Abstract
Initiation of a cell cycle in an adult neuron leads to cell death, placing great importance on the mechanisms that normally suppress the neuronal cell cycle. We have previously shown that the cyclin-dependent kinase Cdk5 is an important part of this process, but only when it is present in the nucleus. We report here that Cdk5 nuclear localization relies on its binding to the cyclin-dependent kinase inhibitor p27. Cdk5 has no intrinsic nuclear localization signal; in the absence of p27, two weak nuclear export signals that bind CRM1 cause it to shuttle to the cytoplasm. When a neuron is subjected to stress, such as exposure to β-amyloid, the Cdk5-p27 interaction is lost, reducing Cdk5 levels in the nucleus and depriving the neuron of a major cell cycle suppression mechanism. Caspase-3 is activated within hours, but death is not immediate; elevated levels of cytoplasmic Cdk5 appear to retard neuronal death by a mechanism that may involve Bcl2. These data suggest a model in which Cdk5 exerts a double protective function in neurons: chronically suppressing the cell cycle when located in the nucleus and transiently delaying cell death in the cytoplasm.
Keywords: Cell/Neuron, Cell/Cycle, Cell/Cyclins, Cell/Division, Diseases/Alzheimer Disease, Neurodegeneration, Nuclear Export, Nuclear Import, Nucleocytoplasmic, p27
Introduction
The Cdks are the catalytic subunits of a family of nine serine/threonine protein kinases: Cdk1–Cdk9. Among all Cdks, Cdk5 is atypical in several ways. First, its activity does not rely on binding to regular cyclins. Instead, Cdk5 is activated by two specific proteins, p35 and p39, that are structurally similar to cyclins yet share no homology at the amino acid level (1). Second, the actions of Cdk5 are not required for cell cycle progression (2); rather, they are critical for neuronal development, migration, and cortical lamination (3). Although Cdk5 does not drive the cell cycle forward, it does hold the cycle in check. As a consequence, the loss of Cdk5 leads to a failure of cell cycle suppression and subsequent neuronal cell death. This is most evident in Cdk5−/− embryonic mouse neocortical neurons, both in vivo and in vitro (4, 5).
Cdk5 is normally located in both nucleus and cytoplasm (5, 6). This distribution changes in neurons that have been shown to re-enter a cell cycle. For example, in the E2f1−/− mouse brain, many neurons in the cerebral cortex have replicated their DNA and continue to express proteins normally found only in cycling cells (7). Similar cell cycle events are found in neurons at risk for death in Alzheimer disease (8, 9). In both of these situations, the “cycling” neurons have lost their nuclear Cdk5 but retain cytoplasmic immunoreactivity (5). These data suggest that nuclear/cytoplasmic transport is important to the cell cycle suppressor function of Cdk5 and stimulated our interest in the mechanisms that control Cdk5 localization in the neuron.
We explore here the role of Cdk5 as a nucleocytoplasmic protein. We show that its nuclear localization is dependent on its binding with p27, whereas its cytoplasmic localization is achieved through the NES-CRM-1 nuclear export mechanism. We show that Cdk5 shuttles between the nucleus and the cytoplasm during the cell cycle. In postmitotic neurons in culture, Cdk5 nuclear export is required for cell cycle re-entry, but once in the cytoplasm, Cdk5 may protect against rapid neuronal death. Thus Cdk5 serves a dual protective function in the highly differentiated postmitotic neuron.
EXPERIMENTAL PROCEDURES
Antibodies and Chemical Regents
Antibodies against β-actin, Cdk4, Cdk5, hnRNP, lamin A/C, lamin B1, GFP,2 Crm-1, HSP90, and hemagglutinin were from Santa Cruz Biotechnology (Santa Cruz, CA). Histone H3 was from Millipore (Billerica, MA). Rat-anti-BrdUrd was bought from Abcam (Cambridge, UK). Cleaved caspase-3 antibody was bought from Cell Signaling (Danvers, MA). Secondary antibodies used for immunocytochemistry were as follows: goat anti-mouse Alexa 488 and 594; goat anti-rat Alexa 488 and 594; goat anti-rabbit Alexa 488 and 594 (Invitrogen). All were used at a dilution of 1:1000. 4′,6′-Diamidino-2-phenylindole was used as a nuclear counterstain at 1 μg/ml. An Amaxa mouse neuron nucleofector kit was bought from Lonza (Köln, Germany).
Animals
A colony of Cdk5−/+ mice were maintained on a mixed (C57BL/6Jx129/S1) background. Homozygous mutant embryos were produced by intercrossing heterozygous Cdk5−/+ mice. Wild type C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Timed pregnancies were established; the date of appearance of a vaginal plug was considered embryonic day 0.5. The embryos were taken at embryonic day 16.5 for either cortical cultures or histology. All of the animal procedures were carried out in accordance with Rutgers University Institutional Animal Care and Use Committee standards. The animal facilities at Rutgers University are fully Association for Assessment and Accreditation of Laboratory Animal Care accredited.
Constructs and Plasmids
Truncated Cdk5 fragments were synthesized by PCR and inserted into pEGFP-C1 or pGEX-4T-2. The fragments of Cdk5 (sequences encoding amino acids 64–83 and 128–147) with insert sites were synthesized and annealed to double-stranded DNA and then inserted into the pEGFP-C1 vector. The caspase-3-sensitive vector was constructed by inserting an NES-Caspase-3 fragment into DsRed-Nuc vector by AgeI/NheI restriction digestion and annealing. The cyclin D1 promoter-luciferase vector (40) was a generous gift of Dr Richard G. Pestell (Thomas Jefferson University), and the cyclin D1 promoter was subcloned into pd2EGFP vector (Clontech). The p27-HA vector was provided by Dr. Elizabeth Nabel (41). The HA-Crm-1 vector was obtained from Addgene.
Transfection
N2a cells were transfected with Lipofectamine 2000 following the instructions of the manufacturer. Primary mouse neurons were transfected with different vectors by the Amaxa mouse neuron nucleofector kit following the manufacturer's instructions.
Cell Culture and Synchronization
Neuroblastoma (Neuro-2a, N2a) and NIH 3T3 cells were purchased from ATCC. The NIH 3T3 (D51) cell line was kindly provided by Dr. Andrew Koff (Memorial Sloan-Kettering Cancer Center, New York). In this cell line, immortalized 3T3 cells were stably transfected with a p27 mutation, p27D51 that removes the N-terminal 51 amino acids of the protein preventing cyclin-Cdk interaction. HCT116-p21−/− cells were kindly provided by Dr. Bert Vogelstein (Johns Hopkins University, Baltimore, Maryland). The cells were synchronized in the G0 phase of the cell cycle by serum deprivation for 48–76 h in medium supplemented with 0.1% fetal bovine serum or 0.1% calf serum. To induce re-entry into the cell cycle, the cells were transferred to medium containing 10% fetal bovine serum or 10% calf serum.
Primary Neuronal Cultures
For Cdk5-deficient cultures, all of the embryos from a Cdk5+/− × Cdk5+/− mating were harvested and treated separately. Isolated embryonic day 16.5 embryonic cerebral cortices from control, Cdk5−/−, or E2f1−/− were treated with 0.25% trypsin-EDTA and dissociated into single cells by gentle trituration. The cells were suspended in neurobasal medium supplemented with B27 and 2 mm glutamine and then plated on coverslips coated with poly-l-lysine (0.05 mg/ml) and laminin (5 μg/ml). All of the cultures were grown for a minimum of 5 days in vitro before treatment. To monitor cultures during treatment, Cdk5−/− or wild type neurons were cultured in glass-bottomed culture chambers (MatTek Corp.). After transfection or drug treatment, the dish to be monitored was placed into a CO2 and temperature-controlled chamber mounted on the motorized stage of an inverted microscope (Leica LTM). Multiple neurons were monitored simultaneously using IP Lab software (BD Biosciences CA). GFP and DsRed were visualized with L5 and N3 filter sets, respectively.
Immunocytochemistry and BrdUrd Incorporation
At the appropriate time, the cultures were rinsed once with PBS and then exposed to 4% paraformaldehyde in 0.1 m phosphate buffer for 30 min at room temperature followed by three rinses with PBS. Immunohistochemistry of cell cultures was done without antigen retrieval. For BrdUrd labeling, the cells were cultured normally or serum-starved for 48 h followed by 12 h of serum add-back. Four hours before the end of the experiment, 10 μm BrdUrd was added to the medium. The cells were then fixed, and DNA was hydrolyzed by exposing the cells to 2 n HCl for 10 min. The specimens were then neutralized in 0.1 m sodium borate (pH 8.6) for 10 min and then rinsed extensively in PBS (three times) for 45 min before treatment with blocking reagent. Nonspecific antibody binding was blocked by exposing the fixed cells to 5% normal goat serum in 0.1% Triton X-100 for 1 h before application of the primary antibody.
Western Blotting and Co-immunoprecipitation
Dissected tissues or harvested cells were homogenized in 1:5 (w/v) ice-cold lysis buffer (1% Triton X-100, 20 mm Tris-HCl, pH 7.5), 150 mm NaCl, with protease inhibitor mix (Roche Applied Science). The samples were centrifuged at 12,000 × g for 20 min at 4 °C. The supernatant was collected, and the total protein levels were measured by a micro bicinchoninic acid protein assay kit (Pierce). Fractionation of cells into cytoplasmic and nuclear components was accomplished with an NER-mammalian kit according to the manufacturer's instructions (Pierce). For Western blots, the lysates were separated with SDS-PAGE and electrophoretically transferred onto nitrocellulose membranes. The membranes were blocked with 5% nonfat milk in TBST and probed with primary antibodies in blocking buffer, followed by treatment with horseradish peroxidase-linked secondary antibodies and ECL Western blotting detection reagents (Pierce). The intensity of immunoreactive bands was quantified using National Institutes of Health ImageJ. For immunoprecipitation, the cell lysates were incubated with immunoprecipitation antibody at 4 °C for 90 min, followed by additional incubation with protein G-Sepharose (GE Healthcare) for 90 min. The beads were washed five times with ice-cold PBS, and the bound proteins were analyzed by SDS-PAGE and immunoblot analysis.
RESULTS
Cdk5 Shuttles between Nucleus and Cytoplasm during the Cell Cycle
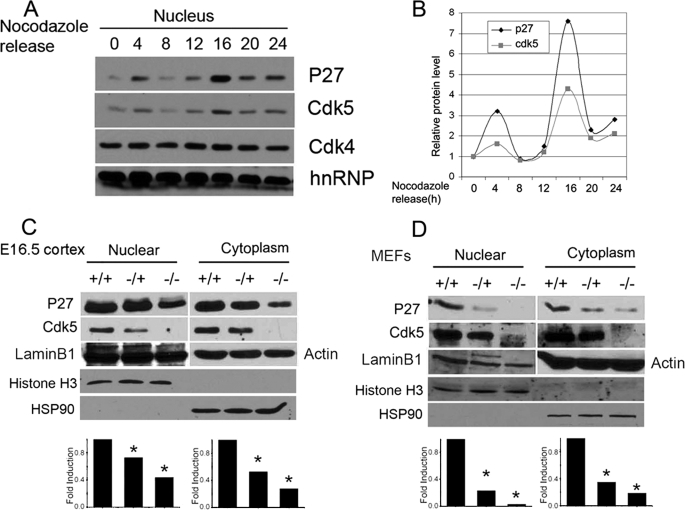
Although the levels of total Cdk5 do not change dramatically during the cell cycle, the nuclear/cytoplasmic ratio does (5). To further explore the movement of Cdk5 during the cell cycle, we used nocodazole to arrest cells at the G2/M phase of the cell cycle, released them, and assayed the location of the Cdk5 at different times. The levels of nuclear Cdk5 show a wave-like pattern, which can be contrasted with nuclear Cdk4 whose levels do not change during the cell cycle (Fig. 1A). Proteins that shuttle between the nucleus and the cytoplasm are termed nucleocytoplasmic, and the movement of Cdk5 is consistent with its identity as a nucleocytoplasmic protein. Cdk5 would thus be predicted to have both an nuclear localization signal (NLS) and a nuclear export signal (NES) to regulate its subcellular localization. A scan of the primary amino acid sequence of Cdk5 did not reveal a canonical NLS. The closest match was found located at amino acids 33–36 (KRVR). This amino acid sequence is similar to the NLS of SV40 (KRKV), but it is unable to drive Cdk5 fragments containing it to the nucleus (supplemental Fig. S1). Without an endogenous NLS, the movement of Cdk5 into the nucleus must depend on binding to an accessory protein. Interestingly, in the course of our experiments, we found that the levels of nuclear Cdk5 during the cell cycle closely followed those of p27 (Fig. 1, A and B). We therefore considered the potential role for this Cdk inhibitor.
FIGURE 1.
A, N2a cells were treated with nocodazole for 48 h and then returned to drug free medium. The cells were harvested at the indicted times. Nuclear fractions were run on Western blots and probed with antibodies against Cdk5, p27, and Cdk4. The RNA binding protein hnRNP was used as loading control. B, the bands in A were quantified by National Institutes of Health ImageJ. Western blots of nuclear and cytoplasm fractions of lysates from embryonic day 16.5 Cdk5+/+, Cdk5+/−, and Cdk5−/− are shown. C and D, embryonic cortex (C) and mouse embryonic fibroblasts (D, MEFs) of the same genotypes. Actin and HSP90 were used as cytoplasmic loading controls. Lamin B1 and Histone H3 were used as nuclear loading controls. *, p < 0.05 by analysis of variance. The error bars represent S.E., n = 3.
Nuclear Localization of Cdk5 Is Dependent on p27 Binding
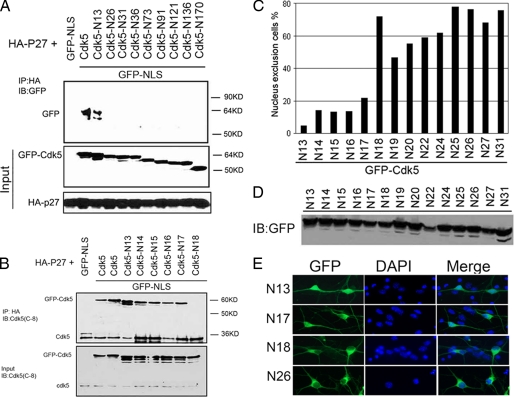
In homozygous Cdk5−/− brain (Fig. 1C) and Cdk5−/− embryonic fibroblasts (Fig. 1D), the levels of p27 are dramatically reduced. This is significant because in Cdk5-deficient cells, cell cycle suppression is lost. To determine whether the two proteins physically interacted with each other, we performed co-immunoprecipitation experiments. Both full-length and GFP-Cdk5N13–292 could be immunoprecipitated by HA-p27 (Fig. 2A; the subscript refers to the amino acid residues remaining in the Cdk5 construct). Successive removal of single amino acids revealed that when the threonine at position 17 was removed, the Cdk5/p27 interaction was eliminated; GFP-Cdk5N18–292 no longer bound to HA-p27 (Fig. 2B, ninth lane). The physical interaction was also confirmed by glutathione S-transferase pulldown experiments (supplemental Fig. S2A). We note that the N-terminal residues around threonine 17 of Cdk5 are highly conserved in species from worm to human (supplemental Fig. S2C).
FIGURE 2.
A, serial Cdk5 truncations with an in-frame NLS were co-expressed with HA-tagged p27. The cell lysates were immunoprecipitated (IP) with HA and blotted (IB) with GFP. Western blots of the lysates show the equal expression levels both within and between runs. B, fine mapping of the p27 binding site on Cdk5. One-by-one deletion of amino acid residues from 12 to 17 identifies the threonine residue at position 17 as crucial for Cdk5 binding to p27. C, N-terminal truncations from N13 to N31 were transfected into N2a cells. The percentage of cells with GFP excluded from the nucleus is shown. D, Western blots of the truncated Cdk5. Note that even the single amino acid differences between two mutations are detectable as a small shift in the migration of the band. E, representative micrographs of the behavior of truncated Cdk5 N13, N17, N18, and N26 in primary mouse cortical neuron.
To determine the correlation between p27 binding and Cdk5 subcellular localization, N-terminal truncation mutations from N13 to N31 of Cdk5 were expressed as GFP fusion proteins, and their localization visualized by GFP fluorescence. Fig. 2C shows that truncations up to residue 17 localized throughout the whole cell; they were rarely excluded from the nucleus (<20%). By contrast, mutations that deleted amino acids Thr17 and beyond localized almost exclusively in cytoplasm (>60%). All of the fusion proteins were expressed at comparable levels (Fig. 2D). Fig. 2E shows representative images of various truncation mutations expressed as GFP fusion proteins in primary neurons (for additional mutations expressed in N2a cells, see supplemental Fig. S3). These data show that the N-terminal 17 residues of Cdk5 are crucial for its interaction with p27 and that the nuclear localization of Cdk5 depends on this interaction.
The Cytoplasmic Localization of Cdk5 Is Dependent on Its Intrinsic NES
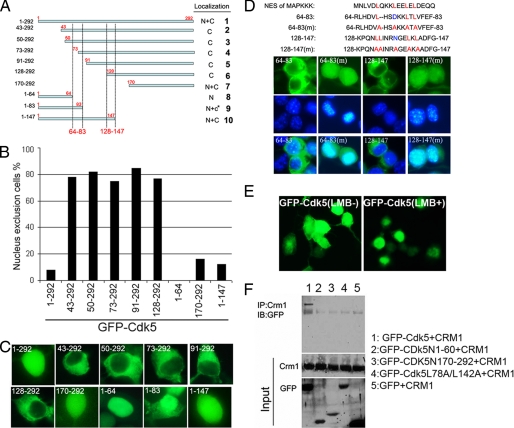
The nuclear exclusion of the N-terminal deleted mutations suggested that the remaining Cdk5 fragment has an intrinsic NES. To determine its location, we designed the truncations shown in Fig. 3A. Wild type Cdk5 is localized throughout the cell. N-terminal truncations from residues 17 to 128 (Fig. 3A, constructs 2–6) localize primarily in the cytoplasm. Further truncations (Fig. 3A, construct 7) lose this cytoplasmic preference. This implies that an NES activity lies between residues 17 and 128. Three C-terminal truncation constructs were made: Cdk5(1–64), Cdk5(1–83), and Cdk5(1–147). Although Cdk5N1–64 localized only in the nucleus; Cdk5N1–83 was mostly nuclear but had some cytoplasmic localization as well; Cdk5N1–147 localized through the cell, similar to full-length Cdk5 (Fig. 3, B and C; see also supplemental Fig. S4). This identified two potential NES, one between amino acids 64 and 83 and another between residues 128 and 147.
FIGURE 3.
A, location of the Cdk5 nuclear export signals determined by truncation mutation of Cdk5. The upper panel shows the position of the mutation. The dashed lines indicate the amino acid fragments with NES activity. B, the percentage of cells with GFP excluded from the nucleus in A. C, each panel shows the behavior of representative truncation mutations after expression in N2a cells. D, top panel is a sequence comparison of the well known NES signal in the MAPKKK protein compared with various fragments in the Cdk5 sequence. The bottom panels illustrate the behavior NES activity of the fragments indicated toward the GFP protein. The middle panels illustrate the 4′,6′-diamino-2-phenylindole counterstain only. E, GFP-Cdk5 was transfected into N2a cells without (left panel, LMB−) or with (right panel, LMB+) leptomycin-B treatment. F, Cdk5, Cdk5(1–64), Cdk5(170–292), and Cdk5(L78A/L142A) were expressed as GFP fusion proteins. A GFP-only vector was expressed as a control. All were co-transfected with HA-CRM-1 into N2a cells. Their interaction was evaluated by immunoprecipitation (IP) with CRM-1 and immunoblotting (IB) with GFP.
To validate the identification of these fragments as NES, we synthesized these fragments and inserted them into a GFP vector. As predicted, both fragments drove GFP into cytoplasm (Fig. 3D). Comparing the sequence of these fragments with the NES of MAPKKK reveals that fragments 64–83 and 128–147 both contain the typical sequence with multiple leucines but are missing the second of four predicted Leu residues (at Asp74 and Asn137; Fig. 3D). Nonetheless, Leu → Ala mutations of the remaining leucines abolished cytoplasmic localization (Fig. 3D). These data show that Cdk5 contains two NES-like signals, each made atypical by the absence of one leucine.
The binding of an NES to CRM1 leads to nuclear export (10), and we wished to determine whether Cdk5 used the CRM1 pathway. GFP-Cdk5 transfected N2a cells were treated with leptomycin B, an inhibitor of CRM1-dependent nuclear export (11, 12). Over 70% of the transfected cells accumulated GFP-Cdk5 in the nucleus (Fig. 3E). A direct interaction between CRM1 and Cdk5 was shown by co-immunoprecipitation (Fig. 3F), and Cdk5 fragments that lacked the NES regions, Cdk5N1–64 and Cdk5N170–292, did not interact with CRM1, nor did the Leu → Ala mutant. An empty GFP vector was used as a negative control.
Cytoplasmic Cdk5 Attenuates the Activation of Caspase-3 Induced by β-Amyloid
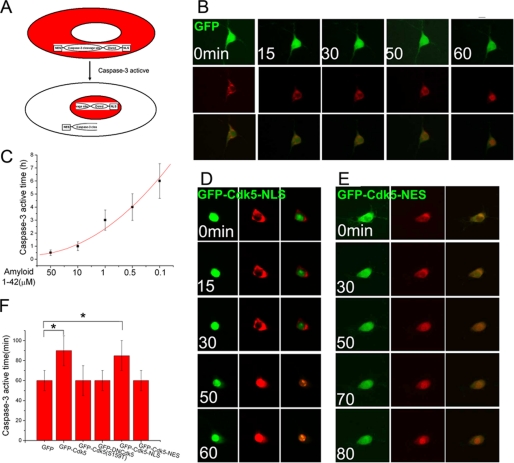
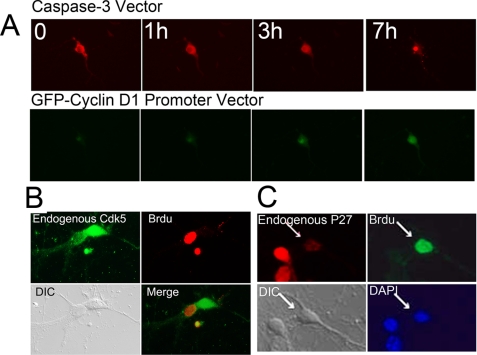
We have previously shown that nuclear Cdk5 functions as a cell cycle suppressor. This raises the question of what happens to Cdk5 once it is shuttled to the cytoplasm. We speculated that this fraction of Cdk5 might play a role in neuronal cell death. To explore this idea, we created a caspase-3-sensitive dsRed fluorescent protein vector in which a caspase-3 cleavage site was placed between an NES and the dsRed coding sequence that included an in-frame NLS. Because NES-based nuclear export is dominant over NLS-based import, a cell expressing our construct will have red fluorescence in the cytoplasm (Fig. 4A, top panel). With the activation of caspase-3, however, the NES is cleaved off. The NLS, now unopposed, moves the dsRed to the nucleus (Fig. 4A, bottom panel). The time required for complete caspase-3 cleavage is defined as the time when the dsRed marker is entirely nuclear. We tested the utility of this construct in Cdk5−/− neurons; within 60 min of exposure to 10 μm fibrillar Aβ1–42 peptide, the dsRed moves from cytoplasm to nucleus (Fig. 4B). The activation time of caspase-3 (dsRed translocation) was dependent on the concentration of Aβ (Fig. 4C).
FIGURE 4.
A, diagram of the behavior of the caspase-3-sensitive dsRed construct during cell death stimulation. B, a GFP-only control vector was co-transfected with caspase-3-sensitive vector into Cdk5−/− neurons, and dsRed localization was monitored at varying times after the administration of 10 μm β-amyloid. Caspase-3 cleavage time is determined by noting the time after Aβ addition when the dsRed marker is completely nuclear. C, decreasing β-amyloid concentrations increase the latency between administration and nuclear import. D and E, GFP-Cdk5-NLS (D) and GFP-Cdk5-NES (E) were co-transfected with caspase-3 vector into Cdk5−/− neurons. After 10 μm β-amyloid administration, caspase-3 cleavage time was monitored. F, the quantification of the time needed for the constructs indicated to lead to nuclear import of dsRed (activate caspase-3). *, p < 0.05 by analysis of variance. The error bars represent S.E., n = 4.
Control GFP, GFP-Cdk5-NLS, or GFP-Cdk5-NES expressing constructs were co-transfected with the caspase-3-sensitive vector into Cdk5−/− neurons. The next day, 10 μm Aβ1–42 was added to the medium, and the location of dsRed was tracked. Although nuclear Cdk5 (GFP-Cdk5-NLS) had no effect on the rate of caspase-3 activation (Fig. 4, D and F), cytoplasmic Cdk5 (GFP-Cdk5-NES) slowed this process significantly (Fig. 4, E and F). These findings suggest that cytoplasmic Cdk5 is neuroprotective. This protection is dependent on kinase activity because both p35 binding-defective and kinase-dead Cdk5 mutants, even if expressed in the cytoplasm, were ineffective at delaying caspase-3 activation.
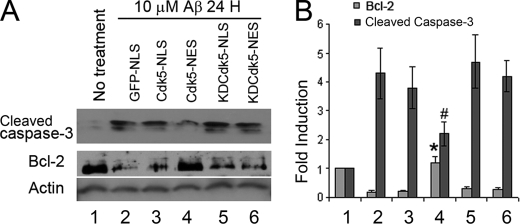
The experiments illustrated in Fig. 4 reflect the short term response of caspase-3 activation to Aβ exposure as measured by an exogenously applied caspase-3 detection peptide. To extend these observations, the endogenous level of cleaved capase-3 was investigated at longer survival times. GFP-NLS, GFP-Cdk5-NLS, GFP-Cdk5-NES, GFP-KDCdk5-NLS, and GFP-KDCdk5-NES were transfected into Cdk5−/− neurons. Twelve hours after Aβ administration, cleaved capase-3 was detected by Western blotting (Fig. 5A). Kinase-active, cytoplasmic Cdk5 (Cdk5-NES) lowered the cleaved caspase-3 level induced by Aβ, whereas none of the other constructs tested (Cdk5-NLS, KDCdk5-NLS, and KDCdk5-NES) were found to have any effect (Fig. 5). The reduced activation of caspase-3 implies an anti-apoptotic action of Cdk5. Aβ is also reported to induce apoptosis through down-regulation of Bcl-2, and Cdk5 has been implicated in this action. Examining the levels of Bcl2 in the Aβ-treated neurons in our cultures revealed that of all of the constructs tested, only cytoplasmic Cdk5 overexpression prevented the decrease of the Bcl-2 level after Aβ administration.
FIGURE 5.
Primary mouse cortical neurons were transfected by Amaxa mouse nucleofector transfection kit. A, cleaved caspase-3 and Bcl-2 levels were detected by Western blotting after 10 μm Aβ1–42 administration for 3 h. B, statistics of the bands in A.
Nuclear Cdk5 Blocks Cell Cycle Re-entry: p27 Dependence
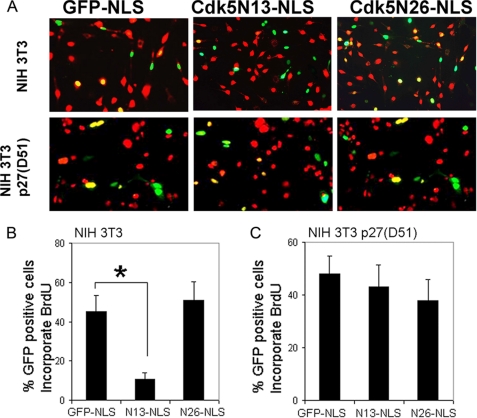
Because nuclear Cdk5 has cell cycle blocking ability (5), and the nuclear localization of Cdk5 depends on its binding with p27, we wondered whether the cell cycle suppression ability of Cdk5 was also dependent on this interaction. To test this idea, two Cdk5 truncation mutations, Cdk5N13–292 and Cdk5N26–292, were forced into the nucleus of NIH 3T3 cells as GFP-NLS fusion proteins. After transfection, cell cycling was arrested by serum starvation followed by reinitiation in the presence of BrdUrd. Cdk5N13–292, which retains p27 binding capacity, was capable of suppressing BrdUrd staining, but Cdk5N26–292, which cannot bind p27, was inert, despite its NLS-directed nuclear location (Fig. 6, A and B). The requirement for p27 was further demonstrated by the inability of GFP-Cdk5-NLS-derived constructs to arrest the cell cycle in the NIH 3T3 p27(D51) cell line, in which a dominant negative p27 that is Cdk/cyclin binding-deficient is constitutively expressed (Fig. 6, A and C). We also investigated a possible role for p21, another member of the Kip/Cip Cdk inhibitor family. We transfected Cdk5-NLS and KDCdk5-NLS into the HCT116 p21−/− cell line. All of the active Cdk5 and KDCdk5 species retain their cell cycle suppressor activity in the absence of p21 in contrast to the results with p27 deficiency (supplemental Fig. S5).
FIGURE 6.
A, BrdUrd staining was performed in NIH 3T3 or NIH 3T3 p27(D51) cells with transfection by GFP-Cdk5N13-NLS and GFP-Cdk5N26-NLS. B and C, the percentages of double-labeled cells percentage in NIH 3T3 cells (B) or NIH 3T3 p27(D51) cells (C). *, p < 0.05 by analysis of variance. The error bars represent S.E., n = 4.
Cdk5 Is a Key Determinant of the Choice between Cell Cycle and Cell Death
It has been proposed that the death of neurons in several different neurodegenerative diseases is preceded by an unscheduled initiation of a cell cycle. We used two marker constructs to determine whether there was a consistent order to the initiation of these two processes in our system. To monitor cell cycle, we inserted the cyclin D promoter in front of a GFP construct. To monitor cell death activity, we used the caspase-3-sensitive vector described above. Cdk5−/− neurons were treated with 1 μm β-amyloid after co-transfection of these two vectors. As shown in Fig. 7A, GFP fluorescence (cyclin D) was seen in neurons ∼3 h after Aβ1–42 addition, marking this as the time of neuronal re-entry into the cell cycle. After 7 h, we observed high expression of cyclin D, and by this time, neurons had also begun the cell death process (nuclear dsRed). Thus cell cycle and cell death are closely linked in cultured neurons, and in this system cell cycle initiation precedes cell death. To validate this conclusion for endogenous Cdk5, neurons were double immunostained with BrdUrd and Cdk5 after β-amyloid exposure. Consistent with our results with the cyclin D-GFP and caspase-sensitive dsRed vector, neurons that had begun DNA synthesis (BrdUrd uptake) showed very low levels of nuclear Cdk5 staining (Fig. 7B) as well as low levels of nuclear p27 staining (Fig. 7C).
FIGURE 7.
A, caspase-3 sensitive vector was co-transfected into Cdk5−/− neurons with cyclin D1-GFP reporter construct. After 1 μm β-amyloid administration, the dsRed and GFP signals were monitored. B and C, wild type primary neurons were treated with 3 μm β-amyloid 24 h. B, double immunostaining for endogenous Cdk5 (green) and BrdUrd (red). C, double immunostaining for endogenous p27 (red) and BrdUrd (Brdu, green). Nuclear Cdk5 and p27 both reveal in inverse relationship with the appearance of BrdUrd incorporation. DAPI, 4′,6′-diamino-2-phenylindole. DIC, differential interference contrast.
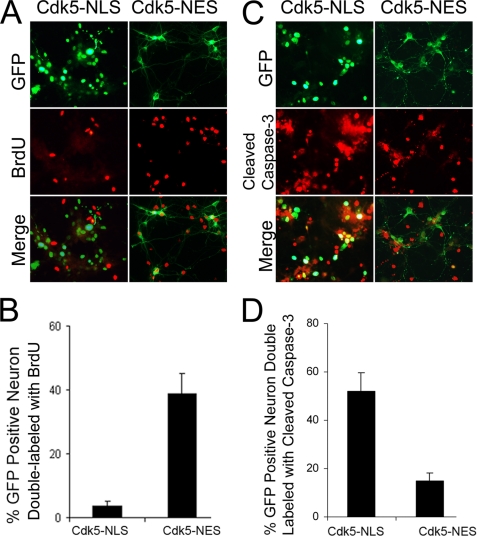
To further validate this ordering of events, GFP-Cdk5-NLS and GFP-Cdk5-NES were transfected into Cdk5−/− neurons. After Aβ administration, BrdUrd incorporation and caspase-3 activation were detected by immunocytochemistry. Only 5% neurons with nuclear Cdk5 (GFP-Cdk5-NLS) incorporated BrdUrd, but more than 40% neurons with cytoplasmic Cdk5 (GFP-Cdk5-NES) incorporated BrdUrd (Fig. 8, A and B). Similarly, ∼50% of the neurons with nuclear Cdk5 was positive for cleaved-caspase-3, but only ∼15% neurons with cytoplasmic Cdk5 was cleaved caspase-3-positive (Fig. 8, C and D). Taken together these data show that Cdk5 is transported into the cytoplasm during or before cell cycle initiation and before caspase-3-dependent cell death begins.
FIGURE 8.
GFP-Cdk5-NLS and GFP-Cdk5-NES were transfected into Cdk5−/− neurons. A and C, after treatment with 3 μm β-amyloid, BrdUrd incorporation (A) and cleaved caspase-3 (C) were detected by immunocytochemistry (red). Note that in the nucleus Cdk5 prevents BrdUrd incorporation (no double-labeling) but permits caspase activation, whereas in the cytoplasm it prevents caspase activation but permits BrdUrd incorporation. B and D, the percentage of double-labeled cells was quantified for BrdUrd (B) and cleaved caspase-3 (D). E, *, p < 0.05 by analysis of variance. The error bars represent S.E.
DISCUSSION
There is now a substantial literature documenting the fact that cell cycle re-entrance by postmitotic central nervous system neurons is tightly correlated with neuronal degeneration (13, 14). It has been observed in human diseases ranging from ataxia-telangiectasia to Parkinson disease to Alzheimer disease (15–17). If developing central nervous system neurons are forced to enter a cell cycle by expression of an oncogene, they will pass into the S phase and synthesize DNA, but rather than divide they will die (18–20). Critically, blocking the cell cycle in these neurons (either pharmacologically or genetically) prevents the cell death (21–23). Despite the frequent correlation of cell cycle and cell death in the same neuronal populations, the mechanistic pathway neurons use to suppress their cell cycle during adult life remains largely undefined. The data presented here begin to address this gap in our knowledge. We have found that cell cycle or cell death stimulation changes the Cdk5 nuclear/cytoplasmic ratio and that this shuttling can actively regulate both the cell cycle and the initiation of the cell death process. This marks the control of Cdk5 transport as a critical part of the maintenance of a normal neuronal homeostasis.
Nucleocytoplasmic proteins usually have both an NLS and an NES signal. During typical nuclear import, an importin family member binds to the NLS of its cargo and moves the importin-cargo complex into the nucleus (24). Although there is a potential NLS located in amino acids 33–36 of Cdk5 (KRVR), our results show that the N31 truncation mutation containing the KRVR residues nonetheless localizes to the cytoplasm (supplemental Fig. S1). In support of this, Fu et al. (25) could not find any significant binding between Cdk5 and any of several different importins. Phosphorylation can also play a role in nuclear localization (26). Indeed we located several potential phosphorylation sites on Cdk5 with the use of the NetPhos web-based program. Mutation of these sites, however, did not change Cdk5 localization (supplemental Fig. S6). Instead, our data show that it is its interaction with p27 that localizes Cdk5 to the nucleus.
As with most nucleocytoplasmic proteins, its subcellular location is also regulated by its ability to bind CRM1, a key mediator of nuclear export (27–29). Truncation mutation studies indicate that the export of Cdk5 relies on two atypical NES motifs, located between amino acids 64 and 83 and amino acids 128 and 147. Their atypical form may help to explain why the 1–83 construct is only partially localized to the cytoplasm (Fig. 3, B and C). Thus unlike its nuclear import, which relies on a second protein (p27), nuclear export of Cdk5 is directed by its endogenous NES signals.
Our cell cycle data illustrate that there are important functional consequences to the loss of the Cdk5-p27 interaction. First, in the absence of p27, the localization of Cdk5 becomes cytoplasmic. Second, this translocation relieves the Cdk5-dependent suppression of the cell cycle. We find it intriguing that if Cdk5 cannot bind p27, even if it is forced into the nucleus with an independent NLS, it cannot suppress the cell cycle. Exactly how the interaction of p27 and Cdk5 serves to arrest the cell cycle is unclear. Further work will be needed to answer this question. The consequences of Cdk5 nuclear export extend beyond the loss of cell cycle suppression. Our caspase-3 data illustrate that cytoplasmic but not nuclear Cdk5 slows the time course of β-amyloid toxicity. Taken together our results suggest that Cdk5 plays a protective function in a nerve cell during the process of cell cycle-related neuronal death. Cdk5 in the nucleus suppresses the cell cycle, whereas Cdk5 in the cytoplasm delays caspase-3 activation and Bcl2 degradation.
Ours is not the only evidence that Cdk5 may have a protective function in neurons. Cdk5 has been shown to protect against excitotoxic death in cerebellar granule neurons (8) and to prevent neuronal apoptosis by negative regulation of c-Jun N-terminal kinase 3 (30). Other reports, including our own, have demonstrated that Cdk5-deficient neurons or cells are more, rather than less, vulnerable to Aβ-induced death than wild type cells (4, 5, 31). The present work broadens this evidence by showing that (i) in our conditions the nuclear and cytoplasmic portions of Cdk5 both contribute to its protective effect and (ii) the cytoplasmic action is independent of its effects on the cell cycle. Questions remain unanswered as to how cytoplasmic Cdk5 serves to protect the neuron. One potential mechanism, suggested by the data in Fig. 5, is that Cdk5 contributes to the stabilization of Bcl-2, which is localized mainly in the cytoplasm. This suggestion is consistent with previous models (32, 33), as well, although other actions may contribute.
This is a complex area of neurobiology because there are numerous reports that excess Cdk5 activity is neurotoxic rather than neuroprotective. For example, Mao and co-workers (34, 35) report that nuclear Cdk5 phosphorylates and destabilizes the anti-apoptotic factor, MEF2, thus contributing to the excitotoxic cell death of cultured neurons. In addition, several labs have argued that the hyperactivation of Cdk5 through the calpain cleavage of p35 to p25 is also responsible for neuronal cell death (36–38). The integration of these disparate findings is crucial to our full understanding of the action of Cdk5 in neurons.
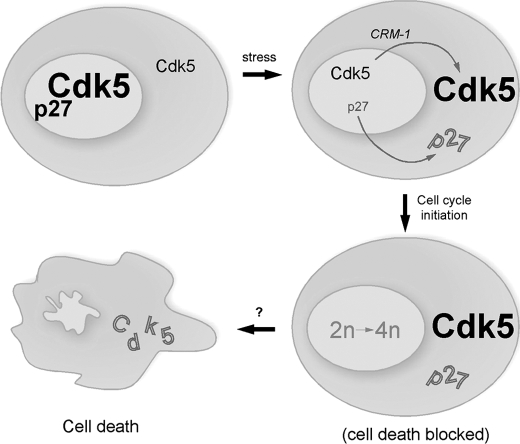
We find that the movement of Cdk5 precedes or is contemporaneous with cell cycle initiation and precedes caspase cleavage. If this is correct, then part of the linkage between cycle and death might occur through Cdk5 itself. Our current model is diagrammed in Fig. 9. When a postmitotic neuron is subjected to cell cycle or cell death stimulation (such as in Alzheimer disease), the physical interaction between Cdk5 and p27 is lost. When this happens, Cdk5 and p27 are both transported into the cytoplasm by CRM1 (Fig. 7, B and C). The reduction in nuclear Cdk5 and p27 deprives the neuron of its cell cycle suppression activity. The stressed neuron re-enters the cell cycle, but with the enhanced levels of cytoplasmic Cdk5, the neurons are temporarily protected from cell death.
FIGURE 9.
When a postmitotic neuron is subjected to cell cycle or cell death stimulation (such as in Alzheimer disease), the interaction between Cdk5 and p27 is lost. When this happens, p27 is transported into the cytoplasm. At the same time, the two atypical NES in the Cdk5 sequence allow its transport to the cytoplasm by the CRM-1 pathway. The reduction in nuclear Cdk5 and p27 deprives the postmitotic neuron of its cell cycle suppression activity. The stressed neurons re-enter the cell cycle, but with the enhanced levels of cytoplasmic Cdk5, the neurons are temporarily protected from cell death. As yet, it is unknown what event causes the ultimate failure of this temporary protection. Driving expression of exogenous nuclear Cdk5 blocks the cell cycle; driving exogenous cytoplasmic Cdk5 attenuates cell death.
An important remaining challenge is to link this model to the situation in the Alzheimer disease brain. Cdk5 protein levels are much higher in adult brain than in developing brain (39) and higher still in the affected regions of the Alzheimer brain (5). We are encouraged that the model shown in Fig. 9 has relevance to AD because, where it has been quantified, death by cell cycle in AD neurons is a slow process (9, 13). We propose that cytoplasmic Cdk5 contributes to this delayed cell death in Alzheimer disease and perhaps other neurodegenerative diseases.
Supplementary Material
Acknowledgments
We acknowledge the generosity of the many laboratories that shared reagents with us. We also thank Gabriella D'Arcangelo, Jianmin Chen, and Jiali Li for thoughtful comments and suggestions during the preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01NS020591.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- GFP
- green fluorescent protein
- BrdUrd
- bromodeoxyuridine
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- NLS
- nuclear localization signal
- NES
- nuclear export signal(s)
- MAPKKK
- mitogen-activated protein kinase kinase kinase.
REFERENCES
- 1.Dhavan R., Tsai L. H. (2001) Nat. Rev. Mol. Cell Biol. 2, 749–759 [DOI] [PubMed] [Google Scholar]
- 2.van den Heuvel S., Harlow E. (1993) Science 262, 2050–2054 [DOI] [PubMed] [Google Scholar]
- 3.Cruz J. C., Tsai L. H. (2004) Trends Mol. Med. 10, 452–458 [DOI] [PubMed] [Google Scholar]
- 4.Cicero S., Herrup K. (2005) J. Neurosci. 25, 9658–9668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J., Cicero S. A., Wang L., Romito-Digiacomo R. R., Yang Y., Herrup K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8772–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikolic M., Dudek H., Kwon Y. T., Ramos Y. F., Tsai L. H. (1996) Genes Dev. 10, 816–825 [DOI] [PubMed] [Google Scholar]
- 7.Wang L., Wang R., Herrup K. (2007) J. Neurosci. 27, 12555–12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Hare M. J., Kushwaha N., Zhang Y., Aleyasin H., Callaghan S. M., Slack R. S., Albert P. R., Vincent I., Park D. S. (2005) J. Neurosci. 25, 8954–8966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busser J., Geldmacher D. S., Herrup K. (1998) J. Neurosci. 18, 2801–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vecchi M., Polo S., Poupon V., van de Loo J. W., Benmerah A., Di Fiore P. P. (2001) J. Cell Biol. 153, 1511–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson B. R., Eleftheriou A. (2000) Exp. Cell Res. 256, 213–224 [DOI] [PubMed] [Google Scholar]
- 12.Engel K., Kotlyarov A., Gaestel M. (1998) EMBO J. 17, 3363–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y., Mufson E. J., Herrup K. (2003) J. Neurosci. 23, 2557–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Höglinger G. U., Breunig J. J., Depboylu C., Rouaux C., Michel P. P., Alvarez-Fischer D., Boutillier A. L., Degregori J., Oertel W. H., Rakic P., Hirsch E. C., Hunot S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3585–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent I., Rosado M., Davies P. (1996) J. Cell Biol. 132, 413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West A. B., Dawson V. L., Dawson T. M. (2005) Trends Neurosci. 28, 348–352 [DOI] [PubMed] [Google Scholar]
- 17.Yang Y., Herrup K. (2005) J. Neurosci. 25, 2522–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feddersen R. M., Clark H. B., Yunis W. S., Orr H. T. (1995) Mol. Cell Neurosci. 6, 153–167 [DOI] [PubMed] [Google Scholar]
- 19.Feddersen R. M., Ehlenfeldt R., Yunis W. S., Clark H. B., Orr H. T. (1992) Neuron 9, 955–966 [DOI] [PubMed] [Google Scholar]
- 20.Feddersen R. M., Yunis W. S., O'Donnell M. A., Ebner T. J., Shen L., Iadecola C., Orr H. T., Clark H. B. (1997) Mol. Cell Neurosci. 9, 42–62 [DOI] [PubMed] [Google Scholar]
- 21.Park D. S., Levine B., Ferrari G., Greene L. A. (1997) J. Neurosci. 17, 8975–8983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park D. S., Morris E. J., Greene L. A., Geller H. M. (1997) J. Neurosci. 17, 1256–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Giovanni S., Movsesyan V., Ahmed F., Cernak I., Schinelli S., Stoica B., Faden A. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8333–8338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Görlich D., Kutay U. (1999) Annu. Rev. Cell Dev. Biol. 15, 607–660 [DOI] [PubMed] [Google Scholar]
- 25.Fu X., Choi Y. K., Qu D., Yu Y., Cheung N. S., Qi R. Z. (2006) J. Biol. Chem. 281, 39014–39021 [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Xiong Y. (2001) Science 292, 1910–1915 [DOI] [PubMed] [Google Scholar]
- 27.Fornerod M., Ohno M., Yoshida M., Mattaj I. W. (1997) Cell 90, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 28.Fukuda M., Asano S., Nakamura T., Adachi M., Yoshida M., Yanagida M., Nishida E. (1997) Nature 390, 308–311 [DOI] [PubMed] [Google Scholar]
- 29.Ossareh-Nazari B., Bachelerie F., Dargemont C. (1997) Science 278, 141–144 [DOI] [PubMed] [Google Scholar]
- 30.Li B. S., Zhang L., Takahashi S., Ma W., Jaffe H., Kulkarni A. B., Pant H. C. (2002) EMBO J. 21, 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner N. C., Lord C. J., Iorns E., Brough R., Swift S., Elliott R., Rayter S., Tutt A. N., Ashworth A. (2008) EMBO J. 27, 1368–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C. X., Song J. H., Song D. K., Yong V. W., Shuaib A., Hao C. (2006) Cell Death Differ. 13, 1203–1212 [DOI] [PubMed] [Google Scholar]
- 33.Cheung Z. H., Gong K., Ip N. Y. (2008) J. Neurosci. 28, 4872–4877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong X., Tang X., Wiedmann M., Wang X., Peng J., Zheng D., Blair L. A., Marshall J., Mao Z. (2003) Neuron 38, 33–46 [DOI] [PubMed] [Google Scholar]
- 35.Tang X., Wang X., Gong X., Tong M., Park D., Xia Z., Mao Z. (2005) J. Neurosci. 25, 4823–4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patrick G. N., Zukerberg L., Nikolic M., de la Monte S., Dikkes P., Tsai L. H. (1999) Nature 402, 615–622 [DOI] [PubMed] [Google Scholar]
- 37.Lee M. S., Kwon Y. T., Li M., Peng J., Friedlander R. M., Tsai L. H. (2000) Nature 405, 360–364 [DOI] [PubMed] [Google Scholar]
- 38.Cruz J. C., Tseng H. C., Goldman J. A., Shih H., Tsai L. H. (2003) Neuron 40, 471–483 [DOI] [PubMed] [Google Scholar]
- 39.Tsai L. H., Takahashi T., Caviness V. S., Jr., Harlow E. (1993) Development 119, 1029–1040 [DOI] [PubMed] [Google Scholar]
- 40.Albanese C., Johnson J., Watanabe G., Eklund N., Vu D., Arnold A., Pestell R. G. (1994) J. Biol. Chem. 270, 23589–23597 [DOI] [PubMed] [Google Scholar]
- 41.Boehm M., Yoshimoto T., Crook M. F., Nallamshetty S., True A., Nabel G. J., Nabel E. G. (2002) EMBO J. 21, 3390–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.