 than by SUMO-1, although this might be attributable to the lower level of expression of SUMO-1 that we saw (data not shown). Subsequent analyses therefore used SUMO-2 to facilitate the detection of Pc2 sumoylation.
To establish whether the SUMO-binding activity is required for Pc2 sumoylation, we analyzed the sumoylation status of a mutant Pc2 construct lacking SIM2 (ΔSIM2). In comparison to wild-type Pc2, the ΔSIM2 mutant exhibited much-reduced levels of sumoylation in vivo (Fig. 3A, lane 2). Importantly, we also tested a more subtle mutant version of Pc2 in which two of the hydrophobic amino acids in SIM2 were replaced by alanine residues, and this protein was also defective in promoting autosumoylation (see Fig. S3B in the supplemental material). Furthermore, the modification of Pc2 by either SUMO-1 or SUMO-2 in vitro was reduced significantly upon the deletion of SIM2 (Fig. 3B). Thus, the sumoylation of Pc2 requires SIM2, which is implicated in noncovalently binding to SUMO.
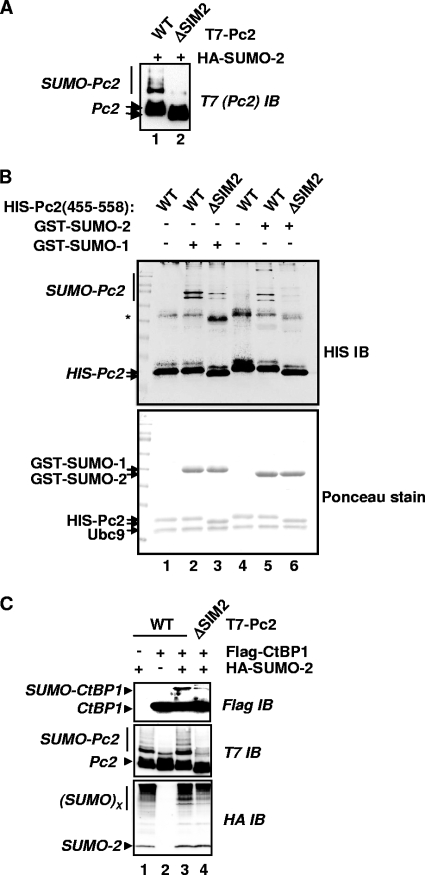
FIG. 3.
The SIM2 motif is important for Pc2 autosumoylation and substrate sumoylation. (A) The indicated WT or ΔSIM2 mutant versions of T7-Pc2(1-558) were coexpressed with HA-SUMO-2. Cell lysates were analyzed by immunoblotting (IB) with an anti-T7 antibody to detect SUMO-modified (bracket) and unmodified (arrows) Pc2. (B) In vitro sumoylation assays were performed with the indicated combinations of GST-SUMO isoforms and either WT or ΔSIM2 mutant versions of Pc2. Pc2 and its modified forms were detected by IB using anti-His antibodies (top), and input proteins were visualized by Ponceau staining of the membrane prior to IB (bottom). The asterisk likely represents anomalously migrating forms of Pc2. (C) Pc2-mediated sumoylation of CtBP1 in vivo. Wild-type or ΔSIM2 mutant versions of T7-Pc2(1-558) were coexpressed with HA-SUMO-2 and Flag-CtBP1 as indicated. Total lysates were analyzed by IB to detect total and sumoylated CtBP1 (Flag) and Pc2 (T7) or total sumoylated cellular proteins (HA).
Next, we tested whether SUMO binding via SIM2 was important in its E3 ligase activity toward other substrates. It was reported previously that CtBP1 is a substrate of Pc2 (15), and we verified that Pc2 acted as an E3 ligase in our system (see Fig. S3C in the supplemental material). We therefore compared the abilities of WT and ΔSIM2 versions of Pc2 to enhance the sumoylation status of CtBP1. In comparison to wild-type Pc2, mutant versions lacking SIM2 exhibited much-reduced E3 ligase activities, as little enhancement of CtBP1 sumoylation was observed (Fig. 3C, lane 4). As expected, a mutant version of CtBP1 lacking its sumoylation site [CtBP(KR)] showed no increases in levels of SUMO modification by Pc2 (data not shown). In addition, Pc2 sumoylation and the overall levels of protein sumoylation in the cell were also substantially reduced upon the mutation of SIM2 (Fig. 3C, middle and bottom), suggesting a wider defect in its E3 ligase activities toward itself and other substrates. Importantly, the sumoylation-defective mutant Pc2(E494A) exhibited wild-type E3 ligase activity, indicating that it was not the lack of SUMO conjugation on Pc2 itself that was the reason for the defect in E3 ligase activity toward CtBP (data not shown). This observation is consistent with data from a previous report demonstrating that the sumoylation of Pc2 is also not required for its E3 ligase activity toward another substrate, HIPK2 (32). The E3 ligase activity of Pc2 toward CtBP in vitro was also analyzed, and consistent with the results seen in vivo, SIM2 played an important role in promoting CtBP sumoylation (see Fig. S3D in the supplemental material).
Taken together, these results suggest that the SUMO-binding activity of Pc2 through SIM2 is a prerequisite for its effective sumoylation. Moreover, these results demonstrate that this SUMO-binding activity is required for the E3 ligase activity of Pc2 toward other substrates.
Pc2 coordinates the colocalization of the SUMO machinery through its SIM2.
Pc2 has been shown to exist in nuclear speckled structures, where it is thought to execute its E3 ligase functions (15, 34). Potentially, a disruption of its subcellular localization would therefore have an impact on its ability to function as an E3 ligase. We therefore tested whether the SIM2 motif is required for the localization of Pc2 into subnuclear foci. As expected, wild-type Pc2 fused to enhanced green fluorescent protein (eGFP) was detected in nuclear speckled structures (see Fig. S4 in the supplemental material). The deletion or mutation of the C-terminal 29 amino acids of Pc2 causes its delocalization from nuclear speckles (14; data not shown). However, the localization of Pc2 into distinct nuclear foci was not affected by the blocking of Pc2 sumoylation by mutating key residues in the core sumoylation motif (K492R and E494A mutants) (see Fig. S4). Similarly, the ΔSIM2 mutant was localized in nuclear speckles in a manner identical to that of the wild-type protein (see Fig. S4), and therefore, the inappropriate subcellular localization of Pc2 is unlikely to be the cause of its defective E3 ligase activity.
A previous report showed that a deletion mutant (Δ381-466) version of Pc2, which lost the ability to bind to Ubc9, failed to stimulate CtBP1 sumoylation (15). Thus, one possible mode of action of SUMO binding by SIM2 might be in facilitating Ubc9 binding and, hence, coordinating substrate-E2 interactions. Such a mode of operation would be similar to that observed for the ubiquitin system, where ubiquitin-binding motifs have been shown to recruit thioester-loaded active ubiquitin E2 enzymes (12). We therefore examined whether Ubc9 might be a potential SIM2-binding target, as Ubc9 was previously shown to itself be a target for SUMO modification and is also transiently linked to SUMO in its active site via a thioester linkage (16). As Pc2 is an E3 SUMO ligase, a mechanistic role in the localization of Ubc9 would be in keeping with its known function. We first used a biochemical approach to examine the effect of Pc2 on the subcellular localization of Ubc9. Pc2 was previously shown to reside mainly in an NP-40-insoluble fraction of the cell (15, 32). Ubc9 was expressed alone or in combination with Pc2 and/or SUMO-2, and its intracellular localization was determined. The majority of Ubc9 was found in the soluble fraction (Fig. 4B, top, lanes 1 and 2). Importantly, the coexpression of Pc2 did not affect this distribution, but in keeping with a potential role for SUMO binding in Ubc9 relocalization, a partial redistribution of Ubc9 to the insoluble compartment was observed when Pc2 and SUMO-2 were coexpressed (Fig. 4B, top, lanes 5 and 6).
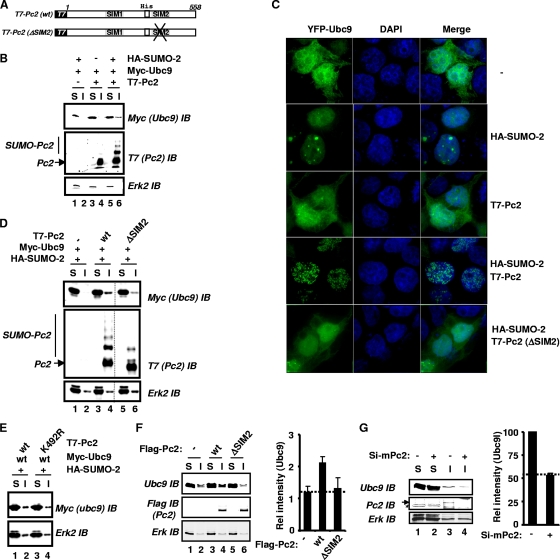
FIG. 4.
SIM2 in Pc2 is essential for the subcellular relocalization of Ubc9. (A) Schematic representation of T7-tagged Pc2 constructs used in subcellular fractionation and fluorescent microscopy studies. (B, D, and E) Wild-type, ΔSIM2, or K492R mutant versions of T7-Pc2(1-558) were expressed in combination with or without HA-SUMO-2 and Myc6-Ubc9 as indicated. Lysates were fractionated into NP-40-soluble (“S”) and insoluble (“I”) fractions, which were further analyzed by IB. The subcellular distribution of Ubc9 was analyzed by IB with an anti-Myc antibody (top panels). Total Pc2 and sumoylated Pc2 were detected by IB with an anti-T7 antibody (middle). Erk2 levels were used as a “loading control” (bottom). The relocalization of Ubc9 in D was reduced to 47.5% ± 13.4% (n = 3) with the ΔSIM2 mutant compared to WT Pc2. (C) Fluorescence images of YFP-Ubc9 fusion proteins. 293 cells were cotransfected with YFP-Ubc9 and vectors expressing the indicated versions of Pc2 and SUMO-2. Fluorescence images of cells expressing YFP-tagged Ubc9 fusion proteins (green channel) are indicated. DNA was stained with DAPI (blue channel). A merge of signals is shown on the right (over 80% of cells exhibited this colocalization). (F and G) Subcellular localization assays of endogenous Ubc9 in 293T cells (F) and mESCs (G). (F) Wild-type or ΔSIM2 mutant versions of T7-Pc2(1-558) were expressed in combination with HA-SUMO-2. (G) Pc2 levels were depleted by treating mESCs with siRNA duplexes against mouse Pc2 (mPc2). Graphs show the quantification of the amount of Ubc9 in the insoluble fraction and represent the averages of data from two independent experiments.
As an alternative way of looking at SUMO-directed Ubc9-Pc2 relocalization, the intracellular localization of a Ubc9-yellow fluorescent protein (YFP) fusion protein was examined, and the results were consistent with the results of the biochemical fractionation experiments. Ubc9 was distributed in a diffuse pattern in both the nuclear and cytoplasmic compartments in the presence or the absence of Pc2 overexpression. An increased nuclear localization of Ubc9 was seen upon the overexpression of SUMO-2, with some evidence of focus production. However, importantly, the redistribution of Ubc9 into multiple nuclear foci was obtained when both SUMO-2 and Pc2 were coexpressed (Fig. 4C), consistent with a role for the SUMO-directed relocalization of Ubc9. This effect was robust and consistently seen in cells where all components were overexpressed (see Fig. S5A in the supplemental material). Collectively, these results demonstrate that the subcellular localization of Ubc9 can be controlled through the combined action of SUMO and Pc2.
To test whether SIM2 in Pc2 might play a role in this intracellular redistribution of Ubc9, we examined the effects of the ΔSIM2 Pc2 mutant. A reduced level of Ubc9 was located in the insoluble fraction when the ΔSIM2 mutant was coexpressed along with SUMO-2 compared to wild-type Pc2 (Fig. 4D, top, lanes 3 to 6). The relocalization of Ubc9 was independent of the sumoylation status of Pc2, as an efficient relocalization of Ubc9 to the insoluble fraction was observed upon the coexpression of Pc2(K492R) (Fig. 4E). Similar effects on the Pc2-dependent redistribution of Ubc9 were observed when the ΔSIM2 mutant was tested in cell imaging experiments, as this mutant exhibited an abrogated ability to redirect the nuclear localization of Ubc9 to punctate structures (Fig. 4C). Importantly, the coexpression of wild-type and ΔSIM2 mutant versions of Pc2 was observed in cells, but despite this coexpression, only the colocalization of wild-type Pc2 and Ubc9 was observed in the subnuclear foci (see Fig. S5A in the supplemental material). The more subtle mutation of the SIM2 motif in Pc2(AILA) also blocked the ability of Pc2 to redistribute Ubc9 into multiple foci (see Fig. S5B). In contrast, a disruption of SIM1 had no effect on the Ubc9-redistributing activity of Pc2 (see Fig. S5B).
Next, we used overexpression and knockdown approaches to establish whether Pc2 was important in determining the levels of endogenous Ubc9 found in the insoluble fraction. The overexpression of wild-type Pc2 in 293T cells caused an increase in the levels of endogenous Ubc9 found in the insoluble fraction, but this increase was not observed with Pc2(ΔSIM2) (Fig. 4F, compare lanes 2, 4, and 6). Conversely, the siRNA-mediated depletion of Pc2 in murine embryonic stem cells (mESCs) caused a reduction in the levels of endogenous Ubc9 found in the insoluble compartment (Fig. 4G).
Taken together, these results demonstrate that the intracellular redistribution of Ubc9 by Pc2 is dependent on SUMO binding through SIM2.
Pc2 relocalizes the active form of Ubc9.
The redistribution of Ubc9 to the insoluble phase by Pc2 was only partial, suggesting that the Pc2 SIM2 region might play a role in partitioning a subpopulation of Ubc9 rather than bulk Ubc9 protein. One such subpopulation of Ubc9 is thioester linked to SUMO (i.e., an active form, the SUMO-loaded Ubc9). To investigate whether Pc2 might function through the active form of Ubc9 via its SIM2, we first mutated the active-site cysteine residue (C93S) in Ubc9 and tested its intracellular redistribution by Pc2 by subcellular fractionation. While wild-type Ubc9 was relocalized to the insoluble fraction by Pc2, no relocalization of Ubc9(C93S) was observed (Fig. 5B, top, lanes 5 to 8). As expected, the sumoylation of Pc2 in the presence of this Ubc9 mutant was diminished (Fig. 5B, middle, lane 8).
FIG. 5.
Thioester-bound and non-covalently-associated SUMO in Ubc9 are important for its redistribution by Pc2. (A) Schematic representation of constructs used in biochemical fractionation assays and cell imaging experiments. (B, C, E, F, and G) Fractionation experiments were performed as described in the legend of Fig. 4B with the exception that they were carried out in the presence and absence of T7-Pc2 and the presence of WT or C93S versions of Ubc9 (B), WT or Δgg versions of HcRed-SUMO-2 (C), WT or R13E/K14E mutant versions (E) or C93S and K14R mutant versions (F) of Ubc9, and combinations of WT or mutant SUMO-2 and Ubc9 (G), as indicated. (D and H) Localization of YFP-Ubc9 fusion proteins. The experiments were performed as described in the legend of Fig. 4C, with the exception that YFP-Ubc9, T7-Pc2(1-558), and WT or Δgg versions of HcRed-SUMO-2 (D) or WT or D62R versions of HA-SUMO-2 (H) were expressed in cells as indicated.
A key prediction from these data is that SUMO mutants that are defective in thioester-bound formation should also be compromised in their ability to cooperate with Pc2 in the redistribution of Ubc9. We therefore compared wild-type SUMO-2 and a diglycine deletion version of SUMO-2 (SUMO-2Δgg) for their abilities to redistribute the intracellular localization of Ubc9 in combination with Pc2. Much-reduced levels of Ubc9 were detectable in the insoluble fractions with this mutant form of SUMO-2 compared to wild-type SUMO-2 (Fig. 5C, top). Similarly, the diglycine residues in SUMO-1 are important for the redistribution of Ubc9 to nuclear speckled structures, as demonstrated by cell imaging experiments (Fig. 5D; see Fig. S6 in the supplemental material). In addition, we also examined the ability of Pc2 to relocate a Ubc9 mutant (R13E/K14E), which exhibits less SUMO loading via active-site thioester bond formation due to a significantly reduced efficiency in transferring SUMO from E1 to E2 (Ubc9) (17). Biochemical fractionation experiments showed that the relocalization of this mutant form of Ubc9 by Pc2 was also much reduced (Fig. 5E, top). As K14 is also the site for isopeptide bond-mediated SUMO conjugation to Ubc9 (16), the absence of isopeptide-linked SUMO might explain the lack of SIM-dependent recruitment of Ubc9 into the insoluble compartment by Pc2. However, this possibility was ruled out by examining the Ubc9(K14R) mutant, which was still efficiently recruited into the insoluble compartment (Fig. 5F, lane 6). Collectively, these results demonstrate that Pc2 selectively relocalizes the active thioester-linked form of Ubc9 to the insoluble compartment and subnuclear speckles.
In addition to forming a thioester linkage, Ubc9 was also found to make a noncovalent interaction with SUMO (4, 17). We therefore probed whether this interaction might also play a role in Ubc9 partitioning by Pc2 by testing two mutants, Ubc9(H20D) and SUMO-2(D62R), that are defective in this noncovalent interaction. Interestingly, a reduced redistribution of Ubc9 by Pc2 was observed when either Ubc9(H20D) was tested or SUMO-2(D62R) was cotransfected with wild-type Ubc9 in place of wild-type SUMO-2 (Fig. 5G, top, lanes 1 and 2 and 5 and 6, respectively). Similarly, the D62R mutant of SUMO-2 also failed to cooperate with Pc2 to promote the formation of nuclear foci containing Ubc9 (Fig. 5H). SUMO chain formation does not appear to play any significant role in Ubc9 partitioning, as in the presence of the SUMO-2(K11R) mutant, it was relocalized normally (Fig. 5G, top, lanes 3 and 4).
Taken together, our results demonstrate that both thioester-linked and noncovalently bound SUMO are important for the relocalization of Ubc9 into the insoluble fraction and nuclear foci by Pc2. This observation is consistent with our finding that SUMO binding via SIM2 is important for the SUMO-dependent relocalization of Ubc9 by Pc2.
Functional role of SIM2 in Pc2 in determining lineage-specific gene expression in mouse embryonic stem cells.
Polycomb-repressive complexes have been shown to be involved in the self-renewal and pluripotent properties of embryonic stem cells (ESCs) (29). As Pc2 is a component of these multiprotein complexes, we sought to investigate whether the E3 ligase activity mediated by the SUMO-binding activities of Pc2 had a role in gene regulation in mouse embryonic stem cells. First, we tested whether Pc2 was important for maintaining the overall protein sumoylation status in E14 mESCs. Notably, a significant reduction of total cellular protein sumoylation by both SUMO-1 and -2 was observed upon Pc2 depletion by siRNA (Fig. 6A). These results indicate that Pc2 indeed functions to control protein sumoylation in mESCs.
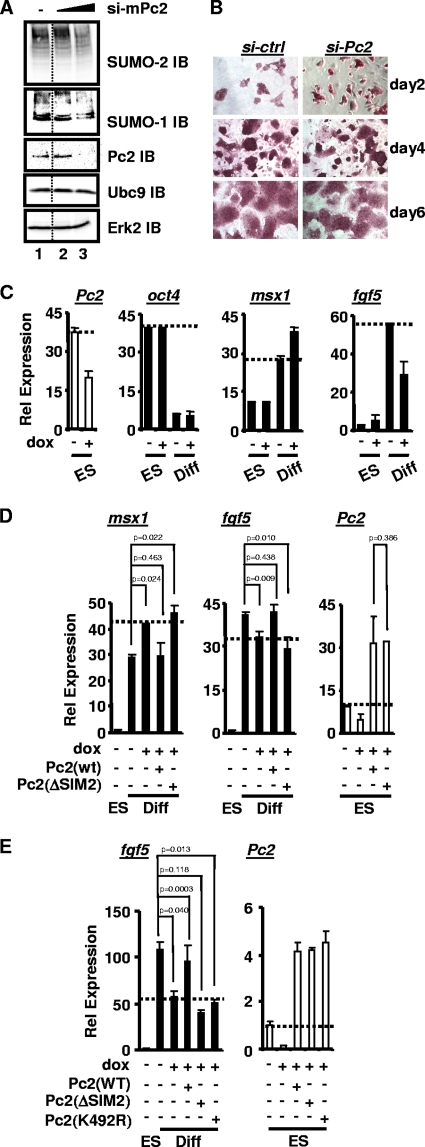
FIG. 6.
SUMO binding via SIM2 is important for Pc2 to regulate lineage-specific gene expression in mouse embryonic stem cells. (A) Effects of Pc2 depletion on total cellular protein sumoylation by either SUMO-1 or SUMO-2 in mESCs. Cells were transfected with increasing concentrations of siRNA (0, 10, and 50 nM) against mouse Pc2 (mPc2) and analyzed by IB with the indicated antibodies. IB with an anti-Erk antibody was used as a loading control (bottom). (B) mESCs were maintained in ES cell medium and stained for alkaline phosphatase activity (red staining) at the indicated times after the transfection of control (si-ctrl) or Pc2 (si-Pc2) RNAi duplexes. (C) RT-PCR analysis of gene expression in a stable mESC line that inducibly expresses RNAi against mPc2 in the presence and absence of doxycycline (dox). The data show the relative mRNA expression levels of the mESC pluripotent marker oct4 and early differentiation markers msx1 and fgf5. “ES” and “diff” indicate that mESCs were cultured in ESC medium for 1 day (ES) or grown in the absence of LIF for 5 days (diff). Data are the averages of data from three independent experiments (SEMs are shown; n = 3). (D and E) Experiments were performed as described above (C) except that siRNA-resistant WT, ΔSIM2, or K492R versions of human Pc2 were transfected. Data are averages of data from three (D) or two (E) independent experiments (SEMs are shown, and statistical significance is shown).
Next, we sought to establish a potential role of Pc2 in mESCs and first focused on their self-renewal properties. However, the depletion of Pc2 did not affect the numbers of alkaline phosphatase-stained mESCs, indicating that Pc2 does not play a significant role in their self-renewal (Fig. 6B). To determine whether Pc2 might instead play a role in mESC differentiation and lineage specification, we established stable mESC lines that inducibly express a doxycycline-inducible RNAi construct targeting Pc2. The expression profiles of a number of lineage-specific markers were monitored (Fig. 6C and data not shown). Decreased Pc2 expression was observed following doxycycline treatment (Fig. 6C). However, no significant changes in the levels of expression of any of the tested markers were observed upon the knockdown of Pc2 in the absence of differentiation signals (ES cell medium) (oct4, msx1, and fgf5) (Fig. 6B). In contrast, aberrant lineage-specific gene expression of both mesendoderm (msx1) and ectoderm (fgf5) markers was observed when Pc2 was depleted under conditions promoting spontaneous differentiation (i.e., LIF withdrawal) (Fig. 6C). The expression of a pluripotent marker (oct4) was not affected by Pc2 depletion (Fig. 6C). Taken together, these results demonstrate that Pc2 plays a key role in lineage-specific gene expression and, hence, contributes to lineage commitment decisions.
To establish whether the SUMO-binding function of Pc2 via SIM2 was important for its role in controlling lineage commitment, we asked whether the defects caused by Pc2 depletion could be rescued by the expression of wild-type human Pc2 and Pc2(ΔSIM2) cDNAs, which are refractory to knockdown by the inducible siRNA construct. The reexpression of wild-type Pc2 rescued the defects in gene expression caused by the depletion of Pc2. In contrast, no rescue was seen in the presence of the Pc2(ΔSIM2) mutant (Fig. 6D). As SIM2 has been shown to be important for the sumoylation of Pc2, we also tested whether the sumoylation site in Pc2 was functionally required by testing Pc2(K492R) in a rescue assay. However, unlike wild-type Pc2, this mutant was unable to rescue fgf5 expression following Pc2 knockdown (Fig. 6E), suggesting that at least one important functional role of SIM2 is likely through promoting Pc2 sumoylation.
Taken together, these results demonstrate that the SUMO-binding activity of Pc2 mediated through SIM2 plays a critical role in its ability to modulate lineage-specific gene expression and potential lineage choices in mESCs.
DISCUSSION
An increasing number of proteins that contain SUMO-binding motifs (SIMs) have been identified (10, 20, 36, 44, 52). These proteins act as potential adapters that recognize sumoylated protein subpopulations and drive the downstream functional consequences of sumoylation. Here, we demonstrate that an E3 ligase, Pc2, is a bona fide SUMO-binding protein that contains two functional SIMs. We focused on the C-terminal SIM, SIM2, and demonstrated that this SIM makes an important contribution to the E3 ligase activity of Pc2. This finding was recently corroborated in another study (23). We find that SIM2 in Pc2 is essential for efficient SUMO binding (Fig. 1 and 2) and is also important for its ability to promote both the sumoylation of its substrate CtBP1 and the sumoylation of Pc2 itself (Fig. 3). SIM2 is also important in determining the subcellular localization of Ubc9 in the insoluble fraction and nuclear foci (Fig. 4 and 5). This relocalizing activity of Pc2 appears to be an important determinant of its E3 ligase activity, as colocalization with Ubc9 is required for both Pc2 sumoylation and the sumoylation of CtBP1. Thus, Pc2 acts as pivotal factor in the SUMO pathway and acts as a bridging factor that binds to SUMO, and this binding activity is then transmitted into further downstream sumoylation events.
An additional spatially distinct motif was identified in the C-terminal region of Pc2, the IIIT motif, which seems to function in part through reinforcing the activities of SIM2. For example, SUMO binding through SIM2 is potentiated by the presence of the IIIT motif (Fig. 2). However, the IIIT motif does not act independently as a SIM (Fig. 2) but instead might act as a functional module with SIM2 (our unpublished data).
SUMO binding through the SIM2 motif is important for the E3 ligase activity of Pc2 both in vitro and in vivo; therefore, either free SUMO or a sumoylated target must be important for this regulatory activity. One such target in vivo appears to be Ubc9, as the level of recruitment of Ubc9 to subnuclear foci by Pc2 is much reduced when a nonconjugatable form of SUMO is introduced (Fig. 5). Thus, it appears that SIM2 functions through conjugated SUMO moieties on targets. The E2 enzyme Ubc9 is covalently linked to SUMO via a thioester linkage in its active form (9, 13). In addition, Ubc9 can noncovalently bind to SUMO and was previously shown to be modified by SUMO through isopeptide linkages (4, 16, 17). Indeed, Pc2 relocalizes Ubc9 to the insoluble compartment and to nuclear foci in a manner that is dependent on both SIM2 and the ability of Ubc9 to be able to be linked to SUMO via thioester linkages and to noncovalently bind to SUMO (Fig. 5). This is highly reminiscent of a similar mechanism seen in the ubiquitin system, where ubiquitin-binding motifs have been shown to be able to recruit ubiquitin-loaded E2 enzymes (12). Our data do, however, suggest a model whereby Pc2 acts via SIM2 to cause the redistribution of the active form of Ubc9 to nuclear foci, which can potentially function as sumoylation centers that ensure the specificity and efficiency of sumoylation (Fig. 7). The simplest model invokes a direct recruitment of SUMO-linked Ubc9, as depicted, but we cannot rule out other potential indirect mechanisms though intermediary proteins that are themselves modified by SUMO. The proposed action of Pc2 in these centers was postulated previously due to its colocalization with SUMO machinery components, but its mechanism of action in this context was not clear (15). The mechanism that we uncover is in keeping with the general model that E3 SUMO ligases act as adapters to bring the E2 enzyme (Ubc9) and substrates into close proximity for efficient SUMO conjugation (9, 13). Moreover, this proposed mode of action is also in agreement with the observation that Pc2 promotes substrate sumoylation subsequent to Ubc9 loading in vitro (14), suggesting a two-step model of relocalization, followed by promoting SUMO transfer. A further prediction of this model is that the efficiency of Pc2 as an E3 ligase promoting substrate sumoylation should be affected by SIM2. This is indeed the case in both in vitro and in vivo experiments and is supported by the observation that the SUMO E3 ligase activity of Pc2 toward CtBP1 is also dependent on the presence of SIM2. Interestingly, the sumoylated form of CtBP1 is found in the insoluble fraction, further emphasizing the potential importance of the Ubc9-relocalizing activity of Pc2 (see Fig. S3 in the supplemental material). Previous studies demonstrated that the phosphorylation of Pc2 at T495 by HIPK2 promoted its E3 ligase activity, and this site is close to SIM2 (32). It is therefore possible that this or other regulatory events might influence Pc2-SUMO interactions and hence control the E3 ligase activity of Pc2.
FIG. 7.
Model illustrating the role of SIM2 (dark gray rectangle) in Pc2 that coordinates the SUMO machinery-substrate association. Pc2 utilizes its SIM2 motif to coordinate substrate (Sub)-Ubc9 interactions in the context of PcG bodies. The SIM2 motif in Pc2 binds to SUMO linked to Ubc9 through a thioester bond and thereby recruits Ubc9 into PcG bodies.
Previous studies have identified a functionally important region of the E3 ligase RanBP2, which resembles the hydrophobic SIM (30). A combination of structural and biochemical assays suggested that mechanistically, a SIM-SUMO interaction is important for the coordination of the thioester-linked Ubc9 with substrate lysine residues. However, no role in determining the subcellular localization of SUMO pathway components was examined. Other E3 ligases in the PIAS family contain SIMs that can bind to SUMO (38). However, recent mutagenesis studies of PIAS1 indicated that these motifs have no role in their E3 ligase activity (39). Thus, although different E3 ligases contain functional SIMs, their molecular roles might differ.
Other studies have shown that SUMO-binding activity and protein sumoylation are linked in several cases, such as with thymine-DNA glycosylase (TDG), Daxx, and SP100 (16, 20, 36). However, the sumoylation status of PML was demonstrated to be independent from noncovalent SUMO interactions. Our results clearly demonstrate that SUMO-binding activity is indispensable for the efficient SUMO conjugation of Pc2 (Fig. 3). In addition, many SUMO E3 ligases are found to be sumoylated, although the function of the sumoylation of these proteins remains unclear (18). Interestingly, we have found that one of the functional consequences of Pc2 sumoylation is in promoting fgf5 expression during mESC differentiation (Fig. 6E), although it is not yet known whether the effect on this target gene is direct or indirect. However, we demonstrate that the sumoylation status of Pc2 plays no roles in (i) its ability to bind to SUMO (see Fig. S1E in the supplemental material), (ii) its subcellular localization (see Fig. S4), (iii) its ability to partition the SUMO machinery (Fig. 4E), and (iv) its E3 ligase activity toward substrates (data not shown).
Pc2 contains two SIMs, which both play a role in SUMO binding. While we have focused on SIM2, SIM1 also appears to be important for Pc2 function as an E3 ligase and also in determining its ability to relocalize other PcG proteins (23), although it still remains unclear how the two SIMs cooperate to promote SUMO binding and subsequent Pc2 functions. While the binding of mono-SUMO is mediated by these two SIMs, it is not clear whether in some instances they might work together to recognize poly- or multisumoylated species. Such a scenario was demonstrated for RNF4, which contains four SIMs that bind weakly to mono- or di-SUMO and show high selectivity for poly-SUMO-2 chains (44). Furthermore, a recent study suggested a difference in the binding of BLM to SUMO paralogues with resulting functional consequences for its own sumoylation (52). However, while BLM discriminates between SUMO paralogues, the SIM2 in Pc2 does not show any obvious discriminatory activity (Fig. 2 and see Fig. S2 in the supplemental material).
Functionally, we demonstrate the importance of SUMO binding via the SIM2 motif in the context of Pc2 function in ESC differentiation (Fig. 6). Polycomb-repressive complexes (PRCs) play an important role in controlling embryonic stem cell properties, and Pc2 is part of these multiprotein complexes (19, 29). The identification of subnuclear foci as potential centers for protein sumoylation through the E3 ligase activity of Pc2 suggests that other components within PRCs may be subjected to SUMO modification and, thus, might lead to the regulation of the activities of PRCs. Indeed, in silico analyses indicate that a number of components of PcG proteins contain SUMO consensus sites. Our data suggest that the sumoylation of Pc2 is important for its role in specifying cell fate, but in addition, it is tempting to speculate that the E3 ligase activity of Pc2 might contribute to the regulation of the activities of other components of the PRCs and, thus, contribute to ESC differentiation. Indeed, we show that lineage-specific markers are aberrantly expressed upon the depletion of Pc2, and this effect is specifically through a loss of the SUMO-binding activity of Pc2 (Fig. 6). One functional consequence of the loss of SUMO-binding activity is the reduced sumoylation of Pc2 itself. This appears likely to be one of the important downstream functional consequences, as a Pc2 protein, which cannot be sumoylated (K492R mutant), is also defective in lineage-specific gene expression (Fig. 6E). Although the role of SIM2 in determining Pc2 function in ESCs is at least partially determined by modulating its E3 ligase activities, we cannot exclude additional roles through interactions with other SUMO-conjugated proteins. Further studies will be needed to identify the potential targets of Pc2 in the context of ESCs, which are subject to SUMO modification.
In summary, our findings demonstrate a novel important role for noncovalent SUMO binding via a SIM in promoting the activity of the E3 ligase Pc2. We provide novel mechanistic insights into how substrate selectivity can be achieved, at least in part, through the partitioning of a specific active E2-E3 complex into specialized subnuclear compartments. It remains to be determined whether the E1 SUMO-activating enzyme is also partitioned in subnuclear structures. The functional importance of SIM-SUMO interactions in promoting the E3 ligase activity of Pc2 is demonstrated in the context of the regulation of lineage-specific gene expression in mouse embryonic stem cells.
Supplementary Material
Acknowledgments
We thank Peter March for the excellent advice in cell imaging experiments; Tom Jowitt for advice and help with fluorescence spectroscopy studies; Alan Whitmarsh, Gino Poulin, and members of our laboratory for comments on the manuscript and stimulating discussions; Karren Palmer, Jane Kott, and Marj Howard for excellent technical support; Ron Hay, David Wotton, and Chris Ward for reagents; Chris Ward for advice on stem cell growth; and David Wotton for discussion of unpublished data.
This work was supported by grants from the Wellcome Trust and a Royal Society-Wolfson award to A.D.S.
Footnotes
Published ahead of print on 22 February 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baba, D., N. Maita, J. G. Jee, Y. Uchimura, H. Saitoh, K. Sugasawa, F. Hanaoka, H. Tochio, H. Hiroaki, and M. Shirakawa. 2005. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435:979-982. [DOI] [PubMed] [Google Scholar]
- 2.Bossis, G., and F. Melchior. 2006. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell 21:349-357. [DOI] [PubMed] [Google Scholar]
- 3.Cão, R., Y. Tsukada, and Y. Zhang. 2005. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20:845-854. [DOI] [PubMed] [Google Scholar]
- 4.Capili, A. D., and C. D. Lima. 2007. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J. Mol. Biol. 369:608-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalkiadaki, A., and I. Talianidis. 2005. SUMO-dependent compartmentalization in promyelocytic leukemia protein nuclear bodies prevents the access of LRH-1 to chromatin. Mol. Cell. Biol. 12:5095-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geiss-Friedlander, R., and F. Melchior. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 12:947-956. [DOI] [PubMed] [Google Scholar]
- 7.Gill, G. 2005. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15:536-541. [DOI] [PubMed] [Google Scholar]
- 8.Guo, B., S. H. Yang, J. Witty, and A. D. Sharrocks. 2007. Signalling pathways and the regulation of SUMO modification. Biochem. Soc. Trans. 35:1414-1418. [DOI] [PubMed] [Google Scholar]
- 9.Hay, R. T. 2005. SUMO: a history of modification. Mol. Cell 18:1-12. [DOI] [PubMed] [Google Scholar]
- 10.Hecker, C. M., M. Rabiller, K. Haglund, P. Bayer, and I. Dikic. 2006. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281:16117-16127. [DOI] [PubMed] [Google Scholar]
- 11.Hietakangas, V., J. Anckar, H. A. Blomster, M. Fujimoto, J. J. Palvimo, A. Nakai, and L. Sistonen. 2006. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U. S. A. 103:45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoeller, D., C. M. Hecker, S. Wagner, V. Rogov, V. Dötsch, and I. Dikic. 2007. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol. Cell 26:891-898. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355-382. [DOI] [PubMed] [Google Scholar]
- 14.Kagey, M. H., T. A. Melhuish, S. E. Powers, and D. Wotton. 2005. Multiple activities contribute to Pc2 E3 function. EMBO J. 24:108-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kagey, M. H., T. A. Melhuish, and D. Wotton. 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113:127-137. [DOI] [PubMed] [Google Scholar]
- 16.Knipscheer, P., A. Flotho, H. Klug, J. V. Olsen, W. J. van Dijk, A. Fish, E. S. Johnson, M. Mann, T. K. Sixma, and A. Pichler. 2008. Ubc9 sumoylation regulates SUMO target discrimination. Mol. Cell 31:371-382. [DOI] [PubMed] [Google Scholar]
- 17.Knipscheer, P., W. J. van Dijk, J. V. Olsen, M. Mann, and T. K. Sixma. 2007. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 26:2797-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotaja, N., U. Karvonen, O. A. Jänne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22:5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine, S. S., I. F. King, and R. E. Kingston. 2004. Division of labor in polycomb group repression. Trends Biochem. Sci. 29:478-485. [DOI] [PubMed] [Google Scholar]
- 20.Lin, D. Y., Y. S. Huang, J. C. Jeng, H. Y. Kuo, C. C. Chang, T. T. Chao, C. C. Ho, Y. C. Chen, T. P. Lin, H. I. Fang, C. C. Hung, C. S. Suen, M. J. Hwang, K. S. Chang, G. G. Maul, and H. M. Shih. 2006. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24:341-354. [DOI] [PubMed] [Google Scholar]
- 21.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 22.Mencia, M., and V. deLorenzo. 2004. Functional transplantation of the sumoylation machinery into Escherichia coli. Protein Expr. Purif. 37:409-418. [DOI] [PubMed] [Google Scholar]
- 23.Merrill, J. C., T. A. Melhuish, M. H. Kagey, S.-H. Yang, A. D. Sharrocks, and D. Wotton. 2010. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 ligase activity. PLoS One 5:e8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minty, A., X. Dumont, M. Kaghad, and D. Caput. 2000. Covalent modification of p73α by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 275:36316-36323. [DOI] [PubMed] [Google Scholar]
- 25.Mohideen, F., A. D. Capili, P. M. Bilimoria, T. Yamada, A. Bonni, and C. D. Lima. 2009. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat. Struct. Mol. Biol. 16:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasini, D., A. P. Bracken, M. R. Jensen, E. Lazzerini-Denchi, and K. Helin. 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23:4061-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perry, J. J., J. A. Tainer, and M. N. Boddy. 2008. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 33:201-208. [DOI] [PubMed] [Google Scholar]
- 28.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 29.Rajasekhar, V. K., and M. Begemann. 2007. Roles of polycomb group proteins in development and disease: a stem cell perspective. Stem Cells 10:2498-2510. [DOI] [PubMed] [Google Scholar]
- 30.Reverter, D., and C. D. Lima. 2005. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435:687-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez, M. S., C. Dargemont, and R. T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276:12654-12659. [DOI] [PubMed] [Google Scholar]
- 32.Roscic, A., A. Möller, M. A. Calzado, F. Renner, V. C. Wimmer, E. Gresko, K. S. Lüdi, and M. L. Schmitz. 2006. Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol. Cell 24:77-89. [DOI] [PubMed] [Google Scholar]
- 33.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satijn, D. P. E., D. J. Olson, J. van der Vlag, K. M. Hamer, C. Lambrechts, H. Masselink, M. J. Gunster, R. G. Sewalt, R. van Driel, and A. P. Otte. 1997. Interference with the expression of a novel human polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol. Cell. Biol. 17:6076-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekiyama, N., T. Ikegami, T. Yamane, M. Ikeguchi, Y. Uchimura, D. Baba, M. Ariyoshi, H. Tochio, H. Saitoh, and M. Shirakawa. 2008. Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J. Biol. Chem. 51:35966-35975. [DOI] [PubMed] [Google Scholar]
- 36.Shen, T. H., H. K. Lin, P. P. Scaglioni, T. M. Yung, and P. P. Pandolfi. 2006. The mechanisms of PML-nuclear body formation. Mol. Cell 24:331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shore, P., and A. D. Sharrocks. 1994. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 14:3283-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song, J., Z. Zhang, W. Hu, and Y. Chen. 2005. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 280:40122-40129. [DOI] [PubMed] [Google Scholar]
- 39.Stehmeier, P., and S. Muller. 2009. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell 33:400-409. [DOI] [PubMed] [Google Scholar]
- 40.Su, H., and S. Li. 2002. Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene 296:65-73. [DOI] [PubMed] [Google Scholar]
- 41.Subramanian, L., M. D. Benson, and J. A. Iñiguez-Lluhí. 2003. A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein alpha inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J. Biol. Chem. 14:9134-9141. [DOI] [PubMed] [Google Scholar]
- 42.Sun, H., J. D. Leverson, and T. Hunter. 2007. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26:4102-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35374. [DOI] [PubMed] [Google Scholar]
- 44.Tatham, M. H., M. C. Geoffroy, L. Shen, A. Plechanovova, N. Hattersley, E. G. Jaffray, J. J. Palvimo, and R. T. Hay. 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10:538-546. [DOI] [PubMed] [Google Scholar]
- 45.Yang, S. H., A. Galanis, J. Witty, and A. D. Sharrocks. 2006. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 25:5083-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, X. J., and S. Grégoire. 2006. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol. Cell 23:779-786. [DOI] [PubMed] [Google Scholar]
- 47.Yang, S. H., E. Jaffray, R. T. Hay, and A. D. Sharrocks. 2003. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell 12:63-74. [DOI] [PubMed] [Google Scholar]
- 48.Yang, S. H., E. Jaffray, B. Senthinathan, R. T. Hay, and A. D. Sharrocks. 2003. SUMO and transcriptional repression: dynamic interactions between the MAP kinase and SUMO pathways. Cell Cycle 2:528-530. [DOI] [PubMed] [Google Scholar]
- 49.Yang, S. H., and A. D. Sharrocks. 2004. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13:611-617. [DOI] [PubMed] [Google Scholar]
- 50.Yang, S. H. and A. D. Sharrocks. 2005. PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J. 24:2161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, S. H., and A. D. Sharrocks. 2006. Interplay of the SUMO and MAP kinase pathways. Ernst Schering Res. Found. Workshop 57:193-209. [DOI] [PubMed] [Google Scholar]
- 52.Zhu, J., S. Zhu, C. M. Guzzo, N. A. Ellis, K. S. Sung, C. Y. Choi, and M. J. Matunis. 2008. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem. 283:29405-29415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.