Abstract
Mimivirus, a virus infecting Acanthamoeba, is the prototype of the Mimiviridae, the latest addition to the nucleocytoplasmic large DNA viruses. The Mimivirus genome encodes close to 1000 proteins, many of them never before encountered in a virus, such as four amino-acyl tRNA synthetases. To explore the physiology of this exceptional virus and identify the genes involved in the building of its characteristic intracytoplasmic “virion factory,” we coupled electron microscopy observations with the massively parallel pyrosequencing of the polyadenylated RNA fractions of Acanthamoeba castellanii cells at various time post-infection. We generated 633,346 reads, of which 322,904 correspond to Mimivirus transcripts. This first application of deep mRNA sequencing (454 Life Sciences [Roche] FLX) to a large DNA virus allowed the precise delineation of the 5′ and 3′ extremities of Mimivirus mRNAs and revealed 75 new transcripts including several noncoding RNAs. Mimivirus genes are expressed across a wide dynamic range, in a finely regulated manner broadly described by three main temporal classes: early, intermediate, and late. This RNA-seq study confirmed the AAAATTGA sequence as an early promoter element, as well as the presence of palindromes at most of the polyadenylation sites. It also revealed a new promoter element correlating with late gene expression, which is also prominent in Sputnik, the recently described Mimivirus “virophage.” These results—validated genome-wide by the hybridization of total RNA extracted from infected Acanthamoeba cells on a tiling array (Agilent)—will constitute the foundation on which to build subsequent functional studies of the Mimivirus/Acanthamoeba system.
Mimivirus, a nucleocytoplasmic large double-stranded DNA (dsDNA) virus (NCLDV) infecting Acanthamoeba hosts, is the largest known virus in particle size (750 nm) and genome complexity (Claverie et al. 2009a). Its 1.2-Mb genome was predicted to encode 911 proteins, among which fewer than 300 have assigned functions (Raoult et al. 2004). Besides exhibiting a gene content larger than that of many intracellular parasitic bacteria, Mimivirus possesses many proteins not encountered in any other viruses, including central components of the protein translation apparatus, thought to be the signature of cellular organisms (Abergel et al. 2007; Claverie et al. 2009a). The unique features of Mimivirus revived the debate about the evolution of DNA viruses (Claverie 2006; Claverie et al. 2006; Iyer et al. 2006) and their position in the Tree of Life (Brüssow 2009; Claverie and Ogata 2009; Moreira and Lopez-Garcia 2009). The complex Mimivirus particle has been the object of detailed proteomic (Renesto et al. 2006) and ultrastructural studies (Zauberman et al. 2008; Xiao et al. 2009), and several individual gene products have been characterized (for review, see Claverie and Abergel 2009); however, a systemic description of the molecular processes at work during the intracellular Mimivirus replication cycle (in particular the eclipse phase) is missing. During this phase, the Acanthamoeba cells are instructed by the infecting Mimivirus to build up a giant organelle-like “virion factory” that appears to centralize most of the metabolic processes leading to the synthesis of new Mimivirus particles (Suzan-Monti et al. 2007; Claverie and Abergel 2009; Claverie et al. 2009b). The functional similarity of these intracytoplasmic virion factories with a cell nucleus (sequestering the DNA replication and transcription apparatus, but devoid of translational and energy producing capacity), is at the origin of fascinating but highly controversial theories on the role that large DNA viruses might have played in the emergence of the eukaryotes by providing the ancestral nucleus to primitive cells (Villarreal and DeFilippis 2000; Takemura 2001; Forterre 2006). Identifying the cellular and viral functions at work during the de novo construction of Mimivirus factories, as well as the functions emulated within them, is thus key in understanding the physiology and the evolutionary origin of this unique virus, as well as of other NCLDVs.
In the present study, we used massively parallel pyrosequencing on the 454 Life Sciences (Roche) FLX platform to sample the transcriptome of infected Acanthamoeba castellanii cells at various time points over the entire replication cycle of Mimivirus. This first application of the deep mRNA sequencing approach to the study of a large DNA virus provided a wealth of information, all at once. First, it confirmed the reality and location of most Mimivirus genes previously annotated using a purely bioinformatic protocol (Raoult et al. 2004), allowed the correction of sequence errors, and refined the exon–intron structure of a few genes. More unexpectedly, it also revealed 75 unpredicted polyadenylated transcripts including several noncoding RNAs. Mimivirus genes were found expressed across a wide dynamic range and according to three main temporal classes: early, intermediate, and late. Finally, the precise delineation of the 5′ and 3′ extremities of Mimivirus mRNAs validated a previously predicted early promoter element (Suhre et al. 2005), confirmed the “hairpin rule” obeyed by Mimivirus mRNA polyadenylation sites (Byrne et al. 2009), and led to the discovery of a late promoter element also prominent in the genome of the recently described Mimivirus “virophage” (La Scola et al. 2008).
Given the richness of this new type of experimental data that can be queried from many different angles, we made a particular effort at providing our entire data set on an interactive public Internet server, offering a genome-wide view of Mimivirus complex transcriptional landscape that can now be used as a basis for all subsequent functional studies of the Mimivirus/Acanthamoeba system (http://www.igs.cnrs-mrs.fr/mimivirus/).
Results
Mapping of Mimivirus transcripts
cDNAs were synthesized using a Clontech SMART protocol optimized for 454 GS FLX sequencing from the polyadenylated RNA fraction of A. castellanii cells at T = −15 min, 0, 1.5 h, 3 h, 6 h, 9 h, and 12 h after their infection by Mimivirus (Supplemental Fig. S1). Our protocol generated small recognizable sequence tags at both the 3′ and 5′ extremities of the cDNAs that were used to determine the likely strand origin of transcripts (see Supplemental Methods). We generated a total of 633,346 reads (average length = 230 ± 8 nucleotides [nt]) of which 322,904 were unambiguously mapped on the Mimivirus genome sequence, while ∼85% of the remaining reads were mapped onto the available (albeit unfinished and unannotated) A. castellanii genome sequence. All previously predicted Mimivirus protein-coding genes (Raoult et al. 2004) were validated by at least one read overlapping the open reading frame (ORF). The number of reads was found to be very unevenly distributed among genes (average number = 203.7 ± 25, median = 45), most likely reflecting large differences in their level of expression (max = 16140, min = 1) (Supplemental Fig. S2). The mapping of the 454 reads allowed the correction of errors in the Mimivirus genome sequence and refinement of the intron–exon structure of a few key genes (such as the major capsid protein L425 and the L244 RNA polymerase small subunit) (Supplemental Fig. S3). Reads matching the predicted Mimivirus six tRNA genes were also found, confirming that all Mimivirus tRNAs are expressed as polyadenylated transcripts (Byrne et al. 2009). The analysis of the genomic positions mapped by the reads exhibiting the 3′ end tags (carefully filtering out the cases corresponding to putative internal priming; Supplemental Fig. S4) led to the precise delineation of the 3′ untranslated region (UTR) of 551 Mimivirus genes. The length of these 3′UTRs was found to be distributed along a nicely bell-shaped distribution (average = 44 ± 20 nt) except for some outliers (N = 60) with lengths ranging from 102 nt to 894 nt (Supplemental Fig. S5). Some of these apparently extra long 3′UTRs correspond to the transcripts of genes not included in the initial genome annotation (see below). As previously reported, 82.3% of these precisely mapped 3′UTRs obey the “hairpin rule,” i.e., end within a palindromic sequence signal (Byrne et al. 2009). Symmetrically, the analysis of the genomic positions mapped by the reads exhibiting the 5′ end tags allowed the precise delineation of the 5′ UTR of 479 Mimivirus genes, thus facilitating the search for promoter elements (see below). The length of most of these 5′ UTRs follows a Poisson-like distribution (average = 16) except for 38 outliers (longer than 70 nt) with lengths ranging from 72 nt to 1507 nt (Supplemental Fig. S5). Again, the longest of these 5′ UTRs encompass the transcripts of genes missed in the initial genome annotation (see below). Combining the 5′ and 3′ end analyses, the complete transcripts of 349 Mimivirus genes were mapped to the genome at a single nucleotide level (Supplemental Fig. S6; http://www.igs.cnrs-mrs.fr/mimivirus/).
Discovery of 75 new genes/transcripts
A number of reads, including some highly abundant, were found to unambiguously match the Mimivirus genome sequence at locations not previously annotated as genes (i.e., intergenic regions or unidentified reading frames [URFs]), or overlapping annotated ORFs but apparently transcribed from the antisense strand. For these unexpected sets of reads to be retained as evidence for bona fide new transcripts, three stringent criteria were applied: (1) the beginning of the new transcript had to be defined by at least five overlapping reads exhibiting the 5′ end tag; (2) the end of the transcript had to be defined by at least five overlapping reads exhibiting the 3′ end tag (i.e., specific for poly(A)+ mRNAs); and (3) the new transcript delimited by these 5′ and 3′ boundaries had to correspond to an uninterrupted tiling of contiguous reads. Using these constraints, 75 new transcriptional units (genes) were defined in the Mimivirus genome (Supplemental Table S1). These new polyadenylated transcripts were mostly located within intergenic regions significantly larger (average length = 631 nt, P < 10−7) than average (i.e., 150 nt), thus, filling up genome segments of previously lower gene density. On the basis of their coding potential (see Methods), 49 of these new transcripts were classified as putative protein coding genes (length = 376 ± 31, median = 303), and 26 as noncoding RNAs (ncRNAs, length = 329 ± 62, median = 168). The hybridization of total RNA extracted from infected Acanthamoeba cells on a tiling array chip (Agilent) covering the full Mimivirus genome sequence unambiguously validated 74 out of 75 new transcripts (Supplemental Fig. S7). Interestingly, one of these newly defined transcripts corresponds to URF277, the product of which was detected in the Mimivirus particle (Claverie et al. 2009a). Taking these newly discovered genes into account, the transcribed fraction of the Mimivirus genome is now established at 95%, and the total gene number at 981 (900 annotated proteins + 6 tRNAs + 75 new transcripts). These results do not rule out additional Poly(A)− transcripts that will be the target of a specific analysis.
The five most expressed new genes (in term of total number of reads; Supplemental Table S2; Supplemental Fig. S7) correspond to five different situations in respect to the Mimivirus genome map. A 360-nt-long transcript (defined by 19,511 reads) is located between the L633 and R634 genes, overlapping and antisense to URF242. Now referred to as R633b, this G + C-rich gene putatively encodes a short 60-amino-acid protein of unknown function. The second most expressed new gene (R549b) is 480-nt long and corresponds to 7677 reads. It is located in between R549 and L550 and, if translated, would correspond to a short 42-amino-acid protein without database homolog. The third most expressed new gene (R750b) is a 1023-nt-long putatively coding transcript, defined by 5248 reads. No functional attributes could be associated with the corresponding putative protein sequence. The fourth most expressed new gene (3671 reads) was detected as a short (127-nt) noncoding transcript (L1b), partially overlapping and antisense to the previously annotated R1 gene. The fifth most expressed new gene (with 2850 reads), R513b, is predicted as coding and precisely overlaps (and thus validates) the previously defined URF197. In summary, highly expressed new transcripts of the following five categories were found: antisense to an ORF; antisense to an URF; validating an URF; or fully intergenic transcripts predicted as coding or noncoding.
Host versus Mimivirus transcriptional activity
The transcriptional activity of each Mimivirus gene was estimated from the proportion of their cognate reads (restricted to those overlapping with the ORF) among the total number of reads (matching or not the Mimivirus genome) generated at each time point from the poly(A)+ RNA content of A. castellanii cells undergoing Mimivirus infection. The variation of these read proportions over the time of infection was used to approximate the transcription profile of the corresponding genes. Only 841 genes totaling at least 10 reads were included into our subsequent analyses of expression patterns and of the correlated regulatory elements.
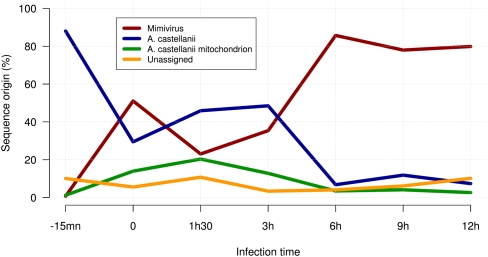
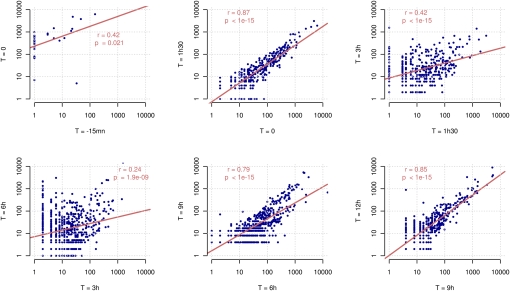
The overall proportion of host versus Mimivirus transcripts is shown in Figure 1. As expected, nearly 90% of the reads generated at T = −15 min correspond to Acanthamoeba genes. At this time, most of the virus particles have not yet delivered their content into the host cytoplasm (Fig. 2A). However, the situation changes dramatically at T = 0, for which more than 50% of the transcripts are already of Mimivirus origin. Another noticeable feature of this very early phase in the infection process is a burst in the transcription of mitochondrial genes that account for nearly half of the transcripts of Acanthamoeba origin. The proportion of Mimivirus transcripts then goes down to about 20% at T = 1.5 h (while the proportion of mitochondrial transcripts continues to increase). Interestingly, this period of lower—but well-detectable—transcriptional activity of Mimivirus genes corresponds to a true eclipse phase, during which no Mimivirus-induced intracytoplasmic structure can be seen by electron microscopy (Fig. 2C). However, after T = 3 h, when virion factories become well visible in infected cells (Fig. 2D), Mimivirus genes start dominating the transcriptional activity, and the proportion of cellular transcripts quickly drops to the 10% range, and even less for mitochondrial transcripts (Fig. 1). The less than 10% of unmapped reads correspond to sequences too short to be confidently aligned and/or to missing parts in the current partial assembly of the A. castellanii genome. The sharp transition that appears to occur around T = 3 h in the transcriptional regime is further confirmed and well visualized by the pairwise comparison of Mimivirus gene transcription levels across the successive time points (Fig. 3). The largest variation in the overall transcription pattern clearly occurs between T = 1.5 h and T = 3 h and between T = 3 h and T = 6 h.
Figure 1.
Host versus Mimivirus reads over infection time. Relative number of reads confidently mapped on Mimivirus and A. castellanii (nuclear and mitochondrial) genomes during the infection.
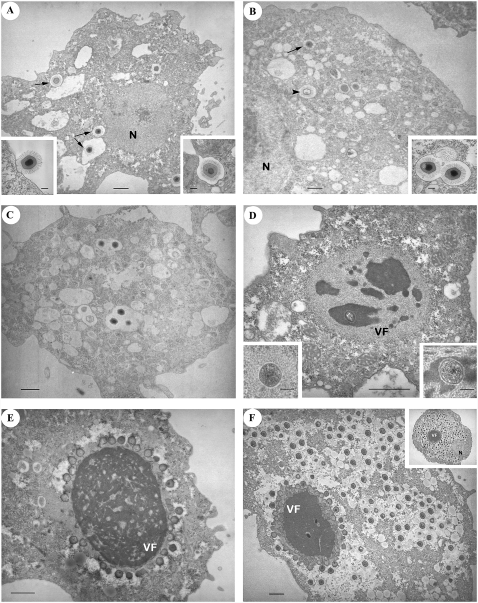
Figure 2.
Progression of A. castellanii infection by Mimivirus over time. (A) T = −15 min: Some virus particles are inside vacuoles in the cytoplasm (arrows). (Left inset) Virus particles are sparsely found in contact with the cytoplasmic membrane. (Right inset) Phagocytosis of a virus particle. (B) T = 0: After 30 min of incubation with a large excess of virus (multiplicity of infection = 1000), the phagocytic vacuoles contain a mixture of empty (arrowheads) and intact (arrows) virus particles (probably not contributing to the measured viral transcripts). (Inset) Several particles can be gathered in the same vacuole. (C) T = 1.5 h. No major change is observed compared with T = 0. Both empty and intact viruses are still visible. (D) T = 3 h: The early virion factory appears as a gray structure, with a fibrous-like aspect, surrounding darker areas. A circular structure (the “seed”) is visible in one of these areas. (Left inset) In some cells, the “seed” is surrounded by the fibrous-like structure only. (Right inset) Higher magnification of the “seed” surrounded by dark matter. (E) T = 6 h: The fully mature virion factory now dominates the picture. Numerous particles are budding from its surface; most capsids are still empty. (F) T = 9 h: A large number of mature (hairy + DNA) virus particles are filling the cytoplasm. New particles are still produced by the virion factory. (Inset) T = 12 h: Ultimate stage of virion production. Panels: bar = 1 μm; insets: bar = 0.2 μm. (N) Nucleus; (VF) virion factory.
Figure 3.
Pairwise comparison of expression profiles between successive time points. Gene expression is measured in normalized read counts. The scatter plots show the expression of genes (blue points) between successive time points during the infection from early phases (top left) to late phases (bottom right). Linear regressions on the log-transformed values, as well as the correlation coefficients and P-values, are shown on each graph.
Transcription profiling of Mimivirus genes
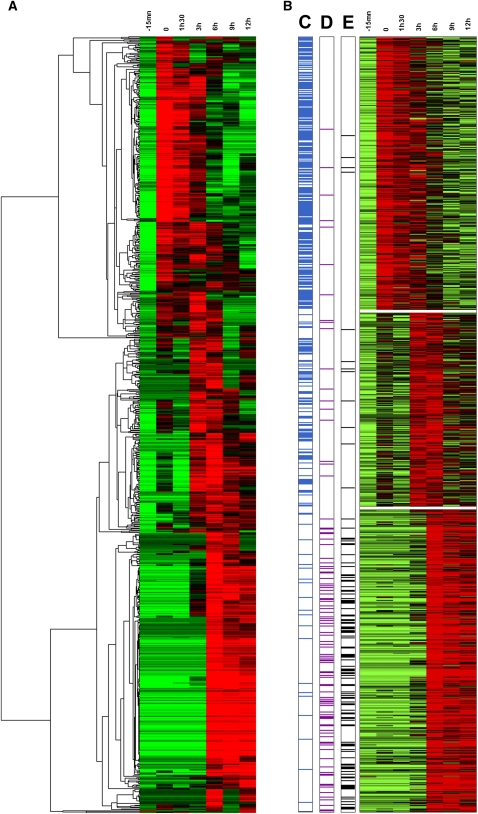
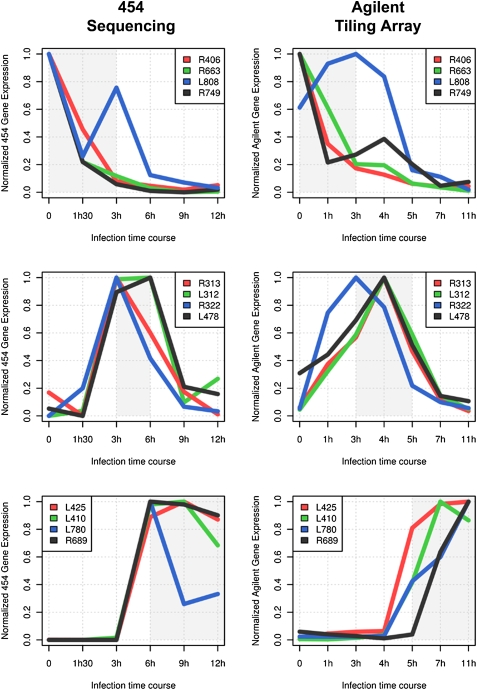
As is customary in sequence tag-based transcriptome studies, a transcription profile for each gene was derived—following a normalization procedure—from the counts of its cognate reads at each time point. These numbers could then be treated as gene “coordinates” in seven-dimensional space and used in a variety of classical statistical methods allowing the pairwise comparison of gene profiles (e.g., distance or correlation indices), their “clustering” into groups sharing similar profiles, as well as the identification and visualization of the dominant transcriptional patterns. Figure 4A presents a combined heat map/hierarchical clustering of 841 Mimivirus genes (with read counts ≥ 10) based on their expression levels at the seven time points. This traditional data-mining procedure was sufficient to clearly display three dominant patterns in the transcriptional program of Mimivirus-infected Acanthamoeba cells: about one third (Fig. 4A, top) of the genes are strongly expressed as soon as Mimivirus enters the cytoplasm (T = 0) and maintain their transcription until T = 3 h, another third (Fig. 4A, middle) begin their expression at T = 3 h and maintain it until T = 6 h, and the remaining third (Fig. 4A, bottom) start to be transcribed at T = 6 h and maintain their activity through the remaining of the virus infection cycle (T = 9 h and T = 12 h). To validate this visual pattern, the same data set was more objectively segmented using the k-means clustering method. Briefly, considering each Mimivirus gene as a seven-dimensional vector, the k-means procedure partition them into k clusters, so as to minimize the sum of squared distances between all vectors assigned to a given cluster and the cluster center. Figure 4B exhibits the transcription profiles of the 841 Mimivirus genes once optimally assigned to three expression classes. These k-means-defined clusters correspond to the three temporal expression classes already visible through a hierarchical clustering and coincide with the traditional classification of viral genes into “early,” “intermediate,” and “late.” The optimality of the above partitioning was confirmed by a variety of quality indices (see Methods). However, a closer inspection of the genes clustered in the same expression class indicates that their individual transcription profiles are not superimposable (Fig. 5; Supplemental Fig. S8), suggesting a finer regulation.
Figure 4.
Main Mimivirus gene expression classes. (A) Heat map of Mimivirus gene expression profiles. Rows correspond to the 841 analyzed genes and columns to the seven infection time points. Expression intensities are displayed from green (low expression) to red (high expression). Expression profiles are clustered using hierarchical clustering (see Methods for details). A dendrogram of the clustering is shown on the left. (B) Heat map of the same expression profiles partitioned into three main classes, “early” (top), “intermediate” (center), and “late” (bottom), by k-means clustering algorithm (see Methods for details). (C) Presence (blue lines) of the AAAATTGA “early” promoter element in the 5′ gene regulatory region; (D) presence (purple lines) of the “late” promoter element (see main text); (E) transcripts corresponding to gene products previously identified in the virus particle proteome (black lines).
Figure 5.
Variability of individual expression profiles. Normalized gene expression profiles are plotted for representative genes from the three main transcript classes (from top to bottom): genes belonging to the “early” class, the “intermediate” class, and the “late” class. The area of highest expression for each class is highlighted in gray (Fig. 4B, red). The expression data obtained from Agilent tiling array experiments are in good agreement with the RNA-seq expression profiles. The absolute number or reads for RNA-seq experiment are shown in Supplemental Fig. S8.
The biological relevance of the three main classes of transcription profiles was confirmed by examining the expression pattern of several genes of known function. The “late” expression class, for instance, includes genes encoding structural components of the Mimivirus particles (such as the main capsid protein L425 or the core protein L410) or genes encoding enzymes carried by the particle (such as the glucose-methanol-choline [GMC]-type oxydoreductase R315) or enzymes most likely involved in the biosynthesis of the lipopolysaccharide (LPS)-like outer layer of the virus particle (Claverie and Abergel 2009; Claverie et al. 2009a; Xiao et al. 2009) (L136, R689, L780) (Fig. 5; Supplemental Fig. S8). Accordingly, the proportion of genes from this class, the products of which were also detected in the particle proteome (Renesto et al. 2006; Claverie et al. 2009a), is much higher than in the other expression classes (Fig. 4E).
The intermediate transcript class corresponds to genes that are transiently expressed in between T = 3 h and T = 6 h, of which a large proportion encode proteins involved in (and presumably necessary to) the replication of the viral DNA. They include enzymes central to the biosynthesis of deoxynucleotides (such as the ribonucleotide reductase small and large subunit L312 and R313) or the deoxynucleotide monophosphate kinase (R512), the DNA polymerase (R322), the proliferating cell nuclear antigen (PCNA) sliding clamp protein (R322), and its five clamp loaders (R395, R411, L478, L499, R510) (Fig. 5; Supplemental Fig. S8). Finally, the early transcript class (mostly detected from T = 0 to T = 3 h) is functionally more diverse as well as enriched in genes of unknown function. Three out of the four viral aminoacyl–tRNA synthetases belong to this class: TyrRS (L124), MetRS (R639), and ArgRS (R663); the CysRS (L164) belongs to the intermediate transcript class.
Correlation with the predicted AAAATTGA early promoter element
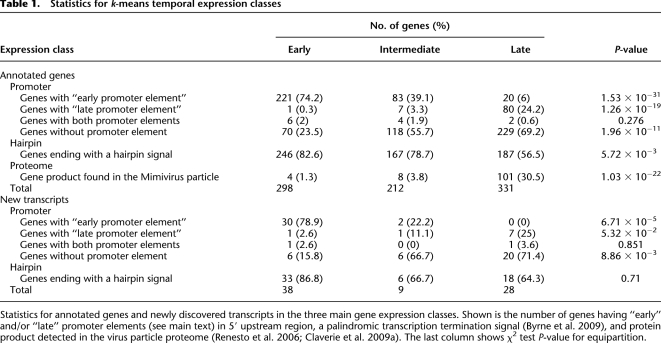
Starting from the predicted gene map, Suhre et al. (2005) identified a strictly conserved AAAATTGA motif as statistically overrepresented in front of 403 Mimivirus ORFs (45% of the total). The nonrandom distribution of this motif and its strong preferential occurrence in the 5′ upstream region of Mimivirus genes was found to be highly significant. Moreover, the type of predicted function associated with the genes exhibiting this motif led to the prediction that AAAATTGA was the promoter signature of early transcripts. This bioinformatic prediction is now experimentally well confirmed by the expression data (Fig. 4; Table 1). A further validation of this strong correlation is provided by the new genes discovered during this study (Table 1B): 78.9% of the newly mapped genes corresponding to early transcripts exhibit the AAAATTGA motif. We also used the 479 transcript start sites (TSSs) precisely mapped in this study to compute the distance distribution between the AAAATTGA motif and the downstream TSS (Supplemental Fig. S9A). As expected from a core promoter element obeying the geometrical constraints of the transcription initiation complex, these distances follow a much narrower distribution (D = −51 ± 5) than when using the downstream ORF start codon as reference.
Table 1.
Statistics for k-means temporal expression classes
Statistics for annotated genes and newly discovered transcripts in the three main gene expression classes. Shown is the number of genes having “early” and/or “late” promoter elements (see main text) in 5′ upstream region, a palindromic transcription termination signal (Byrne et al. 2009), and protein product detected in the virus particle proteome (Renesto et al. 2006; Claverie et al. 2009a). The last column shows χ2 test P-value for equipartition.
Discovery of a promoter element associated with late expression
After the identification of the core promoter element associated with Mimivirus early transcripts, we searched for other motifs that might be associated with genes classified in the other temporal expression classes. Without the gene transcription profiles at hand, our previous attempts at defining significant promoter motifs other than AAAATTGA had failed. Using our new expression data, we could now focus on ORFs corresponding to late transcripts and specifically scrutinize their 5′ upstream sequence using various motif search programs (see Methods). A degenerate but significant motif was thus identified, the logo representation of which is shown in Supplemental Figure S10. This AT-rich motif comprises two 10-nt informative segments separated by a highly degenerated 4-nt sequence. This new motif was then searched in front of all Mimivirus genes. When authorizing a single mismatch, this candidate promoter element was found in front of 24.2% of the gene classified in the late class, compared with only 3.3% and 0.3% for the genes of the intermediate and early classes, respectively (Table 1). As expected from a core promoter element, the distance separating this late motif from the downstream TSS was narrowly distributed (D = −21 ± 5) (Supplemental Fig. S9B).
Mimivirus late promoter element is highly prevalent in the Sputnik “virophage” genome
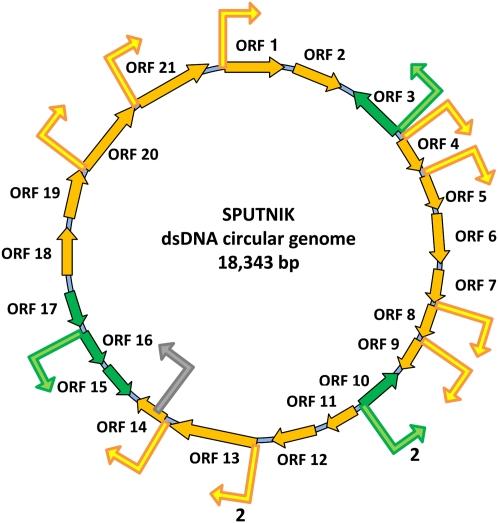
Sputnik is a recently described “satellite” virus only able to replicate in Acanthamoeba cells infected by Mimivirus or its close relative Mamavirus (La Scola et al. 2008). Based on the colocalization of neo-formed Sputnik particles with Mimivirus-induced cytoplasmic virion factory, La Scola et al. (2008) coined the word “virophage” to designate a new type of satellite virus that would be a bona fide parasite of Mimivirus, rather than a mere defective virus requiring the help of Mimivirus to productively infect Acanthamoeba cells. In accordance with this hypothesis, the Sputnik genome, a 18-kb dsDNA molecule encoding 21 proteins but no RNA- or DNA-polymerase of its own, was predicted to be expressed via the Mimivirus-encoded transcription machinery (presumably confined within the virion factories) rather than by the amoeba nuclear transcription apparatus (Claverie and Abergel 2009). A first validation of this concept was the finding that many Sputnik genes share the 3′-end palindromic signal characteristic of Mimivirus mRNA polyadenylation sites (Byrne et al. 2009; Claverie and Abergel 2009). If the completion of Sputnik replication cycle requires fully functional Mimivirus virion factories, one would expect a number of Sputnik genes to be expressed in a late manner. We thus scanned the Sputnik genome for the presence of the newly characterized Mimivirus late promoter element (see above). The motif was found upstream of 12 different genes, for a total of 14 times within the intergenic regions and only once within an ORF (Fig. 6). Given the relative proportion of the intergenic (3759 nt) versus coding (14,584 nt) moieties, this distribution is highly significant (Fisher's exact test, P < 3 × 10−9) of the preferential location of this element in the putative promoter region of Sputnik genes. The distance separating this late motif from the downstream ORF start codon was narrowly distributed (D = −29 ± 2.5) and similar to the distance found for Mimivirus (Supplemental Fig. S9B). Hence, this strongly suggests that these Sputnik genes are transcribed by the Mimivirus-encoded machinery dedicated to late genes. For comparison, the early Mimivirus promoter element AAAATTGA was found only twice in the intergenic region compared with three times within ORFs, thus showing no significant bias (Fisher's exact test, P > 0.26).
Figure 6.
Mimivirus late promoter element in Sputnik genome. Late promoter element locations are depicted by bent arrows in intergenic (yellow and green) and coding regions (gray).
The newly discovered genes obey Mimivirus transcriptional signals
A total of 75 new genes were predicted from the clustering and tiling of reads at genomic positions not corresponding to previously annotated ORFs or tRNAs. Once classified according to the previously used k-means method, they were again partitioned into the three classes of transcripts: early, intermediate, and late. The 5′ and 3′ flanking sequences of these transcripts were then searched for the early and late promoter elements as well as a hairpin signal at their polyadenylation sites. The statistics summarized in Table 1 indicate that the newly identified genes obey the same transcription signals as previously annotated genes, suggesting that they are equally functional.
Functional hints from expression profiles
Interpreting the variation in transcript numbers as being proportional to the variation in the corresponding protein concentrations or enzymatic activities is a common pitfall of transcriptomic studies. Many factors, including mRNA stability and accessibility to the translation machinery, as well as the stability and the eventual post-translational modification of protein products, conjugate to blur the correlation between transcription levels and the change they cause in the cellular metabolism. In the case of Mimivirus, this task is further complicated by the fact that the particle itself incorporates a large number of nonstructural proteins (such as many enzymes and a functional transcription apparatus), the activity of which is presumably available immediately upon the delivery of the particle content into the host cytoplasm, and thus prior to the transcription of the corresponding gene. In the same context, genes expressed as late transcripts may (and some do) correspond to proteins incorporated into the Mimivirus particle and destined to perform “early functions” in the infectious cycle. With these caveats in mind, we analyzed the expression profiles observed for several broad functional categories, as follows (Supplemental Table S3).
Transcription
Most of the genes encoding the Mimivirus transcription apparatus component belong to the “late” temporal class of expression, the abundance of their transcripts reaching their peaks at T ≥ 6 h. One thus expects the corresponding proteins to be translated right in time to be loaded into the newly synthesized particles. Accordingly, with the exception of the R453 gene product (a TATA-box binding protein homolog with a predicted pI of 9.8), all of them were detected in the particle proteome (Renesto et al. 2006; Claverie et al. 2009a). This expression pattern also suggests that the early and intermediate Mimivirus transcripts detected in abundance before T = 3 h (i.e., prior to the appearance of fully mature cytoplasmic virion factories) (Fig. 2) are generated by (1) the transcription apparatus released from the infecting particles, (2) the host transcription apparatus (thus implying an unlikely “nuclear phase”), or (3) proteins encoded by early Mimivirus messengers bearing no detectable similarity with known RNA polymerases.
DNA repair
The transcript abundance of two putative DNA repair enzymes (L386, R406) exhibits a peak at T = 0, suggesting that they are involved in early (possibly transcription-coupled) DNA repair mechanisms. The other DNA repair enzymes correspond to typical “intermediate” or “late” transcripts, as most of the functions involved in the viral DNA replication.
Topoisomerases
The three topoisomerase homologs encoded by the Mimivirus genome exhibit maximal expression at T = 3 h (L221, R480) or T = 6 h (R194). They are thus likely to play a role at the time of DNA replication and encapsidation. However, the corresponding proteins are associated with the particle and may also play a role in helping unload the large Mimivirus genome from the infecting particles.
Translation and tRNA modification
Most of the components of the translation apparatus found encoded in the Mimivirus genome, a unique feature among viruses, are expressed at an average or high level, confirming their role in the replication cycle. Two predicted tRNA methyltransferases (R405, R407) as well as three of the four aminoacyl-tRNA synthetases exhibit an early (T = 0) expression, suggesting that they are involved in the protein translation process right from the beginning of the replication cycle. In contrast, the mRNA cap-binding protein (L496), the peptide chain release factor (R726), and the translation initiation factor R464 exhibit a late temporal expression pattern (T ≥ 6 h), suggesting that they only come into play after that Mimivirus cytoplasmic factories become fully mature.
Chaperonin
The whole set of HSP-70 and DnaJ homologs encoded in the Mimivirus genome exhibit a high and maximal expression at T = 6 h, suggesting that they perform an essential function in the capsid assembly process. Interestingly, despite maintaining a high level of expression until the very end of the replication cycle (T = 12 h), none of these proteins have been detected in the virus particle.
Nucleotide synthesis and DNA replication
Genes in this functional class exhibit a homogeneous expression pattern, all of the corresponding transcripts belonging to the intermediate temporal class. This is consistent with these functions being essential to the viral DNA replication process that only actively begins at T ≥ 6 h in the mature Mimivirus factories. In particular, the expression profile of the Mimivirus homolog (L276) of the mitochondrial ADP/ATP carrier protein concurs with the previous suggestion that the virus targets the host mitochondria as a source for dATP and dTTP (Monne et al. 2007).
Viral metabolism
The Mimivirus genome contains a wealth of putative enzyme homologs predicted to be involved in the biosynthesis of a complex LPS-like particle outer layer and in numerous post-translational protein modifications such as glycosylation, acetylation, or the addition of lipid anchors (Raoult et al. 2004; Claverie et al. 2009a). The fact that an overwhelming proportion of these genes are maximally expressed at T ≥ 6 h strongly suggests that these enzymatic activities are not targeting host proteins but are rather involved in the building of Mimivirus particles (the proteomic analysis of which indicated many post-translational modifications) (Renesto et al. 2006).
Particle structure
As expected, the genes encoding typical virion structural components, such as the four capsid protein paralogs and the major coat protein, are not significantly expressed before T = 6 h. Our data confirm that the three minor capsid protein genes, the products of which were not detected in the particle proteome, are indeed expressed. Interestingly, the same late expression pattern is shared by the six Mimivirus genes encoding collagen-domain proteins that we predicted to be part of the particle structure (Raoult et al. 2004; Claverie et al. 2009a). The nondetection of these proteins—as well as of the minor capsid proteins—in the particle proteome might be due to extensive post-translational modifications or to their covalent association with an insoluble LPS-like moiety (Raoult et al. 2004; Claverie and Abergel 2009; Claverie et al. 2009a; Xiao et al. 2009) conferring its exceptional solidity to the Mimivirus particle.
Discussion
We used massively parallel pyrosequencing to obtain a detailed picture of the transcriptional program of Mimivirus infecting Acanthamoeba castellanii, the first such study applied to a large DNA virus. Compared with conventional ORF-based microarrays, our approach provided several different types of results.
First, we generated a single-nucleotide resolution map of Mimivirus transcripts, confirming the location of most predicted genes. This transcript map was then used to validate the two previously predicted transcription signals (the early promoter element AAAATTGA [Suhre et al. 2005] and the putative hairpin found at most Mimivirus mRNA polyadenylation sites [Byrne et al. 2009]).
Furthermore, the precise mapping of the TSS made possible the identification of a new promoter element strongly correlated with genes starting to be expressed 6 h post-infection. Interestingly, this late promoter element was also found upstream of 12 of the 21 Sputnik genes, in agreement with the notion that the replication cycle of this “virophage” requires the availability of the fully mature Mimivirus intracytoplasmic factories that are readily observed in electron microscopy pictures, starting 6 h post-infection (Fig. 2E,F).
Another main result of our study is the identification of 75 new genes not previously predicted by traditional bioinformatic analysis. About one half of these transcripts do not appear to encode a protein product, thus suggesting that ncRNAs, a handful of which have been described in other large DNA viruses (Sullivan 2008), may constitute a significant component of the gene expression regulatory network of Mimivirus. Following the correction of several sequence errors and the precise mapping of the intron–exon structure of two genes, the Mimivirus genome now exhibits 900 protein coding genes, six tRNA, and 75 “new” genes, for a total of 981 transcription units.
The quantitative analysis of transcript abundances at various times post-infection provided the first global, systemic picture of the transcriptional program of a large DNA virus infecting its host. These data are extremely rich and can be queried from many different angles. As a first step, hierarchical clustering and the k-means clustering approaches were used to objectively classify the various types of expression profiles. This resulted in dividing Mimivirus genes in the three broad classes—early (mostly expressed 3 h post-infection), intermediate (mostly expressed 3 to 6 h post-infection), and late (peaking after 6 h)—that were found to correlate with the presence of two different promoter elements. However, a closer inspection of the individual profiles of genes revealed a significant—within-class—variability (both in terms of intensity as well as timing), suggesting that the expression of Mimivirus genes obeys a much finer level of regulation, the mechanisms of which remain to be elucidated.
In this context, we noticed that an overwhelming fraction (85%) of the most abundant transcripts (Supplemental Table S2) presumably encoding functions essential for the replication cycle correspond to anonymous genes. The only three highly expressed genes with a predicted function encode the major capsid protein (L425), a choline dehydrogenase homolog (R135) associated to the virus particle, and a DNA-interacting protein (R545). Among the top 20 most abundant transcripts, 15 exhibit maximal expression at T ≥ 6 h, and five belong to the early expression class. Five of the new genes identified in this study were found to exhibit similarly high transcript numbers (Supplemental Table S2). We also noticed that the class of early expressed genes—presumably central to the building of the cytoplasmic factories—is particularly enriched in genes of unknown function. Altogether, these findings illustrate our high level of ignorance about the detailed cellular processes at work during Mimivirus infection, while simultaneously pointing out genes to be targeted in priority for future studies.
Another interesting category of Mimivirus genes are those exhibiting a significant number of transcripts at T = −15 min (Supplemental Table S4), which is when a large majority of virus particles are still found unopened in vacuoles (Fig. 2A). Among them, those exhibiting a lower transcript count at T = 0 and corresponding to abundant transcripts at T = 12 h are prime candidates for being incorporated into the viral particle as polyadenylated messengers, which could function as pretranscriptional templates for an early translation immediately after their delivery into the host cytoplasm. An analysis of the RNA content of the Mimivirus particles is ongoing, guided by these results.
The data presented in this study were made fully available on an interactive web server (http://www.igs.cnrs-mrs.fr/mimivirus/) to serve as a blueprint for all kinds of follow-up studies on the unique physiology of Mimivirus.
Methods
A. castellanii infection by Mimivirus
Virus production and purification were performed as previously described (Byrne et al. 2009). The experimental protocols are detailed in the Supplemental material.
RNA and cDNA production
RNA extraction, quantification, and cDNA production were performed as previously described (Byrne et al. 2009). See Supplemental material for details.
Genome mapping of the reads
We generated 633,346 reads that were screened for 5′ and 3′ sequence tags (hereafter called p5 and p3, respectively) using Cross_match (P. Green, unpubl.; http://www.phrap.org/phredphrapconsed.html). A total of 312,641 reads contained a p5 tag and 252,788 a p3 tag. We subsequently aligned the read sequences on the Mimivirus genome (RefSeq ID NC_006450) using the BLAT program (Kent 2002) with the following parameters: query type “rna” and maxIntron = 5000. We only kept the best hit for each read. We were finally able to accurately map 322,904 reads on the Mimivirus genome sequence. The remaining reads were aligned on the available A. castellanii host genomic sequences: mitochondrion DNA sequence (RefSeq ID NC_001637) and draft nuclear genome contigs (Acast_1.0 initial assembly from the Baylor College of Medicine Human Genome Sequencing Center: http://www.hgsc.bcm.tmc.edu). We were able to successfully align 51,910 reads on the mitochondrion genome and 213,479 reads on the nuclear genome. Among the remaining 45,053 unmapped sequences (7%), half were too short to be accurately aligned (length < 30 nt), and the other half did not match the nonredundant (NR) sequence database (BLASTN and BLASTX (Altschul et al. 1990; E-value < 1 × 10−5). Therefore, the remaining sequences presumably originate from unfinished parts of the A. castellanii genome and not from contaminations.
Internal priming filtering
Since our experimental procedure for cDNA production uses 3′ poly(A) sequence for priming, one could expect some internal priming artifacts producing some truncated cDNAs sequences. To minimize this problem, potential internal priming events were filtered using the following procedure. First we extracted p3 reads ending up within ORFs (Supplemental Fig. S11, cases Q and O) as a positive data set, and p3 reads overlapping ORFs but ending in 3′ intergenic regions (Supplemental Fig. S11, case S) as a negative data set. We then analyzed various sequence features to separate the two datasets (poly(A) track length, total number of A, base composition, duplex folding energy). The most discriminating criterion was found to correspond to the hybridization free energy of the duplex formed between the poly(dT)-like p3 tag (5′-TAGAGACCGAGGCGGCCGACATGTTTTGTTTTTTTTTCTTTTTTTTTTN-3′) and its cognate genomic region (i.e., right downstream of the 3′ end of the transcript) (Supplemental Fig. S4). The duplex free energy was calculated by the hybrid-min program from Unafold Package (with default parameters and T = 42) (Markham and Zuker 2008). P3 reads were flagged as potentially arising from internal priming when free energy < −11 kcal/mol.
Read assignment, count, and normalization
To associate reads to their cognate gene we first classified them based on (1) the presence of a p5 or p3 tag, (2) their overlap to an ORF, and (3) their orientation (i.e., sense or antisense). The addition of the p5/p3 tags at the time of cDNA production provided information about transcript strandness. We then took into account the genomic context (i.e., positions relative to the genes) to classify the reads using a decision tree, exhaustively listing all the possible cases (Supplemental Fig. S11).
Transcript abundances were estimated by counting the reads overlapping a given ORF in the same orientation (i.e., read classes B, D, E, I, and S in Supplemental Fig. S11). We were able to assign at least one read to each Mimivirus gene (Supplemental Fig. S2). The counts for redundant time points (6 h-a and 6 h-b) were summed. Data normalization was then performed by dividing raw read counts by the total number of reads obtained for each time point. We multiplied this value by the maximum number of reads and rounded the result to the nearest integer to obtain normalized read counts. Normalized read counts ranged from 1 to 18,102. We only retained the 841 genes supported by at least 10 reads for subsequent analysis.
Gene expression data clustering
To reveal the different transcriptional patterns at play during Mimivirus infection, we clustered gene transcription profiles using various clustering methods. We first log-transformed the normalized read count profiles (see above) and centered this data by the mean. We then used Cluster 3.0 program (de Hoon et al. 2004) to apply a hierarchical clustering on the genes (Fig. 4A), with a Pearson centered correlation similarity metric. Data visualization was done using Java TreeView program (Saldanha 2004).
We then applied a partitioning clustering algorithm (k-means) using the same data and the same metric to classify the expression profiles into k clusters. To define the optimal number of clusters without using any a priori biological information, we used the following procedure. We first computed clustering on the data for different numbers of clusters (k = 2, 3, 4, 5, 10, 20, 50, 100, 200) using the R function “pam” with a Pearson correlation distance. We then calculated various clustering quality indices (as defined in Ray and Turi 1999), namely the Dunn index, the Davies-Bouldin index, and the Validity index, for each of the resulting clusters. According to the Dunn index, the optimal number of clusters in our data set is three. The Davies-Bouldin index gives two clusters as the optimal number, but the Validity index also gives three as the best number of clusters. Therefore, we performed the k-means clustering with k = 3 classes (Fig. 4B).
New gene discovery
Mapping of p5/p3 reads allowed us to accurately identify a number of transcriptional unit starts and ends. We used the following procedure to stringently characterize transcript boundaries. We first converted p5 read genomic coordinates into a profile for each genome strand, so that each single base in the genome was assigned a number of p5 reads. We then scanned the profiles and identified increases of at least five p5 reads in a window of 3 nt. Those genomic positions were defined as transcriptional start sites. We used the same procedure with p3 reads and identified transcript ends by looking at drops of p3 reads (at least five in a window of 3 nt). Only reads predicted not to be derived from internal priming were taken into account (see above). Finally, full transcripts were defined as uninterrupted tiling of reads in between the identified transcript starts and ends. We discovered 75 transcripts not corresponding to previously annotated genes (ORFs and tRNAs) (Supplemental Table S1).
The 75 new genes were subsequently classified as protein coding or noncoding based on the longest ORF they encompass. We determined the longest coding sequence (from START to STOP codon) and shuffled the sequence 1000 times while maintaining the mono- and dinucleotide compositions. The longest ORF was determined for each of these shuffled sequences, and the relative frequency of ORFs equal or longer than the actual ORF was taken as an empirical coding propensity. We classified as potentially coding all genes with a propensity value > 0.9.
For each of these polyadenylated transcript sequences, a homology search was performed using BLASTX (Altschul et al. 1990) against the RefSeq protein sequence database. Only hits in the same orientation as the predicted transcript were retained (E-value < 1 × 10−5). Only three of the 75 sequences exhibited a significant match: L67b with the ankyrin repeat containing Mimivirus protein L62 (E-value = 2 × 10−13), L685b with the uncharacterized Mimivirus protein L781 (E-value = 6 × 10−12), and R776b with the chaperon protein DnaJ protein ZP_06244800.1 (E-value = 7 × 10−6). The longest ORF within each transcript was also searched for protein motifs using RPS-Blast against the Conserved Domain Database (CDD) on (E-value < 1 × 10−5). Only R776b exhibited a significant match to the DnaJ motif (COG0484, E-value < 2 × 10−10).
Promoter regulatory element discovery and assignment
A search for statistically overrepresented motifs in 5′ upstream gene sequences was performed with different algorithms, namely MEME (Bailey and Elkan 1994) and MotifSampler (Thijs et al. 2001). We first extracted intergenic sequences (from 100 bp upstream ORF to START codon) of all genes and grouped the sequences in the three main transcription patterns (see above). MEME algorithm was used with a background model trained on intergenic sequences and parameter w = 25. A new significantly overrepresented motif was found in the “late” cluster (Supplemental Fig. S10A). The same data set was searched with MotifSampler (default parameters, same background model, and w = 25) (Supplemental Fig. S10B).
Mimivirus genome was searched for promoter elements using the Fuzznuc program from the EMBOSS Package (Rice et al. 2000). The “early” AAAATTGA promoter element was searched with no mismatch imposing the following spatial constraints: 110 nt to 50 nt upstream of the START codon for annotated genes or 70 nt to 40 nt upstream of the TSS for new transcripts. The “late” promoter element was searched using the following consensus sequence allowing one mismatch: [AT]{8}T[AC]TN{4}[AT]{5}[AG]TA[TG]A, 50 nt to 10 nt upstream of the START codon for annotated genes or 30 nt to 0 nt upstream of the TSS for new transcripts.
Agilent tiling array validation experiment
RNA production, tiling array design, hybridizations, and quantification of Mimivirus gene expression are described in the Supplemental material.
Acknowledgments
We acknowledge the use of the PACA-Bioinformatics platform and the cellular imaging platform of the Mediterranean Institute of Microbiology (IFR-88). The microarray validation was performed on the Hotel-Express transcriptomic platform. We thank Bruce Roe for very helpful technical suggestions, Jean Weissenbach for providing a speedy access to the Genoscope 454 sequencing platform, Nicolas Boulanger for the tiling array experiment, and Adrien Jeanniard for a preliminary survey of Acanthamoeba transcripts. This work was partially funded by CNRS, ANR grant no. ANR-08-BLAN-0089, IBISA, and the Provence-Alpes-Côte d'Azur region.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequencing data from this study have been submitted to the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA010763. The microarray data from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under accession nos. A-MEXP-1782 and E-MTAB-187.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.102582.109.
References
- Abergel C, Rudinger-Thirion J, Giegé R, Claverie J-M 2007. Virus-encoded aminoacyl-tRNA synthetases: Structural and functional characterization of Mimivirus TyrRS and MetRS. J Virol 81: 12406–12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Brüssow H 2009. The not so universal tree of life or the place of viruses in the living world. Philos Trans R Soc Lond B Biol Sci 364: 2263–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne D, Grzela R, Lartigue A, Audic S, Chenivesse S, Encinas S, Claverie J-M, Abergel C 2009. The polyadenylation site of Mimivirus transcripts obeys a stringent ‘hairpin rule.’. Genome Res 19: 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J-M 2006. Viruses take center stage in cellular evolution. Genome Biol 7: 110. doi: 10.1186/gb-2006-7-6-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J-M, Abergel C 2009. Mimivirus and its virophage. Annu Rev Genet 43: 49–66 [DOI] [PubMed] [Google Scholar]
- Claverie J-M, Ogata H 2009. Ten good reasons not to exclude giruses from the evolutionary picture. Nat Rev Microbiol 7: 615. doi: 10.1038/nrmicro2108-c3 [DOI] [PubMed] [Google Scholar]
- Claverie J-M, Ogata H, Audic S, Abergel C, Suhre K, Fournier P 2006. Mimivirus and the emerging concept of “giant” virus. Virus Res 117: 133–144 [DOI] [PubMed] [Google Scholar]
- Claverie J-M, Abergel C, Ogata H 2009a. Mimivirus. Curr Top Microbiol Immunol 328: 89–121 [DOI] [PubMed] [Google Scholar]
- Claverie J-M, Grzela R, Lartigue A, Bernadac A, Nitsche S, Vacelet J, Ogata H, Abergel C 2009b. Mimivirus and Mimiviridae: Giant viruses with an increasing number of potential hosts, including corals and sponges. J Invertebr Pathol 101: 172–180 [DOI] [PubMed] [Google Scholar]
- de Hoon M, Imoto S, Nolan J, Miyano S 2004. Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Forterre P 2006. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain. Proc Natl Acad Sci 103: 3669–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EV, Aravind L 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res 117: 156–184 [DOI] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—The BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P, Koonin E, et al. 2008. The virophage as a unique parasite of the giant mimivirus. Nature 455: 100–104 [DOI] [PubMed] [Google Scholar]
- Markham NR, Zuker M 2008. UNAFold: Software for nucleic acid folding and hybriziation. Methods Mol Biol 453: 3–31 [DOI] [PubMed] [Google Scholar]
- Monne M, Robinson AJ, Boes C, Harbour ME, Fearnley IM, Kunji ERS 2007. The Mimivirus genome encodes a mitochondrial carrier that transports dATP and dTTP. J Virol 81: 3181–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Lopez-Garcia P 2009. Ten reasons to exclude viruses from the tree of life. Nat Rev Microbiol 7: 306–311 [DOI] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie J-M 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306: 1344–1350 [DOI] [PubMed] [Google Scholar]
- Ray S, Turi RH 1999. Determination of number of clusters in k-means clustering and application in colour image segmentation. Proceedings of the 4th International Conference on Advances in Pattern Recognition and Digital Techniques (ICAPRDT’99, Calcutta), pp. 137–143 Narosa Publishing House, New Delhi, India [Google Scholar]
- Renesto P, Abergel C, Decloquement P, Moinier D, Azza S, Ogata H, Fourquet P, Gorvel J, Claverie J-M 2006. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J Virol 80: 11678–11685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Saldanha AJ 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Suhre K, Audic S, Claverie J-M 2005. Mimivirus gene promoters exhibit an unprecedented conservation among all eukaryotes. Proc Natl Acad Sci 102: 14689–14693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS 2008. New roles for large and small viral RNAs in evading host defences. Nat Rev Genet 9: 503–507 [DOI] [PubMed] [Google Scholar]
- Suzan-Monti M, Scola BL, Barrassi L, Espinosa L, Raoult D 2007. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS One 2: e328. doi: 10.1371/journal.pone.0000328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura M 2001. Poxviruses and the origin of the eukaryotic nucleus. J Mol Evol 52: 419–425 [DOI] [PubMed] [Google Scholar]
- Thijs G, Lescot M, Marchal K, Rombauts S, De Moor B, Rouzé P, Moreau Y 2001. A higher-order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics 17: 1113–1122 [DOI] [PubMed] [Google Scholar]
- Villarreal LP, DeFilippis VR 2000. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J Virol 74: 7079–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Kuznetsov YG, Sun S, Hafenstein SL, Kostyuchenko VA, Chipman PR, Suzan-Monti M, Raoult D, MacPherson A, Rossmann MG 2009. Structural studies of the giant Mimivirus. PLoS Biol 7: e92. doi: 10.1371/journal.pbio.1000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauberman N, Mutsafi Y, Halevy DB, Shimoni E, Klein E, Xiao C, Sun S, Minsky A 2008. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol 6: e114. doi: 10.1371/journal.pbio.0060114 [DOI] [PMC free article] [PubMed] [Google Scholar]